Abstract
The Saccharomyces cerevisiae Spo11 protein catalyses DNA double-strand breaks (DSBs) that initiate meiotic recombination. The model plant Arabidopsis thaliana possesses at least three SPO11 homologues. T-DNA and ethyl-methane sulfonate mutagenesis allowed us to show that meiotic progression is altered in plants in which the AtSPO11-1 gene is disrupted. Both male and female meiocytes formed very few bivalents. Furthermore, no fully synapsed chromosomes were observed during prophase I. Later, in meiosis I, we observed that chromosomes segregated randomly, leading to the production of a large proportion of non-functional gametes. These meiotic aberrations were associated with a drastic reduction in meiotic recombination. Thus, our data show that initiation of meiotic recombination by SPO11- induced DSBs is a mechanism conserved in plants. Furthermore, unlike Drosophila and Caenorhabditis elegans, but like fungi, SPO11 is necessary for normal synapsis in plants.
Keywords: Arabidopsis thaliana/meiosis/mutants/recombination/SPO11
Introduction
In contrast to animals, plant organogenesis is a continuous process, and somatic recombination can therefore lead to transmittable genetic variations. Nevertheless, even in plants, meiosis remains the major source of genetic variability. Cross-overs and genic conversions generate recombination between homologous chromosomes, reorganizing the allelic combinations among them. Cross-overs also establish physical links between homologous chromosomes (chiasmata) so that these remain linked together even when the synaptonemal complex (SC) disappears. This mechanism thus ensures proper segregation of chromosomes at anaphase I when chiasmata are released (for a review see Roeder, 1997), allowing random segregation of paternal and maternal chromosomes (interchromosomal recombination).
In budding yeast, meiotic recombination is initiated by double-strand breaks (DSBs) (Sun et al., 1989; Cao et al., 1990), which will undergo strand exchange interactions with an intact partner duplex. The formation of DSBs requires at least nine different genes (SPO11, RAD50, MRE11, XRS2, MER2, MEI4, REC102, REC104, REC114) (reviewed in Smith and Nicolas, 1998; Pâques and Haber, 1999). There are now several indications that DSBs in Saccharomyces cerevisiae are initiated by the Spo11 protein, which has been found linked to the 5′ termini of the broken DNA molecules in rad50S yeast mutants (Keeney et al., 1997). Spo11 belongs to a novel family of type II topoisomerase (topoisomerase VI, subunit A), suggesting that Spo11 is the protein that catalyses the formation of DSBs through a 5′-phosphotyrosyl linkage (Bergerat et al., 1997). Sequence comparison of the members of the topoisomerase VI/SPO11 family, as well as structural analyses of one of its members (topoisomerase VI A of Methanococcus jannaschii; Nichols et al., 1999), permitted the definition of several conserved domains involved in DNA binding and cleavage (Bergerat et al., 1997; Aravind et al., 1998; Nichols et al., 1999). Evidence for the generation of DSBs during meiosis of other organisms has only been obtained for Schizosaccharomyces pombe (Cervantes et al., 2000; Zenvirth and Simchen, 2000), but SPO11 homologues have been identified in Drosophila melanogaster (mei-W68; McKim and Hayashi-Hagihara, 1998), Caenorhabditis elegans (Dernburg et al., 1998), S.pombe (REC12; Lin and Smith, 1994), Coprinus cinereus (Cummings et al., 1999; Celerin et al., 2000) and mammals (Keeney et al., 1999; Romanienko and Camerini-Otero, 1999; Shannon et al., 1999; Metzler-Guillemain and de Massy, 2000), suggesting that enzymatic generation of DSBs is conserved among all these organisms. As a matter of fact, meiotic defects (including absence of recombination) were found to be associated with SPO11 disruption not only in yeasts (Klapholz et al., 1985; Atcheson et al., 1987; Giroux et al., 1989; Lin and Smith, 1994), but also in Drosophila (McKim and Hayashi-Hagihara, 1998), C.elegans (Dernburg et al., 1998) and Coprinus (Celerin et al., 2000). Differences between species appeared when authors investigated the relationship between synapsis (homologous chromosome association through the SC) and SPO11 disruption. spo11 mutations in fungi co-ordinately affect recombination and SC formation (Giroux et al., 1989; Celerin et al., 2000), whereas SC formation in Drosophila and C.elegans was found to be independent of SPO11 (Dernburg et al., 1998; McKim et al., 1998). These results underline the importance of studying several independent models to understand the relationship between recombination and synapsis.
In addition to DSB formation and synapsis, additional functions in early chromosome pairing (close juxtaposition of homologous chromosomes; Loidl et al., 1994; Weiner and Kleckner, 1994) and S-phase replication rates (Cha et al., 2000) have been assigned to Spo11 in S.cerevisiae. The recent study of Cha et al. (2000) showed that early roles of Spo11 (chromosome pairing and replication control) can be uncoupled from later ones (DSB formation) in mutants where the tyrosine involved in DSB cleavage is replaced by a phenylalanine (spo11Y135F).
In plants, very few molecular data have been obtained on meiosis. Even less is known about meiotic recombination: the only functional data come from the analyses on the Arabidopsis dmc1 mutant that was shown to be deficient in bivalent formation during male and female meiosis (Couteau et al., 1999). The Lilium RecA homologues (LIM15 and RAD51) (Terasawa et al., 1995; Anderson et al., 1997) as well as the maize RAD51 protein (Franklin et al., 1999) have been studied by immunolocalization on meiotic chromosomes, implicating these proteins in several aspects of meiosis in plants, including homology searches between chromosomes, pairing and recombination. In addition, several synaptic mutants have been described in Arabidopsis (Ross et al., 1997) that could lead to the characterization of other genes potentially involved in recombination since most of these mutations are thought to reflect defects in recombination (Dawe, 1998). Until now, the only report of Arabidopsis mutants affected in meiotic recombination comes from Masson and Paszkowski (1997), who selected several lines according to their hypersensitivity to X-ray irradiation (Masson et al., 1997). Two of them were found to present defects in somatic and meiotic recombination (Masson and Paszkowski, 1997).
