Abstract
The common cushion moss Grimmia laevigata (Bridel) Bridel grows on bare rock in a broad range of environments on every continent except Antarctica. As such, it must harbor adaptations to a remarkably broad set of environmental stresses, the extremes of which can include very high temperatures, prolonged nearly complete desiccation, and high ultraviolet B (UVB) exposure. Yet, like many mosses, G. laevigata shows very little morphological variability across its cosmopolitan range. This presents an evolutionary puzzle, the solution to which lies in understanding the phylogeographic structure of this morphologically simple organism. Here we report the results of an analysis of amplified fragment length polymorphisms (AFLPs) in G. laevigata, focusing on individuals from the California Floristic Province. We found evidence that populations within California constitute two distinct geographically overlapping cryptic species. Each clade harbors multiple private alleles, indicating they have been genetically isolated for some time. We suggest that the existence of cryptic species within G. laevigata, in combination with its life history, growth habits, and extreme desiccation tolerance, makes this moss an ideal research tool and a candidate for a biological indicator of climate change and pollution.
Keywords: amplified fragment length polymorphisms, bioindication, California Floristic Province, Grimmiaceae, phylogeography
The broad and, in some cases, cosmopolitan distribution of many moss species suggests that these gametophyte-dominant plants are among Earth's most adaptable taxa. Mosses can be found on every continent and in every terrestrial ecosystem, from tropical rain forests (1) to arid deserts (2) to polar tundra (3). To survive in such diverse and often extreme environments, these sedentary organisms must possess an equally diverse set of physiological adaptations. Mosses have been shown to be capable of surviving complete desiccation (4) and temperatures as extreme as 110°C (5). As climate change causes increased aridity worldwide, and with global temperatures predicted to rise, it will become increasingly important to understand how organisms solve the problem of surviving extremes of temperature and humidity.
The common cushion moss Grimmia laevigata (Bridel) Bridel can be found on every continent except Antarctica, surviving in a diverse set of habitats and with little morphological variability (6–10). It is also one of the classic examples of a drought-resistant bryophyte (11), possessing “apparently extravagant levels of desiccation tolerance” (4). Individuals of G. laevigata have the remarkable ability to survive as dried herbarium specimens for at least 10 years (12), restoring photosynthetic and metabolic activity upon rehydration. To survive even brief periods of desiccation, plants must protect themselves from many forms of damage, including the mechanical strain of dramatic changes in cell volume, disruption of metabolism, damage to DNA, and fusion of unrelated membrane systems (4, 13). G. laevigata therefore presents an evolutionary puzzle as to how such a widespread morphologically uniform species could have adapted to such a broad range of environments.
One possible solution to this puzzle is that G. laevigata consists of a group of morphologically indistinguishable cryptic species adapted to different habitats. Many moss species are characterized as having cosmopolitan distributions, often with little morphological differentiation among individuals from different continents (14–17). However, analysis of molecular markers or isozymes in some moss species has revealed genetic divergence among populations from different regions (17–20) and provided evidence of cryptic species (16, 21). In fact, there is a growing sense that cryptic species with overlapping geographic ranges may be common in mosses (22).
Alternatively, a lack of genetic divergence among G. laevigata from very different habitats would raise interesting questions about the physiological basis of this plant's versatility. In a panmictic population, for example, the ability to live in different environments may be attributable to phenotypic plasticity and to induced rather than constitutive stress responses.
As a first step in solving this puzzle, we undertook a study of the phylogeographic structure of G. laevigata from a diverse set of habitats, focusing on specimens primarily from the California Floristic Province. This region is noted for high levels of endemism in many taxa and is considered one of Earth's biodiversity hotspots (23, 24). Studies of multiple taxa within the region have shown evidence of phylogeographic structure, much of which appears to coincide with boundaries established by past geologic events (25–27). The bryoflora of California is diverse and species-rich, including nearly 50% of the mosses documented for North America (2). The genus Grimmia is frequently encountered in California, and it is the most widespread petricolous genus occurring in all ecological regions of the state (9, 10).
The G. laevigata specimens described here were collected from locations that differ profoundly in mean temperature, extremes of temperature, average humidity, and light intensity, as well as in the nature of the rock substrate and nearby vegetation (Table 1, which is published as supporting information on the PNAS web site). We used analysis of amplified fragment length polymorphisms (AFLPs) to evaluate whether G. laevigata may be a complex of cryptic species. These molecular markers allow assessment of a large number of DNA fragment polymorphisms throughout the genome in a large number of individuals and are becoming an increasingly important tool for identifying early stages of divergence in a wide range of taxa (28–31).
Results
PCR Screen for Contamination of DNA. Fungal endophytes are common in many plants and, because they cannot be removed by surface sterilization, genomic DNA extraction from plants may sometimes produce a mixture of plant and fungal DNA. We therefore initially screened for fungal DNA by performing internal transcribed spacer 2 (ITS-2) region PCR on extractions from all 54 collected individuals [50 G. laevigata, two Grimmia ovalis (Hedwig) Lindberg, and two Ptychomitrium gardneri Lesquereux]. All reactions resulted in products with lengths between 421 and 465 bp. Sequencing of these products from five individuals showed similarity to published bryophyte sequences. A subset of the reactions resulted also in fragments in the range of 323 to 360 bp, the expected size range for fungal ITS-2 regions (32), and sequencing of one of these bands showed similarity to published fungal sequences. Five of the G. laevigata individuals were excluded from further analysis, because >25% of total ITS-2 product was in the fungal size range. One additional sample was excluded because four distinct bands amplified in the plant size range, indicating that the gametophytes of this sample came from more than one individual.
