Abstract
Drosophila Pumilio (Pum) and Caenorhabditis elegans FBF bind to the 3′-untranslated region (3′-UTR) of their target mRNAs and repress translation. Pum and FBF are members of a large and evolutionarily conserved protein family, the Puf family, found in Drosophila, C.elegans, humans and yeasts. Budding yeast, Saccharomyces cerevisiae, has five proteins with conserved Puf motifs: Mpt5/Uth4, Ygl014w, Yll013c, Jsn1 and Ypr042c. Here we report that Mpt5 negatively regulates expression of the HO gene. Loss of MPT5 increased expression of reporter genes integrated into the ho locus, whereas overexpression of MPT5 decreased expression. Repression required the 3′-UTR of HO, which contains a tetranucleotide, UUGU, also found in the binding sites of Pum and FBF. Mutation of UUGU to UACU in the HO 3′-UTR abolished Mpt5-mediated repression. Studies using a three-hybrid assay for RNA binding indicate that Mpt5 binds to the 3′-UTR of HO mRNA containing a UUGU sequence but not a UACU sequence. These observations suggest that the yeast Puf homolog, Mpt5, negatively regulates HO expression post- transcriptionally.
Keywords: HO/MPT5/Puf/3′-UTR
Introduction
Post-transcriptional regulation plays an important role during development in a wide variety of organisms (Gray and Wickens, 1998). For example, the asymmetric patterns of gene expression in Xenopus and Drosophila embryos are determined by the differential localization, stabilization and translation of maternally synthesized mRNA. Translational regulation also plays a key role in germline sex determination in Caenorhabditis elegans (Goodwin et al., 1993; Zhang et al., 1997; Jan et al., 1999). In most cases studied in detail, post-transcriptional regulation is mediated by sites in the 3′-untranslated region (3′-UTR) of the regulated mRNAs. Although regulatory proteins have been identified that bind specifically to such elements, key questions about how their binding mediates regulation remain unanswered.
Drosophila Pumilio protein (Pum) and C.elegans FBF (fem-3 mRNA-binding factor) are RNA-binding proteins which bind to the 3′-UTR of target mRNAs and repress their translation. In Drosophila, Pum binds to a pair of 32 nucleotide sequences (Nanos response elements, NREs) in the 3′-UTR of maternal hunchback (hb) mRNA and represses its translation in the posterior portion of the embryo (Wharton and Struhl, 1991; Murata and Wharton, 1995). In C.elegans, FBF binds to the 3′-UTR of the fem-3 mRNA, thereby promoting the switch from spermatogenesis to oogenesis (Zhang et al., 1997). Pum and FBF are members of a large and evolutionarily widespread protein family, the Puf family (for Pumilio and FBF), found in Drosophila, C.elegans, humans and yeasts (Zhang et al., 1997; Zamore et al., 1999). Pum and FBF contain eight Puf repeats, each of ∼36 amino acids (Zhang et al., 1997). RNA targets of Puf proteins other than Pum and FBF have not yet been identified. It is not known whether these other Puf proteins regulate translation or some other aspect of RNA metabolism.
The budding yeast, Saccharomyces cerevisiae, has five proteins that contain three to eight Puf repeats: Mpt5, Ygl014w, Yll013c, Jsn1 and Ypr042c (Zhang et al., 1997). MPT5, also known as UTH4, was first identified as a multicopy suppressor of pop2 mutants, which are defective in glucose derepression (Hata et al., 1998). Mpt5/Uth4 and another Puf protein, Ygl014w, regulate relocalization of Sir3p and Sir4p from telomeres to the nucleolus and are required for the normal life span of yeast cells (Kennedy et al., 1995, 1997). Mpt5 interacts with the RGS (regulator of G-protein signaling) protein, Sst2, and plays a role in recovery from pheromone-induced G1 arrest (Chen and Kurjan, 1997). JSN1 was identified as a gene whose overexpression supresses a tub2 mutation, which causes a defect in spindle elongation (Machin et al., 1995). Deletion of JSN1 causes no discernible phenotype. YPR042C null mutants are viable and display increased resistance to cycloheximide and paramomycin, both of which inhibit translation (Waskiewicz et al., 1998). The molecular mechanisms by which these Puf homologs function in yeast are not known.
The HO gene codes for an endonuclease that stimulates mating-type switching in budding yeast (Herskowitz, 1988). It is transcribed only in cells that have budded previously (mother cells) but not in newly born cells (daughter cells). This asymmetric transcription of the HO gene results from the preferential accumulation of a negative regulatory protein, Ash1p, in daughter nuclei (Bobola et al., 1996; Sil and Herskowitz, 1996). ASH1 mRNA is targeted to the distal tip of daughter buds in post-anaphase cells (Long et al., 1997; Takizawa et al., 1997). Localization of ASH1 mRNA depends on an intact actin cytoskeleton and five SHE genes (Jansen et al., 1996).
Here we describe the characterization of one of the Puf homologs, Mpt5, and show that it is involved in the regulation of HO expression. We found that Mpt5 binds to the 3′-UTR of HO mRNA and represses HO expression. Our results suggest that Mpt5 operates in the same manner as Pum and FBF, by binding to the 3′-UTR of its target mRNA. An mpt5Δ mutation allows mating-type switching in daughter cells, suggesting that Mpt5 provides a second mechanism for preventing synthesis of HO protein in daughter cells.
Results
Mpt5 represses expression of HO
Because asymmetric expression of HO is determined ultimately by localization of ASH1 mRNA (Long et al., 1997; Takizawa et al., 1997), we carried out a systematic survey of the effect of different candidate RNA-binding proteins on expression of HO. We examined HO expression and ASH1 mRNA localization in yeast mutants lacking each of the five genes coding for members of the Puf family of RNA-binding proteins: MPT5, YGL014w, YLL013c, JSN1 and YPR039c. We constructed disruptants for each of these Puf genes in a strain harboring a HOp-ADE2 reporter gene to monitor expression of HO (see Materials and methods). HOp-ADE2 was constructed by replacing the ho open reading frame (ORF) with the ADE2 ORF at the ho locus (as also described by Jansen et al., 1996). Expression of the reporter can be assayed in an ade2Δ background by growth on medium lacking adenine (SC-Ade). In these strains, the ash1Δ mutant grew faster than wild-type cells on SC-Ade plates, whereas cells overexpressing ASH1 did not grow under these conditions (data not shown). Furthermore, inactivation of SHE genes (SHE1/MYO4, SHE2, SHE3, SHE4 and SHE5), which leads to delocalization of ASH1 mRNA, prevented growth of these strains on SC-Ade plates. These results demonstrate that ASH1 negatively regulates the HOp-ADE2 reporter. mpt5 disruptants carrying this reporter grew faster than wild-type strains on SC-Ade plates; overexpression of MPT5 in the HOp-ADE2 strain completely inhibited growth on SC-Ade plates (see below). In contrast, disruptions of any of the four other Puf genes, YGL014w, YLL013c, JSN1 or YPR039c, had no effect on growth on SC-Ade plates. Therefore, we further characterized the role of MPT5 in the regulation of HO.
In order to quantitate the effect of MPT5 on HO expression, we next used a HOp-lacZ reporter, in which the ho ORF was replaced with the lacZ ORF at the ho locus. Using this reporter, we observed a 2-fold increase in HO expression in mpt5Δ mutants compared with wild-type cells (Figure 1A). In contrast, HO expression decreased ∼40% when MPT5 was overexpressed from a multicopy plasmid (YEpMPT5) (Figure 1B). These results suggest that MPT5 negatively regulates HO expression.
