Abstract
The sulfonylureas and imidazolinones are potent commercial herbicide families. They are among the most popular choices for farmers worldwide, because they are nontoxic to animals and highly selective. These herbicides inhibit branched-chain amino acid biosynthesis in plants by targeting acetohydroxyacid synthase (AHAS, EC 2.2.1.6). This report describes the 3D structure of Arabidopsis thaliana AHAS in complex with five sulfonylureas (to 2.5 Å resolution) and with the imidazolinone, imazaquin (IQ; 2.8 Å). Neither class of molecule has a structure that mimics the substrates for the enzyme, but both inhibit by blocking a channel through which access to the active site is gained. The sulfonylureas approach within 5 Å of the catalytic center, which is the C2 atom of the cofactor thiamin diphosphate, whereas IQ is at least 7 Å from this atom. Ten of the amino acid residues that bind the sulfonylureas also bind IQ. Six additional residues interact only with the sulfonylureas, whereas there are two residues that bind IQ but not the sulfonylureas. Thus, the two classes of inhibitor occupy partially overlapping sites but adopt different modes of binding. The increasing emergence of resistant weeds due to the appearance of mutations that interfere with the inhibition of AHAS is now a worldwide problem. The structures described here provide a rational molecular basis for understanding these mutations, thus allowing more sophisticated AHAS inhibitors to be developed. There is no previously described structure for any plant protein in complex with a commercial herbicide.
Keywords: inhibition, sulfonylurea, x-ray crystallography, imidazolinone, thiamin diphosphate
The sulfonylurea and imidazolinone herbicides are an essential part of the multibillion-dollar weed-control market. There are now >30 herbicides from these families registered for worldwide use. A major advantage of these compounds is that they are nontoxic to animals, highly selective, and very potent, thereby allowing low application rates. These herbicides act by inhibiting acetohydroxyacid synthase [AHAS; also known as acetolactate synthase; EC 2.2.1.6 (1)], the first common enzyme in the biosynthetic pathway of the branched-chain amino acids. The reaction carried out by this enzyme is the synthesis of either (S)-2-acetolactate from two molecules of pyruvate or (S)-2-aceto-2-hydroxybutyrate from a molecule each of pyruvate and 2-ketobutyrate.
AHAS belongs to a superfamily of thiamin diphosphate (ThDP)-dependent enzymes that are capable of catalyzing a variety of reactions, including both the oxidative and nonoxidative decarboxylation of 2-ketoacids. This cofactor is bound by a divalent metal ion such as Mg2+, which coordinates to the diphosphate group of ThDP and to two highly conserved residues (2) in these proteins. AHAS also binds a molecule of FAD, although this cofactor does not participate in the principal reactions. To date, most AHAS enzymes that have been characterized have both a catalytic subunit (≈65 kDa) and a smaller regulatory subunit, which varies in size between 9 and 54 kDa, depending on the species of origin. In plant AHAS, this regulatory subunit stimulates activity and confers sensitivity to inhibition by the branched-chain amino acids (3).
In 2002, we solved the crystal structure of the catalytic subunit of Saccharomyces cerevisiae AHAS (ScAHAS) in the absence of any inhibitor (4). Subsequently, the structure of this enzyme in complex with five sulfonylurea herbicides was determined (5, 6). Although ScAHAS is a reasonable model of the plant enzyme, there are some puzzling differences. For example, the mutation A122V in Arabidopsis thaliana AHAS (AtAHAS) causes only a small (4-fold) decrease in the sensitivity to sulfometuron methyl [SM (7)], whereas the equivalent mutation (A117V) in ScAHAS (8) decreases sensitivity to this inhibitor by nearly 3,000-fold. ScAHAS is inhibited by imidazolinones in the millimolar range (8), whereas micromolar concentrations are sufficient to inhibit AtAHAS (7). Thus, to advance understanding of the binding of these herbicides to their natural target in plants, we initiated studies on the structure of AtAHAS. Moreover, for the rational design of herbicides, the structure of the plant enzyme in complex with a herbicide is far more relevant. Earlier, we reported crystallization of the enzyme in the presence of chlorimuron ethyl [CE (9)]. Here we present the crystal structure of the catalytic subunit of AtAHAS in complex with CE and four other sulfonylurea herbicides and with IQ, a member of the imidazolinone family.