AFLP Reliability. Four primer pairs produced 209 scorable polymorphic fragments (Table 2, which is published as supporting information on the PNAS web site), and each of the 48 individuals had a unique AFLP profile. Blind scoring of duplicated electropherograms from two individuals resulted in a 100% genotype match between duplicates, demonstrating the reliability of manual scoring. The repeatability of the AFLP procedure was confirmed by low mismatch rates in six samples, for which the entire procedure was replicated from the same extraction, with a maximum rate of 0.059 for the MseI-CAG/EcoRI-ACT primer pair (Table 2).
Of the extractions not excluded because of high levels of fungal contamination, some nonetheless contained low levels of fungal DNA (at most 6% of total ITS-2 product). AFLP profiles from extractions of two unwashed samples demonstrated that this level of contamination did not influence the results. In both cases, the washed extraction resulted in no detectable ITS-2 product in the fungal size range, whereas the extraction without the washes resulted in 3% (for sample no. 26) and 5% (for no. 20) fungal ITS. Contaminated and uncontaminated genotypes showed 97% (for no. 26) and 91% (for no. 20) correspondence.
We also tested the robustness of the analyses by including, in a separate principal coordinates analysis and parsimony analysis, the duplicate reaction with the highest mismatch rate (sample no. 36) as well as the contaminated samples nos. 20 and 26 and found that in all cases, duplicate individuals fell within the same clusters. The conclusions of these analyses are therefore robust to the error rates we encountered.
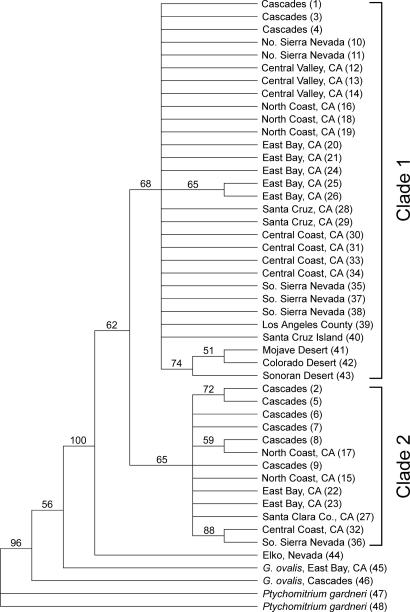
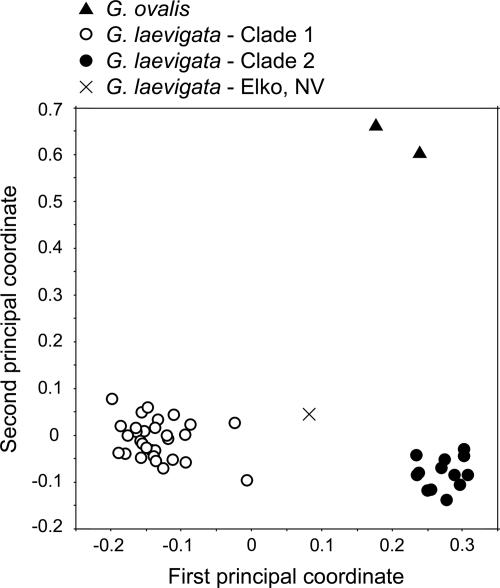
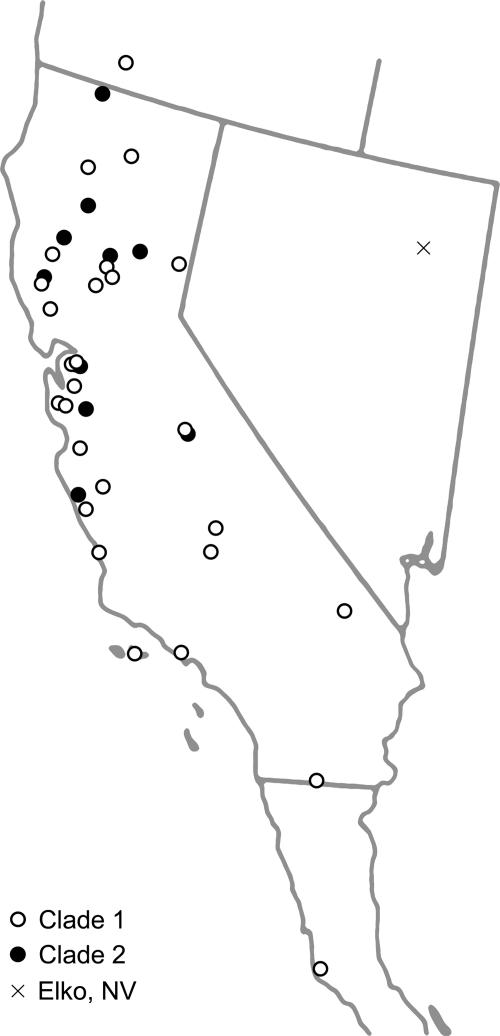
Phylogeography. Phylogenetic analysis indicated the existence of two clades within G. laevigata (Fig. 1), with the single individual from Elko, NV, sister to both clades. Very little phylogenetic structure was found within either clade, with the exception of a group within clade 1 that contained specimens from the Mojave Desert, the Colorado Desert, and the Sonoran Desert. Seventy-five fragments were polymorphic within clade 1 but were absent from clade 2, whereas 19 such “private alleles” were shared only among individuals in clade 2. Clade 1 also contained one diagnostic fragment, present in all individuals in this clade but absent from clade 2. Although boostrap support was weak for each clade, all samples clustered in the same way as found in the principal coordinate (PCO) analysis (Fig. 2). Results are displayed for the PCO in which the P. gardneri samples were excluded, but identical clustering was obtained when P. gardneri was included. The first axis explained 20.0% of the genetic variation, and the second axis explained 11.5%. The mean genetic distance between individuals from clades 1 and 2 was 0.608 (Jaccard index). Each of the two G. laevigata clades included samples from throughout California, with no clear division of clades along geographic boundaries (Fig. 3). The overlapping distributions include some areas of true or near sympatry. Of the five specimens collected in Mt. Diablo State Park, three are in clade 1 (samples 20, 21, and 24), and two are in clade 2 (samples 22 and 23).