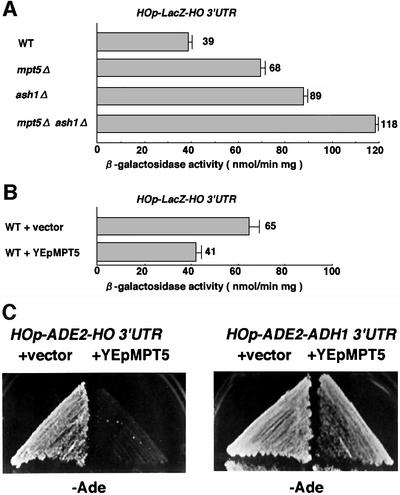
Fig. 1. MPT5 negatively regulates HO expression. (A) Effect of the mpt5Δ mutation on expression of the HOp-lacZ reporter. Yeast strains K1107 (WT), TTC2 (mpt5Δ), TTC91 (ash1Δ) and TTC191 (mpt5Δ ash1Δ), harboring the HOp-lacZ reporter gene, were grown in YPD medium and assayed for β-galactosidase activity. Units shown are averages of five experiments. (B) Effect of MPT5 overexpression on the HOp-lacZ reporter. Yeast strains K1107 carrying HOp-lacZ transformed with vector YEp13 or YEpMPT5 were cultured in SC-Leu medium and assayed for β-galactosidase activity. Units shown are averages of three experiments for independent transformants. (C) The HO 3′-UTR is required for MPT5-mediated repression of HO. Yeast strains 10B (HOp-ADE2-HO 3′-UTR) and TTC47 (HOp-ADE2-ADH1 3′-UTR) were transformed with YEp13 or YEpMPT5. The resulting transformants were streaked on SC-Ade Leu (–Ade) plates and incubated for 3 days at 30°C.
We next examined whether MPT5 affects HO expression by acting on ASH1 mRNA. Compared with wild-type cells, ASH1 mRNA was found to be somewhat delocalized in an mpt5Δ mutant (Figure 2A): 22% of mpt5Δ cells and 87% of wild-type cells localized ASH1 mRNA to the distal cortex of the bud; 62% of mpt5Δ cells and 12% of wild-type cells localized ASH1 mRNA diffusely within the bud; and 16% of mpt5Δ cells and 1% of wild-type cells exhibited substantial ASH1 mRNA in both mother and daughter cells. However, Ash1 protein was still localized asymmetrically in the mpt5Δ cells, similarly to that observed in the wild-type cells (84% for mpt5Δ cells versus 92% for wild-type cells; Figure 2B). Therefore, delocalized mRNA in the bud of the mpt5Δ strain does not affect the daughter-specific localization of Ash1 protein. Since delocalization of ASH1 message in she mutants causes symmetric localization of Ash1 protein and a reduction in HO expression (Jansen et al., 1996), the partial delocalization of ASH1 mRNA in mpt5Δ mutants cannot be responsible for enhancing HO expression. Furthermore, overexpression of MPT5 had no effect on ASH1 mRNA localization (data not shown).
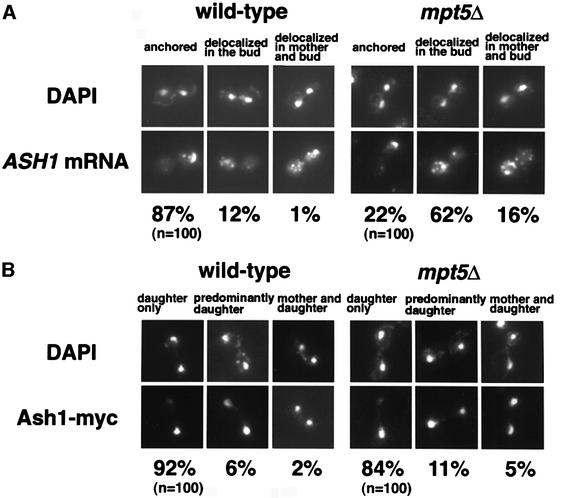
Fig. 2. Effect of the mpt5Δ mutation on the localization of ASH1 mRNA and Ash1 protein. (A) Comparison of ASH1 mRNA localization in K5552 (ASH1-myc; wild type) and in TTC219 (ASH1-myc mpt5Δ). The percentages of cells that showed each pattern of ASH1 mRNA localization were determined by RNA in situ hybridization: anchored = tightly localized ASH1 mRNA at the distal tip; delocalized in the bud = delocalized ASH1 mRNA confined to the bud; delocalized in mother and bud = ASH1 mRNA in both mother cell and bud. (B) Comparison of Ash1 protein localization in K5552 and TTC219. The percentages of cells that showed each pattern of Ash1 protein localization were determined by staining with anti-myc in cells expressing Ash1-myc: daughter only = visible Ash1 only in the daughter; predominantly daughter = Ash1 staining predominantly in the daughter nucleus with an intermediate amount of staining in the mother nucleus; mother and daughter = equivalent levels of Ash1 in both mother and daughter nuclei.
To confirm that MPT5 does not affect HO expression by acting on ASH1 mRNA, we examined HO expression in an mpt5Δ ash1Δ double mutant using a HOp-lacZ reporter. The mpt5Δ ash1Δ double mutant showed a higher level of expression than either an mpt5Δ or ash1Δ single mutant (Figure 1A), indicating that MPT5 and ASH1 act on HO expression independently. Their independent action is supported further by the observation that overexpression of MPT5 still repressed HO in ash1Δ mutants (data not shown).
The HO 3′ UTR is required for MPT5-mediated repression of HO
The Puf proteins, Pum and FBF, bind to the 3′-UTR of their target mRNAs to repress translation (Zhang et al., 1997; Sonoda and Wharton, 1999). Mpt5 has a Puf motif, raising the possibility that it represses HO expression via the 3′-UTR of HO. To test this possibility, we used two reporters that differ only in their 3′-UTR, HOp-ADE2-HO 3′-UTR and HOp-ADE2-ADH1 3′-UTR. In the latter reporter, the HO 3′-UTR was replaced with the ADH1 3′-UTR (see Materials and methods). An ade2Δ strain harboring either HOp-ADE2-HO 3′-UTR or HOp-ADE2-ADH1 3′-UTR grew on SC-Ade plates in a manner similar to the ADE2 strain, indicating that both reporters are expressed. We found that overexpression of MPT5 from YEpMPT5 clearly inhibited growth of the HOp-ADE2-HO 3′-UTR strain and slightly inhibited that of the HOp-ADE2-ADH1 3′-UTR strain on SC-Ade plates (Figure 1C). The partial inhibition of the HOp-ADE2-ADH1 3′-UTR strain by YEpMPT5 apparently is due to non-specific inhibition of growth by MPT5 overexpression, because growth of cells harboring YEpMPT5 was slightly inhibited on SC plates containing adenine when compared with that of cells carrying the empty vector (data not shown). These results indicate that the HO 3′-UTR is required for repression of HO mediated by Mpt5.