Results and Discussion
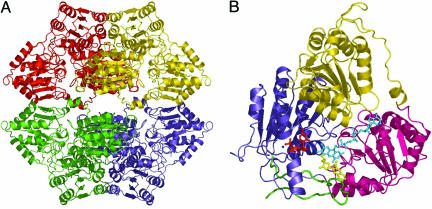
AtAHAS Structure. AtAHAS was crystallized in complex with each of six herbicides, five sulfonylureas and one imidazolinone. The AtAHAS tetramer (Fig. 1A) consists of four identical subunits with an overall fold (Fig. 1B) that resembles that of other ThDP-dependent enzymes such as pyruvate oxidase (10), benzoylformate decarboxylase (11), and ScAHAS (5). AtAHAS and ScAHAS have a rms deviation of 1.00 Å when 525 Cα atoms are superimposed. Each subunit consists of three domains, α (residues 86–280), β (281–451), and γ (463–639), plus a C-terminal tail (646–668) that loops over the active site (Fig. 1B). With the exception of an additional small two-stranded antiparallel β sheet in the α domain, each domain consists of a six-stranded parallel β sheet surrounded by six to nine α helices. The fold of the C-terminal tail closely resembles that of ScAHAS when in complex with the sulfonylureas (5, 6). It is anticipated that this region of AtAHAS is disordered in the absence of herbicide, as it is for the free structure of ScAHAS (4). However, we have not been able to crystallize this form of AtAHAS. A prolyl cis peptide bond is observed between L648 and P649, which also exists in the ScAHAS herbicide complexes but not in the free structure of ScAHAS. This proline is completely conserved across AHAS from 21 species (1), suggesting a critical function for this residue, perhaps as a pivot point where the C-terminal tail changes from a disordered region in the free structure to one that is ordered during the catalytic cycle. Associated with each subunit is FAD, Mg-ThDP, 2-(N-cyclohexylamino)ethanesulfonic acid (CHES) from the crystallization buffer, >200 water molecules, and either one molecule of the appropriate sulfonylurea (Fig. 2 A and B) or two of IQ (Fig. 2 C and D). The electron density is complete for most of the polypeptide, all of the sulfonylureas, one of the IQ molecules, and all of the cofactors, except for the thiazolium and/or pyrimidine rings of ThDP in the sulfonylurea complexes, suggesting that this class of herbicide can directly perturb the stability of ThDP when both are bound to AHAS.
Fig. 1.
The overall fold of AtAHAS. (A) The tetrameric structure with each monomer colored separately. (B) A single subunit. The individual domains α (86–280), β (281–451), and γ (463–639) are colored gold, red, and blue, respectively. The C-terminal tail (646–668) is colored green. ThDP, Mg2+, FAD, and IQ are shown as ball-and-stick models and are colored red, dark blue, cyan, and yellow, respectively.
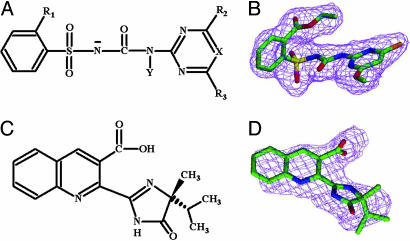
Fig. 2.
Structures of sulfonylureas and IQ and electron densities for CE and IQ. (A) The chemical structure for the sulfonylureas. R1 is CO-OC2H5 for CE, Cl for CS, and CO-OCH3 for SM, MM, and TB. R2 is OCH3 for CE, MM, CS, and TB or CH3 for SM. R3 is CH3 for MM, SM, TB, and CS or Cl for CE. X is N for CS, MM, TB, or CH for SM and CE. Y is H for CE, CS, SM, and MM or CH3 for TB. (B) 2Fo – Fc electron density for CE. (C) The chemical structure of (R)-IQ. (D) 2Fo – Fc electron density for (R)-IQ bound within the active site access channel. Carbon is green; nitrogen, blue; sulfur, yellow; oxygen, red; and chlorine is orange.
