Abstract
Premature translation termination codons are common causes of genetic disorders. mRNAs with such mutations are degraded by a surveillance mechanism termed nonsense-mediated decay (NMD), which represents a phylogenetically widely conserved post-transcriptional mechanism for the quality control of gene expression. How NMD-competent mRNPs are formed and specified remains a central question. Here, we have used human β-globin mRNA as a model system to address the role of splicing and polyadenylation for human NMD. We show that (i) splicing is an indispensable component of the human β-globin NMD pathway, which cannot be compensated for by exonic β-globin ‘failsafe’ sequences; (ii) the spatial requirements of human β-globin NMD, as signified by the maximal distance of the nonsense mutation to the final exon–exon junction, are less constrained than in yeast; and (iii) non-polyadenylated mRNAs with a histone 3′ end are NMD competent. Thus, the formation of NMD-competent mRNP particles critically depends on splicing but does not require the presence of a poly(A) tail.
Keywords: failsafe sequences/histone 3′ end processing/human nonsense-mediated decay/polyadenylation/splicing
Introduction
Accurate gene function depends on a low error rate of each step of the gene expression pathway. The fidelity of gene expression is enhanced further by a phylogenetically widely conserved quality control mechanism against faulty mRNAs with premature translation termination codons, which has been termed nonsense-mediated mRNA decay (NMD). The importance of NMD in man is signified by an estimated frequency of nonsense and frameshift mutations in genetic disorders of ∼25% (Culbertson, 1999). After the phenomenon of NMD was first described in a patient with β-thalassaemia (Chang and Kan, 1979), the human β-globin gene has been central in both elucidating the mechanism (Maquat et al., 1981; Maquat, 1995; Thermann et al., 1998) and documenting the medical significance of NMD. A specific beneficial effect of NMD in the context of genetic diseases is exemplified by β-thalassaemia. The commonly occurring nonsense mutations in the 5′ part of the β-globin gene activate NMD and are associated with a recessive mode of inheritance, whereas the rarely occurring nonsense mutations in the 3′ part that escape NMD cause symptoms of thalassaemia in heterozygous carriers and a dominant mode of inheritance (Thein et al., 1990; Hall and Thein, 1994).
The mechanism that specifies termination codons as premature appears to vary in different classes of organisms. In yeast, the discrimination between the correct and prematurely occurring termination codons requires the presence of exonic cis-acting sequences that have been termed downstream sequence elements (DSEs) (Peltz et al., 1993). Some of the trans-acting factors have also been recognized in yeast, Caenorhabditis elegans and humans. They represent components of a translation-dependent complex that functions to identify and to degrade nonsense-mutated mRNAs (Peltz et al., 1993; Pulak and Anderson, 1993; Cui et al., 1995; Perlick et al., 1996; Applequist et al., 1997; Cali and Anderson, 1998; Czaplinski et al., 1998, 1999; Ruiz-Ecchevarria et al., 1998a,b; Cali et al., 1999; González et al., 2000). Although cis-acting DSE-like sequence elements have also been suggested to occur in mammalian genes (Cheng et al., 1994; Zhang et al., 1998a,b), mammalian NMD differs from that in yeast in that splicing clearly plays a crucial role in the former but not in the latter (Carter et al., 1996; Nagy and Maquat, 1998; Thermann et al., 1998). According to the post-termination surveillance model, splicing is thought to position a factor(s) at the exon–exon junction of the mRNA, which may represent a functional equivalent of the DSE-binding complex in yeast. Within an open reading frame (ORF), such a factor would be encountered by the elongating ribosome, which results in a stable mRNA. In mRNAs with a premature translation termination codon at a position upstream of a critical boundary in the penultimate exon, the factor(s) at the downstream exon–exon junction (Le Hir et al., 2000) would be recognized by the hypothetical surveillance complex that scans the mRNA after termination has occurred. As a result of this interaction, the mRNA will be degraded (Carter et al., 1996; Thermann et al., 1998; Zhang et al., 1998b; Hentze and Kulozik, 1999). In the alternative termination/mRNP context model, splicing also plays a critical role (Hilleren and Parker, 1999). According to this model, a termination event is defined as improper when it occurs outside of an RNP domain that has been recruited by a notional property of the process that defines the final exon. Subsequent to the discrimination between a premature and a correct termination event, the actual degradation pathways of wild-type and mutant mRNAs appear to differ. Poly(A) tail shortening is a central event in the turnover of mRNAs with termination codons at a proper position. Deadenylation occurs before decapping and subsequent 5′→3′ or 3′→5′ exonucleolytic degradation (Beelman and Parker, 1995). In contrast, nonsense-mutated yeast mRNAs have been shown to be decapped and degraded without prior deadenylation (Muhlrad and Parker, 1994), raising the possibility that the preservation of the poly(A) tail may play a pivotal role in NMD. Furthermore, the poly(A) tail enhances the efficiency of splicing and translation (Tarun and Sachs, 1996; Gray and Wickens, 1998; Preiss and Hentze, 1998; Cooke et al., 1999; Minvielle-Sebastia and Keller, 1999; Gray et al., 2000), both of which are known functional components of the NMD pathway. Therefore, one might suspect that the poly(A) tail may be required to direct nonsense-mutated transcripts to NMD.
Here, we have studied the importance of splicing and polyadenylation for NMD in the human β-globin model system.
Results
Splicing is indispensable for human β-globin NMD
There are multiple lines of evidence supporting an important role for splicing in mammalian NMD (Cheng et al., 1994; Carter et al., 1996; Thermann et al., 1998; Zhang et al., 1998a,b). Specifically, the insertion of an intron into the 3′-untranslated region (3′-UTR) of a human β-globin mRNA with a completely normal ORF redefines the physiological stop codon operationally as premature and subjects the mRNA to the NMD pathway (Thermann et al., 1998), thus establishing that the translational sense of a given ORF per se does not represent the relevant signal. Rather, under conditions of enabled translation, the termination event appears to be sufficient to activate NMD if an exon–exon junction exists at least 50 nucleotides downstream from the termination codon (Thermann et al., 1998; Zhang et al., 1998a). This interpretation is consistent with the observation that introns in the 3′-UTR are rare and, if they do occur, are mostly located close to the translation termination codon where they are expected not to induce NMD (Nagy and Maquat, 1998).
