Abstract
Nucleotide-microarray technology, which allows the simultaneous measurement of the expression of tens of thousands of genes, has become an important tool in the study of disease. In disorders such as malignancy, gene expression often undergoes broad changes of sizable magnitude, whereas in many common multifactorial diseases, such as diabetes, obesity, and atherosclerosis, the changes in gene expression are modest. In the latter circumstance, it is therefore challenging to distinguish the truly changing from nonchanging genes, especially because statistical significance must be considered in the context of multiple hypothesis testing. Here, we present a balanced probability analysis (BPA), which provides the biologist with an approach to interpret results in the context of the total number of genes truly differentially expressed and false discovery and false negative rates for the list of genes reaching any significance threshold. In situations where the changes are of modest magnitude, sole consideration of the false discovery rate can result in poor power to detect genes truly differentially expressed. Concomitant analysis of the rate of truly differentially expressed genes not identified, i.e., the false negative rate, allows balancing of the two error rates and a more thorough insight into the data. To this end, we have developed a unique, model-based procedure for the estimation of false negative rates, which allows application of BPA to real data in which changes are modest.
Keywords: metabolic disease, microarray analysis, multiple hypothesis testing, statistics
Changes in gene expression may occur as part of the primary pathogenesis of a disease or secondary to other factors involved in disease progression. Identifying the changes in gene expression in common, multifactorial diseases, such as obesity, type 2 diabetes, hypertension, dyslipidemia, and the metabolic syndrome is particularly challenging, because the changes are often of small magnitude and involve multiple pathways and multiple tissues (1–3). Nonetheless, existing work using nucleotide-microarray technology, which allows the simultaneous measurement of the expression of tens of thousands of genes, suggests that modest changes in gene expression in metabolic disease can be reproducible and of fundamental importance in unraveling new aspects of the pathogenesis of disease (4, 5).
Many of the existing statistical methods for microarray analysis have been fashioned by using data sets from diseases such as cancer, where the changes in gene expression are abundant, with many genes having a high magnitude of change that far exceeds the observed variability in expression (6). Under these conditions, existing algorithms are able to detect numerous genes that survive statistical correction for the number of hypotheses tested. However, for many physiological and metabolic conditions, the changes in gene expression are often moderate compared to the variability in expression leading to modest p values, such that existing statistical models often miss most of the real changes (4, 5).
We therefore sought to develop an analytical approach that provides both the biologist and the bioinformatician with a more thorough understanding of the statistical significance of any list of genes produced from conditions where the real changes are of modest magnitude. Through the creation of a unique, model-based procedure that allows estimation of the false negative rate (FNR) and by adapting existing statistical methods, we have developed a balanced probability analysis that should be useful in addressing this challenging situation.
Results
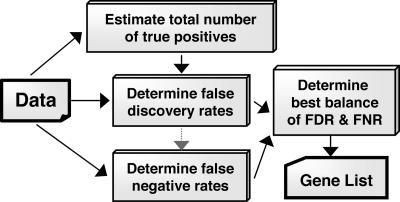
The strategy for a balanced approach to understanding the statistical significance of a list of genes produced from a microarray analysis, i.e., balanced probability analysis (BPA), is shown in Fig. 1. We reasoned that three variables would be of fundamental importance to the investigator. (i) The total number of true positives (TTP), i.e., the number of genes that truly are undergoing a change. (ii) The false discovery rate (FDR), defined as the aggregate chance that any gene listed is truly not changing and is, thus, on the list by statistical accident. (iii) The FNR, defined as the fraction of truly changing genes that are not on the list at hand. This rate is highly intuitive, simply the fraction of truly changed genes on the microarray that remain unselected. Other definitions of type 2 error rates, such as the false nondiscovery rate (the ratio of nondiscovered truly changing genes to the number of unselected genes), although statistically useful, are difficult to intuit for the nonstatistician.
Fig. 1.
Approach for balanced probability analysis.