Sequencing of the Arabidopsis thaliana genome is nearly complete and allowed Hartung and Puchta (2000) to describe two SPO11 homologues in this model plant. A third SPO11-like coding sequence can be found in the DDBJ/EMBL/GenBank databases (accession No. CAB86036). Until now, SPO11 genes have been found in every eukaryote tested, but only plants possess more than one homologue. The meaning of such sequence redundancy is not yet understood. The study presented here shows that mechanisms by which recombination initiates are conserved between yeast, animals and plants, and that disruption of the AtSPO11-1 gene is sufficient to drastically reduce meiotic recombination and pachytene-stage homologue juxtaposition.
Results
Isolation of spo11-1 mutants of A.thaliana
To identify mutants affected in meiosis, we screened the Versailles collection of T-DNA-transformed lines (Bechtold et al., 1993) as well as ethyl-methane sulfonate (EMS)-mutagenized lines for reduced fertility. Screening for sterility among mutagenized plants allowed two allelic mutations conferring a semi-sterile phenotype (Figure 1) to be isolated.

Fig. 1. spo11-1-1 mutant phenotype. Comparison of 6-week-old Ws (left) and spo11-1-1 (right) plants. Higher magnification of one inflorescence of each is also presented to focus on fruit development (arrows). The scale bar represents 3 cm.
The semi-sterile phenotype segregated 3:1 as a single recessive mutation in both lines (see Materials and methods).
According to the results shown later, the T-DNA and EMS alleles were called spo11-1-1 and spo11-1-2, respectively.
Male and female fertilities are affected in spo11-1 mutants
Plants heterozygous for one of the spo11-1 mutations could not be distinguished from wild type. Homozygous spo11-1 mutant plants exhibited an overall morphology similar to wild type, except for seed production (Figure 1). Both mutant alleles produced very reduced amounts of seeds (on average 2 seeds per silique instead of 45 ± 5 for wild-type plants). Furthermore, 32% of these seeds were abnormal (dark, shrunken and non-viable).
Seeds counted on spo11-1 mutants come from self-fertilization of the plants. In order to examine the effects of spo11-1 mutation on male and female fertilities individually, a spo11-1-1 homozygous mutant plant was used as either the female or the male parent in crosses with a wild-type strain of the same line [ecotype Wassilewskija (Ws)]. When the mutant was used as the female parent, an average of 3.2 seeds per silique were recovered (counted from 51 siliques) and among them very few abnormal seeds were observed (0.2 per silique). When reciprocal crosses were performed (using the spo11-1-1 mutant as male parent), an average of 10.1 seeds per silique were obtained (counted from 46 siliques). Among these, 33% were abnormal (dark and shrunken). These data show that abnormal seeds observed in selfing progeny probably come from male gamete defects. Furthermore, these crosses indicate that both male and female fertilities are impaired by the spo11-1-1 mutation, explaining the low level of seed production observed in spo11-1 selfing progeny.
To investigate the basis of these gametic defects, we examined mature gametophytes in mutant plants (Figure 2). The life cycle of flowering plants is predominantly diploid (sporophytic). The production of gametes occurs within haploid gametophytes (male pollen grains and female embryo sacs) differentiated within the sporophyte after meiosis.
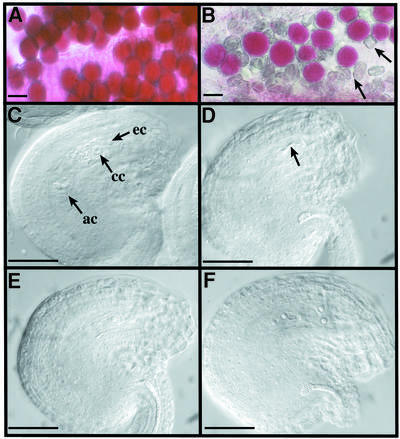
Fig. 2. Male and female gametophyte observation. Viability of mature pollen grains of Ws (A) or spo11-1-1 (B) plants after Alexander staining. Cytoplasm from viable pollen grains is coloured pink. Arrows point to dead pollen grains. The scale bar represents 25 µm in (A) and (B). (C–F) DIC observation of mature ovules of Ws (C) or spo11-1 plants (D–F) after clearing. In a wild-type mature ovule (C), we can observe some of the seven cells of a mature embryo sac (ec, egg cell; cc, central cell; ac, antipodal cells; synergids are out of focus). At the same stage of development, we observe various differentiation stages in the mutant. As examples, in (D) no embryo sac has developed and only degenerated cells can be seen (arrow), (E) shows a one-nucleus embryo sac and (F) shows a four-nuclei embryo sac. For (C–F), the scale bar represents 35 µm.
Mature pollen grains scored for viability by Alexander staining (Figure 2A and B) showed that spo11-1 mutants produced a large proportion of dead pollen in their anthers (shown by arrows for spo11-1-1 in Figure 2B) when compared with wild type (Figure 2A), where 100% of the mature pollen was stained pink. Furthermore, remaining ‘viable’ pollen grains in mutants (monitored by the colouration of the cytoplasm) were variable in size and shape. In vitro germination of spo11-1-1 pollen showed that ∼11% of the pollen grains (among 2969 investigated) germinated, whereas 97% germination of wild-type pollen was observed (data not shown).
On the female side, embryo sacs (mature female gametophytes) develop within ovules. In every ovule, a single nucellar cell differentiates into the megaspore mother cell and undergoes meiosis to produce a tetrad of haploid megaspores. Only one of these survives, and after three mitotic divisions gives rise to an eight-nuclei coenocytic embryo sac, which will then cellularize into a seven-cell embryo sac (Figure 2C). As the embryo sac develops, surrounding ovule tissues (nucellus, integuments and funiculus) evolve to produce a fully curved (anatrope) mature ovule (for a review see Schneitz et al., 1995). Wild-type 2-mm pistils were cleared and observed by differential interference contrast microscopy (DIC). The mature embryo sac composed of seven mature cells could be observed (Figure 2C). In spo11-1-1 plants, however, only 3% (among 1358 observed) of the ovules showed such differentiated female gametophytes. In the remaining ovules, we observed fully developed ovules with no distinguishable embryo sac in 74% of the cases (Figure 2D). However, some intermediate stages of development were sometimes observed, as shown in Figure 2E and F, where apparently mature ovules (as defined by the anatrope shape) contain embryo sacs with one, two or four nuclei, suggesting that meiosis occurred but differentiation of the embryo sac has stopped at different stages of development.