The reason for the presence of FAD in AHAS has been controversial, but it is thought to be an evolutionary remnant from a common ancestor of AHAS and pyruvate oxidase (12). The position and conformation of FAD are similar in the two enzymes (6), and the isoalloxazine ring has a bend across the N5–N10 axis that is indistinguishable from that in pyruvate oxidase. Moreover, it has been shown that the FAD can act as an electron acceptor in a slow side reaction catalyzed by AHAS (13).
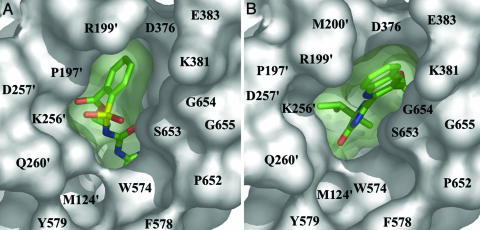
Sulfonylurea and Imidazolinone Binding to AtAHAS. We have shown how sulfonylureas bind to and inhibit ScAHAS (5–7). In binding to plant AHAS, they adopt a similar structure, with a bend at the sulfonyl group that positions the two rings almost orthogonal to each other. The sulfonyl group and the adjacent aromatic ring are situated at the entrance to a channel leading to the active site with the rest of the molecule inserting into the channel (Fig. 3A). A significant difference between sulfonylurea-bound AtAHAS and ScAHAS structures is that residues 652–660 are 3 Å closer to the herbicide in the plant enzyme, giving rise to the additional herbicide contact, S653.
Fig. 3.
Connolly surface and herbicide blocking of the active site channel of AtAHAS. (A) CE. (B) IQ. Both herbicides are shown as stick models. The color scheme for the herbicide atoms is as described in Fig. 2. The residues that line the channel are depicted as a gray surface. ′ indicates residues from the neighboring subunit.
The structure of ScAHAS with a bound imidazolinone herbicide has not been determined. Moreover, the inhibition constant for the yeast enzyme is several-hundred-fold higher than that of plant AHAS. This difference may be due to the positioning of S653 mentioned above, because it is known that mutation of this residue in plant AHAS decreases imidazolinone sensitivity substantially (7). This emphasizes the importance of the structure of the AtAHAS–IQ complex reported here, and that there are significant differences in the active sites of the enzyme in these two species. In this complex, there are two herbicide molecules bound to each subunit. One of these is within the channel leading to the active site, whereas a second is located ≈20 Å from the active site, in a pocket that also contains the CHES molecule. The presence of this second IQ molecule is attributed to the high concentration of herbicide used for crystallization. IQ both activates and inhibits imidazolinone-resistant mutants of ScAHAS, thus implying the existence of two binding sites (8). The IQ molecule in the channel leading to the active site is situated so that the dihydroimidazolone ring is directed toward the C2 center of ThDP, whereas the quinoline ring protrudes out toward the surface of the protein (Fig. 3B). IQ forms extensive noncovalent interactions across 12 amino acids, including a salt bridge between the side chain of R377 and the carboxylate group of IQ. The numerous contacts between AtAHAS and the isopropyl and methyl groups are important for anchoring the herbicide to the protein and help to explain why these substituents are required for good herbicidal activity (14, 15). Racemic IQ was used for crystallization, but it is the (R) enantiomer, a 10-fold more effective herbicide than the (S) isomer (14, 15), which is bound to AtAHAS (Fig. 2 C and D). If the (S) isomer is positioned in the active-site channel, three unfavorable contacts form between the isopropyl substituent of IQ and R377, W574, and G121. Thus, to accommodate (S)-IQ, each of these residues would have to move, although binding is feasible.
The Ki(app) for IQ with AtAHAS is 3.0 μM, whereas the Ki(app) values for the sulfonylureas with AtAHAS range from 10.8 nM (CE) to 253 nM [tribenuron methyl (TB)] (7). This difference in affinity is reflected in the structure in two ways. First, there are 28 van der Waals contacts and only one hydrogen bond between IQ and the enzyme, whereas for the sulfonylureas, there are at least 50 van der Waals contacts and six hydrogen bonds. Second, the sulfonylureas are buried deeper into the active site ≈2 Å closer to the C2 center of ThDP than IQ. The overall orientation and differing chemical structures of the herbicides put IQ ≈6 Å closer to the surface of the protein than any of the sulfonylureas.