As a first step toward a BPA, we assessed the capability to determine the three parameters of interest, TTP, FDR, and FNR, under real-life conditions. We used the modeling approach outlined in Methods to create synthetic data sets whereby we could track the distribution of the true positives in the significance list and, thus, determine the actual FDR and FNR. Simultaneously, we applied a “real-world” analysis by assuming no knowledge of any of the parameters or distributions used to create the synthetic data set. This approach allowed us to explore the capacities of algorithms to estimate the values of interest. By varying the conditions used to create the synthetic data sets, we studied the impact on algorithm performance of three parameters: the magnitude of the fold changes, the percentage of genes affected, and the number of experimental replicates.
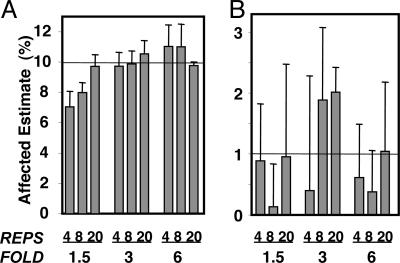
Estimation of the Total Number of True Positives. Only with perfect knowledge of the total number of true positives can the FDR be accurately determined (7). However, for real-life data, the total number of true positives cannot be directly observed but, rather, only estimated. By using an adaptation of the algorithm of Storey and Tibshirani (7) coupled with the modeling approach outlined above, we explored the ability to estimate the total number of true positives at varying percentages of genes actually altered in their expression. When a larger fraction of the genes in the sample was affected, the estimates tended to be fairly accurate and precise, even at a low number of replicates (Fig. 2A). However, when the fold change was modest (1.5-fold), with lower numbers of replicates (four to eight), the algorithm tended to underestimate the number of total changing genes. As fewer genes were affected, the estimates exhibited markedly increased variability (Fig. 2B) and sometimes resulted in negative estimates of the number of true positive genes. This behavior is not unexpected, and other algorithms for estimating the number of total truly changing genes also perform with increased relative variability when the effect and population affected becomes small (8).
Fig. 2.
Estimation of TTP under different fold changes (FOLD) and number of replicates (REPS). Actual TTP was set to 10% (A) or 1% (B). The mean estimate produced from four independent simulations is plotted.
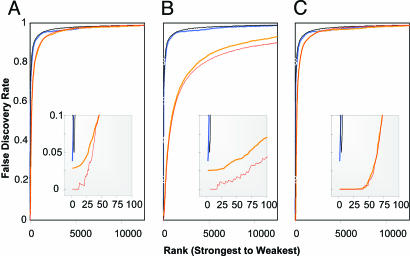
Estimation of the False Discovery Rate. The false discovery rate has been well described as a suitable means to detail type 1 statistical errors for microarray analysis and has become one of the standards to which gene-significance lists derived from microarray analyses are held. We modeled a standard procedure for estimation of the FDR (9). This procedure performed very well over a range of population conditions and replicates (Fig. 3). The procedure discerned the adverse consequences on the FDR of a reduction in fold change (Fig. 3A). At strong changes, such as 3-fold, the FDR curve stays low (i.e., highly significant) initially and moves upward to higher error rates only after the first 0.25% top-scoring genes. In contrast, for fold changes of modest effect, such as 1.5-fold, the FDR curve takes on an entirely different shape, climbing steeply from the onset to higher error rates, even when one focuses on the genes that have the best p values (Fig. 3A Inset). Likewise, the algorithm discerned that, as a smaller percentage of genes are affected, there is an adverse effect on the FDR (Fig. 3B) and that increasing the number of experimental replicates improves the FDR curve (Fig. 3C). However, although the FDR estimates were accurate across the majority of the FDR curves, the estimates produced were sometimes overly conservative for the genes with the largest significance, potentially missing some of the truly changing genes (Fig. 3 A and B Insets).
Fig. 3.
FDR was estimated (thicker lines) under real-life conditions, i.e., TTP was unknown. Base conditions were 1.5-fold change with 1% of genes changing and six replicates (A–C, blue lines). The effects of 3-fold change (A, orange line), 10% of genes changing (B, orange line), or 20 replicates (C, orange line) are shown. Actual FDR is represented in thin lines of related color. All lines represent the average of 10 simulations, each using 500 permutations. (Insets) Magnified view of the bottom left portion of the graph.