Data are shown for spo11-1-1, but were also observed for the spo11-1-2 allele.
Both male and female meiosis are altered in spo11-1 mutants
To further determine what stages of sporogenesis or gametogenesis are impaired in spo11-1 mutants, we observed the developing male and female gametophytes by DIC after clearing treatments.
Comparison of early steps of male sporogenesis did not reveal any difference between wild-type and mutant plants (Figure 3A and B): round pollen mother cells (PMCs) could be distinguished within the anther loculus. Later, when these cells had performed the two meiotic divisions, we could observe, in wild-type anthers, the characteristic tetrad of microspores encased in a callose wall (Figure 3C). In mutant plants (data are shown only for spo11-1-1), meiosis products encased in a callose wall were observed (Figure 3D), but instead of a regular tetrahedral structure, either asymmetric tetrads or ‘polyads’ containing more than four meiotic products could be distinguished.

Fig. 3. DIC observation of developing pollen grains. (A and B) PMCs at meiosis. Round meiocytes can be seen in wild-type anthers (A) as well as in mutant ones (shown for spo11-1-1; B). (C and D) After meiosis, microspores encased in callose are visible in anthers. The four regular products of meiosis arranged in tetrahedra are observed in wild type (C), whereas irregular tetrads and polyads containing more than four cells are seen in the mutant (D). Scale bar = 10 µm.
Male meiosis was therefore investigated, staining chromosomes with 4′,6-diamidino-2-phenylindole (DAPI) (Figures 4, 5, 7 and 8). Wild-type Arabidopsis meiosis has already been described in detail by Ross et al. (1996) and Figure 4 summarizes its major stages. At metaphase I, the five bivalents of Arabidopsis are easily distinguishable (Figure 4A), each of them being composed of two homologous chromosomes linked together by chiasmata (see also Figure 7F). Anaphase I leads to the formation of dyads containing two pools of five chromosomes (Figure 4B). A partial decondensation of the chromosomes (Figure 4C) occurs before meiosis II, during which segregation of sister chromatids happens, producing four pools of five chromosomes (Figure 4D). Decondensation of chromosomes (Figure 4E) and cytokinesis then take place, producing tetrads of microspores encased in callose (Figures 4F and 3C).

Fig. 4. DAPI staining of wild-type (Ws) PMCs during meiosis. The five bivalents of homologous chromosomes are located at the centre of the cell at metaphase I (A). After the first anaphase, two pools of five chromosomes can be observed (B). They will partially decondense (C) before the second meiotic division, which produces four pools of five chromosomes (D). Decondensation of each of them occurs (E) before cytokinesis (F). Scale bar = 10 µm.

Fig. 7. DAPI staining of wild-type PMCs during meiotic prophase. (A) Leptotene stage. (B) Partial homologous pairing (arrows). (C and D) Pachytene stages showing fully ‘synapsed’ homologous chromosomes. Some bright pericentromeric regions are indicated by arrows. (E) Diplotene stage. (F) Diakinesis stage. Scale bar = 10 µm.
In spo11-1 mutant PMCs, the 10 Arabidopsis chromosomes were condensed, but bivalents were rarely formed (Figure 5A). However, some chromosome bridges were observed (Figure 5B, arrow), showing that chromosomes can still sometimes form bivalents. Out of 84 spo11-1 cells in metaphase I or in the beginning of anaphase I, 36 contained chromosomes showing evidence of some cross-over (39 chromosome bridges and 8 bivalents). These data suggest that ∼7% of chiasma are formed compared with wild type since an average of nine cross-overs per wild-type meiosis was reported by Copenhaver et al. (1998). Anaphase I proceeded, but because most homologous chromosomes were not linked together by chiasmata, segregation appeared to be random. Instead of the two pools of five chromosomes observed at metaphase II in wild-type cells (Figure 4B), variable partitioning of the 10 chromosomes was seen in mutant meiocytes (Figure 5C, D and E). Sister chromatids were then separated and cells with 20 chromosomes were observed (Figure 5F). Decondensation (Figure 5G) and cytokinesis occurred normally, producing the ‘polyads’ already described (Figure 3D) with variable number of cells (Figure 5H), reflecting the variation in numbers of pools of chromosomes obtained at the end of meiosis.

Fig. 5. DAPI staining of spo11-1-1 PMCs during meiosis. At the end of the prophase, cells containing 10 fully condensed chromosomes most of the time are observed (A). When anaphase migration occurs, chromosome bridges can sometimes be seen (B, arrow). Segregation of unpaired chromosomes at anaphase I leads to random partitioning of chromosomes (C–E). The second meiotic division produces cells with more than four pools of DNA, corresponding to a variable number of chromosomes (F). Decondensation (G) and cytokinesis occur, producing polyads (H). Scale bar = 10 µm.
Female meiosis was also investigated after chromosome staining by propidium iodide and observation by confocal microscopy. Univalents were also observed at the end of prophase I (compare Figure 6C and A), leading to random segregation of the chromosomes at anaphase I (compare Figure 6B and D). Therefore, gametophyte development deficiencies described earlier appeared to be a consequence of aberrant meiosis in the mutants.
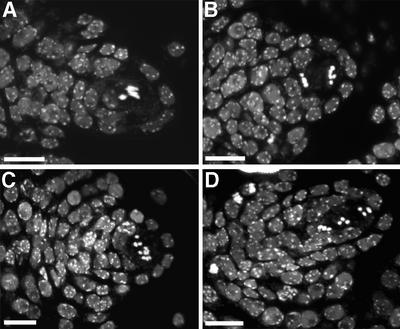
Fig. 6. Propidium iodide staining of developing ovules at meiosis, observed with a confocal laser scanning microscope. Metaphase I of wild type (A) or spo11-1-1 mutant (C). Metaphase II of wild type (B) or spo11-1-1 mutant (D). Scale bar = 10 µm.