To determine whether the HO 3′-UTR is sufficient for repression by Mpt5, we replaced the ADE2 3′-UTR of the chromosomal ADE2 locus with the HO 3′-UTR or ADH1 3′-UTR. We also added three copies of the hemagglutinin (HA) epitope to the 3′ end of the ADE2 coding sequence to monitor the levels of Ade2 protein by western blotting. Replacement of the ADE2 allele with the ADE2-3HA-HO 3′-UTR or ADE2-3HA-ADH1 3′-UTR constructs did not affect growth on SC-Ade plates, indicating that replacement of the 3′-UTR and addition of the HA tag did not inactivate the ADE2 gene. When MPT5 was overexpressed from the GAL1 promoter in these strains, growth of the ADE2-HO 3′-UTR strain, but not of the ADE2-ADH1 3′-UTR strain, was inhibited on SG-Ade plates (Figure 3A). Thus, the HO 3′-UTR confers MPT5-mediated repression.
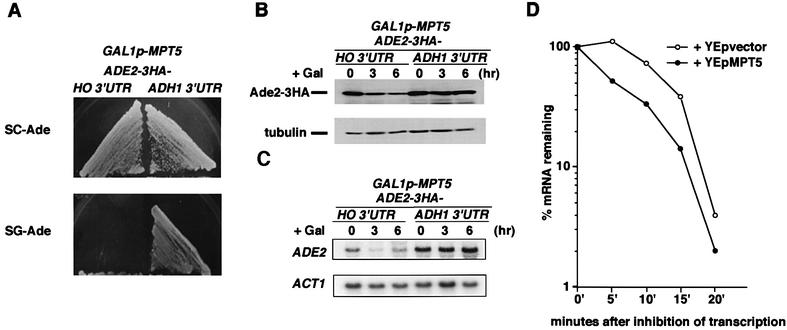
Fig. 3. The HO 3′-UTR is sufficient for MPT5-mediated repression of HO. (A) Effect of MPT5 overexpression on ADE2-3HA reporter strains. Yeast strains TTC59 (ADE2-3HA-HO 3′-UTR, GAL1p-MPT5) and TTC62 (ADE2-3HA-ADH1 3′-UTR, GAL1p-MPT5) were streaked on the SC (glucose)-Ade or SG (galactose)-Ade plates and incubated for 3 days at 30°C. (B) Effect of MPT5 overexpression on Ade2-3HA protein levels. Yeast cells were cultured in 2% raffinose medium at 30°C and treated with galactose (2%) to induce MPT5 expression from GAL1p-MPT5. At the times indicated, cells were harvested and western blot analysis was performed to assay the level of Ade2-3HA protein (top). Tubulin protein (bottom) was included as a quantity control. (C) Effect of MPT5 overexpression on ADE2-3HA mRNA levels. Yeast cells were harvested as in (B), and northern blot analysis was performed to assay the level of ADE2-3HA mRNA (top). ACT1 mRNA (bottom) was included as a quantity control. (D) Effect of MPT5 overexpression on degradation of mRNA. Yeast strain TTC181 (GAL1p-ADE2-3HA-HO 3′-UTR) harboring YEp195-MPT5 or empty vector was incubated in SR-Ura. Transcription was induced by adding galactose to a final concentration of 2%. The culture was then transferred to medium containing 2% glucose. Aliquots were removed at various times, and the amounts of the ADE2 transcript were quantitated by dot blotting as described in Materials and methods. Decay rates were determined from semilog plots of the percentage of hybridizing material remaining at different times after the inhibition of transcription.
We confirmed that this growth inhibition by MPT5 reflected a decrease in Ade2 protein levels. Western blotting analysis revealed that the levels of Ade2-3HA protein were reduced in the ADE2-3HA-HO 3′-UTR strain but not in the ADE2-3HA-ADH1 3′-UTR strain after MPT5 was overexpressed (Figure 3B). Overexpression of MPT5 also altered the levels of ADE2-3HA mRNA in a manner dependent upon the HO 3′-UTR (Figure 3C). The decrease of ADE2-3HA-HO 3′-UTR mRNA in the cells overexpressing MPT5 suggested that MPT5 affects mRNA turnover. To test this possibility, we measured the decay rates of mRNA by performing transcriptional pulse–chase experiments using the regulated GAL1 promoter. For this purpose, we constructed a reporter, GAL1p-ADE2-HO 3′-UTR. Transcription was first induced in the presence of galactose and then repressed by shifting the medium from galactose to glucose. The mRNA produced by GAL1p-ADE2-HO 3′-UTR showed a faster decay rate in the strain harboring YEpMPT5 as compared with the strain harboring the control vector (Figure 3D). The decay rates of mRNA produced by GAL1p-ADE2-ADH1 3′-UTR were not affected by MPT5 overexpression (data not shown). These results suggest that overexpression of MPT5 promotes degradation of mRNA carrying the HO 3′-UTR.
A region within the HO 3′-UTR required for repression by Mpt5
To determine which region of the HO 3′-UTR is required for repression by Mpt5, we first analyzed the 3′ end of HO mRNA using the 3′-RACE method (see Materials and methods). The HO transcript extends 67 nucleotides from the stop codon (Figure 4B). This segment contains a positioning element (+32 to +37 from the stop codon), which is involved in the addition of poly(A), and an efficiency element UAUAUA (+7 to +12 from the stop codon), which enhances the efficiency of downstream positioning elements (Guo and Sherman, 1996).
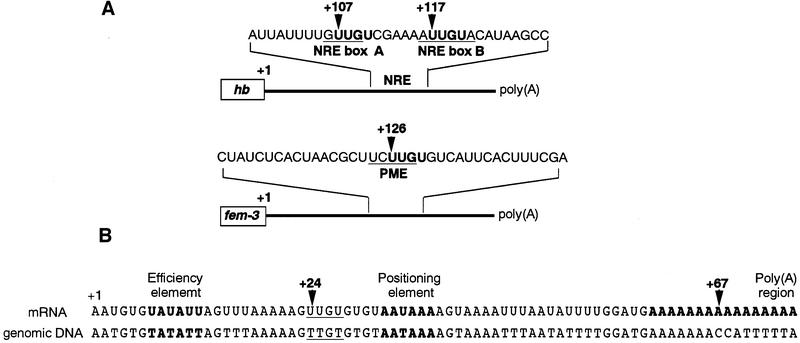
Fig. 4. Sequence of the 3′ end of HO mRNA. (A) Schematic drawing of the 3′-UTR structures of hunchback (hb) and fem-3 mRNAs. Numbers indicate the number of nucleotides between the stop codon and UUGU sites. (B) The 3′ end of HO mRNA was determined by 3′-RACE analysis. The putative efficiency element, positioning element and poly(A) region are shown in bold as indicated. The UUGU site is underlined. Numbers indicate the number of nucleotides from the stop codon.
Pum binds to a pair of 32 nucleotide sequences, called NREs, in the 3′-UTR of hb mRNA (Wharton and Struhl, 1991; Murata and Wharton, 1995). Binding of FBF to the 3′-UTR of fem-3 mRNA requires a five nucleotide segment in the center of the 3′-UTR, called the point mutation element (PME) (Zhang et al., 1997). Although NREs and the PME have little overall similarity to each other, they both contain the tetranucleotide UUGU (Figure 4A), suggesting that this tetranucleotide might be a conserved binding sequence for Puf proteins. The 3′-UTR of HO mRNA contains only one UUGU site, which is located 24 nucleotides downstream of the stop codon (Figure 4B). No UUGU site is present in the ADH1 3′-UTR region, which is not repressed by Mpt5. We thus examined whether this UUGU is important for repression by Mpt5 by changing its sequence to UACU. In NREs, this change eliminates Pum regulation (Murata and Wharton, 1995; Sonoda and Wharton, 1999). Changing UUGU to UACU in the HOp-ADE2 reporter greatly reduced its repression by MPT5 (Figure 5), suggesting that the UUGU sequence in the HO 3′-UTR is indeed required for repression by Mpt5.