We had expected that the binding sites for the sulfonylureas and IQ would overlap and had anticipated that either the dihydroimidazolone or the quinoline ring would superimpose on either the aromatic or heterocyclic ring of the sulfonylureas. Although it is observed that the two sites overlap with the dihydroimidazolone ring and part of the quinoline ring of IQ sharing part of the sulfonylurea binding site, no pair of rings coincide. Furthermore, despite these differing orientations, of the 12 residues involved in securing IQ to the enzyme, G654 and R199 are the only two residues that do not make contact with the sulfonylureas. Therefore, it is impossible that both herbicides could bind to AtAHAS simultaneously in their preferred orientations.
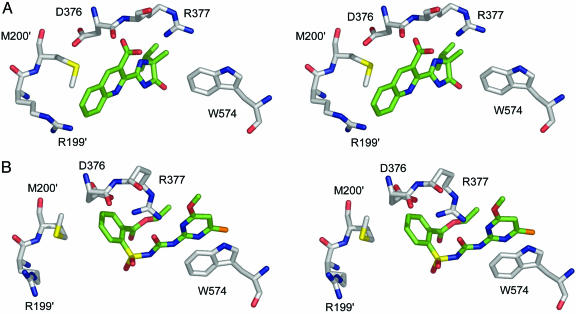
The positions of several key residues move to accommodate either CE or IQ (Fig. 4). R199, at the entrance to the active-site channel, has a different conformation, and the side chains of M200 and W574 are also in different orientations, so that both are closer to IQ. Although the same conformation of M200 would allow for either CE or IQ binding, the orientation of W574 must change. In addition, due to repulsive forces between the carboxylate group of IQ and the side chain of D376, this residue is forced into a different conformation, so that the salt bridge with R377, which exists for the sulfonylurea complexes, cannot be made. In an accompanying change, R377 rotates back and away from D376 to establish an ionic interaction with IQ.
Fig. 4.
Stereoview of the conformational adjustments in the AtAHAS herbicide-binding sites. (A) IQ. (B) CE. Herbicide carbon atoms are colored green. AtAHAS carbon atoms are colored gray, whereas the color scheme for noncarbon atoms is as described in Fig. 2. ′ indicates that these residues are from the neighboring subunit.
Herbicide Resistance. Only a few years after the introduction of sulfonylureas and imidazolinones to the herbicide market, resistant weeds began to emerge. Since that time, there has been an explosion in the appearance of resistant plants, most commonly due to single point mutations resulting in amino acid substitutions for A122, P197, W574, or S653 [using the A. thaliana numbering favored by Tranel and Wright (16)]. The 3D structure of AtAHAS allows us to explain how these amino acid substitutions result in resistance to one or both classes of herbicide. The most comprehensively characterized mutations are those of W574, which result in tolerance to both classes of herbicide in several plants (1, 16, 17). Not only is this residue important for defining the shape of the active-site channel, but it also serves to anchor both classes of herbicide to the enzyme. Consequently, the commonly observed mutation of this residue to leucine changes the shape of the herbicide binding site and results in the loss of several interactions.
Eight different amino acid substitutions for P197 are known to confer herbicide resistance (1, 16). Whereas only one of these, P197L, has been implicated in strong resistance to imidazolinones (18), all known mutations of this residue impart resistance to sulfonylureas. P197 is observed at one end of an α-helix at the entrance to the active-site access channel. Although this residue always contacts the aromatic ring of the sulfonylureas, it does not interact directly with IQ, which lies more toward the middle of the opening. For this reason, it is likely that only bulky amino acid substitutions for P197 will impede the entry of imidazolinones, whereas almost any substitution will prevent sulfonylurea access.