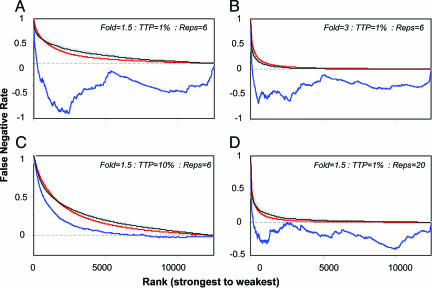
Estimation of the False Negative Rate. We sought an FNR algorithm that would, likewise, produce overall accurate estimates across a range of conditions. Existing procedures for calculation of the FNR depend on estimates of the FDR and TTP (10, 11). We modeled one such standard procedure under a variety of conditions. When knowledge of TTP is perfect, such an approach can produce accurate estimates of the FNR. However, in the analysis of real data, TTP is not known and can only be estimated. When this estimate is even modestly incorrect, the calculated FNR often markedly deviates from the true FNR (Fig. 4 A–D, blue versus black curves). This instability becomes more marked as fewer genes in the sample are affected.
Fig. 4.
FNR was estimated under real-life conditions, i.e., TTP was unknown. Base conditions were 1.5-fold change with 1% of genes changing and six replicates (A). The effects of 3-fold change (B), 10% of genes changing (C), or 20 replicates (D) are shown. Blue lines represent the customary approach to estimation of the FNR, whereas the red lines show the proposed approach. Actual FNR is represented in thin black lines with gray error bars. All lines represent the average of 10 simulations, each using 500 permutations.
We therefore created a unique procedure for estimating the FNR. Rather than depending on TTP, our procedure calculates the FNR directly from the data, by using resampling to estimate the null and alternate distributions for each gene. Comparison of these distributions allowed the estimation of the number of truly changing genes on any given “significance list,” leading directly to calculation of the FNR. The procedure weakly depends on the estimated FDR, and requires one model-dependent step to optimize a single parameter, α. In simulation trials, we found that, for any given condition, a suitable α could be chosen to allow the resultant FNR estimate to closely approximate the actual FNR (Fig. 4 A–D, compare red versus black curves). This result was true, even under conditions where TTP tends to be substantially misestimated (Fig. 4A). In general, the best fitting α took on low values in simulations containing strong effects or higher numbers of replicates (Fig. 4 B and D) and required higher values when the effect under study was small (Fig. 4 A and C).
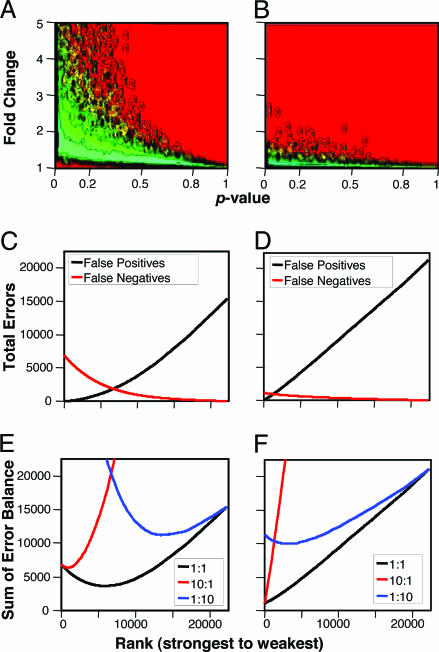
Exemplary Analysis of Real Data. To demonstrate the insight allowed by BPA, we performed this analysis on two illustrative public data sets: one data set representing malignant disease (bone marrow from individuals with two types of acute lymphoblastic leukemia) and the other representing metabolic disease (skeletal muscle from morbidly obese versus normal-weight individuals). To visualize the differences in the magnitude of change between these two data sets, we created contour plots of the observed p values versus the absolute fold changes across all genes. As can be seen for malignancy (Fig. 5A), many genes have experienced large fold changes with p values shifted toward significance, whereas, in metabolic disease, only a few genes have fold changes >1.5–2 (Fig. 5B). As a further marker of the changes in gene expression experienced in these data sets, we calculated the median absolute fold changes of the top-1%-scoring genes for both models (Table 1). Note that the median fold change in malignant disease was almost as great as the largest fold change experienced in the metabolic disease.