Chromosome behaviour during the earlier stage of meiosis was then analysed, comparing meiotic prophase I in wild type (Figure 7) and in the spo11-1-1 mutant (Figure 8). Figure 7A shows a leptotene nucleus with very thin chromosome threads. Later, synapsis progresses and can be identified in some nuclei where loops of paired and unpaired chromosomes alternate (Figure 7B, arrows). Synapsis is maximum at pachytene (Figure 7C and D), during which bivalents appear thick. Later, each member of the bivalent tends to separate (Figure 7E), and chiasmata become visible at diakinesis (Figure 7F). Out of 90 cells in prophase I in wild type, we found six in leptotene (Figure 7A), 21 in diakinesis (Figure 7F) and 43 in pachytene (Figure 7C and D). The remaining cells (20) showed intermediate synapsis. The same analyses on the spo11-1-1 mutant did not allow the identification of any pachytene stage among 160 cells observed. Nevertheless, condensation progressed until diakinesis [eight cells were observed in leptotene (Figure 8A), 27 in diakinesis (Figure 8D) and 125 at intermediate stages of condensation like the one shown in Figure 8B and C], but fully synapsed chromosomes were never observed.

Fig. 8. DAPI staining of spo11-1-1 PMCs during meiotic prophase. (A) A cell at leptotene. In (B and C), chromosomes are condensing but no formation of bivalents is observed. (D) Diakinesis. Scale bar = 10 µm.
Molecular characterization of spo11-1 mutations
Plant DNA flanking the T-DNA insertion in the spo11-1-1 allele was cloned either by plasmid rescue or tail PCR. PCR mapping on the CIC YAC library showed that the two borders map on the same part of chromosome 3 on CIC clones 6H6 and 5G8. The Columbia allele of this gene was found in the database as part of a P1 clone sequenced on chromosome three of Arabidopsis (clone MJG9, DDBJ/EMBL/GenBank accession No. AP000375). The T-DNA insertion occurred between nucleotides 43553 and 43555. The EMS mutation (spo11-1-2) was mapped to chromosome 3, between morphological markers hy2 (12 cM) and gl1 (30 cM), confirming the location of spo11-1 mutations on the top arm of chromosome 3 between 15 and 25 cM.
Southern analyses (not shown) showed that spo11-1-1 possessed two linked left borders (LB) and no right border (RB). Indeed, cloning and sequencing of the T-DNA flanking sequences showed that a recombined insertion had occurred (Figure 9). A first LB was truncated within the 24 bp repeat, which is observed frequently in T-DNA transformants (LB1; Figure 9). The RB region and the promotorless GUS coding sequence of the T-DNA were deleted, and replaced by a piece of the binary vector pGKB5 and a second LB, complementary to the first one (LB2; Figure 9).
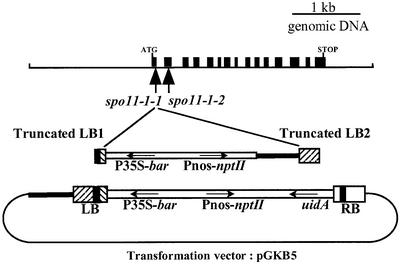
Fig. 9. AtSPO11-1 coding region and spo11-1 mutations. Schematic representations of AtSPO11-1, spo11-1-1 T-DNA insertion and pGKB5 transformation vector. Black boxes represent exons [from the start of translation (ATG) to the stop codon (STOP)] of the main transcript of the AtSPO11-1 gene. The location of the T-DNA insertion in the spo11-1-1 allele as well as the location of the point mutation that occurred in the spo11-1-2 allele are indicated by vertical arrows. The pGKB5 transformation vector used to generate the T-DNA transformant collection and the recombined T-DNA structure of the spo11-1-1 allele are indicated underneath. Boxes represent the T-DNA borders (RB and LB), where the 24 bp repeats are indicated in black. The orientation of the T-DNA coding sequences (bar gene conferring basta resistance, nptII gene conferring kanamycin resistance and uidA gene encoding the GUS enzyme) is indicated by horizontal arrows.
We found that T-DNA insertion occurred in the first exon of AtSPO11-1 recently described by Hartung and Puchta (2000). T-DNA insertion in the spo11-1-1 allele disrupted the gene as early as the first exon (Figures 9 and 10), probably leading to a null mutant. The Ws sequence for AtSPO11-1 was obtained (DDBJ/EMBL/GenBank accession No. AF302928) and compared with the Columbia sequence, showing 96% identity at the nucleotide level. cDNA was obtained through RT–PCR (data not shown) and encodes a protein showing 98.6% identity with the Columbia sequence (Figure 10). Expression studies performed by RT–PCR showed that AtSPO11-1 is expressed at a low level and mainly, but not exclusively, in reproductive tissues (data not shown). Interestingly, alternate splicing as described previously by Hartung and Puchta (2000) was constantly observed. It is not clear yet how this pattern of intron retention is developmentally regulated. Sequencing of the AtSPO11-1 gene in the EMS allele (spo11-1-2) revealed a C to T transition in the second exon of AtSPO11-1, transforming a glutamine codon CAA into a stop codon TAA (Figures 9 and 10), confirming that mutant phenotypes observed in both mutants are the result of a non-functional AtSPO11-1 gene.

Fig. 10. Clustal_X alignment of the three Arabidopsis SPO11-like proteins (Columbia ecotype) (AtSPO11-1, DDBJ/EMBL/GenBank accession No. CAB81544; AtSPO11-2, accession No. CAB81545; AtSPO11-3, accession No. CAB86036) with S.cerevisiae (Sc; accession No. AAA65532) and mouse (accession No. AAD52563) SPO11 proteins as well as S.shibatae topoisomerase VI A protein (accession No. O05208). Conserved motifs I–V, as defined by Bergerat et al. (1997), are indicated in black boxes and conserved amino acids are shadowed in grey. The location of the T-DNA insertion in the spo11-1-1 allele and the stop mutation of the spo11-1-2 allele are indicated by arrowheads. An asterisk shows the conserved tyrosine (in position 135 in S.cerevisiae) thought to be responsible for DNA cleavage (Bergerat et al., 1997). Amino acids that differ between Ws and Columbia AtSPO11-1 are underlined. An isoleucine replaces the phenylalanine (position 5), a glutamic acid replaces the valine (position 26), a serine replaces the asparagine (position 39), a leucine replaces the phenylalanine (position 65), and there is an insertion of a valine in position 213 between the underlined threonine and glycine.
