Abstract
Development of the Caenorhabditis elegans vulva serves as a paradigm for intercellular signaling during animal development. In wild-type animals, the somatic gonadal anchor cell generates the LIN-3/EGF ligand to induce vulval fates in the underlying hypodermis, whereas FBF, FOG-1, and FOG-3 control germ-line development. Here we report that FBF functions redundantly with FOG-1 and FOG-3 to control vulval induction: animals lacking FBF and either FOG-1 or FOG-3 have multiple vulvae, the Muv phenotype. The fog; fbf Muv phenotype is generated by aberrant induction of vulval precursor cells (VPCs): in wild-type animals, three VPCs are induced to form a single vulva, but, in fog; fbf mutants, four or five VPCs are typically induced, resulting in ectopic vulvae. Laser ablation experiments and mosaic analyses demonstrate that the germ line is critical for the fog; fbf Muv phenotype. Consistent with that site of action, we detect FBF and FOG-1 in the germ line but not in the VPCs. The simplest interpretation is that FOG-1, FOG-3, and FBF act in the germ line to influence vulval fates. The LIN-3/EGF ligand may be the germ-line signal to the VPCs: the fog; fbf Muv phenotype depends on LIN-3 activity, and the lin-3 3′ UTR possesses an FBF binding element. Our findings reveal new insights into germ line-to-soma signals and the role of PUF proteins in animal development.
Keywords: fem-3 binding factor, FOG-1, FOG-3, LIN-3
The Caenorhabditis elegans vulva serves as a paradigm for cell–cell interactions in animal development. Intensive studies over many years have converged on an elegant and detailed model for how multiple signaling pathways are integrated to establish the precise pattern of cell division and differentiation necessary to form the mature vulva, a well defined and tightly organized multicellular structure (1). Given this wealth of information from C. elegans, vulval development is now being investigated in other nematode species to dissect the evolution of pathways controlling organogenesis (2). Here we report that a signal from the germ line can induce vulval development and that fem-3 binding factor (FBF), FOG-1, and FOG-3 normally repress that signal.
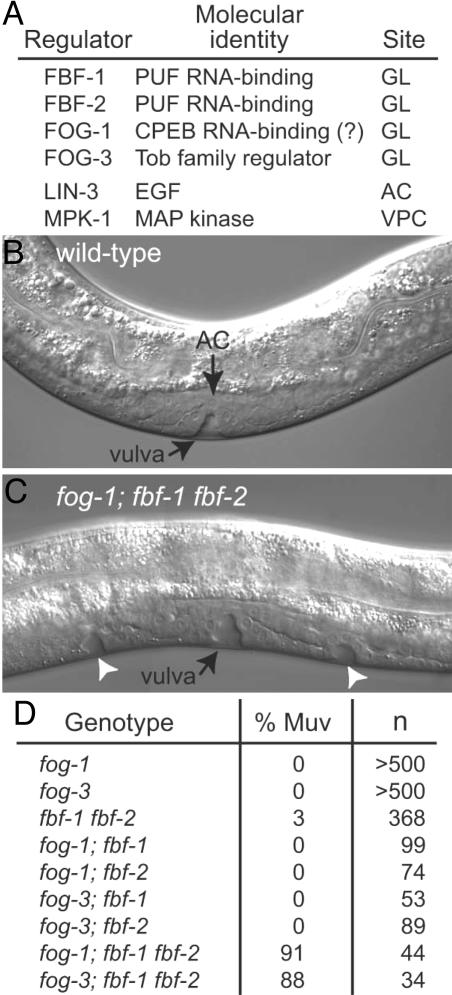
We began this work by analyzing the molecular regulation of germ-line development in C. elegans. Regulatory proteins central to our work are summarized in Fig. 1A. FBF is encoded by two nearly identical genes, fbf-1 and fbf-2 (3). FBF-1 and FBF-2 are largely redundant and biochemically interchangeable (3–6). FBF belongs to the PUF family of RNA-binding proteins, and it binds specifically to FBF binding elements (FBEs) within the 3′ UTR of target mRNAs to repress their expression (3–10). Both FBF-1 and FBF-2 are expressed in the germ line throughout postembryonic development (3–5) and control stem cell maintenance and sex determination in that tissue (3, 4).
Fig. 1.
FBF and FOG-1 or FOG-3 control vulval development. (A) Summary of regulators relevant to this work. (B) Wild-type L4, Nomarski micrograph. A single vulva is induced. (C) fog-1; fbf-1 fbf-2 L4, Nomarski micrograph. Ectopic vulvae (arrowheads) are induced in addition to the main vulva. (D) Penetrance of Muv defects. Genotype, all mutants are homozygous for null mutations of the indicated loci; % Muv, percentage of animals with at least one ectopic pseudovulva; n, number of animals scored.
FOG-1 (for feminization of the germ line) is a homolog of CPEB (for cytoplasmic polyadenylation element binding protein) (11, 12), and FOG-3 is a Tob family protein (13). Vertebrate CPEB binds specifically to cytoplasmic polyadenylation elements (CPEs) in target mRNA 3′ UTRs and either represses or activates their expression (reviewed in ref. 14). Vertebrate Tob proteins are antiproliferative (reviewed in ref. 15) and have been implicated in both transcriptional and posttranscriptional regulation (16–19). FOG-1 protein is present in the germ line (9); fog-3 mRNA is expressed in the germ line (13), but anti-FOG-3 antibodies are not available. Phenotypically, fog-1 and fog-3 mutants are essentially indistinguishable, although the two regulators are molecularly distinct.