Fig. 1.
Strict consensus cladogram showing maximum-parsimony relationships among the G. laevigata samples. P. gardneri and G. ovalis were used to root trees. Numbers above branches are boostrap values. Individuals are labeled with region and sample identification in parentheses, corresponding to Table 1.
Fig. 2.
Principal coordinates analysis of Jaccard's distance among the 46 Grimmia individuals. The first two axes explain 31.5% of total variation.
Fig. 3.
Map of collection sites of the 44 G. laevigata individuals.
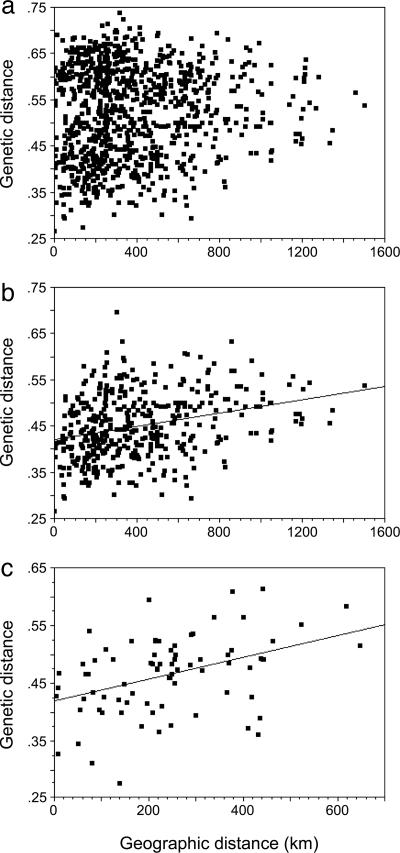
The degree of genetic distance between pairs of individuals was not related to geographic distance when all G. laevigata samples were included in the analysis (Fig. 4a). The same analysis performed separately for each clade, however, indicated significant isolation by distance (Fig. 4 b and c). Mantel tests using Jaccard's index, Nei's index, and Cavalli–Sforza's chord values all showed significant isolation by distance within both clades (P < 0.05). Only the Mantel tests using Euclidean distance deviated from this pattern, indicating significant isolation by distance for clade 2 (P < 0.01) but not clade 1. All statistical inferences remained the same if the analyses were performed by using logarithmic rather than arithmetic distances. The consistent results among most measures of isolation by distance suggest that these two Grimmia clades spread independently across California.
Fig. 4.
Isolation-by-distance analysis among G. laevigata individuals. (a) All samples analyzed together. (b) Clade 1 analyzed separately. (c) Clade 2 analyzed separately. Each point shows Jaccard's index of genetic distance between a pair of individuals and the geographic distance between that pair.
Discussion
Overlapping Cryptic Species Within G. laevigata. Results suggest that the cosmopolitan G. laevigata may comprise sympatric cryptic species at least within the California Floristic Province. Both clades identified by AFLPs occur throughout a broad region of overlap in California, and there is no clear phylogeographic structure within either clade. The allelic differentiation found between these two clades suggests they may have long been genetically isolated within California. Worldwide sampling would be needed to determine whether each of these two clades has a cosmopolitan distribution, whether one of the two clades may be a California endemic, or whether the taxon known as G. laevigata is in fact a mosaic of geographically restricted cryptic species.
Some regions, like California, are known as biodiversity hotspots and may be especially conducive to phylogeographic differentiation and cryptic speciation. Studies of multiple taxa within the region have shown evidence of phylogeographic structure (e.g., refs. 33–35). Moreover, multiple studies have shown higher levels of phylogeographic differentiation in California than in the Pacific Northwest, where populations of many species became established only after the retreat of Pleistocene glaciers (26, 36, 37). Recent comparative analyses have indicated that patterns of molecular differentiation within many California taxa coincide with geographic barriers, such as those created by the Transverse Range, the Monterey Bay, and the Los Angeles Basin (25, 26). Together, these studies indicate that such phylogeographic structure evolves over time due in part to reduction of gene flow across geographic barriers.