Hartung and Puchta (2000) described a second Arabidopsis SPO11-like gene located on chromosome 1 (DDBJ/EMBL/GenBank accession No. AJ251990). In addition, a third sequence putatively encoding a protein with strong similarity to Spo11 was detected in the Arabidopsis genome (chromosome 5, DDBJ/EMBL/GenBank accession Nos AL162973, CAB86036). We checked that this third gene is transcribed (DDBJ/EMBL/GenBank accession number No. AF323679). Figure 10 shows an alignment of several SPO11/Topo VI A orthologues including the three Arabidopsis homologues. The level of amino acid identity between AtSPO11-1 and S.cerevisiae Spo11 is 30%. Conserved regions I–V, as defined by Bergerat et al. (1997), are indicated. Region I contains the active tyrosine site (indicated by an asterisk), which is invariant among all the proteins of this family and which has been shown to be necessary for DSB induction in S.cerevisiae (Bergerat et al., 1997).
AtSPO11-1 disruption drastically decreases meiotic recombination
The S.cerevisiae Spo11 protein is thought to catalyse DNA DSBs that initiate meiotic recombination. To examine recombination frequencies in a spo11-1 mutant background, lines heterozygous for the spo11-1-1 mutation (Ws ecotype) were crossed to Columbia (Col-0). Meiotic recombination rates were investigated in the progeny of F2 plants either homozygous for the mutation spo11-1-1, heterozygous or wild type. Microsatellites showing polymorphisms between the two ecotypes used (Bell and Ecker, 1994) were chosen to measure recombination frequencies on two chromosome arms: the bottom of chromosome 1 between markers nga280 and nga111, and the top of chromosome V between markers nga106 and nga76. MAPMAKER (Lander et al., 1987) was used to convert data into genetic distance in centimorgans (Table I). According to the recent RI map (http://nasc.nott.ac.uk/), markers nga280 and nga111 are separated by 31.72 cM, and markers nga76 and nga106 by 35.05 cM. These data fit with the distances we calculated from our results on either heterozygous or wild-type plants (Table I; spo11-1/SPO11-1 and SPO11-1/SPO11-1 plants). In contrast, distances between the same markers calculated in the spo11-1 background were ∼10 times lower, since the calculated map distance between nga111 and nga280 has fallen from 31.72 to 4.3 cM, and the distance between nga76 and 106 from 35.05 to 2.4 cM. We checked that this effect is not plant dependent since three different homozygous spo11-1/spo11-1 plants were used to collect these data (sister plants 15, 27 and 39; Table I).
Table I. Effect of spo11-1 mutation on meiotic recombination.
| Genotype | No. | Map distanceanga111/280 | Map distanceanga76/106 |
|---|---|---|---|
| spo11-1/spo11-1 | 15 | nd | 1.4 (70) |
| 27 | 4.2 (86) | nd | |
| 39 | 4.4 (71) | 3.3 (77) | |
| average | 4.3 (157) | 2.4 (147) | |
| spo11-1/SPO11-1 | 48 | 37.5 (45) | 30.7 (45) |
| SPO11-1/SPO11-1 | 6 | nd | 20.6 (78) |
| 10 | 36.1 (71) | nd | |
| average | 36.8 (116) | 23.8 (123) |
aMap distance calculated in centimorgans in the progeny of sister plants either homozygous for the spo11-1-1 mutation (plants 15, 27 and 39), heterozygotes (plant 48) or wild type (plants 6 and 10). Total numbers of plants tested for each marker are indicated in parentheses. nd, not done.
Discussion
Initiation of meiotic recombination in plants via DSBs
In this paper, we show that disruption of AtSPO11-1 induces perturbation of male and female meiosis, with the formation of very few bivalents at the end of prophase I. This absence of association between homologous chromosomes can be explained by an absence of crossing over, prerequisite to any chiasmata and therefore to bivalent stabilization. Another possibility could be that recombination proceeds normally, but premature release of the chiasmata leads to univalent formation, as has already been reported for the dy and dsy mutants of maize (Maguire, 1978; Maguire et al., 1993). The drastic decrease of meiotic recombination observed in a spo11-1 background indicates that the first hypothesis is the correct one.
There is now quite a lot of evidence that Spo11 is the protein involved in the induction of DSBs in yeast, necessary for cross-overs and therefore chiasmata formation. Even if the formation of DSBs has only been demonstrated in yeasts, recent analyses showed that induction of DSBs by Spo11 is probably conserved in C.elegans (Dernburg et al., 1998), D.melanogaster (McKim and Hayashi-Hagihara, 1998) and C.cinereus (Celerin et al., 2000). In plants, it has been shown that artificially induced DSBs (via γ-irradiation) of Lilium longiflorum buds can increase the number of chiasmata per bivalent (Lawrence, 1961). Irradiation can also partially bypass the requirement for Spo11 in C.elegans (Dernburg et al., 1998) and C.cinereus (Celerin et al., 2000), as in S.cerevisiae (Thorne and Byers, 1993). These data, taken together with the fact that Arabidopsis spo11-1 mutants are defective in recombination, suggest that initiation of recombination via SPO11-induced DSBs is conserved in plants, even if there is no biochemical proof so far that any SPO11 can make DSBs in vitro.
Why are there three SPO11-like genes in Arabidopsis?
It is likely that events initiating meiotic recombination are conserved between plants and other eukaryotes, but the existence of at least three SPO11-like genes in the Arabidopsis genome remains to be explained. It has emerged from Arabidopsis sequencing project data that this genome shows an unexpectedly high level of genetic redundancy, since as many as 75% of gene products show significant similarity to another protein coded in the genome (Bancroft, 2000). Our study proves that AtSPO11-1 is a functional homologue of other eukaryote SPO11 genes. The severe phenotype of both spo11-1 mutants we isolated shows that the other two AtSPO11 genes are not functionally redundant with AtSPO11-1. Nevertheless, recombination is not null in spo11-1-1 mutant (see Table I), and we obtained cytological evidence for the formation of some bivalents (Figure 5B). These results are different from what was obtained for yeast, Drososphila and C.elegans spo11 mutants, where no evidence for any meiotic recombination events was obtained (Klapholz et al., 1985; Dernburg et al., 1998; McKim et al., 1998). Even if a T-DNA insertion can sometimes lead to a leaky mutation, we have good reason to believe that spo11-1-1 is null since the T-DNA insertion occurred within the first exon of the gene. Therefore, it is plausible that at least one of the remaining AtSPO11-like genes is responsible for the low level of recombination we measured in the spo11-1-1 mutant, restoring (very partially) AtSPO11-1 function. However, we cannot exclude the possibility that other protein(s) partially responsible for DSB formation exist in plants and not in other eukaryotes. Another possibility is that environmental factors might be able to induce DSBs in plants and not in other eukaryotes where reproductive cells are less exposed to external factors.