During early larval development, FBF functions redundantly with FOG-1 and FOG-3 to control germ-line proliferation (9). Later, FBF maintains germ-line stem cells (4), FOG-1 and FOG-3 specify the sperm fate (20, 21), and FBF promotes the hermaphrodite switch from spermatogenesis to oogenesis (3). FBF controls germ-line development by repression of a set of target mRNAs that themselves are key regulators of germ-line development, including fbf-1, fbf-2, fog-1, gld-1, gld-3, fem-3, and lip-1 (3–5, 8–10). Little is known about FOG-1 binding specificity or target mRNAs (22), and virtually nothing is known about the FOG-3 molecular mechanism of control.
The C. elegans vulva is a specialization of the body wall, an opening in the hypodermis used for egg laying and sperm entry. A somatic regulatory cell in the gonad, the anchor cell (AC), induces vulval fates in vulval precursor cells (VPCs) of the underlying hypodermis (reviewed in ref. 1). Normally, only three VPCs (P5.p–P7.p) are induced, although all six (P3.p–P8.p) are competent to adopt vulval fates. Briefly, the AC signals by LIN-3/EGF to the underlying VPCs, which express the LET-23/EGF receptor. The activated LET-23/EGF receptor in turn stimulates Ras/mitogen-activated protein kinase signaling to promote vulval fates. In the absence of LIN-3/EGF signaling, VPCs adopt a nonvulval fate, and their daughters fuse with the syncytial hypodermis. Here we show that FBF acts redundantly with FOG-1 and FOG-3 to repress ectopic vulval induction.
Results and Discussion
Germ-Line Regulators Control Vulval Development. Wild-type hermaphrodites possess a single, centrally located vulva (Fig. 1B). Similarly, all fog-1 and fog-3 single mutants have a single vulva, as do most fbf-1 fbf-2 double mutants (Fig. 1D). By contrast, most fog-1; fbf-1 fbf-2 and fog-3; fbf-1 fbf-2 triple mutants have multiple vulvae, the Muv phenotype (Fig. 1 C and D). This effect requires removal of both fbf genes: fog; fbf-1 and fog; fbf-2 double mutants are not Muv (fog refers to either fog-1 or fog-3) (Fig. 1D). We conclude that fog-1 and fog-3 function redundantly with fbf to inhibit ectopic vulval formation.
To learn the cellular basis of the fog; fbf Muv defect, we examined VPC induction in both fog-1; fbf and fog-3; fbf mutants (fbf refers to the fbf-1 fbf-2 double mutant). In the wild type, P5.p–P7.p adopt vulval fates, whereas P3.p, P4.p, and P8.p adopt nonvulval fates, and their daughters fuse with the hypodermis. By contrast, in fog-1; fbf and fog-3; fbf mutants, P4.p and P8.p daughters often failed to fuse with the hypodermis; instead, they continued to divide and generated ectopic vulvae (for P4.p 50% and for P8.p 60% adopted a vulval fate in both fog-1; fbf and fog-3; fbf mutants; n = 10 for each genotype); P3.p was not induced and fused with the hypodermis. However, we occasionally observed two ectopic vulvae anterior to the main vulva, suggesting that P3.p can adopt a vulval fate. To determine whether ectopically induced VPCs in fog; fbf mutants followed a primary or secondary vulval fate, we examined their lineage and found that P4.p descendants divided in a manner characteristic of the primary vulval fate (n = 2 for fog-1, and n = 1 for fog-3). In contrast, P3.p was not induced; either P8.p was not induced or its complete lineage was not determined. We conclude that the presence of FOG-1 and FOG-3, or FBF, normally inhibits vulval fates in the descendents of P3.p, P4.p, and P8.p.
FBF, FOG-1, and FOG-3 control germ-line development by repressing the activities of gld genes (4, 8, 9). Whereas fbf-1 fbf-2 double mutant adults possess no germ-line stem cells and make only sperm (4), gld-1 gld-2; fbf-1 fbf-2 quadruple mutant adults have a germ-line tumor (8); similarly, whereas fog; fbf-1 fbf-2 triple mutants possess very few germ cells that are oogenic, gld-1 gld-2; fbf-1 fbf-2; fog-1(RNAi) germ lines are tumorous (9). We therefore depleted gld-1 and gld-2 by RNA interference (RNAi) to ask whether a similar suppression was observed for the fog; fbf Muv defect. In these experiments, RNAi-treated animals generated half fog/+; fbf-1 fbf-2 and half fog; fbf-1 fbf-2 triple homozygotes (fog refers to either fog-1 or fog-3). In animals not treated with RNAi, half were Muv, as expected (fog-1, 47%, n = 17; fog-3, 42%, n = 113). By contrast, in all gld-1(RNAi) gld-2(RNAi) animals, vulval development was normal (n = 14 for fog-1, and n = 31 for fog-3). Similarly, vulval development was normal when fog-1 was depleted by RNAi in gld-1 gld-2; fbf-1 fbf-2 quadruple mutants (data not shown). Therefore, depletion of gld-1 and gld-2 abolishes both germ-line and vulval defects in fog-1; fbf-1 fbf-2 and fog-3; fbf-1 fbf-2 animals.
We conclude that fbf-1 and fbf-2 function redundantly with either fog-1 or fog-3 to affect vulval development. It is important to note that fog-1 and fog-3 were indistinguishable in all Muv assays, which was also true for their effects on germ-line development (refs. 9, 20, and 21 and B.E.T., unpublished observations). We therefore refer to fog-1; fbf-1 fbf-2 and fog-3; fbf-1 fbf-2 triple mutants as fog; fbf for simplicity, and we name the specific fog gene when relevant.