Both S653N (19, 20) and A122T (17) confer strong resistance to the imidazolinones but not to the sulfonylureas. A122 makes important hydrophobic contacts to the isopropyl, and methyl substituents of the dihydroimidazolone ring and mutation to a larger polar residue, such as threonine, would tend to dislodge the herbicide from its site. Because A122 interacts only with the large ethyl ester substituent of CE, but not other sulfonylureas, it is plausible that the slightly larger threonine could be accommodated without seriously compromising sulfonylurea binding. Substitution of S653 with asparagine would not displace the sulfonylureas, provided the side chain is oriented correctly. In contrast, replacement of S653 with almost any other larger residue would impair imidazolinone binding, because it would obstruct the space where the aromatic ring is situated.
It has been suggested (21) that the herbicide-binding site in AHAS may have originated from the binding site for the quinone substrate of pyruvate oxidase. This idea is consistent with the finding that the residues equivalent to R377 and W574 in Escherichia coli AHAS interact with the carboxylate and alkyl side chains, respectively, of the second substrate of AHAS (22). Further, almost all of the residues at herbicide-resistance mutation sites are highly conserved across species, suggesting they may play a role in the normal function of the enzyme. Conflicting with this proposal is the finding that the activity of most herbicide-resistant mutants is similar to that of the wild type (7, 8). However, it should be understood that the selection conditions under which these mutations arise make it probable that mutants with low or no activity will not be isolated. Thus R377 mutations have never been found in herbicide-resistant organisms, because such mutants have little or no activity (22, 23).
Conclusion
To date, most crystal structures of herbicides in complex with their protein target are of microbial origin. The only crystal structures of herbicides in complex with their plant protein target are of experimental herbicides that are not used commercially; DAS 869 and DAS 645 (Dow AgroSciences; PDB ID codes 1TFZ and 1TG5) bound to 4-hydroxyphenylpyruvate dioxygenase from A. thaliana (24), and IpOHA (PDB ID code 1YVE) bound to Spinacia oleracea ketol acid reductoisomerase (25). This study reports structural data at atomic resolution for two common classes of commercial herbicides in complex with their actual target protein. Both the sulfonylureas and imidazolinones, which have partially overlapping binding sites, inhibit the enzyme by binding within and obstructing the channel leading to the active site. These structures now enable us to offer explanations as to how mutant weeds/plants have developed herbicide resistance and will aid in the design of future herbicides.
Materials and Methods
Crystal Structure Determination. The catalytic subunit of recombinant AtAHAS was expressed, purified, and crystallized in the presence of herbicides, as described (9). X-ray data (Table 1) were collected from cryoprotected crystals (30% vol/vol ethylene glycol) at 100 K on Beam-Line 14BMD at the Advanced Photon Source, Argonne National Laboratory, Chicago. The data were indexed, integrated, and scaled by using denzo and scalepack (26). The crystal structure of the catalytic subunit of AtAHAS in complex with CE (2.5 Å) was solved by molecular replacement with amore (27) by using the protein component of the ScAHAS–CE complex as the starting model. The other structures were determined from the AtAHAS–CE complex by difference Fourier methods. Model building and refinement of the structures were performed by using o (28) and cns (29), respectively. Figures were generated with setor (30), chemsketch (ACD Labs, Toronto), and pymol (DeLano Scientific, South San Francisco, CA).
Table 1. Data collection and refinement statistics for the AtAHAS herbicide complexes.