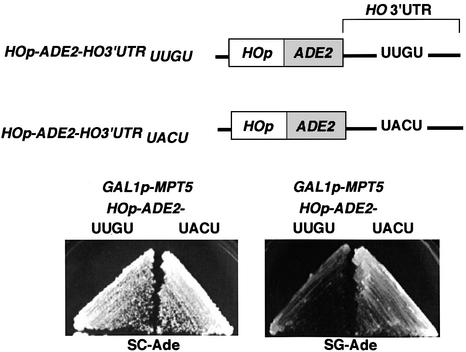
Fig. 5. The UUGU sequence in the HO 3′-UTR is required for MPT5-mediated repression of HO. Yeast strain TTC74 (GAL1p-MPT5) was transformed with YCplac33 (HOp-ADE2-HO 3′-UTRUUGU) or YCplac33 (HOp-ADE2-HO 3′-UTRUACU). Resulting transformants were plated on SC (glucose)-Ade Leu or SG (galactose)-Ade Leu plates and incubated for 3 days at 30°C.
Analysis of Mpt5 by the three-hybrid system
To determine whether Mpt5 acts through the HO 3′-UTR, we used the three-hybrid RNA-binding assay (described in Figure 6A) (SenGupta et al., 1996). The reporter strain, L40c, contains a HIS3 gene with LexA-binding sites inserted in place of the normal upstream promoter elements, and expresses a fusion bewteen LexA and the MS2 coat protein (LexA-CP). This strain was transformed with plasmids coding for two additional hybrids: plasmid pGAD-MPT5 codes for a fusion of Mpt5p to the Gal4 transcriptional activation domain; plasmid pMS2-HO 3′-UTR produces a chimeric nuclear RNA containing 134 bp of the region 3′ of the HO stop codon and a binding site for the bacteriophage MS2 coat protein. The resulting transformants grew on SC-His plates. In contrast, transformants carrying pGAD-MPT5 and a control plasmid (MS2) that contains only the MS2 RNA segment failed to grow on SC-His plates (Figure 6B). Similarly, transformants carrying the pGAD control vector and pMS2-HO 3′-UTR were also unable to grow on SC-His plates (data not shown). These results suggest that Mpt5 binds to the HO 3′-UTR. As described above, mutation of a single UUGU site in the 3′-UTR of HO abolishes repression by Mpt5. We thus generated a derivative of plasmid pMS2-HO 3′-UTR containing the UUGU to UACU change and tested it in the three-hybrid system. This mutation did not affect the expression of MS2-HO 3′-UTR reporter RNA (data not shown). These transformants were unable to grow on SC-His plates (Figure 6B), suggesting that the UUGU site is indeed essential for Mpt5 binding.
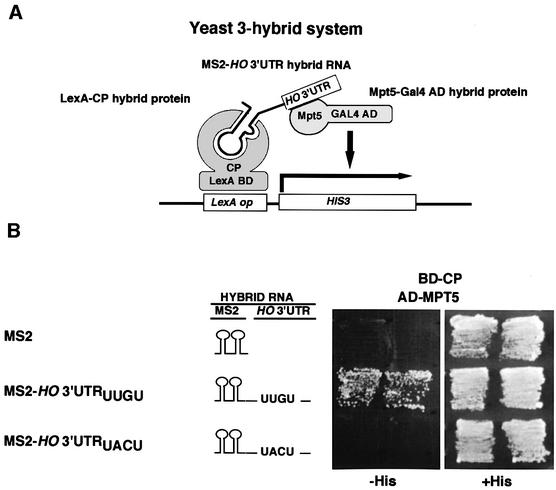
Fig. 6. Requirement for the HO mRNA 3′-UTR for functioning of Mpt5 in the three-hybrid RNA-binding assay. (A) Schematic drawing of the yeast three-hybrid assay. The LexA-CP hybrid protein (BD-CP) contains the LexA DNA-binding domain joined to the MS2 coat protein and binds to the LexA operator of the HIS3 reporter gene. The Gal4 AD-Mpt5 hybrid protein (AD-MPT5) is a fusion of Mpt5 and the yeast Gal4 activation domain. The hybrid RNA (MS2-HO 3′-UTR) contains the binding site for MS2 coat protein and the HO 3′-UTR. If the Mpt5 segment of the Gal4 AD-Mpt5 hybrid binds to the HO 3′-UTR, then the MS2-HO 3′-UTR hybrid RNA will link the two hybrid proteins to each other and activate the HIS3 reporter gene. (B) Yeast strain L40c, which expresses a LexA-MS2 coat protein fusion, was transformed with the indicated plasmids, and the transformants were streaked on SC-Ura Leu His (–His) and SC-Ura Leu (+His) plates and incubated for 2 (+His plates) or 3 days (–His plates) at 30°C. Plasmids were MS2, MS2-HO 3′-UTRUUGU, MS2-HO 3′-UTRUACU and pGAD-MPT5.
MPT5 affects mating-type switching in daughter cells
The ability of cells in a cell lineage to undergo mating-type switching has a very precise pattern in which mother cells switch efficiently (typically ∼65% of cell divisions), but daughter cells do not (<0.1% of cell divisions) (Strathern and Herskowitz, 1979). The basis for this difference is the presence of HO mRNA in mother cells but not in daughter cells (Nasmyth, 1983). Ash1 protein, located in daughter cells, limits HO transcription (Bobola et al., 1996; Sil and Herskowitz, 1996). To test whether Mpt5-mediated repression of HO affects the regulation of mating-type switching, we disrupted the MPT5 gene in an HO background and measured its effect on mating-type switching by pedigree analysis. In wild-type cells, we observed that 59% of mother cells and <1.5% of daughters switched mating type. In contrast, in the mpt5Δ mutant, 60% of mother cells and 27% of daughters switched mating type. Thus, the mpt5Δ mutation greatly increased the frequency of mating-type switching in daughter cells, whereas it had no effect in mothers. Although the effect of mpt5Δ on mating-type switching is not as strong as that of the ash1Δ mutation, in which 85–95% of daughter cells switch mating type (Bobola et al., 1996; Sil and Herskowitz, 1996), Mpt5 is clearly required for proper regulation of mating-type switching. The mpt5Δ mutation had no effect in mothers, suggesting that Mpt5-mediated repression might function only in daughter cells (see Discussion).
The observation that the mpt5Δ mutation allows mating-type switching in a substantial fraction of daughter cells suggests that some residual expression of HO may occur in daughter cells even in the presence of Ash1p and that this expression is blocked by Mpt5. To test this possibility, we examined the effect of MPT5 on HO expression in myo4Δ mutants, in which Ash1p is delocalized and is thus present in both mother and daughter nuclei (Bobola et al., 1996). myo4Δ mutants containing the HOp-ADE2 reporter failed to grow on SC-Ade plates (Figure 7). In contrast, the mpt5Δ myo4Δ double mutant containing the HOp-ADE2 reporter grew on SC-Ade plates (Figure 7). These results suggest that Mpt5p inhibits the utilization of HO mRNA that has escaped from repression by Ash1.