Concerning the functions of AtSPO11-2 and -3, we do not possess much data yet. The specificity of expression of these genes, as well as their disruption, will give more information on their role in plants. Several hypotheses can already be raised. First, they might be involved in recombination within organs other than meiocytes since most recombination events are known to be induced by DSBs (Pâques and Haber, 1999). Another explanation might be that these other two sequences have topoisomerase functions independent of recombination. Spo11 function in S.cerevisiae was elucidated thanks to the isolation of a new type of topoisomerase II from Sulfolobus shibatae (topoisomerase VI; Bergerat et al., 1997). In the Archaea, this topoisomerase is composed of two subunits: A and B. Subunit A is homologous to Spo11 and subunit B has ATP binding activity. Homologues of topoisomerase VI subunit A were found in every eukaryote studied and all have meiotic functions similar to yeast Spo11. Until now, no B subunit homologue has been found, even in the fully sequenced S.cerevisiae and C.elegans genomes. However, a BLAST search revealed topoisomerase VI B subunit-like sequences in several plant genomes, including Arabidopsis (DDBJ/EMBL/GenBank accession Nos AB025629, BAB02486). The fact that plants are the only eukaryotes to possess a topoVIB gene and more than one topoVIA coding sequence might indicate that plants have conserved topoisomerase VI function in addition to a specialized meiotic SPO11-topoisomerase VI-related function.
What is the link between synapsis and recombination in plants?
The relationship and timing between different events in meiotic prophase, including chromosome synapsis and recombination, are very striking and subject to debate. Historically, it was proposed that chromosomes synapse before they recombine. In yeast, however, it is now obvious that DSBs occur during the early stages of meiotic prophase (Padmore et al., 1991), well before the SC assembles and triggers synapsis between homologues. In this organism, there is also some evidence that recombination and synapsis are tightly coupled events (reviewed in Zickler and Kleckner, 1999). The yeast spo11Δ and spo11Y135F mutants do not form any SC (Giroux et al., 1989; Cha et al., 2000), showing that SC formation in yeast is dependent upon Spo11 function and possibly upon DSB formation. The discovery that in C.elegans (Dernburg et al., 1998) and Drosophila (McKim et al., 1998) spo11 mutant chromosomes can pair and synapse without any recombination is very intriguing, but these may be exceptions. Indeed, the C.cinereus spo11 mutant was recently found to be unable to form mature SC (Celerin et al., 2000). The analyses we present in this paper show that, in plants also, synapsis is affected when SPO11 function is absent, since no pachytene stages were observed in the Arabidopsis spo11-1 mutant. Therefore, it seems that compared with other eukaryotes, Drosophila and C.elegans have different control mechanisms for SC formation.
Meiosis in spo11-1 mutants is abnormal but not prevented, leading to chromosomal missegregations and production of unbalanced gametes
Male and female meiosis in spo11-1 mutants is perturbed, with little formation of bivalents at the end of prophase I; nevertheless, chromosome segregation occurs. Male meiosis of spo11-1 mutants produces aberrant ‘tetrads’ composed of more than four cells, having different sizes. In Arabidopsis, all the meiotic mutants described so far produce this type of polyad (He et al., 1996; Peirson et al., 1996; Ross et al., 1997; Bhatt et al., 1999; Couteau et al., 1999; Yang et al., 1999). Some of the genes affected have been cloned and they concern mutations that affect recombination steps (dmc1; Couteau et al., 1999), chromosome synapsis (asy1; Caryl et al., 2000), chromosome or chromatid segregation (syn1, Bai et al., 1999; dif1, Bhatt et al., 1999; ask1, Yang et al., 1999) or late abnormalities of division (ms5/tdm; Glover et al., 1998).
Gametogenesis in spo11-1 mutants, like for many other plant meiotic mutants, is not abolished since differentiation of male and female gametophytes is observed. Microspore aneuploidy, followed by chromosome staining with DAPI, results in a large proportion of aborted or abnormal pollen grains (only 11% of spo11-1 pollen grains germinate in vitro). Studies on chromosomal rearrangements and pollen grain irradiation have shown that although both male and female gametophytes are fairly tolerant to additions to the single set of chromosomes (trisomics, duplications), the gametophytes are particularly sensitive to deficiencies, therefore most unbalanced gametes are lethal. On the female side, we mainly observed mature ovules that failed to differentiate a functional embryo sac, and stopped at different stages of development (no nucleus, one, two or four nuclei embryo sacs), probably according to the level of aneuploidy of the meiosis products. Therefore, female fertility (3% of mature embryo sacs) fits with what is expected from random segregation of the 10 unpaired chromosomes of Arabidopsis, as was suggested by Couteau et al. (1999) for the At-dmc1 mutant phenotype [(1/2)5 = 0.03]. We obtained approximately the same amount of seeds per silique in the progeny of self-pollinated homozygous mutant plants (2 seeds per silique) or in crosses using the mutant as female (3.2 seeds per silique). Therefore, the number of pollen grains able to fertilize is probably not a limiting parameter in spo11-1 mutant fertility.
Among the few seeds produced in spo11-1 mutant selfing progeny, 32% are abnormal. The analysis of reciprocal crosses using spo11-1 mutants as male or female parent showed that the large proportion of aborted seeds observed in selfing progeny comes from a male defect and not a female one. These data are interesting since most of the data obtained on the consequences of unbalanced gametes on seed development come from studies on irradiated pollen used as pollinator on wild-type ovules (see e.g. Cave and Brown, 1954; Vizir et al., 1994). Therefore, spo11-1 and other synaptic mutants represent new tools to study the impact of unbalanced macro- or microspores on gamete development, fertilization and seed development.