Investigations of NMD of the human triose phosphate isomerase gene (tpi) and a mouse–human β-globin hybrid gene led to the conclusion that cis-acting exonic sequences may activate a splicing-independent failsafe mechanism to activate NMD in the absence of 3′ splicing (Zhang et al., 1998a,b). We first tested a non-hybrid human β-globin mRNA for the existence of exonic failsafe sequences that could specify NMD for mutant mRNAs lacking an intron.
We generated intron-containing and intronless human β-globin genes with or without a nonsense codon at position 39 (NS 39) (Figure 1A). Following the transfection of these constructs together with an internal control (CAT) into HeLa cells, mRNA accumulation was assessed by northern blotting. Long (top panel) and short exposures (bottom panel) of the northern blot are shown (Figure 1B), because the signals generated by the intronless genes are much weaker than those generated by the intron-containing genes. The mRNA level of the intronless minigene with the normal ORF is reduced to ∼5% in comparison with the intron-containing gene (Figure 1B, top and bottom panel, compare lanes 1 and 3). Whereas the mRNA expressed from the intron-containing NS 39 gene is clearly subjected to NMD (Figure 1B, bottom panel, compare lanes 1 and 2), the mRNA expressed from the intronless NS 39 gene is immune to NMD (Figure 1B, top panel, compare lanes 3 and 4). Since cytoplasmic translation is necessary for NMD of the NS 39 mRNA (Thermann et al., 1998), the immunity to NMD of the intronless NS 39 mRNA may result from the lack of translatability of the intronless mRNA. As shown in the western blot in Figure 1C, the mRNA expressed from the intronless cWT construct is translated and the amount of synthesized β-globin protein correlates with the mRNA expression levels (Figure 1B and C, lanes 1 and 3). This result demonstrates that the intronless β-globin mRNA is translated and that the NMD immunity of the intronless β-globin gene is not an indirect consequence of the lack of translation.


Fig. 1. mRNAs from intronless β-globin genes escape NMD. (A) Human β-globin gene constructs used for transfection. The ORF is represented by boxes; introns and the UTRs by lines. AUG, initiation codon; NS 39, nonsense mutation at codon 39; Ter, termination codon. (B) Northern blot of cytoplasmic HeLa cell RNA and hybridization with β-globin- and CAT-specific cRNA probes. The percentage values refer to the mean of the phosphoimaging data of two independent experiments after normalization for transfection efficiency. For clarity, the top panel shows a 6 h and the bottom panel a 2 h exposure. (C) β-globin immunoblot of lysates of transfected HeLa cells (lanes 1–4). Three times more of the intronless (lanes 3 and 4) than the genomic β-globin genes (lanes 1 and 2) were transfected, because the intronless genes were expressed at low levels.
To control for the possibility that the lack of NMD of the intronless construct could be a function of its low level of mRNA expression, we inserted a spliceable MINX intron into the 3′-UTR of the intronless cWT gene (Figure 2A) at a position that had previously been documented to activate NMD in a genomic gene (Thermann et al., 1998). The expression of the constructs with the normal or the NS 39-mutated ORF without an intron in the 3′-UTR (Figure 2A, constructs cWT-UTR-1 and cNS 39-UTR-1) is low and does not display an NMD phenotype (Figure 2B, lanes 1 and 2), confirming the data shown in Figure 1. When a spliceable intron is too close to the termination codon to trigger NMD (construct cWT-UTR-4), its insertion per se does not increase the expression of the encoded mRNA compared with the intronless wild-type construct (compare lane 5 with lane 1). In contrast, the insertion of a spliceable MINX intron into the 3′-UTR of cWT at a distance of 62 nucleotides downstream from the termination codon (construct cWT-UTR-2, lane 3) resulted in the redefinition of the physiological termination codon as premature (Thermann et al., 1998) and reduced the expression level of this mRNA to 25%. As expected, mRNAs expressed from the control constructs with a spliceable MINX intron and a Ter→Gln mutation (construct cWT-UTR-3, lane 4) or a non-spliceable MINX intron in the 3′-UTR (construct cWT-UTR-5, lane 6) are expressed at high levels and are not subjected to NMD that typically down-regulates mRNA expression levels ∼4- to 5-fold.
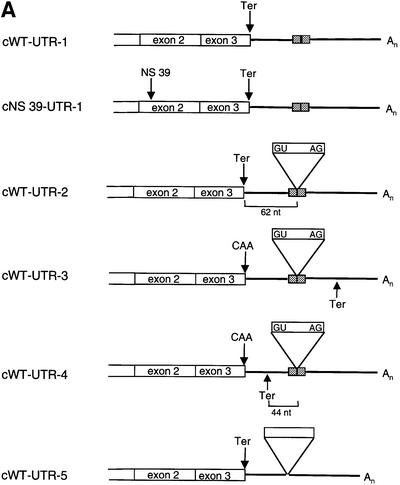
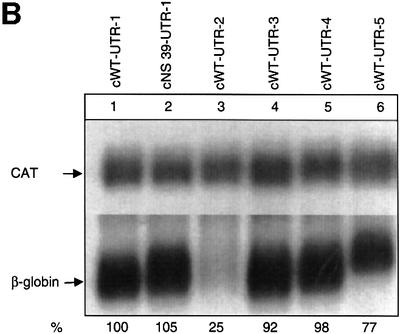
Fig. 2. The reduced expression of intronless β-globin genes does not result in NMD incompetence. (A) Human β-globin gene constructs with modified 3′-UTRs. (B) Northern blot of cytoplasmic HeLa cell RNA and hybridization with β-globin- and CAT-specific cRNA probes. The percentage values refer to the mean of the phosphoimaging data of three independent experiments after normalization for transfection efficiency.