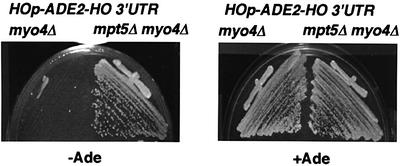
Fig. 7. Effect of MPT5 on Ash1-mediated repression of HO. Yeast strains TTC120 (myo4Δ HOp-ADE2-HO 3′-UTR) and TTC121 (mpt5Δ myo4Δ HOp-ADE2-HO 3′-UTR) were streaked on SC-Ade (–Ade) or SC (+Ade) plates and incubated for 3 days at 30°C.
Discussion
We show that one of the Puf homologs of S.cerevisiae, Mpt5, is involved in the regulation of HO expression. mpt5Δ mutants showed increased HO expression compared with wild-type cells, whereas cells overexpressing MPT5 from a multicopy plasmid showed a decrease in HO expression. We have presented several pieces of evidence suggesting that Mpt5, like Pumilio and FBF, binds to the 3′-UTR of the HO mRNA and regulates HO expression post-transcriptionally. First, the HO 3′-UTR is required for Mpt5-mediated repression of HO expression. Secondly, the 3′-UTR of HO mRNA contains a UUGU site which might be a conserved binding sequence for Puf proteins. This site is essential for repression by Mpt5. Finally, studies using the three-hybrid assay indicate that Mpt5 binds to the HO 3′-UTR and that the UUGU site is required for its binding. We thus propose that Mpt5 regulates HO expression post-transcriptionally by binding to the 3′-UTR of HO.
Mpt5 is a Puf protein with functional similarity to Pum and FBF
Mpt5 has structural and functional similarities to the founding members of the Puf family. Like Pum and FBF, which contain eight Puf repeats, Mpt5 contains seven or eight repeats. All three of these proteins are negative regulators that are known or inferred to act by binding to specific elements in the 3′-UTRs of their respective RNA targets (Murata and Wharton, 1995; Zhang et al., 1997; Sonoda and Wharton, 1999). The structural similarities of these proteins suggest that they may recognize related RNA sequences, and indeed their binding to 3′-UTRs appears to require the UUGU sequence. Changing UUGU to UACU eliminates regulation by Pum and Mpt5, indicating that UUGU is a conserved binding sequence for Puf family proteins. Mpt5, Pum and FBF all regulate expression post-transcriptionally. It is clear that Pum represses translation in early embryos (Wreden et al., 1997). In some cases, it appears to do so by promoting deadenylation of poly(A) RNA, leading to mRNA turnover (Wickens et al., 1997). The observation that overexpression of MPT5 promotes degradation of ADE2-3HA-HO 3′-UTR mRNA suggests that Mpt5 may affect mRNA stability or turnover via binding to the HO 3′-UTR. Recently, Olivas and Parker reported that another Puf protein, Puf3/Yll013c, specifically promotes deadenyl ation and degradation of COX17 mRNA (Olivas and Parker, 2000). Thus, Puf proteins regulate mRNA turnover in yeast, although previous work with Puf proteins from Drosophila and C.elegans has demonstrated a role in translational inhibition. Pum and FBF function with companion proteins, Nanos and Nos-3, respectively (Wreden et al., 1997; Wharton et al., 1998; Kraemer et al., 1999; Sonoda and Wharton, 1999; Subramaniam and Seydoux, 1999). It would not be surprising if Mpt5 also functioned with a companion protein, although there is no obvious homolog of Nanos in S.cerevisiae. The observation that inactivation of MPT5 leads to only a 2-fold increase in HO expression could reflect the limits of regulation by Mpt5, or it could suggest the existence of a functionally redundant product. Inactivation of other Puf proteins, Ygl014w and Yll013c, in an mpt5Δ background did not increase HO expression further, indicating that Ygl014w and Yll013c are not redundant with Mpt5 for this function.
Mpt5 may provide a second mechanism to prevent synthesis of HO protein in daughter cells
Prior studies of mating-type interconversion focused on the regulation of HO by the transcriptional activator proteins Swi1–Swi6 and the negative regulators Sin1, Sin3 and Ash1. The Ash1 protein was shown recently to play a key role in generating the asymmetric pattern of mating-type switching as it is a daughter cell-specific repressor of HO transcription (Bobola et al., 1996; Sil and Herskowitz, 1996). Our studies on Mpt5 reveal an unexpected aspect of HO regulation, in particular that HO expression is also regulated post-transcriptionally by Mpt5 through the 3′-UTR of the HO transcript. Surprisingly, in mpt5Δ mutants, a substantial fraction (27%) of daughter cells switched mating type, whereas in wild-type strains, daughter cells are never observed to switch mating type. Thus, Mpt5-mediated repression is required for proper regulation of mating-type switching. Why does the mpt5Δ mutation allow mating-type switching in daughter cells, where Ash1p is thought to repress HO transcription? One possibility is that HO is still transcribed at a low level in daughter cells, even in the presence of Ash1p, and that use of this residual HO transcript is prevented by Mpt5. This explanation is supported further by the observation that expression of HO in myo4Δ mutants, in which Ash1p localizes to both mother and daughter, is enhanced by deletion of MPT5.
Does Mpt5 inhibit HO expression in mother cells as well as in daughter cells? Because the mpt5Δ mutation does not noticeably affect the frequency of mating-type switching in mother cells, Mpt5-mediated repression might, in principle, function only in daughter cells. However, using an Mpt5–Myc fusion protein, we have observed that Mpt5p is present in both mother and daughter cells (data not shown). Perhaps Mpt5 function requires a companion protein whose expression is restricted to daughter cells.
Possible roles of Mpt5p in regulating other processes
MPT5, also known as UTH4, was first identified as a multicopy suppressor of the growth defect of POP2-deficient strains (Hata et al., 1998). POP2 encodes a component of the Ccr4 transcription regulatory complex required for glucose derepression (Sakai et al., 1992; Liu et al., 1998). pop2Δ mutants exhibit temperature-sensitive growth, perhaps due to overproduction of some protein. Overexpression of Mpt5 might restore growth to pop2Δ mutants by reducing the level of this growth-inhibitory product. Although we do not know the mechanism by which Mpt5 negatively regulates utilization of HO and other mRNAs, it should be possible to identify Mpt5-regulated transcripts other than HO by use of DNA microarrays to assess the level of poly(A) RNA species or by isolation of RNA from polysomes in wild-type versus mpt5Δ strains (Diehn et al., 2000).
mpt5Δ mutants exhibit many defects, including temperature-sensitive growth, shortened life span, a slight increase in sensitivity to mating pheromone and a defect in recovery from pheromone arrest (Kennedy et al., 1995, 1997; Chen and Kurjan, 1997). Can this pleiotropic phenotype be explained by the ability of Mpt5 to negatively regulate gene expression post-transcriptionally? Mpt5 might down-regulate synthesis of proteins that control growth at high temperature, life span and response to mating pheromones. Accordingly, the 3′-UTRs of the encoding mRNAs might contain one or more UUGU sites. However, the finding that Mpt5 protein interacts with Sst2, Fus3, Kss1 and Cdc28 proteins, as assayed by the two-hybrid system and by co-immunoprecipitation (Chen and Kurjan, 1997), suggests that Mpt5 could also affect pheromone response by a mechanism unrelated to post-transcriptional regulation.
Materials and methods
Strains and general methods
Escherichia coli DH5α was used for DNA manipulations. The yeast strains used in this study are described in Table I. Standard procedures were followed for yeast manipulations (Kaiser et al., 1994). The media used in this study included rich medium, synthetic complete medium (SC), synthetic minimal medium (SD) and sporulation medium (Kaiser et al., 1994). SC lacking amino acids or other nutrients (e.g. SC-Leu lacks leucine, SC-Ade lacks adenine) was used to select transformants and to score ADE2 reporter activity. SG and SR were identical to SC except that they contained 2% galactose and raffinose, respectively, instead of 2% glucose. Recombinant DNA procedures were carried out as described previously (Sambrook et al., 1989).