Germ-Line Expression of FBF-1, FBF-2, and FOG-1. The FOG-1, FOG-3, and FBF proteins are key regulators of germ-line development (23); a role in somatic development was unexpected. Antibodies specific for FOG-1, FBF-1, and FBF-2 have shown that each of these regulators is present in the germ line (3, 5, 9), but no somatic staining has been reported. Furthermore, although anti-FOG-3 antibodies have not been made, fog-3 mRNA was not detected in animals lacking a germ line, and mosaic analysis indicated that fog-3 functions in the germ line to promote spermatogenesis (13). If FOG-1, FOG-3, FBF-1, and FBF-2 act cell-autonomously in the VPCs to control vulval development, we would expect them to be expressed in the VPCs. We therefore attempted to detect FBF-1, FBF-2, or FOG-1 in the VPCs or their daughters during larval development. However, we could not detect them in the ventral hypodermis of L1, L2, or L3 larvae (Fig. 5, which is published as supporting information on the PNAS web site, and data not shown). However, we cannot exclude the possibility that these regulators are present in the VPCs at a level below detection.
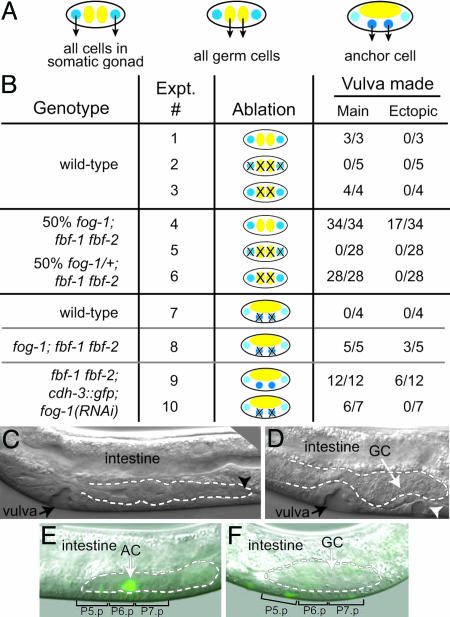
Germ-Line Induction of Vulval Fates: Laser Ablation Studies. The presence of FBF-1, FBF-2, and FOG-1 in the germ line and their apparent absence from VPCs suggested that they might function in the germ line to control vulval development. To address this idea, we used laser ablation in fog; fbf mutants to assess the importance of specific gonadal precursor cells for vulval development. Specifically, we ablated individual precursor cells in early L1 larvae and scored vulvae in L4s; our results are summarized in Fig. 2B, with each experiment assigned a number. The gonadal primordium contains four cells, two somatic precursor cells that give rise to the entire somatic gonad, including the vulva-inducing AC, and two germ-line precursor cells that generate the entire germ line (Fig. 2 A Left and Center). We first examined wild-type controls (Fig. 2B, Exps. 1–3). Unablated wild-type animals generated a single main vulva and no ectopic vulvae (Fig. 2B, Exp. 1), ablation of all four gonadal precursor cells abolished vulval induction (Fig. 2B, Exp. 2), and ablation of the two germ-line precursor cells had no effect on vulval development (Fig. 2B, Exp. 3) (also see ref. 24). To do these same experiments in fog; fbf mutants, we used L1s derived from a mating that segregates half fog-1; fbf homozygotes and half fog-1/+; fbf animals. In unablated controls, all progeny made a main vulva, and the expected half made ectopic vulvae (Fig. 2B, Exp. 4). When all four gonadal precursor cells were ablated, no vulval tissue was made (Fig. 2B, Exp. 5). Therefore, generation of ectopic vulvae indeed depends on the presence of the gonad. Finally, and most importantly, when the two germ-line precursor cells were ablated, all progeny made a main vulva, but none made ectopic vulvae (Fig. 2 B, Exp. 6, and C). We conclude that the generation of ectopic vulvae depends not only on the gonad but more specifically on the germ line.
Fig. 2.
Germ cells induce ectopic vulvae in fog-1; fbf-1 fbf-2. (A) Gonadal precursor cells at time of laser ablation. (Left and Center) Gonadal primordium. Blue, somatic gonadal precursor cells; yellow, germ-line precursor cells. (Right) Somatic gonadal precursor cells give rise to Z1.p and Z4.a daughters with AC potential. (B) Summary of ablation results. See text for details. (C–E) Broken lines indicate gonad. GC, germ cells. (C) Representative L4 from an experiment in which germ-line precursor cells, Z2 and Z3, were ablated in L1s from a mating that generates half fog-1/+; fbf-1 fbf-2 and half fog-1; fbf-1 fbf-2 progeny. The main vulva and somatic gonad are both present and normal, but no germ cells are present. The black arrowhead indicates the distal end of the gonad. (D) fog-1; fbf-1 fbf-2 mutant in which AC precursors Z1.p and Z4.a were ablated. Both main vulva and an ectopic pseudovulva (white arrowhead) have been induced despite the absence of an AC. Germ cells are present directly above the induced VPCs. (E) fbf-1 fbf-2; cdh-3::gfp; fog-1(RNAi) animal, unablated. AC is readily detectable (green), and P5.p–P7.p have been induced. Gonad visible in this focal plane (outlined by white dashed line) consists primarily of somatic cells. (F) fbf-1 fbf-2; cdh-3::gfp; fog-1(RNAi) animal with Z1.p and Z4.a ablated; no AC is detectable (lack of green). Nonetheless, P5.p–P7.p have been induced. Gonad visible in this focal plane (outlined by white dashed line) consists primarily of germ cells.