These data establish that the NMD of NS 39-mutated human β-globin RNA is strictly dependent on splicing. Furthermore, these findings provide direct evidence against the existence of a splicing-independent mechanism for the degradation of this mRNA, as previously suggested for human tpi and a hybrid mouse–human β-globin gene (Zhang et al., 1998a,b). It is difficult to account for these seemingly controversial results, but it is perfectly conceivable that cis-acting sequences that are capable of activating NMD in a splicing-independent manner may be present in some mammalian genes and not in others.
The spatial requirements of human β-globin NMD are less constrained than in yeast
The identification of exon–exon junctions downstream from a termination codon has been suggested to be mediated by a so-called post-termination surveillance complex. In yeast, the assembly of such a complex involves interactions between the translation termination factors and the proteins Upf1, Upf2 and Upf3 (Czaplinski et al., 1998). The yeast surveillance complex is thought to examine the region 3′ of translation termination codons for the presence of a DSE, which may represent a functional analogue of the mammalian exon–exon junction. In yeast, the maximal distance between the translation termination codon and the DSE appears to be ∼150–200 nucleotides (Ruiz-Echevarria et al., 1998a), and the function of the surveillance complex hence appears to be spatially constrained. In human tpi, a nonsense mutation that is located 416 nucleotides 5′ of the only 3′ splice junction was reported to induce NMD, whereas a nonsense mutation 559 nucleotides upstream of the 3′-most splice junction did not induce NMD (Zhang et al., 1998b; Sun and Maquat, 2000). This observation suggested that human NMD may also be constrained in a similar albeit more relaxed way.
To study the effect of the distance between a nonsense codon and the 3′ splicing event in human β-globin mRNA systematically, the genomic distance between the NS 39 mutation and the 3′ exon–exon junction was increased from the normal 180 nucleotides to up to 654 nucleotides by in-frame insertions of homologous β-globin sequences of 102, 204, 300 and 474 nucleotides between the NS 39 mutation and the intron (Figure 3A). These fragments were chosen because they have been shown directly in the experiments described above not to contain any notional cis-acting failsafe sequences that might be capable of activating NMD in the absence of splicing (Figures 1 and 2). The analysis of RNA accumulation in cells transfected with these constructs demonstrates that even the nonsense-mutated mRNA with the longest distance of 654 nucleotides between the termination event and the 3′ splice junction is NMD competent (Figure 3B). Similar results were obtained with human β-globin gene constructs without introns in the ORF but with a spliceable MINX intron in their natural 3′-UTR (data not shown). In these constructs, the natural termination codon was mutated and the distance between the NS 39 mutation and the intron was extended from 395 to 695 nucleotides. Even the construct with the longest distance exhibited NMD competence (not shown). These data demonstrate that human β-globin NMD is capable of recognizing the necessary exon–exon junction at much longer distances than its yeast counterpart, as has been reported previously for the human tpi mRNA. The spatial requirements of human β-globin NMD are therefore less constrained than those observed in yeast and appear different from those of human tpi. These results also show that, in principle, the human NMD machinery can accommodate distances between stop codons and exon–exon junctions of at least 695 nucleotides. This long distance surveillance ability may result from a more progressive linear scanning activity of the mammalian surveillance complex than in yeast or, alternatively, may be a reflection of a non-linear interaction of the functional components of the mammalian NMD pathway as the termination/mRNP context model would predict (Hilleren and Parker, 1999).
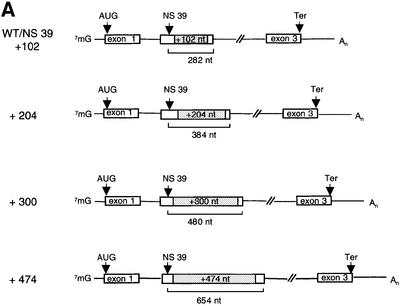
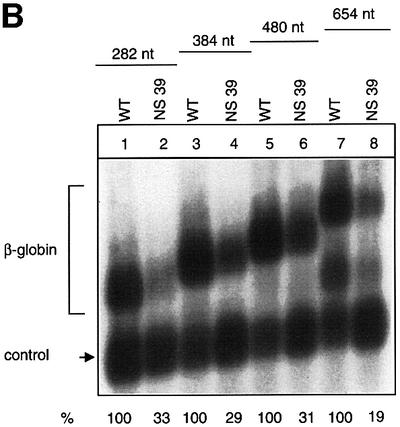
Fig. 3. Recognition of the exon–exon junction can occur at long distances between the translation termination codon and a 3′ intron. (A) Human β-globin gene constructs with insertions of homologous β-globin sequences of various lengths (shaded). (B) Northern blot of cytoplasmic HeLa cell RNA and hybridization with β-globin- and CAT-specific cRNA probes. The additional fragment detected in lanes 7 and 8 probably represents atypically spliced mRNAs. Nucleotide (nt) numbers refer to the distance between the nonsense codon and the 3′ exon–exon junction. The percentage values refer to the mean of the phosphoimaging data of three independent experiments after normalization to transfection efficiency.
Human NMD is poly(A) independent
The poly(A) tail enhances the efficiency of splicing and translation (Tarun and Sachs, 1996; Gray and Wickens, 1998; Preiss and Hentze, 1998; Cooke et al., 1999; Minvielle-Sebastia and Keller, 1999; Gray et al., 2000), both of which are important for NMD. It might therefore be expected that the poly(A) tail represents an important cis-acting element within NMD-competent mRNPs. We directly tested this hypothesis in an experimental system that was designed to analyse the expression of non-polyadenylated nonsense-mutated human β-globin genes with a histone 3′-UTR. Histone mRNAs do not carry a poly(A) tail. Instead, histone mRNAs terminate in a stem–loop structure that is bound by a specific protein termed stem–loop-binding protein (SLBP) (Wang et al., 1996) or histone hairpin-binding protein (HBP) (Martin et al., 1997; Müller and Schümperli, 1997).