Fig. 5.
Analysis exemplifying circumstances where expression has undergone widespread changes of high magnitude (malignant disease) (A, C, and E) versus changes of less proportion and magnitude (metabolic disease) (B, D, F). A and B show a density contour plot of the observed p values versus fold changes for these two data sets. Red represents areas that are not populated with any genes, whereas colors increasing toward turquoise represent increasing densities of genes, with each contour representing an increase in density of 10-fold. C and D show the number of estimated false positive and false negative genes determined by BPA, and E and F show the sum of false positive and false negative errors at the relative weights indicated.
Table 1. Characteristics of exemplary real data.
| Data set
|
||
|---|---|---|
| Parameter | Malignant | Metabolic |
| Total features on microarray | 22,283 | 22,283 |
| Estimated total true positives | 6,865 | 1,135 |
| Median fold change among top 1% | 2.48 | 1.19 |
| Greatest fold change | 293.4 | 2.66 |
The malignant data set analysis represents comparison of RNA expression in bone marrow between the T cell and B precursor forms of acute lymphoblastic leukemia. The metabolic-data set analysis represents comparison of RNA expression in abdominal-wall muscle between morbidly obese versus nonobese individuals.
We then subjected each data set to BPA. For malignant disease, a large number of genes were estimated to be changing, whereas in the metabolic disorder, a smaller number of genes were estimated (Table 1). The FDR and FNR curves determined by BPA were used to predict the total number of false positive and false negative genes. For the malignant disease data set, the number of false positive genes stays low until one has chosen several thousand genes (Fig. 5C). Likewise, the number of false negatives drops rapidly across this early range. In contrast, for the metabolic disease data set, the number of false positive genes increases almost linearly from the start, and the number of false negatives drops slowly (Fig. 5D). For the malignancy data set, sole consideration of the FDR at the traditional cutoff of <0.05 selects several hundred genes of which only a few dozen are expected to be false positives (Table 2). By contrast, at this cutoff for the metabolic data set, no genes would be selected, a problematic result in some sense, given that the TTP algorithm estimates that >1,000 genes are truly affected.
Table 2. Balanced probability analysis of exemplary data.
| Data set
|
||
|---|---|---|
| Analysis | Malignant | Metabolic |
| FDR < 0.05 | ||
| Total on list | 643 | 0 |
| Falsely on list | 32 | 0 |
| FNR < 0.20 | ||
| Total on list | 8,159 | 13,525 |
| Falsely on list | 2,601 | 12,417 |
| BPA 10:1 | ||
| Total on list | 978 | 1 |
| Falsely on list | 66 | 0 |
| BPA 1:1 | ||
| Total on list | 5,818 | 1 |
| Falsely on list | 1,427 | 0 |
| BPA 1:10 | ||
| Total on list | 13,433 | 2,803 |
| Falsely on list | 6,482 | 2,180 |
Data sets described in Table 1 were analyzed by BPA. The number of resampling iterations was 500. Optimal α levels were determined by simulation, assuming that the expected number of changing genes were experiencing the median fold changes shown in Table 1: For the malignancy data set, α was set to 0.1; for the morbid obesity data set, 0.8.
The opposite approach, sole consideration of the FNR by choosing enough genes to obtain an expected aggregate power of discovery of 80% (i.e., FNR <0.2), is also problematic. For the malignancy data set, selection of >8,000 genes is required to obtain a FNR <0.2, and, of these, >2,000 genes would be false positives (Table 2). The situation, however, is far worse for metabolic disease, because one must choose >13,000 genes to discover 80% of the truly changing genes, but this comes at the expense of having concomitantly selected >12,000 false positives.
Balancing False Discovery and False Negative Rates. Over the past few years, most microarray investigations have focused on only the FDR when trying to determine which genes in a nucleotide-array analysis have altered expression. This approach is parallel to solely considering an ordinary p value in the context of testing a single hypothesis. This approach is extremely valuable, because it allows the researcher an estimate of the chances that a gene on a significance list is there accidentally. In many contexts, this is a wise approach, although it neglects statistical power.