We next asked whether the AC is critical for vulval induction in fog; fbf mutants. Specifically, we ablated Z1.p and Z4.a, the two somatic gonadal cells capable of producing an AC (Fig. 2 A Right). Ablation of Z1.p and Z4.a in wild-type animals abolished vulval induction (Fig. 2B, Exp. 7) (also see ref. 24). By contrast, when Z1.p and Z4.a were ablated in fog-1; fbf triple mutants, all animals generated a main vulva and most generated ectopic vulvae (Fig. 2 B, Exp. 8, and D). To confirm AC removal, we ablated Z1.p and Z4.a in mutants carrying an AC marker, cdh-3::gfp (25). Thus, we generated fbf-1 fbf-2 double mutants carrying the integrated cdh-3::gfp transgene, and we treated these animals with fog-1 RNAi. All intact animals made a GFP-expressing AC that induced a main vulva (Fig. 2E), and half of them also made ectopic vulvae (Fig. 2B, Exp. 9). The lower penetrance of the Muv phenotype is likely a result of using RNAi to deplete fog-1 instead of using a fog-1-null mutant. When Z1.p and Z4.a were ablated in fbf-1 fbf-2; cdh-3::gfp; fog-1(RNAi) animals, no GFP-expressing AC was seen (Fig. 2F), but a vulva was nonetheless induced (Fig. 2 B, Exp. 10, and F). Although vulval induction was less robust than in the triple mutant, this is likely due to variability in the effectiveness of RNAi. The key result is that vulval induction occurred in the absence of the AC. Interestingly, in other nematode species, vulval induction can occur independent of the AC (26, 27). We conclude that the Muv phenotype in fog-1; fbf mutants is dependent on the germ line and independent of the AC. Therefore, we favor the model that the germ line is capable of vulval induction.
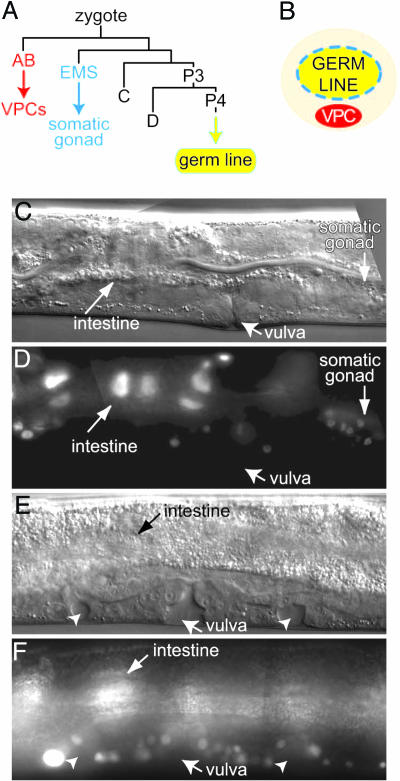
Germ-Line Induction of Vulval Fates: Mosaic Analysis. To further assess the apparent germ cell focus of the fog; fbf Muv defect, we turned to a genetic mosaic analysis. The first step in making mosaic animals in C. elegans is generation of an extrachromosomal array carrying both the gene of interest (e.g., fog-3) and a ubiquitously expressed and easily detectable marker (e.g., sur-5::gfp). Mosaics are identified as animals with some cells expressing the ubiquitous marker and other cells not expressing the marker. Given the invariant lineage of C. elegans (28), it is possible to identify mosaic animals in which the extrachromosomal array was lost from one of the major founder cells during early embryogenesis. For example, the AB founder cell produces all six VPCs and most of the hypodermis, EMS generates the entire somatic gonad, and P4 gives rise to the entire germ line (Fig. 3A). Thus, although the VPCs, somatic gonad, and germ line are in close contact (Fig. 3B), each derives from a distinct founder cell.
Fig. 3.
Mosaic analysis of fog; fbf Muv phenotype. (A) Lineage of embryonic founder cells. The AB founder cell generates all VPCs as well as most neurons and hypodermis; EMS generates the somatic gonad; P4 generates all germ cells. Modified from Sulston et al. (28). (B) Schematic of tissues relevant to this work. The gonad includes both somatic cells (blue) that partially encapsulate the germ line and germ cells (yellow); VPCs (red) lie under the gonad. (C and D) Genetic mosaic in which qEx555 [fog-3(+); sur-5::gfp] was lost from the AB lineage but was retained in P3; the main vulva was made, but no ectopic vulvae are seen. The same animal is shown in C and D.(C) Nomarski micrograph. (D) GFP is expressed in the intestine and somatic gonad, but not in the vulval cells, neurons, or hypodermis. See text for further explanation. (E and F) Genetic mosaic in which qEx555 [fog-3(+); sur-5::gfp] was lost from the P3 lineage, which includes both D and P4, but retained in the AB lineage; both the main vulva and ectopic pseudovulvae (arrowheads) are made. The same animal is shown in E and F.(E) Nomarski micrograph. (F) GFP is expressed in vulval cells and intestine; GFP is also expressed in neurons and hypodermis, but not in muscle cells derived from the D founder cell (data not shown).
To generate mosaics, we focused on the fog-3 gene for several reasons. First, an array carrying the fog-3 genomic region can rescue most fog-3 mutants to fertility (13); by contrast, fbf transgenes are not capable of efficient rescue (B.E.T., unpublished observations). Second, the fog-3 genomic region is contained on a single cosmid; by contrast, fog-1 is much larger. Third, fog-3 is in a better genetic location than fog-1 for chromosomal balancing. Given these various technical constraints, the fog-3 gene was selected for mosaic analysis.