In light of these common phylogeographic patterns, the genetic structure of G. laevigata is surprising in two ways. First, there is no clear phylogeographic structure within either clade. Possible explanations include insufficient time since the two clades split or extensive gene flow among populations within each clade. Second, the split between the two Grimmia clades does not correspond to any obvious geographic barriers. The two clades may have originated in allopatry in California or elsewhere, later expanding to their current overlapping distributions, or the two clades could have diverged subsequent to ecological specialization. Populations are often more ecologically specialized than the species or species complex as a whole (38, 39). In G. laevigata, the existence of a desert clade within clade 1 suggests the potential for divergence due to habitat specialization, but there was no clear indication that the two main clades are specialized to different environments or types of rock substrate. Additional sampling and detailed habitat evaluation will be needed to test the hypothesis of ecological specialization.
The few other studies of molecular differentiation in mosses within the California Floristic Province have shown little evidence of geographic structure. Shaw (16), for example, found only a single nuclear ribosomal ITS clade in California populations of the Mielichhoferia elongata/mielichhoferiana species group, and there was little differentiation among individuals collected over hundreds of kilometers in California. Yet this group, which ranges across the Northern Hemisphere, harbors multiple ITS clades that overlap in other regions, and morphologically indistinguishable but phylogenetically distant populations have been found within a few meters of each other.
Our findings corroborate the developing view that widely distributed generalist taxa are often collections of genetically distinct populations or sibling species. Cryptic species are being revealed at an increasing rate as studies of molecular variation include more individuals, more populations, and samples over broader geographic areas. Some widespread taxa once thought to be ecological generalists are now known to be collections of genetically distinct and sometimes broadly sympatric populations specialized to different habitats, microhabitats, or species interactions. Recent examples, using analysis of DNA sequences or fragments, span a wide range of taxa, including sympatric fig wasps that use the same fig species (40), the widespread neotropical skipper butterfly Astraptes fulgerator that is now recognized as 10 species (41), the millipede Anadenobolus excisus in Jamaica that includes three deeply divergent clades (42), the black mould Stachybotrys chartarum (43) that includes two distinct lineages, and cooccurring clades of marine calcifying algae called coccolithophores that may differ in their ecological preferences (44).
Too few species of moss have been analyzed by any single molecular technique to draw robust conclusions about the geographic scale of molecular differentiation within moss species or species complexes. Molecular phylogeographic and phylogenetic studies of mosses have analyzed sequence variation in the nuclear ribosomal ITS region (e.g., refs. 17 and 19), chloroplast genes such as trnL-trnF or rps4 (e.g., refs. 20 and 45), or microsatellites (e.g., ref. 46). No previous phylogeographic study of moss has used AFLPs to evaluate patterns of differentiation among populations across a large geographic region, but multiple laboratories are now undertaking similar studies using AFLPs or related genome-wide marker techniques (e.g., ref. 47).
These results emphasize the need to make molecular characterization of species a standard part of ecological analyses of populations and communities. The extent to which these clades differ in life history or other nonmorphological traits is unknown, but they appear to function as reproductively isolated units within California. In that sense, they are part of the rich but hidden biodiversity that is becoming increasingly apparent as broadly distributed taxa are analyzed in more molecular detail.
Implications for Future Research. The cryptic species of G. laevigata, each found in both desert and temperate areas of California, provide a unique opportunity to investigate how desiccation tolerance may evolve in different ways in reproductively isolated lineages. It has been suggested that some G. laevigata specimens are capable of surviving even longer and more intense drought than they ever encounter in the wild, raising the question of how such extreme tolerance could have evolved (11). Desiccation tolerance is constitutive in some moss species and inducible in others. In the well studied Tortula ruralis [=Syntrichia] (48), stable ubiquitin transcripts survive dehydration and can be translated upon rehydration to initiate rapid cellular repair (49). In several other moss species, desiccation tolerance is controlled by the growth regulator abscisic acid (4), which induces cellular protection and repair mechanisms by turning on the expression of genes encoding the so-called dehydrin proteins (50). The morphologically indistinguishable but genetically divergent clades of G. laevigata could be viewed as replicated experiments in which selection has shaped physiology and life histories while holding morphology constant.
The molecular genetics of desiccation tolerance is a field still in its infancy and in need of a genetic model system (51). Characterizing the genes involved has obvious biotechnological potential in an era of increasing global aridity and temperatures. In mosses, the desiccation-tolerant tissues are haploid, opening up a rare opportunity for functional gene analysis through gene replacement using efficient homologous recombination techniques (51).