We first designed an experimental system that enabled an analysis of the NMD competence of transcripts lacking a poly(A) tail. The constructs (Figure 4) contain a genomic human β-globin gene with or without NS 39. The β-globin 3′-UTR was replaced by the 3′-UTR of the histone H1.3 gene that contains the histone 3′ end processing signals. This stabilizes the mRNA, and supports nucleo-cytoplasmic transport and translation in a poly(A) tail-independent manner (Müller and Schümperli, 1997). Two additional modifications enable the identification of NMD-specific events. First, an iron-responsive element (IRE) or its non-functional derivative (IREΔC) in the 5′-UTR allowed the effect of cytoplasmic translation on the accumulation of non-polyadenylated human β-globin mRNA to be analysed specifically (Hentze et al., 1987; Thermann et al., 1998). Secondly, the introns of the genomic human β-globin gene were removed to study the influence of splicing on NMD.
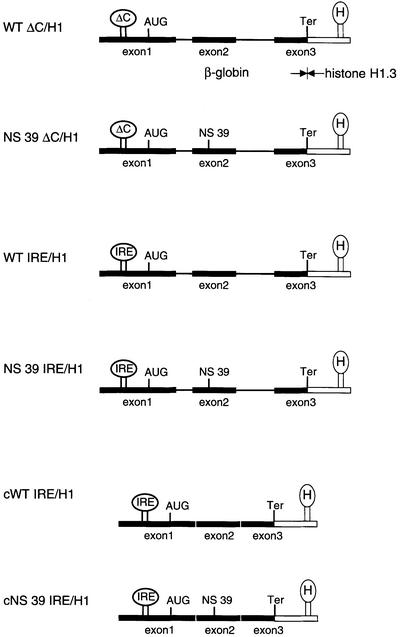
Fig. 4. Hybrid human β-globin constructs with modified 5′- and 3′-UTRs. All constructs contain the genomic human β-globin gene or a corresponding intronless ORF from the transcription start site to the XhoI site introduced immediately upstream of the physiological termination codon. The termination codon (Ter) and the 3′-UTR were derived from the human H1.3 gene (boxed) containing the histone stem–loop and 918 nucleotides downstream to include all sequences essential for histone 3′ end processing. IRE, iron-responsive element; H1, histone H1.3; ΔC, IREΔC; H, histone stem–loop.
Non-polyadenylated RNAs with a histone 3′ end tend not to be spliced efficiently and spliced mRNAs tend to be polyadenylated even at cryptic poly(A) sites (Pandey et al., 1990). Therefore, it was crucial for this analysis to establish directly that the hybrid mRNAs are processed correctly. Poly(A)+/poly(A)– fractionation of cytoplasmic RNA obtained from cells transfected with the hybrid gene constructs and a human β-globin construct with its natural 3′-UTR shows that the unfractionated cytoplasmic RNA contains both the globin control and the hybrid mRNAs (Figure 5A, lane 1). In contrast, the poly(A)+ and the poly(A)– fractions contain almost exclusively the control or the hybrid mRNA, respectively (Figure 5A, lanes 2 and 3). Moreover, the probe was designed to assess the correct length of the mRNA with a histone 3′ end, because vertebrate histone 3′ end processing entails cleavage downstream of the sequence ACCCA that is directly juxtaposed to the histone hairpin. In the RNase protection assay with a probe spanning parts of the final intron, the entire final exon and parts of 3′-flanking sequences, the spliced and correctly 3′-end-processed hybrid and control mRNAs protect fragments of 184 and 125 nucleotides, respectively. The lengths of the fragments protected by this probe enable the mapping of the 3′ ends of the non-polyadenylated hybrid mRNA and the polyadenylated control mRNA at single nucleotide resolution, which confirms the expected respective positions of the 3′ ends (Figure 5B). The correct removal of both introns of the hybrid RNA is confirmed in an RNase protection assay with a probe spanning the entire mRNA. The correctly spliced hybrid and control mRNAs protect fragments of 497 and 438 nucleotides, respectively. There is only a trace of 313 and 610 nucleotide mismatch bands that have probably arisen from partial splicing of intron 2 and atypical 3′ end processing, respectively (Figure 5C). These findings establish that both introns are spliced correctly and that the hybrid mRNA is not polyadenylated.
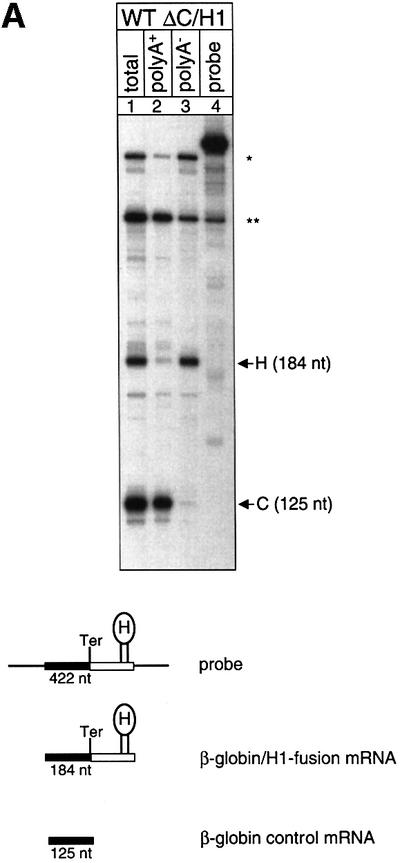
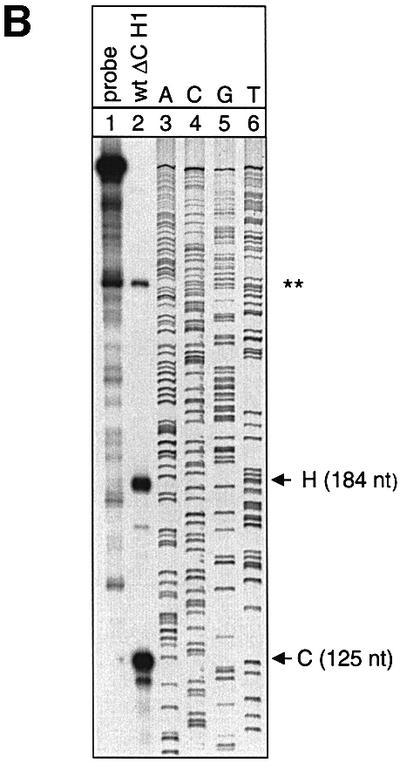
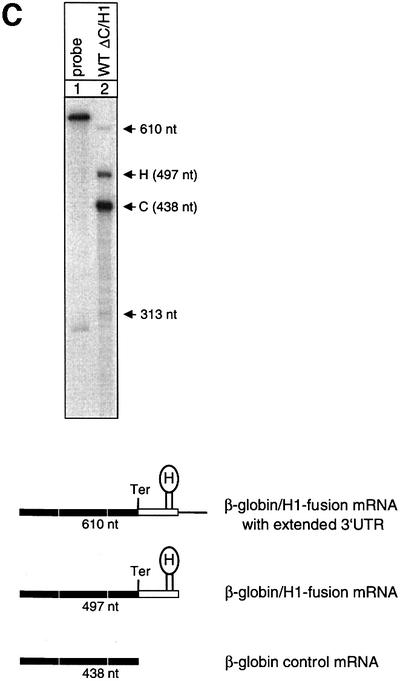
Fig. 5. Hybrid human β-globin–histone H1.3 mRNAs are spliced and 3′ end processed correctly. (A) RNase protection analysis of HeLa cells that were co-transfected with construct WT ΔC/H1 and the β-globin control plasmid. The total cytoplasmic RNA (lane 1) was separated into poly(A)+ (lane 2) and poly(A)– (lane 3) fractions. Lane 4: undigested probe. The fragments protected by either the correctly spliced and 3′-end-processed β-globin–histone hybrid mRNA (H) or the β-globin control mRNA (C) are shown schematically at the bottom. The 422 nucleotide probe encompasses the 3′ 120 nucleotides of human β-globin intron 2, the entire exon 3 of the hybrid constructs and 113 nucleotides of histone H1.3 downstream sequences. Correctly spliced and 3′-end-processed