Mosaics were generated by using the extrachromosomal array qEx555 [fog-3(+); sur-5::gfp], which was introduced into an animal heterozygous for both fog-3 and fbf-1 fbf-2: fog-3/Balancer; fbf-1 fbf-2/GFPBalancer, where the GFP balancer carries an integrated GFP transgene (see Methods). Without the array, this heterozygous mother produced three types of sterile progeny: (i) fog-3; fbf-1 fbf-2/GFPBal, which made only oocytes; (ii) fog-3/Bal; fbf-1 fbf-2, which made only sperm; and (iii) fog-3; fbf-1 fbf-2 triple homozygotes, which made only a few oogenic germ cells (9). We assessed function of the array in multiple ways. First, most fog-3; fbf-1 fbf-2/GFPBal; qEx555 animals were fertile (62%, n = 163). Also, fog-3; fbf-1 fbf-2; qEx555 animals made >100 germ cells instead of the 10 germ cells typical of the triple mutant. The rescue was therefore not complete, but it was sufficient for mosaic analysis.
We identified mosaics that had lost the qEx555 array from the AB founder cell to ask whether fog-3 might act in the VPCs or other AB-derived tissues (e.g., hyp7 is also largely derived from AB); in addition, we found other mosaics that had lost the array from the P3 founder cell to assess its action in the germ line. AB loss of the array was scored by loss of GFP from cells in the ventral nerve cord, as well as head and pharyngeal neurons; AB retention of the array was scored by expression of GFP in most of those same cells. P3 loss of the array was scored by loss of GFP from muscle cells specific to the D founder cell as well as the presence of only a few oogenic germ cells, which is typical of fog-3; fbf triple mutants; P3 retention of the array was scored by GFP expression in D muscle cells as well as production of >100 germ cells, a feature typical of fog-3; fbf-1 fbf-2; qEx555 germ lines.
We found five fog-3; fbf-1 fbf-2 mosaics that had lost the array from the AB lineage but retained it in the P3 lineage. All of these “AB-loss” mosaics possessed a main vulva that appeared normal and did not have ectopic vulvae (Fig. 3 C and D). This result suggests that wild-type fog-3 activity expressed in cells outside the AB lineage was sufficient to prevent a Muv phenotype. We also found 14 fog-3; fbf-1 fbf-2 mosaics that had lost the array from the P3 lineage but maintained it in most descendants of the AB lineage. Most of these “P3-loss” mosaics had one or two ectopic vulvae in addition to a main vulva (85%, n = 14) (Fig. 3 E and F). The high percentage of Muv animals among P3-loss mosaics was similar to the percentage of Muv animals among fog-3; fbf-1 fbf-2 triple mutants (85% vs. 88%). This result suggests that loss of wild-type fog-3 activity from the P3 lineage results in ectopic vulvae. The simplest explanation is that fog-3 functions in the germ line to control vulval development.
The Germ Line Can Induce Vulval Fates. In-depth analyses of vulval development have been conducted for years (1), but induction by the germ line became apparent only after removal of redundant regulators (FOG-1 or FOG-3 in addition to FBF). A low-penetrance Muv phenotype had been noted in fbf-1 fbf-2 double mutants and fbf(RNAi) animals for several years (J.K., unpublished observations), but that effect was not practical to analyze. The germ line might have a role in vulval development in other nematode species: depletion of the germ-line regulator glp-1 by RNAi in Caenorhabditis briggsae also results in a Muv defect (29). However, the site of GLP-1 action was not assessed in those experiments.
Our experiments distinguish between two simple models to explain the Muv phenotype in fog; fbf mutants. The first model is that the fog and fbf genes act in the soma to specify vulval fates, with the simplest version of that model being that they act in the VPCs. The second model is that the fog and fbf genes act in the germ line to affect vulval fates. Other models can be envisioned: for example, FOG-1 and FOG-3 might act in the germ line, and FBF might act in the soma. Regardless, we conclude that the germ line is capable of inducing vulval fates and that the germ line does not accomplish this by modulating an AC signal (laser ablation studies). We also conclude that FOG-3 functions in the germ line to antagonize production of ectopic vulvae (mosaic analysis). Finally, we failed to detect FBF-1, FBF-2, or FOG-1 in the VPCs during larval development, but we did detect all three proteins in the germ line (antibody staining). The simplest interpretation of these data, when they are considered together, is that FBF, FOG-1, and FOG-3 function together in the germ line to affect vulval fates.
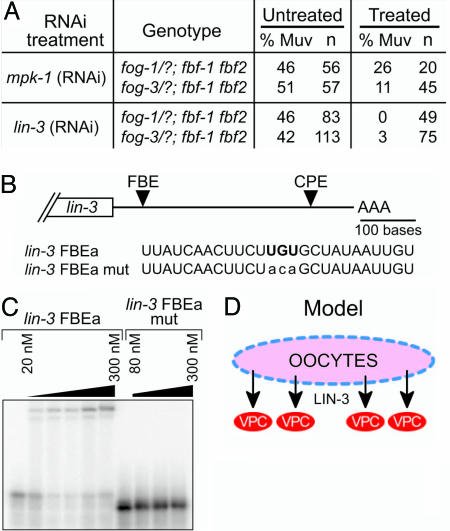
lin-3 mRNA Is a Candidate Target of FBF Repression. The AC normally induces vulval fates by expression of the LIN-3/EGF ligand, which activates the LET-23/EGF receptor and the Ras/mitogen-activated protein kinase pathway in the VPCs (1). We next asked whether the fog; fbf Muv defect operates via this same pathway. We first tested mpk-1, the C. elegans mitogen-activated protein kinase that normally controls vulval fates in the VPCs (30). If mpk-1 activity were activated aberrantly in fog; fbf mutants, we would expect depletion of mpk-1 by RNAi to alleviate the fog; fbf Muv defect. In these experiments (as described above), we examined populations comprised of half fog; fbf homozygotes and half fog/+; fbf animals. In controls not treated with mpk-1(RNAi), Muv animals were observed at the expected ratio; by contrast, in fog; fbf; mpk-1(RNAi) animals, the Muv defect was reduced (Fig. 4A). Because mpk-1(RNAi) defects were less severe than those of mpk-1(0) mutants (data not shown), the Muv animals remaining in the fog; fbf; mpk-1(RNAi) experiments may reflect incomplete mpk-1 depletion. We suggest that mpk-1 functions downstream of fog; fbf to control vulval development.