Toward that end, we have refined and optimized an extraction protocol that produces DNA of high-enough quality for genotyping, and that could also be useful for studies of DNA damage and repair under the high UVB levels to which most G. laevigata are routinely exposed. Storage of some of the specimens for many years in herbaria did not compromise DNA extraction or the quality of AFLP profiles. We have also demonstrated the utility in this organism of using ITS-2 PCR to screen for fungal contamination, a method reviewed and advocated by Saar et al. (32) for angiosperm DNA. Our use of this PCR screen resulted in the exclusion of five contaminated individuals but also provided the useful information that G. laevigata does not harbor obligate endophytic fungi of appreciable quantity to interfere with future molecular studies. Moreover, the existence of a single ITS-2 band in the moss size range in all but one of the 54 collected individuals suggests that collecting gametophytes from a single cushion in the field usually generates a sample with a single genotype.
G. laevigata has numerous general attributes that make it a tractable experimental system. First, it is a sedentary organism, common in many geographic regions across the globe, easy to find and identify. Individuals can be nondestructively sampled, marked, and monitored in the field over long periods of time, because individual cushions can persist for decades. Samples can be dried and stored in herbaria for many years and later rehydrated and propagated (ref. 12; confirmed in our laboratory), paving the way for a panoply of genomic, transcriptional, translational, and proteomic analyses of mosses subjected to precisely known environmental conditions.
Bioindication. As a cosmopolitan lineage, G. laevigata also has potential as a biological indicator of climate change and pollution. A bioindicator is an organism that contains information on the quality of the environment, because a change in the organism can be linked to a change in its habitat (52). Phylogeographic information provides a template for evaluation of the “correlative complexes” (53) that exist between environmental (i.e., pollution, temperature, and aridity) changes and biotic (genetic, morphological, and physiological) changes. If the putative cryptic species of G. laevigata respond in similar ways to environmental change, they could be treated as a single widely distributed bioindicator system. If they do not, the variability in response may provide more environmental information than a genetically uniform set of bioindicator organisms could.
The cosmopolitan distribution of G. laevigata offers an important advantage. Many of the moss species that have been developed as bioindicators of heavy-metal pollution are species that can be found only in specific regions, such as Northern Europe or the northeastern U.S., necessitating the development of new bioindicator systems when pollution threatens an area outside these regions (54). Because G. laevigata is found on every continent except Antarctica, and in such a diversity of habitats, an investment into thoroughly researching the utility of this species complex as a bioindicator system is likely to be rewarded with global applicability.
Yet another advantage of G. laevigata is the relative simplicity of the rock substrates on which they grow. Bryophytes are known to be good indicators of changes in temperature and humidity (55) and may be more useful than many plants in which responses to climate change can be complicated by indirect influences acting through the soil (56). Petricolous organisms like G. laevigata are also particularly useful as pollution bioindicators, because they cannot pick up metals from the soil, a process that confounds the correlation between air pollution and metal accumulation in other plants, even other mosses (57). Finally, the life history and growth habits of G. laevigata suggest that it may be ideal for biomonitoring. Individual cushions can persist in the same spot for decades and are easy to find, mark, monitor, and correlate with local environmental change.
Overall, analysis of G. laevigata reveals hidden complexity within this remarkable drought-tolerant cosmopolitan species complex. Moreover, it provides a basis for using this species in multiple forms of research on adaptation to extreme or changing environments.
Methods
Distribution of Samples. We analyzed leaves (phyllids) from 50 G. laevigata individuals, 47 of which were collected throughout California, plus one each from southern Oregon, Nevada, and Baja California (Table 1, which is published as supporting information on the PNAS web site). Collection sites ranged in elevation from 30 to 2,010 m above sea level, representing a broad range of habitats and rock substrates. The sites spanned the major common phylogeographic breaks that have been found in molecular studies of a wide range of taxa within this region (25, 26). We also analyzed leaves of two G. ovalis and two P. gardneri individuals to use as outgroups in clustering analyses. Ptychomitrium in the Ptychomitriaceae is a sister family to the Grimmiaceae in the order Grimmiales (58).
DNA Extraction. The protocol for DNA extraction was based upon that of Schlink and Reski (59), with substantial modification. Dried gametophytes were rehydrated in distilled water, and green tissue for extraction was manually separated from brown tissue and inorganic matter. Samples were washed several times in distilled water and vacuum-dried. Approximately 40 mg of clean dried tissue was placed in a microcentrifuge tube with 1-mm glass beads, frozen in liquid nitrogen and ground to a fine powder by using a Biospec MiniBeadbeater (Biospec Products, Bartlesville, OK). Samples were incubated in extraction buffer (59) for 1 h at 60°C in a shaking water bath. One volume of 5 M ammonium acetate was added to the liquid phase of each sample for a 5-min incubation at room temperature.
Centrifugation for 10 min at 13,000×g caused the separation of an upper pigmented layer from a lower clear layer. Gel quantification and PCR trials revealed that removal of this pigmented layer increased PCR success rate >3-fold but did not noticeably decrease DNA yield. Consequently, the pigmented layer was removed from all analyzed samples before isopropanol precipitation. Pellets were washed in 70% ethanol, dried down, resuspended in TE buffer, and treated with 0.6 μg/ml RNase A at 37°C for 15 min. Two chloroform extractions were followed by a final isopropanol precipitation. Typical yields were 400 ng of DNA per sample.