Materials and methods
Plant material
Arabidopsis thaliana (L.), ecotype Wassilewskija (Ws), was used to generate a collection of T-DNA insertional mutants by infiltration of adult plants (Bechtold et al., 1993). Three per cent of the T2 lines showed reduced fertility (scored on the basis of limited elongation of the siliques). Among them, spo11-1-1 was isolated.
spo11-1-2 was isolated from Arabidopsis ecotype Columbia (Col-O) seed stocks that were mutagenized by EMS, essentially as described by Chory et al. (1989). After individual screening of 300 M2 progenies, 17 lines affected in their fertility were found, among which spo11-1-2 was characterized. This EMS line was backcrossed twice by the wild-type parent (Col-O ecotype).
Genetic analyses
The T-DNA mutant (in a Ws background) was initially found to segregate 63:1 for kanamycin resistance (one of the T-DNA markers), suggesting the presence of at least three unlinked inserts. After crossing to wild type, lines were obtained segregating 3:1 for kanamycin resistance and for the semi-sterile mutation. Linkage between the remaining T-DNA insert and the semi-sterile phenotype was checked by analysing the progeny of 100 plants resulting from self-pollination of a plant heterozygous for the remaining kanamycin insertion. Among these 100 plants, 25 exhibited the mutant phenotype and were resistant to kanamycin. For the remaining fertile plants, it was checked that each plant heterozygous for the kanamycin resistance gene (segregating 3:1 in vitro for the kanamycin resistance) was also producing 25% semi-sterile plants in its selfed progeny, when every wild-type plant (100% of progeny sensitive to kanamycin) did not generate any semi-sterile plants after selfing. Three backcrosses with the Ws wild-type line were performed on several independent heterozygotes, checking the expected 1:1 segregation of the kanamycin resistance gene every generation.
In order to test for allelism, crosses using as female a plant heterozygous for the EMS mutation (25% of sterile plants in its progeny) and as male a plant heterozygous for the T-DNA mutation (progeny segregating 3:1 for the kanamycin resistance in vitro) yielded 13 sterile plants among 69 F1 examined, indicating that the two mutations are allelic.
Mapping of spo11-1-1. Oligonucleotides were deduced from plant genomic sequences flanking T-DNA insert. They were used for PCR screening of the CIC YAC library as described by Creusot et al. (1995) on three-dimensional YAC pools.
Mapping of spo11-1-2. Heterozygous plants were crossed with tester lines (NW, provided by NASC, Nottingham Arabidopsis Stock Center) in Landsberg erecta ecotype, carrying 2 or 3 different phenotypic markers per chromosome (Koornneef et al., 1983). The recombined F2 plants were checked and the data analysed with MAPMAKER (version 3.0; SAS Institute, 1988).
Measurement of meiotic recombination in a spo11-1-1 background. Lines heterozygous for the spo11-1-1 mutation (Ws ecotype) were crossed to a Columbia strain (Col-0). F1 plants heterozygous for the spo11-1-1 mutation were selected according to their resistance to kanamycin. F2 plants were generated by selfing and characterized for several loci including AtSPO11-1, and microsatellite markers showing polymorphisms between the two ecotypes Ws and Col-0 (Bell and Ecker, 1994). Levels of recombination were estimated choosing plants heterozygous for markers nga280 and nga111 or heterozygous for markers nga106 and nga76.
Isolation of plant T-DNA flanking sequences
One of the LB flanking genomic sequences (LB1) was obtained with thermal asymmetric interlaced PCR (TAIL PCR) according to Liu et al. (1995), using the arbitrary degenerate primer AD3 [5′-(A/T)GTGNAG(A/T)ANCANAGA-3′]. The three specific primers of the LB of the pGKB5 vector (Bouchez et al., 1993) are TAG5 (5′-ATTGCCTTTTCTTATCGACC-3′), TailB (5′-CGGCTATTGGTAATAGGACACTGG-3′) and LB-Bar1 (5′-CAACCCTCAACTGGAAACGGGCCGGA-3′).
The other T-DNA border was isolated by performing kanamycin rescue experiments according to Bouchez et al. (1996). A 6 kbp PstI insert was cloned in pResc38; 3.1 kb of the cloned insert were found to be plant DNA.
Light and confocal microscopy
Observation of developing ovules and embryo sacs by DIC was performed as described by Motamayor et al. (1999).
Development of PMCs was observed under DIC after clearing in Herr’s buffer (phenol:chloral hydrate: 85% lactic acid:xylene:oil of clove; 1:1:1:0.5:1; w:w:w:w:w) of fresh buds of different sizes.
Mature pollen grain viability was estimated as described by Alexander (1969).
DAPI staining of meiotic chromosomes was performed either according to the technique described by Maluszynska and Heslop-Harrison (1991) or according to that described by Ross et al. (1996).
For confocal laser scanning microscopy, anthers and pistils of different stages were observed as described by Motamayor et al. (1999).
Acknowledgments
Acknowledgements
We are grateful to Françoise Budar, Herman Höfte, Raphaël Mercier, Christine Mezard and Ian Small for helpful discussions and critical reading of the manuscript. We also thank Christine Camilleri for her great help in genetic experiments and Gareth Jones for his advice on DAPI staining of prophase chromosomes.