Fig. 4.
FBF binds FBE in lin-3 3′ UTR. (A) Summary of mpk-1(RNAi) and lin-3(RNAi) experiments. RNAi-treated animals are half fog/+; fbf and half fog; fbf homozygotes. (B) Putative FBE in the lin-3 3′ UTR, beneath which is shown the nucleotide sequence of predicted FBE (FBEa) with the boldface UGU motif in the core of the conserved consensus. In FBEa mut, this conserved UGU is mutated to ACA (lowercase). (C) FBF-2 binds the wild-type FBE from the lin-3 3′ UTR but not to a sequence with a mutated site (FBE mut). Protein concentrations are 20, 40, 80, 150, and 300 nM for wild-type RNA and 80, 160, and 300 nM for mutant RNA. (D) Model for induction of vulval fates by LIN-3 signal from the germ line.
To refine our placement of fog and fbf in the vulval induction pathway, we next tested lin-3, the EGF ligand that normally induces vulval induction by expression in the AC (31). Again, half the animals tested were fog; fbf homozygotes and half were fog/+; fbf animals. In controls not treated with lin-3(RNAi), Muv animals were observed at the expected ratio, but the Muv defect was sharply reduced in fog; fbf; lin-3(RNAi) animals (Fig. 4A). Therefore, LIN-3/EGF is required for vulval induction in fog; fbf animals. This result was unexpected because the fog; fbf Muv defect appears to be independent of the AC, the source of LIN-3/EGF in wild-type animals. We suggest that the fog; fbf mutant germ line signals to the soma by the LIN-3/EGF ligand.
We next postulated that FBF might affect vulval development by repressing lin-3 mRNA in the germ line. This idea was based on the following: (i) FBF represses expression of target mRNAs (7); (ii) the fog; fbf Muv defect relies on induction from the germ line; (iii) the fog; fbf Muv defect is alleviated by lin-3 RNAi. To explore the idea that FBF might directly regulate lin-3 mRNA, we examined the lin-3 3′ UTR for putative FBF regulatory elements and found a putative FBE that conforms to the consensus UGURHHAUW (where H is A, C, or U and W is A or U) (6) (Fig. 4B). We next tested whether FBF could bind this putative FBE. Specifically, we incubated purified FBF-2 (a kind gift of Brad Hook, University of Wisconsin, Madison, WI) with 32P-labeled synthetic RNA containing the potential FBE plus surrounding sequences from the lin-3 3′ UTR, and then we separated products by gel electrophoresis. FBF-2 retarded the mobility of an RNA containing the wild-type FBE (Fig. 4C) but did not retard a similar RNA in which the central UGU of the FBE had been mutated to ACA (Fig. 4C). We conclude that FBF-2 can bind directly to the FBE within the lin-3 3′ UTR and that this interaction is specific. Because FBF-1 and FBF-2 are biochemically indistinguishable with respect to binding specificity (ref. 6 and references therein), we suggest that either FBF-1 or FBF-2 can bind the lin-3 3′ UTR and repress lin-3 expression in the germ line. Recently, it has been shown that LIN-3 signals from the VPCs in addition to the AC and that this VPC signaling must be fine-tuned to allow proper development (32). FBF repression of lin-3 may similarly prevent a broad and unlocalized induction and therefore be part of a larger mechanism that localizes LIN-3 activity.
What Are the Roles of FOG-1 and FOG-3? FBF repression of lin-3 expression cannot fully explain the fog; fbf Muv defect. What are the roles of FOG-1 and FOG-3? We do not know the answer to this question, but we suggest two possibilities. One idea is that FOG-1 and FOG-3 also repress lin-3 expression. FOG-1 is a homolog of vertebrate CPEB, which can act as an mRNA repressor (reviewed in ref. 14), and FOG-3 is a Tob family protein, members of which physically interact with known RNA regulators (16–19). The lin-3 3′ UTR contains a putative CPE that conforms to the consensus UUUUUAU for vertebrate CPEs (reviewed in ref. 14), but little is known about FOG-1 binding sites. A second idea is that FOG-1 and FOG-3 act indirectly, perhaps by feminizing the germ line. In the wild-type germ line, it has been suggested that LIN-3 from oocytes might stimulate sheath cell contraction (33, 34). LIN-3 production by oocytes in wild-type adults (or in fog single mutants) is not expected to interfere with vulval induction, because oocytes are made only after the vulva has been induced and completed maturation. However, in fog; fbf triple mutants, oogenesis occurs prematurely, during the second or third larval stage, just when vulval induction by the AC normally occurs (9). Suppression of the fog; fbf Muv phenotype by removal of gld genes (e.g., gld-1 and gld-2) is consistent with this explanation because germ cells no longer enter oogenesis prematurely: the gld-1 gld-2; fbf-1 fbf-2; fog-1(RNAi) germ lines are tumorous.