PCR Screen for Contamination of DNA. The ITS-2 region was amplified with the primers ITS4 and ITS3 (32). We labeled the ITS3 primer with the blue fluorophore 6-FAM on the 5′ end. PCR was performed in 25-μl reaction volumes containing 0.2 mM each dNTP, 0.2 mM each oligonucleotide primer, 0.05 mg/ml BSA, 3.5 mM MgCl2, ≈5 ng of a DNA template, and 1 unit of TaqDNA polymerase (Qiagen, Valencia, CA). An Applied Biosystems GeneAmp 9700 thermal cycler was used to perform the amplifications, programmed for one cycle for 3 min at 94°C, 1 min at 50°C, and 2 min at 72°C; followed by 30 cycles for 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C; and finished with 10 min at 72°C. Amplified products were sized and quantified on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Before sample loading, 1 μl of a 1:10 dilution of each PCR product was combined with 1 μl of ROX-500 size standard and 10 μl of Hi-Di formamide and denatured at 95°C for 5 min. Relative fluorescence intensity was used as a measure of the relative abundance of ITS-2 regions of different sizes. Fungal ITS-2 regions are expected to be 50–100 bp smaller than plant ITS-2 regions (32).
AFLPs. AFLP methods largely followed the procedures described by Vos et al. (60) and the Applied Biosystems protocol (AFLP Plant Mapping Kit). For each sample, ≈50 ng of total genomic DNA was digested to completion with restriction enzymes EcoRI and MseI. AFLP adapters for both restriction enzymes were then ligated to the restriction fragments. Preamplification of template DNA was carried out using an EcoRI-complementary primer with a 3′ A and an MseI-complementary primer with a 3′ C. Selective amplification reactions were performed with four primer combinations: MseI-CAT with EcoRI-ACT-FAM (blue fluorophore), MseI-CAG with EcoRI-AAG-JOE (green fluorophore), MseI-CAG with EcoRI-ACT-FAM, and MseI-CAC with EcoRI-AAG-JOE. Detection and sizing of PCR products were performed with internal size standards on an ABI Prism 3100 Genetic Analyzer. All electropherograms were visualized with genotyper Ver. 3.7 from Applied Biosystems and inspected manually. Distinct polymorphic peaks in the 50- to 500-bp range were scored as present or absent, and the only characters included in the analysis were those for which unambiguous determinations could be made in all electropherograms.
Assessment of AFLP Error Rate. We used three methods to assess the error rate of AFLP genotyping, each of which involved creating duplicates at different stages of the process. All electropherograms were scored blind, i.e., without knowledge of collection site or of which samples were unique and which were duplicates. The first method assessed the repeatability of allele scoring by duplicating two electropherograms before blind scoring. The second method assessed the repeatability of the AFLP reactions by replicating the entire procedure, from restrictionligation reactions through capillary electrophoresis, on genomic DNA from a single extraction of each of six individuals. The third method assessed the robustness of AFLP profiles to fungal contamination by performing AFLP reactions on replicate extractions from the same samples, one of which showed fungal contamination, and one of which did not. This was accomplished for two individuals by performing one extraction as described above and one extraction omitting the initial gametophyte washes.
Analysis. Phylogenetic analysis was accomplished by using maximum parsimony in paup Ver. 4.0 (61). Heuristic searches were run from 1,000 initial trees obtained with a random addition sequence for stepwise addition, followed by tree bisection–reconnection branch swapping. Multiple most-parsimonious trees were summarized with a strict consensus tree, and boostrap analyses included 100 bootstrap replicates. Trees were rooted by using G. ovalis and P. gardneri as outgroups. The same internal topology resulted when the P. gardneri samples were excluded from parsimony analysis.
The presence/absence matrix of AFLP fragments was used to generate genetic distance and similarity matrices by using the r package for multivariate and spatial analysis Ver. 4.0 (Philippe Casgrain, University of Montreal, Montreal, PQ, Canada). Clustering of individuals based upon these genetic matrices was then summarized with PCO in the r package. We present PCO results (Fig. 2) based upon Jaccard similarities, which ignore the shared absence of AFLP fragments, but similar results were obtained by using either Nei's genetic distance or Euclidean distance.
Mantel tests were performed by using the r package to test for patterns of isolation by distance. Three different combinations of individuals were evaluated based upon the groupings revealed by parsimony and PCO analyses, and separate Mantel tests were performed for each combination. Each Mantel test was repeated by using four measures of genetic distance (Cavalli–Sforza and Edward's chord distance, Jaccard's index, Nei's index, and Euclidean distance) and two measures of geographic distance (arithmetic distance in kilometers, correcting for the curvature of the Earth, and log distance) in all possible combinations.
Supplementary Material
Acknowledgments
We are grateful to Pamela Soltis and Jonathan Shaw for reviewing and greatly improving the manuscript. We also thank Mariana Cuautle-Arenas, Sarah Dwiggins, Samantha Forde, Jason Hoeksema, Katherine Horjus, and Bridget Piculell for helpful comments. We thank the curators at the California Academy of Sciences and University of California, Berkeley, for access to specimens and Lowell Ahart, Colin Dillingham, Judy Harpel, and David Toren for providing additional G. laevigata collections for this study. This work was supported by National Science Foundation Grant DEB-344147.