| CE | MM | CS | SM | TB | IQ | |
|---|---|---|---|---|---|---|
| Crystal parameters | ||||||
| Unit cell length, Å, a = b, c | 178.52, 184.78 | 178.23, 185.25 | 178.52, 185.68 | 178.44, 185.24 | 179.06, 186.15 | 179.08, 186.08 |
| Space group | P6422 | P6422 | P6422 | P6422 | P6422 | P6422 |
| Crystal dimensions, mm | 0.35 × 0.35 × 0.3 | 0.3 × 0.3 × 0.2 | 0.3 × 0.3 × 0.3 | 0.3 × 0.3 × 0.2 | 0.3 × 0.3 × 0.2 | 0.3 × 0.2 × 0.2 |
| Diffraction data* | ||||||
| Temperature, K | 100 | 100 | 100 | 100 | 100 | 100 |
| Resolution range, Å | 50-2.5 | 50-2.7 | 50-2.7 | 50-2.7 | 50-2.9 | 50-2.8 |
| Observations [I>0(I)] | 492,411 (25,429) | 170,699 (18,987) | 320,581 (31,026) | 284,055 (27,517) | 168,939 (11,584) | 227,297 (7,150) |
| Unique reflections [I>0(I)] | 59,390 (5,770) | 45,178 (4,506) | 48,379 (4,700) | 45,961 (4,484) | 35,505 (3,276) | 40,076 (2,424) |
| Completeness, % | 98.9 (97.2) | 93.8 (95.5) | 99.5 (98.3) | 95.4 (94.9) | 90.7 (85.5) | 90.8 (56.0) |
| Rsym† | 0.056 (0.297) | 0.099 (0.239) | 0.076 (0.207) | 0.099 (0.254) | 0.095 (0.237) | 0.060 (0.286) |
| 〈I 〉/ 〈σ(I) 〉 | 24.3 (4.1) | 7.3 (3.1) | 12.0 (5.7) | 7.2 (3.0) | 8.4 (3.2) | 23.0 (3.3) |
| Refinement statistic | ||||||
| Resolution limits, Å | 50.0-2.5 | 50.0-2.7 | 50.0-2.7 | 50.0-2.7 | 50.0-2.9 | 50.0-2.8 |
| Rfactor | 0.1920 | 0.1928 | 0.1766 | 0.1767 | 0.1868 | 0.1987 |
| Rfree | 0.2226 | 0.2235 | 0.2011 | 0.2032 | 0.2250 | 0.2157 |
| rmsd bond lengths, Å | 0.0060 | 0.0063 | 0.0059 | 0.0061 | 0.0064 | 0.0065 |
| rmsd bond angles, ° | 1.255 | 1.251 | 1.250 | 1.249 | 1.279 | 1.187 |
| Ramachandran plot, % | ||||||
| Most favored | 90.4 | 90.2 | 89.6 | 88.4 | 86.9 | 86.9 |
| Additionally allowed | 9.2 | 9.4 | 10.0 | 11.0 | 12.4 | 12.7 |
| Generously allowed | 0.4 | 0.4 | 0.4 | 0.6 | 0.6 | 0.4 |
| Disallowed | 0 | 0 | 0 | 0 | 0 | 0 |
rmsd, rms deviation.
Values in parentheses are for the outer-resolution shells: 2.5-2.6 Å for CE; 2.7-2.8 Å for MM, CS, and SM; 2.9-3.0 Å for TB; and 2.8-2.9 Å for IQ.
Rsym = Σ|I- 〈I 〉|/Σ 〈I 〉, where I is the intensity of an individual measurement of each reflection, and I is the mean intensity of that reflection.
Acknowledgments
We thank Harry Tong and Keith Brister for assistance with data collection at the Advanced Photon Source, Argonne National Laboratory, Chicago. This work was supported by Grant DP0450275 from the Australian Research Council. Financial assistance from Badische Anilin und Soda Fabrik is gratefully acknowledged. The use of the BioCARS sector was supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program. Use of BioCARS Sector 14 was also supported by the National Institutes of Health, National Center for Research Resources, under Grant RR07707. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research, under Contract W-31-109-Eng-38.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AHAS, acetohydroxyacid synthase; ScAHAS, Saccharomyces cerevisiae AHAS; ThDP, thiamin diphosphate; AtAHAS, Arabidopsis thaliana AHAS; SM, sulfometuron methyl; CE, chlorimuron ethyl; IQ, imazaquin; TB, tribenuron methyl; MM, metsulfuron methyl; CS, chlorsulfuron.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdg.org (PDB ID codes 1YBH, 1YHY, 1YHZ, 1YI0, 1YI1, and 1Z8N for the CE, MM, CS, SM, TB, and IQ complexes, respectively).