Conclusions and Implications for Animal Development
Fig. 4D presents a simple model to summarize our findings. Our discovery that the germ line can induce the vulva provides insights into diverse areas of biological regulation: vulval development, germ line-to-soma signals, and the roles of PUF proteins in animal development. First, our findings underscore the importance of coordinating the regulation of potent signaling molecules during animal development. To ensure normal vulval development, expression of LIN-3/EGF in the germ line must be repressed or delayed. Second, we note that Drosophila oocytes use a LIN-3/EGF homolog, known as GURKEN, to signal surrounding follicle cells and establish both anteroposterior and dorsoventral axes (35, 36). It is plausible, although of course speculative, that LIN-3/EGF ligands are used broadly for signaling from germ line to soma. Third and most speculative is the idea that FBF, and indeed PUF proteins more generally, repress a broader spectrum of target mRNAs than previously appreciated. The known FBF target mRNAs to date all encode proteins that direct germ-line differentiation, and their repression by FBF made sense for maintenance of stem cells (3, 4, 8–10). However, lin-3 has no obvious role in the immature germ line. Therefore, FBF repression of lin-3 mRNA may represent a general repression of potent developmental regulators in stem cells.
Methods
Nematode Methods. All strains were maintained at 20°C (37). Mutations include the following: LGI, fog-1(q250) (20, 22) balanced by sep-1(e2406) and fog-3(q520) (13, 21) balanced by sys-1(q544); LGII, fbf-1(ok91) (4), fbf-2(q738) (5), and fbf-1(ok91) fbf-2(q704) (4), all balanced by mIn1[mIs14 dpy-10(e128)]. syIs50 [cdh-3::gfp] (25) was an AC marker. RNAi was carried out by feeding L4s bacteria expressing dsRNA and scoring F1 progeny (38, 39). Homozygous fog; fbf animals were generated for RNAi and ablations by using a cross that was designed to produce half fog/+; fbf and half fog; fbf homozygotes.
Immunocytochemistry. Mixed-stage worms were fixed by the Finney–Ruvkun method (40). Antibodies used included rat anti-FBF-1 (1:5), rabbit anti-FBF-2 (1:5), and rat anti-FOG-1 (1:5) (4, 5, 9). Mouse anti-MH27 was used as a permeability control and to stage larvae (41). DNA was visualized by DAPI. Confocal images were collected on a Bio-Rad MRC1024 confocal microscope.
Laser Microsurgery. Laser ablations were done as described in ref. 42. Ablations were validated either by the absence of the entire gonad during L4 (Z1–Z4 ablations) or by remounting 3–8 h after surgery (Z2/Z3 and Z1.p/Z4.a ablations). Nonablated controls were grown in parallel with experimental animals. Killing AC precursors was confirmed in cdh-3::gfp transgenic animals by scoring L3s for lack of gonadal GFP.
Mosaic Analysis. The extrachromosomal array, qEx555, was generated by microinjection of fog-3 genomic DNA (pJK1098) (0.25 ng/μl) with sur-5::gfp (pTG96) (25 ng/μl) (43) into fog-3(q520)/sys-1(q544); fbf-1(ok91) fbf-2(q704)/mIn1[mIs14 dpy-10(e128)]. qEx555 rescued 62% of fog-3; fbf-1 fbf-2/mIn1[mIs14 dpy-10(e128)] to fertility (n = 163). Transgenic L4s were scored for mosaicism and vulval phenotype.
FBE Analysis. Candidate FBEs were identified, and gel shifts were done as described in ref. 6. Briefly, GST-fused FBF-2 (amino acids 121–634) was combined with 100 fmol 32P-end-labeled RNA oligoribonucleotides (Integrated DNA Technologies, Coralville, IA).
Supplementary Material
Acknowledgments
We are grateful to Min Han (University of Colorado, Boulder), Jonathan Pettitt (University of Aberdeen, Aberdeen, U.K.), and Robert Waterston (University of Washington, Seattle) for reagents; Brad Hook for preparation of the FBF-2 protein; and members of the J.K. and M. Wickens laboratories for helpful discussions during the course of this work. We specifically thank Mike Chesney for comments on the manuscript. We thank Anne Helsley-Marchbanks and Laura Vanderploeg for help preparing the manuscript and figures. J.K. is an investigator with the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grant GM069454 (to J.K.). L.B.L. was supported by National Institutes of Health Predoctoral Training Grant T32 GM07215 in Molecular Biosciences.
Conflict of interest statement: No conflicts declared.
Abbreviations: FBF, fem-3 binding factor; VPC, vulval precursor cell; AC, anchor cell; FBE, FBF binding element; RNAi, RNA interference.
References
- 1.Sternberg, P. W. (June 25, 2005) WormBook, 10.1895/wormbook.1.6.1, www.wormbook.org.
- 2.Sommer, R. J. (2005) WormBook, www.wormbook.org, in press.