Conflict of interest statement: No conflicts declared.
Abbreviations: AFLP, amplified fragment length polymorphisms; PCO, principal coordinate.
References
- 1.Pócs, T. (1982) in Bryophyte Ecology, ed. Smith, A. J. E. (Chapman & Hall, London), pp. 59–104.
- 2.Shevock, J. R. (2003) Fremontia 31, 12–20. [Google Scholar]
- 3.Longton, R. E. (1988) The Biology of Polar Bryophytes and Lichens (Cambridge Univ. Press, Cambridge, U.K.).
- 4.Proctor, M. C. F. & Pence, V. C. (2002) in Desiccation and Survival in Plants: Drying without Dying, eds. Black, M. & Pritchard, H. W. (CABI, Wallingford, U.K.), pp. 207–237.
- 5.Liu, Y., Cao, T. & Glime, J. M. (2003) Bryologist 106, 53–60. [Google Scholar]
- 6.Alpert, P. (1988) J. Bryol. 15, 219–227. [Google Scholar]
- 7.Muñoz, J. & Pando, F. (2000) Monogr. Syst. Bot. 82, 1–133. [Google Scholar]
- 8.Greven, H. C. (2003) Grimmias of the World (Backhuys, Leiden, The Netherlands).
- 9.Norris, D. H. & Shevock, J. R. (2004) Madroño 51, 1–131. [Google Scholar]
- 10.Norris, D. H. & Shevock, J. R. (2004) Madroño 51, 133–269. [Google Scholar]
- 11.Alpert, P. & Oliver, M. J. (2002) in Desiccation and Survival in Plants: Drying without Dying, eds. Black, M. & Pritchard, H. W. (CABI, Wallingford, U.K.), pp. 3–43.
- 12.Keever, C. (1957) Ecology 38, 422–429. [Google Scholar]
- 13.Walters, C., Farrant, J. M., Pammenter, N. W. & Berjak, P. (2002) in Desiccation and Survival in Plants: Drying without Dying, eds. Black, M. & Pritchard, H. W. (CABI, Wallingford, U.K.), pp. 263–291.
- 14.Schofield, W. B. (1980) in The Mosses of North America, eds. Taylor, R. J. & Levitan, A. E. (Pacific Division, American Association for the Advancement of Science, California Academy of Sciences, San Francisco), pp. 131–170.
- 15.Schofield, W. B. (1984) J. Hattori Bot. Lab. 55, 35–43. [Google Scholar]
- 16.Shaw, A. J. (2000) Mol. Ecol. 9, 595–608. [DOI] [PubMed] [Google Scholar]
- 17.Shaw, A. J., Werner, O. & Ros, R. M. (2003) Am. J. Bot. 90, 540–550. [DOI] [PubMed] [Google Scholar]
- 18.Wyatt, R., Staneburner, A. & Odrzykoski, I. J. (1989) in Isozymes in Plant Biology, eds. Soltis, D. E. & Soltis, P. S. (Dioscorides, Portland, OR), pp. 221–240.
- 19.Chiang, T. Y. & Schaal, B. A. (1999) Mol. Ecol. 8, 1037–1042. [Google Scholar]
- 20.Werner, O. & Guerra, J. (2004) Plant Biol. 6, 147–157. [DOI] [PubMed] [Google Scholar]
- 21.McDaniel, S. F. & Shaw, A. J. (2003) Evolution (Lawrence, KS) 57, 205–215. [DOI] [PubMed] [Google Scholar]
- 22.Shaw, A. J. (2001) J. Biogeogr. 28, 253–261. [Google Scholar]
- 23.Mittermeier, R. A., Myers, N., Gil, P. R. & Mittermeier, G. C. (1999) Hotspots: Earth's Biologically Richest and Most Endangered Terrestrial Ecoregions (CEMEX Conservation, Mexico City, Mexico).
- 24.Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B. & Kent, J. (2000) Nature 403, 853–858. [DOI] [PubMed] [Google Scholar]
- 25.Calsbeek, R., Thompson, J. N. & Richardson, J. E. (2003) Mol. Ecol. 12, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. N. & Calsbeek, R. (2005) in Insect Evolutionary Ecology, eds. Fellowes, M., Holloway, G. & Rolff, J. (Royal Entomological Society, London), pp. 405–421.