Table I. Strains used in this study.
| Strain | Genotype | Source | |
|---|---|---|---|
| W303 | a | ade2 trp1 can1 leu2 his3 ura3 GAL psi+ | Sil and Herskowitz (1996) |
| K1107 | a | HOp-LacZ-HO 3′-UTR | Nasmyth (1987) |
| K5552 | a | ASH1-myc | Jansen et al. (1996) |
| 10B | α | HOp-ADE2-HO 3′-UTR | this study |
| TTC2 | a | HOp-LacZ-HO 3′-UTR mpt5Δ::CgHIS3 | this study |
| TTC28 | α | HOp-ADE2-HO 3′-UTR His3MX6::GAL1p-MPT5 | this study |
| TTC47 | α | HOp-ADE2-ADH1 3′-UTR | this study |
| TTC59 | a | ADE2-3HA-HO 3′-UTR::His3MX6 kanMX6::GAL1p-MPT5 | this study |
| TTC62 | a | ADE2-3HA-ADH1 3′-UTR::His3MX6 kanMX6::GAL1p-MPT5 | this study |
| TTC74 | a | kanMX6::GAL1p-MPT5 | this study |
| TTC85 | a/α | HO/HO | Sil and Herskowitz (1996) |
| TTC87 | a/α | HO/HO mpt5Δ::CgHIS3 mpt5Δ::CgHIS3 | this study |
| TTC91 | a | HOp-LacZ-HO 3′-UTR ash1Δ::hisG URA3 hisG | this study |
| TTC120 | α | HOp-ADE2-HO 3′-UTR myo4Δ::CgTRP1 | this study |
| TTC121 | α | HOp-ADE2-HO 3′-UTR mpt5Δ::CgHIS3 myo4Δ::CgTRP1 | this study |
| TTC181 | a | kanMX6::GAL1p-ADE2-3HA-HO 3′-UTR::His3MX6 | this study |
| TTC191 | a | HOp-LacZ-HO 3′-UTR mpt5Δ::CgLEU2 ash1Δ::hisG URA3HisG | this study |
| TTC219 | a | ASH1-myc mpt5Δ::CgHIS3 | this study |
| L40c | a | ura3 leu2 his3 trp1 ade2 LYS2::(lexAop)-HIS3 ura3::(lexAop)-LacZ TRP1-LexA-coat | SenGupta et al. (1996) |
All strains except L40c were isogenic derivatives of W303.
Plasmids
Plasmids used in this study are described in Table II. Plasmid YEpMPT5 (kindly provided by A.Sakai) is YEp13 carrying MPT5 (Hata et al., 1998). Plasmid YEp195-MPT5 is YEplac195 carrying MPT5. Plasmid pFA6a-3HA-HO 3′-UTR-His3MX6 was constructed as follows. The HO 3′-UTR was amplified by PCR using a 5′ primer (5′-CTCGGCGCGCCAATGTGTATATTAGTTTAAAAAG-3′, incorporating an AscI site) and a 3′ primer (5′-CTCAGATCTGAATTCGACTTGAAGAACATCCC-3′, incorporating a BglII site). The resulting fragment was inserted into the AscI–BglII gap of pFA6a-3HA-His3MX6 (Longtine et al., 1998) to generate pFA6a-3HA-HO 3′-UTR-His3MX6. Plasmids YCplac33 (HOp-ADE2-HO 3′-UTRUUGU) and YCplac33 (HOp-ADE2- HO 3′-UTRUACU) are YCplac33 carrying HOp-ADE2-HO 3′-UTR reporter genes. YCplac33 (HOp-ADE2-HO 3′-UTRUACU) contains two base changes, from TTGT to TACT. Plasmids pIIIA/MS2-1-HO 3′-UTRUUGU and pIIIA/MS2-1-HO 3′-UTRUACU express chimeric nuclear RNAs bearing binding sites for the MS2 coat protein and 134 bp 3′ of the HO stop codon. pIIIA/MS2-1-HO 3′-UTRUACU contains two base changes, from TTGT to TACT. The DNA fragments of the HO 3′-UTR containing nucleotides +1 to +134 from the stop codon were amplified by PCR and inserted into the SmaI gap of pIIIA/MS2-1 vector (SenGupta et al., 1996) to generate pIIIA/MS2-1-HO 3′-UTRUUGU and pIIIA/MS2-1-HO 3′-UTRUACU. The hybrid RNAs were transcribed by RNA polymerase III. Plasmid pGAD-MPT5 expresses MPT5 fused to the GAL4 transcriptional activation domain. The N-terminal portion of the MPT5 coding sequence was amplified by PCR using a 5′ primer (5′-CTCGGATCCATGATCAATAACGAACCATTTCC-3′, incorporating a BamHI site) and a 3′ primer (5′-GAGTTAGCAGAATTCGATG-3′). An ∼0.3 kb BamHI–EcoRI fragment generated by PCR and a 2.5 kb EcoRI–XhoI fragment containing the C-terminal portion of MPT5 were inserted into the BamHI–SalI gap of pGAD-C1 vector (James et al., 1996) to generate pGAD-MPT5. Plasmid pCgHIS3 is pUC19 carrying the Candida glabrata HIS3 gene (Sakumoto et al., 1999).
Table II. Plasmids used in this study.
| Plasmid | Relevant markers | Source |
|---|---|---|
| YEp13 | LEU2, 2 µm | |
| YEp13MPT5 | MPT5 sequence in YEp13 | Sakai et al. (1992) |
| YEp195 | URA3, 2 µm | |
| YEp195MPT5 | MPT5 sequence in YEp195 | this study |
| YCplac33 | URA3, CEN4 | |
| YCplac33HOp-ADE2-HO 3′-UTRUUGU | HOp-ADE2-HO 3′-UTRUUGU in YCplac33 | this study |
| YCplac33HOp-ADE2-HO 3′-UTRUACU | HOp-ADE2-HO 3′-UTRUACU in YCplac33 | this study |
| pIIIA/MS2-1 | URA3, 2 µm, MS2 sequence behind pol III promoter | SenGupta et al. (1996) |
| pIIIA/MS2-1-HO 3′-UTRUUGU | MS2-HO 3′-UTRUUGU sequence in pIIIA/MS2-1 | this study |
| pIIIA/MS2-1-HO 3′-UTRUACU | MS2-HO 3′-UTRUACU sequence in pIIIA/MS2-1 | this study |
| pGAD-C1 | LEU2, 2 µm, GAL4-AD sequence behind ADH1 promoter | James et al. (1996) |
| pGAD-MPT5 | GAL4-AD-MPT5 sequence in pGAD-C1 | this study |
| pFA6a-3HA-His3MX6 | 3HA-ADH1 3′-UTR-His3 sequence | Longtine et al. (1998) |
| pFA6a-3HA-HO 3′ UTR-His3MX6 | 3HA-HO 3′-UTR-His3 sequence | this study |
| pFA6a-kanMX6-GAL1p-3HA | KanMX6-GAL1p-3HA sequence | Longtine et al. (1998) |
| pCgHIS3 | Candida glabrata HIS3 sequence in pUC19 | Sakumoto et al. (1999) |
3′-RACE
Total RNA was isolated from wild-type yeast cells, W303, as described previously (Sambrook et al., 1989), and treated with RQ1 DNase (Promega). RT–PCR was performed using an Access RT–PCR kit (Promega) and 0.1 µg of total RNA sample as template. Primers were 5′-GGGAACCCGTATATTTCAGC-3′ (5′ primer) and 5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′ (3′ primer). Cycling conditions were 45 min at 48°C, followed by 30 cycles of 30 min at 94°C, 1 min at 60°C, 2 min at 68°C, and then 15 min at 68°C. Nested PCR was performed using a 5′ primer (5′-CTGTCCGCGGGCCTCATAAGAG-3′), a 3′ primer (5′-GCCACGCGTCGACTAGTAC-3′) and Ex Taq polymerase (Takara). The resultant fragment (∼150 bp) was subcloned into the pCR TOPO 2.1 vector (Invitrogen) and sequenced.