However, when the effect under study is modest, as in the case of metabolic disorders, or when the percentage of genes truly changing is small, as in perturbations involving only a single pathway, stringent attention solely to the FDR can mask the majority, and sometimes even all, of the truly changing genes. Under these conditions, discovery of the truly changing genes requires consideration of the FNR in addition to the FDR.
The notion of balancing false positive and false negative errors has long existed in the single-hypothesis-testing context (12, 13). Recently, these ideas have been extended to the multiple-hypothesis-testing scenario (10, 11, 14). For BPA, we used the simple, informative method of assigning separate penalties for false discoveries and false negatives (15). The total penalty is then calculated across all genes, and the point that minimizes the total penalty is taken as the best cutoff.
To illustrate this approach, we performed alternate analyses on the exemplary data sets emphasizing balanced (i) control of false positives, (ii) control of false negatives, or (iii) a more equal temperament, by setting the relative “penalties” for false positives and false negatives at 10:1, 1:10, or 1:1, respectively. The resultant curves (Fig. 5 E and F) show the total penalty as one chooses an increasingly larger portion of the genes. For each data set, the point that minimizes the error sum for each discovery curve is furthest left for the 10:1 weighting, furthest right for the 1:10 weighting, and in between for the equally weighted approach (1:1). The resultant expected number of false positives and false negatives for the optimal balance at each weighting condition are listed in Table 2.
Discussion
Defining the set of genes undergoing small fold changes in a microarray analysis, as often occurs in the process of normal physiologic change or metabolic disease, is statistically challenging. Although having a large number of replicates helps improve the statistics, it is not a full remedy for this situation. To create the most useful gene list for further analysis of the pathophysiology of disease, the FDR is best interpreted in context with the FNR. To this end, the BPA procedure should aid in the choice of rational statistical cutoffs for significance. This approach may enjoy the most benefit when combined with other microarray analysis techniques, such as gene-set-enrichment analysis (4) or clustering, which provide additional statistical power by considering groupings of genes. Furthermore, the FNR component can define truly nonchanging genes, an essential step in defining functional networks (16).
The ultimate intent of the analysis at hand is a very important consideration in striking the ideal balance between the FDR and FNR. When the intended use of the microarray data is exploratory in nature, it may be desirous to accept a larger FDR for the purpose of minimizing the FNR. For example, such a balanced approach could be used as prefiltering for subsequent clustering analysis. By assigning a high relative penalty to false negatives, the researcher would be able to capture a larger portion of the truly changing genes in an informed fashion. Such prefiltering would have an advantage over use of the entire array, because clustering tends to perform better with smaller samples enriched for truly changing genes (17) but would maintain a larger portion of the biology at hand. Alternatively, there are times when minimization of false positives must be the driving force in deciding which genes to call significant, for example, use of the analysis to define a pathologic diagnosis of grave consequences. In such cases, sole attention to the FDR may be desirable.
In this study, we provide an FNR-determining procedure that can be applied to real-world data. Our FNR approach is a microarray algorithm that uses resampling to define the alternate distribution for each gene, thus allowing estimation of statistical power. Further refinement and broader application of this technique to multiple hypothesis-testing can be envisioned. Although our procedure provided robust estimates of the FNR in challenging simulations that emulated real-world conditions, the limitation of our approach is that it requires the optimization of a single modeling-parameter α for each data set studied. Thus, the deduced FNR will depend on the suitability of the modeling assumptions, including the chosen data distribution and range of affected fold changes. For example, we used a Gaussian data distribution for our synthetic data sets, but expression data for a given gene may not assume this distribution. Although the algorithms used in our approach make no assumptions about normality or data distribution, it is possible that synthetic modeling choices will affect the determined FNR.