hybrid mRNAs are identified as a protected 184 nucleotide fragment. The mRNA of the co-transfected human β-globin control construct protects a fragment of 125 nucleotides. One of the large protected fragments corresponds in size to the plasmid DNA (*). The other large fragment is also present in the lane loaded with the probe alone and is therefore not derived from the cellular RNA (**). Both of these fragments occur with variable intensities in different experiments. (B) The length of the protected fragments of the β-globin–histone H1.3 hybrid mRNA and the β-globin control mRNA was confirmed (lane 2) at the resolution of a DNA sequence ladder (lanes 3–6). Lane 1: undigested probe. (C) RNase protection analysis of HeLa cells that were co-transfected with construct WT ΔC/H1 and the β-globin control plasmid. The probe used in this experiment encompasses the β-globin–hybrid construct from the ATG in exon 1 to the histone 3′ end processing site and 113 nucleotides of histone H1.3 downstream sequences, and enables the determination of correct splicing of both introns of the β-globin–histone hybrid construct. The fragments protected by either the correctly spliced and 3′-end-processed β-globin–histone hybrid mRNA (H) or the β-globin control mRNA (C) are shown schematically at the bottom. Correctly spliced and 3′-end-processed hybrid mRNAs are identified as a protected 497 nucleotide fragment. The mRNA of the co-transfected human β-globin control construct protects a fragment of 438 nucleotides. The 610 nucleotide fragment corresponds in size to atypically 3′-end-processed hybrid mRNAs. The 313 nucleotide fragment represents mRNAs that contain an abnormally processed or unprocessed intron 2.
Next we analysed whether nonsense-mutated mRNAs with a histone 3′ end are subjected to NMD. The comparison of the wild-type and NS 39 mRNAs (WT ΔC/H1 versus NS 39 ΔC/H1; Figure 4) shows that the nonsense-mutated hybrid gene is expressed at a level of 18% of that of the wild type (Figure 6, compare lanes 2 and 3). Translation dependence represents one of the key features of bona fide NMD. Therefore, we next examined the specific influence of cytoplasmic translation on the accumulation of NS 39 hybrid mRNA. The IRE–iron regulatory protein (IRP) interaction has previously been shown by ourselves and others to represent a robust experimental system to control translation specifically in an iron-dependent manner and to assess the role of translation in NMD (Hentze et al., 1987; Gray et al., 1993; Rouault and Klausner, 1997; Thermann et al., 1998).

Fig. 6. Degradation of nonsense-mutated mRNAs with a histone 3′ end is translation dependent. RNase protection analysis of HeLa cells that were co-transfected with constructs WT ΔC/H1 (lanes 2, 4 and 6), NS 39 ΔC/H1 (lanes 3, 5 and 7), WT IRE/H1 (lanes 8 and 10) or NS 39 IRE/H1 (lanes 9 and 11) and the genomic β-globin control plasmid. Translation of the IRE-containing transcripts was either blocked or enabled specifically by iron depletion with deferoxamine (lanes 8 and 9) or iron repletion with haem-arginate (lanes 10 and 11), respectively. Treatment of cells transfected with the non-functional IREΔC constructs with either deferoxamine (lanes 6 and 7) or haem-arginate (lanes 4 and 5) had no influence on the mRNA expression level of the hybrid mRNAs with respect to untreated cells (lanes 2 and 3). The relative mRNA levels of wild-type and NS 39 mRNAs were measured by phosphoimaging and normalized for transfection efficiency. The protected fragments (H, C and **) are indicated as in Figure 5. Lane 1, undigested probe; lane 12, β-globin control mRNA; lane 13, untransfected control.
The cells were transfected with the hybrid genes containing a functional IRE and were either iron depleted with deferoxamine (DFO) or iron repleted with haem-arginate, thus specifically disabling or enabling translation of the mRNAs. Under conditions of enabled translation, the NS 39 mRNA is expressed at a level of 26% of wild type (Figure 6, compare lanes 10 and 11), which confirms the results shown in lanes 2 and 3. In contrast, when translation was disabled by DFO treatment, the NS 39 and the wild-type mRNAs display similar mRNA levels (Figure 6, compare lanes 8 and 9). Hybrid mRNAs with a non-functional IRE in the 5′-UTR display an NMD phenotype regardless of the iron status of the cells (Figure 6, compare lanes 4–7). These findings demonstrate that the reduction in NS 39-mutated hybrid mRNA accumulation requires cytoplasmic translation.