- 27.Lapointe, F.-J. & Rissler, L. J. (2005) Am. Nat. 166, 290–299. [DOI] [PubMed] [Google Scholar]
- 28.Douek, J., Barki, Y., Gateño, D. & Rinkevich, B. (2002) Zool. J. Linn. Soc. 136, 315–320. [Google Scholar]
- 29.Whittall, J. B., Hellquist, C. B., Schneider, E. L. & Hodges, S. A. (2004) Am. J. Bot. 91, 2022–2029. [DOI] [PubMed] [Google Scholar]
- 30.Irwin, D. E., Bensch, S., Irwin, J. H. & Price, T. D. (2005) Science 307, 414–416. [DOI] [PubMed] [Google Scholar]
- 31.Mendelson, T. C. & Shaw, K. L. (2005) Nature 433, 375–376. [DOI] [PubMed] [Google Scholar]
- 32.Saar, D. E., Polans, N. O., Sørensen, P. D. & Duvall, M. R. (2001) Plant Mol. Biol. Rep. 19, 249–260. [Google Scholar]
- 33.Alexandrino, J., Baird, S. J. E., Lawson, L., Macey, J. R., Moritz, C. & Wake, D. B. (2005) Evolution (Lawrence, KS) 59, 1334–1347. [PubMed] [Google Scholar]
- 34.Kuchta, S. R. & Tan, A.-M. (2004) Mol. Ecol. 14, 225–244. [DOI] [PubMed] [Google Scholar]
- 35.Schaffer, B., Pauly, G. B., Oliver, J. C. & Trenham, P. C. (2004) Mol. Ecol. 13, 3033–3049. [DOI] [PubMed] [Google Scholar]
- 36.Brown, J. M., Leebens-Mack, J. H., Thompson, J. N., Pellmyr, O. & Harrison, R. G. (1997) Mol. Ecol. 6, 215–224. [DOI] [PubMed] [Google Scholar]
- 37.Soltis, D. E., Gitzendanner, M. A., Strenge, D. D. & Soltis, P. S. (1999) Plant Syst. Evol. 206, 353–373. [Google Scholar]
- 38.Sáez, A. G. & Lozano, E. (2005) Nature 433, 111. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. N. (2005) The Geographic Mosaic of Coevolution (Univ. of Chicago Press, Chicago).
- 40.Molbo, D., Machado, C. A., Sevenster, J. G., Keller, L. & Herre, E. A. (2003) Proc. Natl. Acad. Sci. USA 100, 5867–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hebert, P. D. N., Penton, E. H., Burns, J. M., Janzen, D. H. & Hallwachs, W. (2004) Proc. Natl. Acad. Sci. USA 101, 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bond, J. E. & Sierwald, P. (2002) Evolution (Lawrence, KS) 56, 1123–1135. [DOI] [PubMed] [Google Scholar]
- 43.Koster, B., Scott, J., Wong, B. Malloch, D. & Straus, N. (2003) Can. J. Bot. 81, 633–643. [Google Scholar]
- 44.Sáez, A. G., Probert, I., Geisen, M., Quinn, P., Yound, J. R. & Medlin, L. K. (2003) Proc. Natl. Acad. Sci. USA 100, 7163–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw, A. J. & Allen, B. (2000) Mol. Phylogenet. Evol. 16, 225–237. [DOI] [PubMed] [Google Scholar]
- 46.Van der Velde, M. & Bijlsma, R. (2000) Heredity 85, 328–337. [DOI] [PubMed] [Google Scholar]
- 47.Hassel, K. & Gunnarsson, U. (2003) Lindbergia 28, 152–157. [Google Scholar]
- 48.Bewley, J. D. & Oliver, M. J. (1992) in Water and Life, eds. Somero, G. N., Osmond, C. B. & Bolis, C. L. (Springer, Berlin), pp. 141–160.
- 49.O'Mahony, P. J. & Oliver, M. J. (1999) Plant Mol. Biol. 41, 657–667. [DOI] [PubMed] [Google Scholar]
- 50.Knight, C. D., Sehgal, A., Atwal, K., Wallace, J. C., Cove, D. J., Coates, D., Quatrano, R. S., Bahadur, S., Stockley, P. G. & Cuming, A. C. (1995) Plant Cell 7, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips, J. R., Oliver, M. J. & Bartels, D. (2002) in Desiccation and Survival in Plants: Drying without Dying, eds. Black, M. & Pritchard, H. W. (CABI, Wallingford, U.K.), pp. 319–341.
- 52.Markert, B. A., Breure, A. M. & Zechmeister, H. G. (2003) Bioindicators and Biomonitors: Principles, Concepts and Applications (Elsevier, Amsterdam).
- 53.Zonneveld, I. S. (1983) Env. Monit. Assess. 3, 207–217. [DOI] [PubMed] [Google Scholar]
- 54.Schintu, M., Cogoni, A., Durante, L., Cantaluppi, C. & Contu, A. (2005) Chemosphere 60, 610–618. [DOI] [PubMed] [Google Scholar]
- 55.Frahm, J. P. & Gradstein, S. R. (1991) J. Biogeogr. 18, 669–678. [Google Scholar]
- 56.Callaghan, T. V., Carlsson, B. Å., Sonesson, M. & Temesváry, A. (1997) Funct. Ecol. 11, 157–165. [Google Scholar]
- 57.Onianwa, P. C. (2001) Env. Monit. Assess. 71, 13–50. [DOI] [PubMed] [Google Scholar]
- 58.Buck, W. R. & Goffinet, B. (2000) in Bryophyte Biology, eds. Shaw, J. & Goffinet, B. (Cambridge Univ. Press, Cambridge, U.K.), pp. 71–123.
- 59.Schlink, K. & Reski, R. (2002) Plant Mol. Biol. Rep. 20, 423a–423f. [Google Scholar]
- 60.Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M. & Zabeau, M. (1995) Nucleic Acids Res. 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swofford, D. L. (2002) paup: Phylogenetic Analysis Using Parsimony (and Other Methods) (Sinauer, Sunderland, MA), 4.0 Beta.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.