The small fold changes incurred in metabolic disease make discovery by other methodologies such as RT-PCR or Northern blotting likewise challenging (18). Thus, the traditional experimental paradigm of confirmation of expression changes found on arrays by these methods may not be an effective means of defining false positives and false negatives. Instead, a strong knowledge of the biology of the system (19) coupled with alternate means of testing the hypotheses generated by the data analysis may be the best approach for maximizing the value of the array analysis. Clearly, as nucleotide array analysis becomes even more widely applied to increasingly more subtle situations, such as defining the differences between two normal populations or the impact of mild environmental variables, further refinements in the algorithms to determine the TTP and FNR as well as complementary scientific approaches will be needed.
Methods
Modeling. Real data were used to drive modeling analysis. Specifically, means and standard deviations were calculated from real data obtained for the 12,488 consensus sequences on Affymetrix U74Av2 microarrays. Source RNA was from skeletal muscle, with replicates obtained from related male mice on a mixed genetic background (control group) (20). Expression values were calculated by using genechip mas v. 5.0 and normalized by using a standard protocol (21). Thus, these means and variances typify the real-world effects of biological variability and experimental noise, including that introduced by a mixed genetic background, on observed expression values.
Model “control” data of any number of replicates can thus be created from the real data's means and standard deviations. Furthermore, replicates for model “experiment” data can likewise be created, altering a portion of the means by a given fold change. In our model simulations, we assumed that half of the changing genes were altered upward by the given fold change and half downward. The standard deviations for the modeled “experimental” data were obtained in parallel from related mice (knockout group) (20) under similar but nonidentical biological conditions.
Actual false discovery and false negative error rates were calculated for the synthetic control and experimental data as follows. All genes were sorted from most to least significant by using the standard t statistic, assuming unequal variance. For any significance list of top-scoring genes, the FDR was defined as the ratio of genes inappropriately called significant to the total number of genes called significant (22). Likewise, the FNR was defined as the number of truly differentially expressed genes missing from the significance list divided by the total number of differentially expressed genes (10, 11, 22).
Balanced Probability Analysis. We adapted established resampling algorithms for the estimation of TTP (7) and FDR (9). We also explored the customary method for computation of FNR: simply FNR = [TTP – #SL× (1 – FDR)]/TTP, where #SL is the total number of genes on the significance list.
A Unique Procedure for the Estimation of the False Negative Rate. Our approach is outlined here and detailed in the supporting information, which is published on the PNAS web site. As defined, FNR = “number of truly changing genes missing from the significance list/TTP.” Because the TTP is invariant across all possible significance lists, the crux is to determine the number of truly changing genes present on any significance list. The contribution of any single gene to this number is simply the probability that the gene is truly changing times the statistical power to observe the gene on the significance list. Thus, from the former probability (estimated as described in supporting information) and the statistical power (estimated as outlined below) calculated for each gene on the significance list, the number of identified truly changing genes can be estimated. The relative growth of this estimate across increasingly larger significance lists then defines (1 – FNR).
To estimate the statistical power to observe any given gene, we used resampling to approximate the distribution under the alternate hypothesis. Specifically, random samples were drawn from the experimental or control replicates, with replacement (a modified bootstrap approach). Class labels were maintained, such that only experimental replicates contributed to the experimental resamples, and only control replicates contributed to the control resamples. Thus, a series of such randomly drawn resamples will recapitulate the range and distribution of possible true effects predicted by the data. By comparing the alternate distribution thus derived to the null distribution determined by resampling by randomization of class labels, the statistical power can be estimated.
Computation. Statistical algorithms were hand-implemented in the C++ programming language, with control scripts created in perl and bash. Contour plots were created with the program gri after density extraction and gridding by using hand-implement scripts. Error bars represent standard deviations (Figs. 2 and 4).
Exemplary Data Analyses. Public data exemplifying clonal and metabolic disease were obtained from the Entrez Gene Expression Omnibus website (www.ncbi.nlm.nih.gov/geo) and were used to explore BPA in the context of additional real-world data, including an example of metabolic disease (accession no. GDS268, skeletal muscle from morbidly obese individuals) and cancer (accession no. GDS760, acute lymphoblastic leukemia). Analysis used eight microarrays per experimental group, chosen randomly in the GDS760 case.