We also analysed the expression of intronless hybrid β-globin gene constructs under conditions of enabled or disabled translation. Under both conditions, an NS 39 mRNA with a histone 3′ end but without introns displays similar expression levels to those of the wild-type transcript (Figure 7A, compare lanes 4 and 5 and lanes 6 and 7), whereas the respective intron-containing constructs display the NMD phenotype (Figure 7A, lanes 2 and 3; Figure 6, lanes 2–7, 10 and 11). Furthermore, poly(A)+/poly(A)– fractionation analysis demonstrates that the correct histone type 3′ end processing without polyadenylation is maintained in the intronless pre-mRNA species (Figure 7B). These findings show that the down-regulation of nonsense mRNAs bearing a histone 3′ end is also splicing dependent.
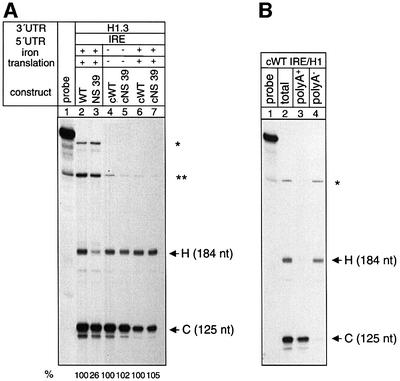
Fig. 7. Degradation of nonsense-mutated mRNAs with a histone 3′ end is splicing dependent. (A) RNase protection analysis of HeLa cells that were co-transfected with the intron-containing constructs WT IRE/H1 (lane 2) and NS 39 IRE/H1 (lane 3) or with the intronless constructs cWT IRE/H1 (lanes 4 and 6) and cNS 39 IRE/H1 (lanes 5 and 7) and the genomic β-globin control plasmid. Lane 1: undigested probe. Translation was either blocked or enabled specifically by iron depletion with deferoxamine (lanes 4 and 5) or iron repletion with haem-arginate, respectively (lanes 2, 3, 6 and 7). The relative mRNA levels of wild-type and NS 39 mRNAs were measured by phosphoimaging and normalized for transfection efficiency. The protected fragments (H, C, * and **) are indicated as in Figure 5. (B) RNase protection analysis of HeLa cells that were co-transfected with construct cWT IRE/H1 and the β-globin control plasmid. The total cytoplasmic RNA (lane 2) was separated into poly(A)+ (lane 3) and poly(A)– (lane 4) fractions. Lane 1: undigested probe. The protected fragments (H and C) are indicated as in Figure 5. The fragment protected by the intronless plasmid DNA (*) is shorter than that protected by the intron-containing DNA (see Figure 5A).
Discussion
NMD depends on the recognition of the improper position of a translation termination codon. In yeast, the relative position of the termination codon with respect to cis-acting DSEs plays a critical role (Peltz et al., 1993). The DSEs bind specific proteins that are thought to interact either with a hypothetical post-termination complex or with the elongating ribosome to induce or not induce mRNA degradation by NMD, respectively (Zhang et al., 1997; Ruiz-Echevarria et al., 1998a,b; Mangus and Jacobson, 1999; González et al., 2000). In mammalian NMD, splicing at a position of more than ∼50 nucleotides 3′ of a termination codon is able to specify NMD, even if the translational sense of the ORF is completely normal (Carter et al., 1996; Thermann et al., 1998). It is very possible that the link between splicing and NMD is represented by one or more of the numerous proteins that have recently been shown to be recruited to the exon–exon junctions of mRNAs in the course of splicing and that are thought to facilitate transport of the mRNA into the cytoplasm (Kataoka et al., 2000; Le Hir et al., 2000; McGarvey et al., 2000; Zhou et al., 2000). The position of these proteins within the mRNP particle may also serve as a reference point for the elongating ribosome or a hypothetical mammalian post-termination surveillance complex to distinguish a proper from an improper translation termination codon. Our finding that an unspliced human β-globin mRNA that is not expected to bind any of these proteins is poorly expressed and escapes NMD (Figure 1) meets the predictions of this model.
The necessity for splicing and the apparent lack of cis-acting failsafe sequences in human β-globin NMD raises the question of similarities and differences between NMD-competent RNPs in mammals and yeast. In yeast, NMD does not occur when the distance between the nonsense mutation and the DSE exceeds 100–150 nucleotides (Ruiz-Echevarria et al., 1998a; Czaplinski et al., 1999). In human β-globin mRNA, the distance between the nonsense mutation and the 3′ exon–exon junction can be extended to at least 695 nucleotides. This indicates a less constrained spatial requirement of the interaction between the mammalian mRNP and the surveillance complex than in yeast. This difference could be explained either by a more processive nature of the mammalian compared with the yeast surveillance complex, or by a non-linear mechanism that may be predicted by the RNP/context model (Hilleren and Parker, 1999). The RNP/context model predicts that alterations of the 3′ end would activate the NMD machinery (Hilleren and Parker, 1999; Sun and Maquat, 2000), as for example an extension of the 3′-UTR beyond the generally short length of ∼100 nucleotides in yeast (Muhlrad and Parker, 1999). It is therefore interesting that mammalian mRNAs with a histone 3′ end and a normal ORF are not subjected to NMD. In contrast, nonsense-mutated transcripts with a histone 3′ end are down-regulated. This shows directly that the definition of an NMD-competent mRNP is independent of the presence of a poly(A) tail, suggesting that the 3′ end structure of the mRNP does not play a dominant role in mammalian NMD.
Materials and methods
Constructs
The β-globin constructs with the wild-type ORF or with the nonsense codon at position 39 (NS 39) contain a genomic 1423 bp β-globin gene fragment extending from the physiological translation initiation codon to the translation termination codon, which was inserted into the pCIneo vector (Promega) at the XhoI–XbaI sites of the polylinker. The wild-type and NS 39 gene sequences were derived from a healthy proband and from a patient with homozygous β-thalassaemia, respectively. Constructs cWT and cNS 39 were derived from RT–PCRs of cytoplasmic RNA from HeLa cells transfected with wild-type and NS 39 constructs and were inserted into the same position of the pCIneo vector.