Deletion of the Puf genes
The deletions of five Puf genes, MPT5, YGL014w, YLL013c, JSN1 and YPR039c, were constructed by the PCR-based gene deletion method (Baudin et al., 1993; Schneider et al., 1996; Sakumoto et al., 1999). Primer sets were designed such that 46 bases at the 5′ end of the primers were complementary to those at the corresponding region of the target gene, and 20 bases at their 3′ end were complementary to the pUC19 sequence outside the polylinker region in plasmid pCgHIS3 containing the C.glabrata HIS3 gene as a selectable marker. Primer sets for PCR were designed to delete the ORF completely. The PCR products were used to transform the strains K1107 and 10B by selection for His+. The disruption was verified by colony-PCR amplification (Huxley et al., 1990) to confirm that replacement had occurred at the expected locus.
Construction of GAL1p-MPT5 strains
The GAL1p-MPT5 strain was constructed by the PCR-based gene modification method (Longtine et al., 1998). PCR was performed using pFA6a-kanMX6-GAL1p-3HA as template, a 5′ primer (5′-TCTACGCAAATTTATAAATCAATTTCGATTTTTCCAGTTTCTCTTGAATTCGAGCTCGTTTAAAC-3′) and a 3′ primer (5′-TCCATTGGCGAAGAAAAATATTGGGAATTAAGAGATAATACATACGCACTGAGCAGCGTAATCTG-3′). The resultant transformation module was used to transform a haploid strain by selection on YPD medium containing 200 µg/ml geneticin (Gibco-BRL). The resultant transformants were verified by colony-PCR amplification using a 5′ primer (5′-GGGTACTGTTGAGTCAACATC-3′, upstream of the MPT5 ORF) and a 3′ primer (5′-GTTTAAACGAGCTCGAATTC-3′, annealing with the pFA6a-kanMX6-GAL1p-3HA module) to confirm that replacement had occurred at the expected locus.
Construction of ADE2-3HA-HO 3′-UTR and ADE2-3HA-ADH1 3′-UTR strains
The ADE2-3HA-HO 3′-UTR and ADE2-3HA-ADH1 3′-UTR strains were constructed as follows. 3HA-HO 3′-UTR and 3HA-ADH1 3′-UTR transformation modules were amplified by PCR using pFA6a-3HA-HO 3′-UTR-His3MX6 and pFA6a-3HA-GFP(S65T)-His3MX6 as templates, a 5′ primer (5′-CAAAAGTTAGAAACTGTCGGTTACGAAGCTTATCTAGAAAACAAGTGAGGCGCGCCACTTCTAAA-3′) and a 3′ primer (5′-ATTTTATAATTATTTGCTGTACAAGTATATCAATAAACTTATATAGAATTCGAGCTCGTTTAAAC-3′). The resultant PCR products were used to transform a haploid strain by selection for His+. Yeast colony-PCR was carried out using a 5′ primer (5′-GTTTAAACGAGCTCGAATTC-3′, annealing with a portion of the templates) and a 3′ primer (5′-ATTGGAAGACCTTCCAAGGG-3′, annealing downstream of the ADE2 ORF) to confirm that replacement had occurred at the expected locus.
Construction of GAL1p-ADE2-3HA-HO 3′-UTR strain
The GAL1p-ADE2-3HA-HO 3′-UTR strain was constructed by the PCR-based gene modification method (Longtine et al., 1998). PCR was performed using pFA6a-kanMX6-GAL1p as template, a 5′ primer (5′-TCTACGCAAATTTATAAATCAATTTCGATTTTTCCAGTTTCTCGTTAATTCGAGCTCGTTTAAAC-3′) and a 3′ primer (5′-TCCATTGGCGAAGAAAAATATTGGGAATTAAGAGATAATACATACGCACTGAGCAGCGTAATCTG-3′). The resultant PCR product was used to transform the ADE2-3HA-HO 3′-UTR strain by selection on YPD medium containing 200 µg/ml geneticin (Gibco-BRL). The resultant transformants were verified by colony-PCR amplification using a 5′ primer (5′-GGGTACTGTTGAGTCAACATC-3′, upstream of the MPT5 ORF) and a 3′ primer (5′-GTTTAAACGAGCTCGAATTC-3′, annealing with the pFA6a-kanMX6-GAL1p module) to confirm that replacement had occurred at the expected locus.
Mating-type switching assays
Pedigree analysis was performed as described previously (Strathern and Herskowitz, 1979) using homothalic strains TTC85 (HO MPT5) and TTC87 (HO mpt5Δ::CgHIS3).
β-galactosidase assays
β-galactosidase assays were performed as described previously (Kaiser et al., 1994).
Preparation of yeast extracts and western blot analysis
Yeast cells were grown to an optical density (600 nm) of 0.5–1.0 and treated with 2% galactose to activate the GAL1 promoter. After treatment, yeast cultures were quickly chilled, and cells were collected by rapid centrifugation. The pellet was washed twice and then suspended in breaking buffer (4% SDS, 40 mM Tris–HCl pH 7.0, 8 M urea, 0.1 mM EDTA, 1% 2-mercaptoethanol). Glass beads (0.4–0.6 mm diameter) were added to this suspension, and cells were broken by vigorous vortexing for 5 min at room temperature. The beads and cell debris were removed by centrifugation at 14 000 r.p.m. at room temperature. Protein concentrations of cell extracts were measured at 280 nm. Cell extracts were subjected to SDS–PAGE on 7% acrylamide gels followed by electroblotting onto Hybond N+ membrane (Amersham). Blots were blocked by incubation for 15 min at room temperature in TBS-M (TBS with 4% non-fat dry milk). Blots were then incubated with monoclonal antibody HA11 (Babco) diluted 1:2000 (to detect Ade2-3HA) or anti-tubulin antibody diluted 1:1000 (to detect tubulin) in TBS-M overnight at 4°C. After three washes with TBS, blots were incubated for 2 h with peroxidase-conjugated secondary antibody (Calbiochem) diluted 1:3000 with TBS-M. After three final washes with TBS, blots were detected using an enhanced chemiluminescence detection kit (Amersham).
Nothern blot analysis
RNA preparation and sample analysis were performed as described previously (Sambrook et al., 1989). The probes used were all gel-purified, PCR-amplified DNA fragments labeled by random-prime labeling using the Random Primer DNA Labeling Kit Ver 2.0 (Takara). The primers used were as follows: ADE2, a 5′ primer (5′-CAGACTCTGACTCTGACTTGCCGGTAATG-3′) and a 3′ primer (5′-CTCGAATTCCTCGGCGCGCCTTACTTGTTTTCTAGATAAGC-3′), amplifying 0.4 kb of ADE2 coding sequence; ACT1, a 5′ primer (5′-GGAATCTGCCGGTATTGACC-3′) and a 3′ primer (5′-ACATACGCGCACAAAAGCAG-3′), amplifying 0.4 kb of ACT1 coding sequence.