References
- 1.Duggleby, R. G. & Pang, S. S. (2000) J. Biochem. Mol. Biol. 33, 1–36. [Google Scholar]
- 2.Hawkins, C. F., Borges, A. & Perham, R.N. (1989) FEBS Lett. 255, 77–82. [DOI] [PubMed] [Google Scholar]
- 3.Lee, Y.-T. & Duggleby, R. G. (2001) Biochemistry 40, 6836–6844. [DOI] [PubMed] [Google Scholar]
- 4.Pang, S. S., Duggleby, R. G. & Guddat, L. W. (2002) J. Mol. Biol. 317, 249–262. [DOI] [PubMed] [Google Scholar]
- 5.Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2003) J. Biol. Chem. 278, 7639–7644. [DOI] [PubMed] [Google Scholar]
- 6.McCourt, J. A., Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2005) Biochemistry 44, 2330–2338. [DOI] [PubMed] [Google Scholar]
- 7.Chang, A. K. & Duggleby, R. G. (1998) Biochem. J. 333, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duggleby, R. G., Pang, S. S., Yu, H. & Guddat, L. W. (2003) Eur. J. Biochem. 270, 2895–2904. [DOI] [PubMed] [Google Scholar]
- 9.Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2004) Acta Crystallogr. D 60, 153–155. [DOI] [PubMed] [Google Scholar]
- 10.Müller, Y. A. & Schulz, G. E. (1993) Science 259, 965–967. [DOI] [PubMed] [Google Scholar]
- 11.Hasson, M. S., Muscate, A., McLeish, M. J., Polovnikova, L. S., Gerlt, J. A., Kenyon, G. L., Petsko, G. A. & Ringe, D. (1998) Biochemistry 37, 9918–9930. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y.-Y. & Cronan, J. E., Jr. (1988) J. Bacteriol. 170, 3937–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tittmann, K., Schröder, K., Golbik, R., McCourt, J., Kaplun, A., Duggleby, R. G., Barak, Z., Chipman, D. M. & Hübner, G. (2004) Biochemistry, 43, 8652–8661. [DOI] [PubMed] [Google Scholar]
- 14.Los, M. (1984) in Pesticide Synthesis Through Rational Approaches, eds. Magee, P. S., Kohn, G. K. & Menn, J. J. (Am. Chem. Soc., Washington, DC), pp. 29–44.
- 15.Stidham, M. A. & Singh, B. K. (1991) in The Imidazolinone Herbicides, eds. Shaner, D. L. & O'Connor, S. L. (CRC, Boca Raton, FL), pp. 71–90.
- 16.Tranel, P. J. & Wright, T. R. (2002) Weed Sci. 50, 700–712. [Google Scholar]
- 17.Bernasconi, P., Woodworth, A. R., Rosen, B. A., Subramanian, M. V. & Siehl, D. L. (1995) J. Biol. Chem. 270, 17381–17385. [DOI] [PubMed] [Google Scholar]
- 18.Sibony, M., Michel, A., Haas, H. U., Rubin, B. & Hurle, K. (2001) Weed Res. 41, 509–522. [Google Scholar]
- 19.Haughn, G. W. & Somerville, C. R. (1990) Plant Physiol. 92, 1081–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattori, J., Rutledge, R., Labbé, H., Brown, D., Sunohara, G. & Miki, B. (1992) Mol. Gen. Genet. 232, 167–173. [DOI] [PubMed] [Google Scholar]
- 21.Schloss, J. V., Ciskanik, L. M. & Van Dyk, D. E. (1988) Nature 331, 360–362. [Google Scholar]
- 22.Tittmann, K., Vyazmensky, M., Hübner, G., Barak, Z. & Chipman, D. (2005) Proc. Natl. Acad. Sci. USA 102, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le, D. T., Yoon, M.-Y., Kim, Y.-T. & Choi, J.-D. (2005) Biochim. Biophys. Acta 1749, 103–112. [DOI] [PubMed] [Google Scholar]
- 24.Yang, C., Pflugrath, J. W., Camper, D. L., Foster, M. L., Pernich, D. J. & Walsh, T. A. (2004) Biochemistry 43, 10414–10423. [DOI] [PubMed] [Google Scholar]
- 25.Biou, V., Dumas, R., Cohen-Addad, C., Douce, R., Job, D. & Pebay-Peyroula, E. (1997) EMBO J. 16, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Navaza, J. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 28.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 29.Brünger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 30.Evans, S. V. (1993) J. Mol. Graphics 11, 134–138. [DOI] [PubMed] [Google Scholar]