The cWT-UTR and the cNS 39-UTR series of constructs was generated by replacement of a fragment spanning from the EcoRI site in the nominal exon 3 to an XhoI site that was inserted by in vitro mutagenesis immediately 3′ of the termination codon by sequences extending from the same EcoRI site to the XhoI site immediately 3′ of the MINX intron- (Zillmann et al., 1988) flanking sequences of construct WT-spf, which was described previously (Thermann et al., 1998). The cWT-UTR-1/cNS 39-UTR-1 constructs contain the MINX-intron flanking sequences only. The cWT-UTR-2 construct contains a spliceable MINX intron together with its flanking sequences at an NMD-competent position 62 nucleotides 3′ of the translation termination codon. The construct cWT-UTR-3 differs from cWT-UTR-2 in that Ter was mutagenized to a CAA codon. The cWT-UTR-4 construct was generated by introducing a novel termination codon into the cWT-UTR-3 construct at an NMD-incompetent position 44 nucleotides 5′ of the inserted intron. Construct cWT-UTR-5 differs from cWT-UTR-2 by the absence of the MINX-flanking sequences including the splice donor and acceptor sites. Construct cNS 39-UTR-3 was created by site-directed mutagenesis of cWT-UTR-3.
For the in-frame introduction of exonic sequences into the WT/NS 39 constructs, SacII and ApaI restriction sites were inserted between 21 and 24 nucleotides 5′ of the exon 2 end by site-directed mutagenesis. Homologous human β-globin fragments starting from codon 1 and extending 102, 204 and 300 nucleotides into the coding sequence, respectively, were generated by PCR. These fragments were also inserted into the EcoRI site in the nominal exon 3 of cWT-UTR-3 or cNS 39-UTR-3 for a second series of constructs (data not shown). The 474 nucleotide insertion was created by PCR of construct cWT-UTR-3 and extends from codon 1 to a position 33 nucleotides 3′ of the CAA that replaces Ter in this construct.
The constructs WT IRE/H1 and NS 39 IRE/H1 were generated by the in-frame insertion of an XhoI site directly upstream of the physiological termination codon of the β-globin gene into the plasmids WT IRE and PTC 39-IRE (Thermann et al., 1998). The complete β-globin 3′-UTR including the canonical and a cryptic AUUAAA poly(A) site, and 1.8 kb of 3′-flanking sequences including the U-rich element were subsequently replaced by inserting a fragment from the human histone H1.3 gene containing the termination codon, the 3′-UTR and 918 downstream nucleotides including the histone-specific purine-rich spacer element for 3′ end processing (Müller and Schümperli, 1997) into the novel XhoI site and the XbaI site in the β-globin 3′-flanking region 1.7 kb 3′ of the poly(A) site. This histone 3′-UTR does not contain any canonical or obvious cryptic poly(A) sites. The histone H1.3 fragment was PCR amplified from human genomic DNA (primer sequences: 5′-TTTTCTCGAGTGAAACTGGCGGGACGTTCCCCTTTG-3′ and 5′-TTTTTCTAGAGAGCCCCTGGGAAAATAAGTCC-3′). Constructs WT ΔC/H1 and NS 39 ΔC/H1 differ from the IRE constructs by a deletion of a single cytosine residue, which functionally inactivates the IRE (Hentze et al., 1987). In the constructs cWT IRE/H1 and cNS 39 IRE/H1, the intron-containing genomic β-globin gene fragment was replaced by an intronless β-globin ORF. All site-directed mutagenesis procedures were performed using standard protocols (Hagemeier, 1996). The identity of all constructs was confirmed by DNA sequence analysis.
Cell culture and transfections
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) under standard conditions. For the experiments described in Figures 2 and 3, cells were transiently transfected by calcium phosphate precipitation (Ausubel et al., 1994) with 20 µg of the test constructs and 15 µg of an Escherichia coli CAT gene cloned into the multiple cloning site of the plasmid pSG5 (Green et al., 1988), which served as a control for transfection efficiency. For the northern blot analyses in Figure 1, cells were transfected with 25 µg of the test and the control constructs. For the analysis of protein expression, 8 µg of the wild-type and NS 39 constructs and 24 µg of the cWT and cNS 39 β-globin variants were transfected to compensate for the low expression of genes without an intron in their ORF. Cells were washed after 20 h and harvested 24 h later.
For the experiments with the hybrid human β-globin–histone genes, 35 µg of the test constructs were co-transfected with 0.75 µg of a wild-type β-globin gene (Thermann et al., 1998), which served as a control for transfection efficiency. Cells were washed 16 h after transfection and harvested 26 h later.
The translation of the IRE-containing transcripts was regulated specifically by supplementing the culture media with either 100 µM iron source haem-arginate (Leiras, Turku, Finland) 20 h after washing (iron repletion; enabled translation), or 100 µM iron chelator DFO 6 h after washing (iron depletion; disabled translation).
RNA analysis
Total cytoplasmic RNA was isolated and northern blotting was performed by methods described previously (Thermann et al., 1998). RNase protection assays were performed according to standard protocols. The 3′ probe for the globin–histone hybrid mRNA was prepared by in vitro transcription of a plasmid containing exon 3 of the construct WT IRE/H1 and 120 nucleotides upstream (intron 2) and 113 nucleotides downstream from the histone 3′ end processing site. Therefore, the RNase protection analysis assays for both correct histone 3′ end formation and splicing of the last intron of the hybrid constructs. Correctly spliced and 3′-end-processed mRNA is expected to protect a fragment of 184 nucleotides. The probe also detects wild-type β-globin that was co-transfected as a control. This results in a protected fragment of 125 nucleotides. The probe used in Figure 5C was prepared by in vitro transcription of a plasmid containing the hybrid β-globin–histone H1.3 cDNA from the ATG in exon 1 to the histone 3′ processing site, and 113 nucleotides of histone H1.3 downstream sequences. Therefore, this probe assays for both correct histone 3′ end formation and splicing of both introns of the hybrid construct. Correctly spliced and 3′-end-processed hybrid mRNA is expected to protect a fragment of 497 nucleotides. The mRNA of the co-transfected human β-globin construct protects a fragment of 438 nucleotides. Abnormal splicing of introns 1 or 2 will result in fragments of 403 or 313 nucleotides, respectively.