- 3.Zhang, B., Gallegos, M., Puoti, A., Durkin, E., Fields, S., Kimble, J. & Wickens, M. P. (1997) Nature 390, 477–484. [DOI] [PubMed] [Google Scholar]
- 4.Crittenden, S. L., Bernstein, D. S., Bachorik, J. L., Thompson, B. E., Gallegos, M., Petcherski, A. G., Moulder, G., Barstead, R., Wickens, M. & Kimble, J. (2002) Nature 417, 660–663. [DOI] [PubMed] [Google Scholar]
- 5.Lamont, L. B., Crittenden, S. L., Bernstein, D., Wickens, M. & Kimble, J. (2004) Dev. Cell 7, 697–707. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, D., Hook, B., Hajarnavis, A., Opperman, L. & Wickens, M. (2005) RNA 11, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickens, M., Bernstein, D. S., Kimble, J. & Parker, R. (2002) Trends Genet. 18, 150–157. [DOI] [PubMed] [Google Scholar]
- 8.Eckmann, C. R., Crittenden, S. L., Suh, N. & Kimble, J. (2004) Genetics 168, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson, B. E., Bernstein, D. S., Bachorik, J. L., Petcherski, A. G., Wickens, M. & Kimble, J. (2005) Development 132, 3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, M.-H., Hook, B., Lamont, L. B., Wickens, M. & Kimble, J. (Dec. 1, 2005) EMBO J., 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed]
- 11.Jin, S.-W., Kimble, J. & Ellis, R. E. (2001) Dev. Biol. 229, 537–553. [DOI] [PubMed] [Google Scholar]
- 12.Luitjens, C., Gallegos, M., Kraemer, B., Kimble, J. & Wickens, M. (2000) Genes Dev. 14, 2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, P.-J., Singal, A., Kimble, J. & Ellis, R. E. (2000) Dev. Biol. 217, 77–90. [DOI] [PubMed] [Google Scholar]
- 14.Mendez, R. & Richter, J. D. (2001) Nat. Rev Mol. Cell Biol. 2, 521–529. [DOI] [PubMed] [Google Scholar]
- 15.Matsuda, S., Rouault, J.-P., Magaud, J.-P. & Berthet, C. (2001) FEBS Lett. 497, 67–72. [DOI] [PubMed] [Google Scholar]
- 16.Rouault, J.-P., Prévôt, D., Berthet, C., Birot, A.-M., Billaud, M., Magaud, J.-P. & Corbo, L. (1998) J. Biol. Chem. 273, 22563–22569. [DOI] [PubMed] [Google Scholar]
- 17.Ikematsu, N., Yoshida, Y., Kawamura-Tsuzuku, J., Ohsugi, M., Onda, M., Hirai, M., Fujimoto, J. & Yamamoto, T. (1999) Oncogene 18, 7432–7441. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida, Y., Hosoda, E., Nakamura, T. & Yamamoto, T. (2001) Jpn. J. Cancer Res. 92, 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okochi, K., Suzuki, T., Inoue, J., Matsuda, S. & Yamamoto, T. (2005) Genes Cells 10, 151–163. [DOI] [PubMed] [Google Scholar]
- 20.Barton, M. K. & Kimble, J. (1990) Genetics 125, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis, R. E. & Kimble, J. (1995) Genetics 139, 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, S. W., Arno, N., Cohen, A., Shah, A., Xu, Q., Chen, N. & Ellis, R. E. (2001) Genetics 159, 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimble, J. & Crittenden, S. L. (August 15, 2005) WormBook, 10.1895/wormbook.1.13.1, www.wormbook.org.
- 24.Kimble, J. (1981) Dev. Biol. 87, 286–300. [DOI] [PubMed] [Google Scholar]
- 25.Pettitt, J., Wood, W. B. & Plasterk, R. H. A. (1996) Development 122, 4149–4157. [DOI] [PubMed] [Google Scholar]
- 26.Sommer, R. J. & Sternberg, P. W. (1994) Science 265, 114–118. [DOI] [PubMed] [Google Scholar]
- 27.Sigrist, C. B. & Sommer, R. J. (1999) Dev. Genes Evol. 209, 451–459. [DOI] [PubMed] [Google Scholar]
- 28.Sulston, J. E., Schierenberg, E., White, J. G. & Thomson, J. N. (1983) Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 29.Rudel, D. & Kimble, J. (2001) Genetics 157, 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lackner, M. R., Kornfeld, K., Miller, L. M., Horvitz, H. R. & Kim, S. K. (1994) Genes Dev. 8, 160–173. [DOI] [PubMed] [Google Scholar]
- 31.Hill, R. J. & Sternberg, P. W. (1992) Nature 358, 470–476. [DOI] [PubMed] [Google Scholar]
- 32.Dutt, A., Canevascini, S., Froehli-Hoier, E. & Hajnal, A. (2004) PLoS Biol. 2, 1799–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clandinin, T. R., DeModena, J. A. & Sternberg, P. W. (1998) Cell 92, 523–533. [DOI] [PubMed] [Google Scholar]
- 34.Bui, Y. K. & Sternberg, P. W. (2002) Mol. Biol. Cell 13, 1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth, S., Neuman-Silberberg, F. S., Barcelo, G. & Schupbach, T. (1995) Cell 81, 967–978. [DOI] [PubMed] [Google Scholar]
- 36.González-Reyes, A., Elliott, H. & St Johnston, D. (1995) Nature 375, 654–658. [DOI] [PubMed] [Google Scholar]
- 37.Brenner, S. (1974) Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 39.Timmons, L. & Fire, A. (1998) Nature 395, 854 (lett.). [DOI] [PubMed] [Google Scholar]
- 40.Finney, M. & Ruvkun, G. (1990) Cell 63, 895–905. [DOI] [PubMed] [Google Scholar]
- 41.Francis, R. & Waterston, R. H. (1991) J. Cell Biol. 114, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bargmann, C. I. & Avery, L. (1995) in Caenorhabditis elegans: Modern Biological Analysis of an Organism, eds. Epstein, H. F. & Shakes, D. C. (Academic, San Diego), pp. 225–250.
- 43.Yochem, J., Gu, T. & Han, M. (1998) Genetics 149, 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.