Supplementary Material
Acknowledgments
We thank Pei Lin for helpful discussion regarding programming. This work was supported by National Institutes of Health Grant K08-DK064906 (to A.W.N.), by Diabetes Genome Anatomy Project Grant R01-DK060837 (to C.R.K.), and by the Mary K. Iacocca Professorship (to C.R.K.).
Conflict of interest statement: No conflicts declared.
Abbreviations: BPA, balanced probability analysis; FDR, false discovery rate; FNR, false negative rate; TTP, number of total true positives.
References
- 1.Yechoor, V. K., Patti, M.-E., Saccone, R. & Kahn, C. R. (2002) Proc. Natl. Acad. Sci. USA 99, 10587–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shannon, M. F., McKenzie, K. U. S., Edgley, A., Rao, S., Peng, K., Shweta, A., Schyvens, C. G., Anderson, W. P., Wilson, S. R., Pittelkow, Y. E., Ohms, S. & Whitworth, J. A. (2005) Kidney Int. 67, 364–370. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, K. H. S., Eckenrode, S. E., Li, Q.-Z., Ruan, Q.-G., Yang, P., Shi, J.-D., Davoodi-Semiromi, A., McIndoe, R. A., Croker, B. P. & She, J.-X. (2003) Diabetes 52, 2151–2159. [DOI] [PubMed] [Google Scholar]
- 4.Mootha, V. K., Lindgren, C. M., Eriksson, K.-F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrle, M., Laurila, E., et al. (2003) Nat. Genet. 34, 267–273. [DOI] [PubMed] [Google Scholar]
- 5.Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramaswamy, S., Tamayo, P., Rifkin, R., Mukherjee, S., Yeang, C. H., Angelo, M., Ladd, C., Reich, M., Latulippe, E., Mesirov, J. P., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storey, J. D. & Tibshirani, R. (2003) Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsueh, H., Chen, J. J. & Kodell, R. L. (2003) J. Biopharm. Stat. 13, 675–689. [DOI] [PubMed] [Google Scholar]
- 9.Ge, Y. C., Dudoit, S. & Speed, T. P. (2003) Test 12, 1–77. [Google Scholar]
- 10.Pawitan, Y., Michiels, S., Koscielny, S., Gusnanto, A. & Ploner, A. (2005) Bioinformatics 21, 3017–3024. [DOI] [PubMed] [Google Scholar]
- 11.Delongchamp, R. R., Bowyer, J. F., Chen, J. J. & Kodell, R. L. (2004) Biometrics 60, 774–782. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, R. A. & Jekel, J. F. (1969) Am. Rev. Respir. Dis. 100, 645–650. [DOI] [PubMed] [Google Scholar]
- 13.Lusted, L. B. (1971) Science 171, 1217–1219. [DOI] [PubMed] [Google Scholar]
- 14.Genovese, C. & Wasserman, L. (2002) J. R. Stat. Soc. B 64, 499–517. [Google Scholar]
- 15.Zweig, M. H. & Campbell, G. (1993) Clin. Chem. 39, 561–577. [PubMed] [Google Scholar]
- 16.Jansen, R. & Gerstein, M. (2004) Curr. Opin. Microbiol. 7, 535–545. [DOI] [PubMed] [Google Scholar]
- 17.Deutsch, J. M. (2003) Bioinformatics 19, 45–52. [DOI] [PubMed] [Google Scholar]
- 18.Souaz, F., Ntodou-Thom, A., Tran, C. Y., Rostne, W. & Forgez, P. (1996) BioTechniques 21, 280–285. [DOI] [PubMed] [Google Scholar]
- 19.Noordewier, M. O. & Warren, P. V. (2001) Trends Biotechnol. 19, 412–415. [DOI] [PubMed] [Google Scholar]
- 20.Norris, A. W., Chen, L., Fisher, S. J., Szanto, I., Ristow, M., Jozsi, A. C., Hirshman, M. F., Rosen, E. D., Goodyear, L. J., Gonzalez, F. J., et al. (2003) J. Clin. Invest. 112, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butte, A. J., Ye, J., Hring, H. U., Stumvoll, M., White, M. F. & Kohane, I. S. (2001) Pac. Symp. Biocomput., pp. 6–17. [DOI] [PubMed]
- 22.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.