Total cytoplasmic RNA was separated into poly(A)+ and poly(A)– fractions by using the poly(A) spin column kit (New England Biolabs). RNA from the unbound poly(A)– fraction and from the final poly(A)+ eluate was recovered and analysed by RNase protection.
The signals were quantitated by imaging in a GS-250 Molecular imager (Bio-Rad).
Protein analysis
Immunoblotting and immunostaining of blots with a β-globin-specific antibody were performed as previously described (Thermann et al., 1998).
Acknowledgments
Acknowledgements
This study was supported financially by the Deutsche Forschungsgemeinschaft and the Fritz Thyssen Stiftung.
References
- Applequist S.E., Selg,M., Raman,C. and Jäck,H.-M. (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res., 25, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.I., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY.
- Beelman C.A. and Parker,R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179–183. [DOI] [PubMed] [Google Scholar]
- Cali B.M. and Anderson,P. (1998) mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet., 260, 176–184. [DOI] [PubMed] [Google Scholar]
- Cali B.M., Kuchma,S.L., Latham,J. and Anderson,P. (1999) smg-7 is required for mRNA surveillance in Caenorabditis elegans. Genetics, 151, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.S., Li,S. and Wilkinson,M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Chang J.C. and Kan,Y.W. (1979) β0-Thalassemia, a nonsense mutation in man. Proc. Natl Acad. Sci. USA, 76, 2886–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng I., Belgrader,P., Zhou,X. and Maquat,L. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C., Hans,H. and Alwine,J.C. (1999) Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol., 19, 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Hagan,K.W., Zhang,S. and Peltz,S.W. (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev., 9, 423–436. [DOI] [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance—unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., González,C.I. and Peltz,S. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- González C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- Gray N.K. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- Gray N.K., Quick,S., Goossen,B., Constable,A., Hirling,H., Kühn,L.C. and Hentze,M.W. (1993) Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. Eur. J. Biochem., 218, 657–667. [DOI] [PubMed] [Google Scholar]
- Gray N.K., Coller,J.M., Dickson,K.S. and Wickens,M. (2000) Multiple portions of the poly(A)-binding protein stimulate translation in vitro. EMBO J., 19, 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Issemann,I. and Sheer,S. (1988) A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res., 16, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier C. (1996) Site-directed mutagenesis using a uracil-containing phagemid template. Methods Mol. Biol., 57, 45–54. [DOI] [PubMed] [Google Scholar]
- Hall G.W. and Thein,S. (1994) Nonsense codon mutations in the terminal exon of the β-globin gene are not associated with a reduction in β-mRNA accumulation: a mechanism for the phenotype of dominant β-thalassemia. Blood, 83, 2013–2017. [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hentze M.W., Rouault,T.A., Caughman,S.W., Dancis,A., Harford,J.B. and Klausner,R.D. (1987) A cis-acting element is necessary and sufficient for translational regulation of human ferritin expression in response to iron. Proc. Natl Acad. Sci. USA, 84, 6730–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Yong,J., Kim,V.N., Velazquez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.J. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable associations of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A. and Jacobson,A. (1999) Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods, 17, 28–37. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. (1995) When cells stop making sense: effect of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E., Kinniburgh,A.J., Rachmilewitz,E.A. and Ross,J. (1981) Unstable β-globin mRNA in mRNA-deficient β0 thalassemia. Cell, 27, 543–553. [DOI] [PubMed] [Google Scholar]
- Martin F., Schaller,A., Eglite,S., Schümperli,D. and Müller,B. (1997) The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J., 16, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T. et al. (2000) The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon–product complexes. J. Cell Biol., 150, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L. and Keller,W. (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1999) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA, 5, 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B. and Schümperli,D. (1997) The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol., 8, 567–576. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Pandey N.B., Chodchoy,N., Liu,T.J. and Marzluff,W.F. (1990) Introns in histone genes alter the distribution of 3′ ends. Nucleic Acids Res., 18, 3161–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S.W., Brown,A.H. and Jacobson,A. (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- Perlick H.A., Medghalchi,S.M., Spencer,F.A., Kendzior,R.J. and Dietz,H.C. (1996) Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc. Natl Acad. Sci. USA, 93, 10928–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T. and Hentze,M.W. (1998) Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature, 392, 516–520. [DOI] [PubMed] [Google Scholar]
- Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- Rouault T. and Klausner,R. (1997) Regulation of iron metabolism in eukaryotes. Curr. Top. Cell Regul., 35, 1–19. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Gonzalez,C.I. and Peltz,S.W. (1998a) Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J., 17, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ecchevarria M.J., Yasenchak,J.M., Han,X., Dinman,J.D. and Peltz,S.W. (1998b) The upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc. Natl Acad. Sci. USA, 95, 8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. and Maquat,L.E. (2000) mRNA surveillance in mammalian cells: the relationship between introns and translation termination. RNA, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1996) Association of yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thein S.L., Hesketh,C., Taylor,P., Temperley,I.J., Hutchinson,R.M., Old,J.M., Wood,W.G., Clegg,J.B. and Weatherall,D.J. (1990) Molecular basis for dominantly inherited inclusion body β-thalassaemia. Proc. Natl Acad. Sci. USA, 87, 3924–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.F., Whitfield,M.L., Ingledue,T.C., Dominski,Z. and Marzluff,W.F. (1996) The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required fo histone pre-mRNA processing. Genes Dev., 10, 3028–3040. [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun,X. and Maquat,L. (1998a) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X. and Maquat,L.E. (1998b) At least one intron is required for nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear pre-mRNA splicing and cytoplasmic mRNA translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Welch,E.M., Hagan,K., Brown,A.H., Peltz,S.W. and Jacobson,A. (1997) Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA, 3, 234–244. [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.-J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger mRNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Zapp,M.L. and Berget,S.M. (1988) Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol., 8, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]