Yeast three-hybrid assays
Three-hybrid assays were performed as described previously (SenGupta et al., 1996). Yeast strain L40c (L40-coat ofSenGupta et al., 1996) carrying the pGAD-MPT5 plasmid was transformed with pIIIA/MS2-1-HO 3′-UTRUUGU, pIIIA/MS2-1-HO 3′-UTRUACU or pIIIA/MS2-1 vector. Transformants were tested for growth on SC-Ura Leu His plates.
Localization of ASH1 mRNA and Ash1 protein
In situ RNA hybridization with digoxigenin-labeled ASH1 antisense probe was performed as described previously (Takizawa et al., 1997). Indirect immunofluorescence microscopy of an Ash1-myc strain was described (Sil and Herskowitz, 1996) using anti-myc antibodies (9E10).
Analysis of mRNA decay rate
The GAL1p-ADE2-3HA-HO 3′-UTR strain harboring YEp195-MPT5 or empty vector was incubated in SR-Ura. Transcription was induced by adding galactose to a final concentration of 2%. An aliquot was removed at 10–15 min after addition of galactose. The culture was then transferred to medium containing 2% glucose. Aliquots were removed at various times, and cells were collected by rapid centrifugation. RNAs were isolated from each sample and the amounts of the ADE2 transcript and the ACT1 transcript as a control were quantitated by dot blotting. Decay rates were determined from semilog plots of the percentage of hybridizing material remaining at different times after the inhibition of transcription.
Acknowledgments
Acknowledgements
We thank R.P.Jansen, A.Sakai, M.P.Wickens, Y.Mukai and S.Harashima for materials; A.Sil, R.Vale, P.Takizawa and S.Shaham for helpful discussions; and R.Parker for communicating results prior to publication. These studies were supported by the Senri Life Science Foundation (K.I.); special grants for CREST, Advanced Research on Cancer from the Ministry of Education, Culture, and Science of Japan, the Uehara Memorial Foundation and the Daiko Foundation (K.M.); and by the United States National Institute of Health (I.H.).
References
- Baudin A., Ozier,K.O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N., Jansen,R.P., Shin,T.H. and Nasmyth,K. (1996) Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell, 84, 699–709. [DOI] [PubMed] [Google Scholar]
- Chen T. and Kurjan,J. (1997) Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol. Cell. Biol., 17, 3429–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Eisen,M.B., Botstein,D. and Brown,P.O. (2000) Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nature Genet., 25, 58–62. [DOI] [PubMed] [Google Scholar]
- Goodwin E.B., Okkema,P.G., Evans,T.C. and Kimble,J. (1993) Translational regulation of tra-2 by its 3′ untranslated region controls sexual identity in C.elegans. Cell, 75, 329–339. [DOI] [PubMed] [Google Scholar]
- Gray N.K. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- Guo Z. and Sherman,F. (1996) Signals sufficient for 3′-end formation of yeast mRNA. Mol. Cell. Biol., 16, 2772–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H., Mitsui,H., Liu,H., Bai,Y., Denis,C.L., Shimizu,Y. and Sakai,A. (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics, 148, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. (1988) Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev., 52, 536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C., Green,E.D. and Dunham,I. (1990) Rapid assessment of S.cerevisiae mating type by PCR. Trends Genet., 6, 236. [DOI] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E., Motzny,C.K., Graves,L.E. and Goodwin,E.B. (1999) The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J., 18, 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.P., Dowzer,C., Michaelis,C., Galova,M. and Nasmyth,K. (1996) Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell, 84, 687–697. [DOI] [PubMed] [Google Scholar]
- Kaiser C.A., Adams,A. and Gottschling,D.E. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kennedy B.K., Austriaco,N.J., Zhang,J. and Guarente,L. (1995) Mutation in the silencing gene SIR4 can delay aging in S.cerevisiae. Cell, 80, 485–496. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K. et al. (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S.cerevisiae. Cell, 89, 381–391. [DOI] [PubMed] [Google Scholar]
- Kraemer B., Rittenden,S., Gallegos,M., Moulder,G., Barstead,R., Kimble,J. and Wickens,M. (1999) NANOS-3 and FBF proteins physically interact to control the sperm–oocyte switch in Caenorhabditis elegans. Curr. Biol., 9, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Liu H.Y., Badarinarayana,V., Audio,D.C., Rappsilber,J., Mann,M. and Denis,C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 18, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Singer,R.H., Meng,X., Gonzalez,I., Nasmyth,K. and Jansen,R.P. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science, 277, 383–387. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.R., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Machin N.A., Lee,J.M. and Barnes,G. (1995) Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell, 6, 1241–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y. and Wharton,R.P. (1995) Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell, 80, 747–756. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (1983) Molecular analysis of a cell lineage. Nature, 302, 670–676. [DOI] [PubMed] [Google Scholar]
- Olivas W. and Parker,R. (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J., 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., Chibazakura,T., Shimizu,Y. and Hishinuma,F. (1992) Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res., 20, 6227–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumoto N. et al. (1999) A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast, 15, 1669–1679. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schneider D., Bruton,C.J. and Chater,K.F. (1996) Characterization of spaA, a Streptomyces coelicolor gene homologous to a gene involved in sensing starvation in Escherichia coli. Gene, 177, 243–251. [DOI] [PubMed] [Google Scholar]
- SenGupta D.J., Zhang,B., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A. and Herskowitz,I. (1996) Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell, 84, 711–722. [DOI] [PubMed] [Google Scholar]
- Sonoda J. and Wharton,R.P. (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev., 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J.N. and Herskowitz,I. (1979) Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell, 17, 371–381. [DOI] [PubMed] [Google Scholar]
- Subramaniam K. and Seydoux,G. (1999) nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development, 126, 4861–4871. [DOI] [PubMed] [Google Scholar]
- Takizawa P.A., Sil,A., Swedlow,J.R., Herskowitz,I. and Vale,R.D. (1997) Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature, 389, 90–93. [DOI] [PubMed] [Google Scholar]
- Waskiewicz S.B., Skala,J., Jasinski,M., Grenson,M., Goffeau,A. and Ulaszewski,S. (1998) Functional analysis of three adjacent open reading frames from the right arm of yeast chromosome XVI. Yeast, 14, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Wharton R.P. and Struhl,G. (1991) RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell, 67, 955–967. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda,J., Lee,T., Patterson,M. and Murata,Y. (1998) The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell, 1, 863–872. [DOI] [PubMed] [Google Scholar]
- Wickens M., Anderson,P. and Jackson,R.J. (1997) Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev., 7, 220–232. [DOI] [PubMed] [Google Scholar]
- Wreden C., Verrotti,A.C., Schisa,J.A., Lieberfarb,M.E. and Strickland,S. (1997) Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development, 124, 3015–3023. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Bartel,D.P., Lehmann,R. and Williamson,J.R. (1999) The PUMILIO–RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry, 38, 596–604. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gallegos,M., Puoti,A., Durkin,E., Fields,S., Kimble,J. and Wickens,M.P. (1997) A conserved RNA-binding protein that regulates sexual fates in the C.elegans hermaphrodite germ line. Nature, 390, 477–484. [DOI] [PubMed] [Google Scholar]