References
- Alexander M.P. (1969) Differential staining of aborted and non aborted pollen. Stain Technol., 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Anderson L.K., Offenberg,H.H., Verkuijlen,W. and Heyting,C. (1997) RecA-like proteins are components of early meiotic nodules in lily. Proc. Natl Acad. Sci. USA, 94, 6868–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Leipe,D.D. and Koonin,E.V. (1998) Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res., 26, 4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atcheson C.L., DiDomenico,B., Frackman,S., Esposito,R.E. and Elder,R.T. (1987) Isolation, DNA sequence and regulation of a meiosis-specific eukaryotic recombination gene. Proc. Natl Acad. Sci. USA, 84, 8035–8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Peirson,B.N., Dong,F., Cai,X., Makaroff,C.A., Bai,X.F., Xue,C. and Dong,F.G. (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell, 11, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I. (2000) Insights into the structural and functional evolution of plant genomes afforded by the nucleotide sequences of chromosomes 2 and 4 of Arabidopsis thaliana. Yeast, 17, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Ellis,J. and Pelletier,G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, 316, 1194–1199. [Google Scholar]
- Bell C.J. and Ecker,J.R. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics, 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bergerat A., de Massy,B., Gadelle,D., Varoutas,P.C., Nicolas,A. and Forterre,P. (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature, 386, 414–417. [DOI] [PubMed] [Google Scholar]
- Bhatt A.M., Lister,C., Page,T., Fransz,P., Findlay,K., Jones,G.H., Dickinson,H.G. and Dean,C. (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J., 19, 463–472. [DOI] [PubMed] [Google Scholar]
- Bouchez D., Camilleri,C. and Caboche,M. (1993) A binary vector based on basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris, 316, 1188–1193. [Google Scholar]
- Bouchez D., Vittorioso,P., Courtial,B. and Camilleri,C. (1996) Kanamycin rescue: a simple technique for the recovery of T-DNA flanking sequences. Plant Mol. Biol. Rep., 14, 115–123. [Google Scholar]
- Cao L., Alani,E. and Kleckner,N. (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell, 61, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong,S.J., Jones,G.H. and Franklin,F.C. (2000) A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma, 109, 62–71. [DOI] [PubMed] [Google Scholar]
- Cave M.S. and Brown,S.W. (1954) The detection and nature of dominant lethals in Lilium. II. Cytological abnormalities in ovules after pollen irradiation. Am. J. Bot., 41, 469–483. [Google Scholar]
- Celerin M., Merino,S.T., Stone,J.E., Menzie,A.M. and Zolan,M.E. (2000) Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J., 19, 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes M.D., Farah,J.A. and Smith,G.R. (2000) Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell, 5, 883–888. [DOI] [PubMed] [Google Scholar]
- Cha R.S., Weiner,B.M., Keeney,S., Dekker,J. and Kleckner,N. (2000) Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev., 14, 493–503. [PMC free article] [PubMed] [Google Scholar]
- Chory J., Peto,C., Feinbaum,R., Pratt,L. and Ausubel,F. (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell, 58, 991–999. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P., Browne,W.E. and Preuss,D. (1998) Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl Acad. Sci. USA, 95, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Belzile,F., Horlow,C., Grandjean,O., Vezon,D. and Doutriaux,M.P. (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell, 11, 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F. and et al. (1995) The CIC library: a large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J., 8, 763–770. [DOI] [PubMed] [Google Scholar]
- Cummings W.J., Celerin,M., Crodian,J., Brunick,L.K. and Zolan,M.E. (1999) Insertional mutagenesis in Coprinus cinereus: use of a dominant selectable marker to generate tagged, sporulation-defective mutants. Curr. Genet., 36, 371–382. [DOI] [PubMed] [Google Scholar]
- Dawe R.K. (1998) Meiotic chromosome organization and segregation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 371–395. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald,K., Moulder,G., Barstead,R., Dresser,M. and Villeneuve,A.M. (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell, 94, 387–398. [DOI] [PubMed] [Google Scholar]
- Franklin A.E., McElver,J., Sunjevaric,I., Rothstein,R., Bowen,B. and Cande,W.Z. (1999) Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell, 11, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux C.N., Dresser,M.E. and Tiano,H.F. (1989) Genetic control of chromosome synapsis in yeast meiosis. Genome, 31, 88–94. [DOI] [PubMed] [Google Scholar]
- Glover J., Grelon,M., Craig,S., Abdul,C., Dennis,E. and Chaudhury,A. (1998) Cloning and characterization of MS5 from Arabidopsis: a gene critical in male meiosis. Plant J., 15, 345–356. [DOI] [PubMed] [Google Scholar]
- Hartung F. and Puchta,H. (2000) Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res., 28, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Tirlapur,U., Cresti,M., Peja,M., Crone,D.E., Mascarenhas,J.P. and He,C.P. (1996) An Arabidopsis mutant showing aberrations in male meiosis. Sexual Plant Reprod., 9, 54–57. [Google Scholar]
- Keeney S., Giroux,C.N. and Kleckner,N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Keeney S., Baudat,F., Angeles,M., Zhou,Z.H., Copeland,N.G., Jenkins,N.A., Manova,K. and Jasin,M. (1999) A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics, 61, 170–182. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Waddell,C.S. and Esposito,R.E. (1985) The role of the SPO11 gene in meiotic recombination in yeast. Genetics, 110, 187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., van Eden,J., Hanhart,C.J., Stam,P., Braaksma,F.J. and Feenstra,W.J. (1983) Linkage map of Arabidopsis thaliana. J. Hered., 74, 265–272. [Google Scholar]
- Lander E.S., Green,P., Abrahamson,J., Barlow,A., Daly,M.J., Lincoln,S.E. and Newburg,L. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics, 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W. (1961) The effect of the irradiation of different stages in microsporogenesis on chiasma frequency. Heredity, 16, 83–89. [Google Scholar]
- Lin Y. and Smith,G.R. (1994) Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics, 136, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa,N., Oosumi,T. and Whittier,R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J., 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Loidl J., Klein,F. and Scherthan,H. (1994) Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J. Cell Biol., 125, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M.P. (1978) Evidence for separate genetic control of crossing over and chiasma maintenance in maize. Chromosoma, 65, 173–183. [Google Scholar]
- Maguire M.P., Riess,R.W. and Paredes,A.M. (1993) Evidence from a maize desynaptic mutant points to a probable role of synaptonemal complex central region components in provision for subsequent chiasma maintenance. Genome, 36, 797–807. [DOI] [PubMed] [Google Scholar]
- Maluszynska J. and Heslop-Harrison,J.S. (1991) Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J., 1, 159–166. [Google Scholar]
- Masson J.E. and Paszkowski,J. (1997) Arabidopsis thaliana mutants altered in homologous recombination. Proc. Natl Acad. Sci. USA, 94, 11731–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J.E., King,P.J. and Paszkowski,J. (1997) Mutants of Arabidopsis thaliana hypersensitive to DNA-damaging treatments. Genetics, 146, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S. and Hayashi-Hagihara,A. (1998) mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev., 12, 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Green-Marroquin,B.L., Sekelsky,J.J., Chin,G., Steinberg,C., Khodosh,R. and Hawley,R.S. (1998) Meiotic synapsis in the absence of recombination. Science, 279, 876–878. [DOI] [PubMed] [Google Scholar]
- Metzler-Guillemain C. and de Massy,B. (2000) Identification and characterization of an SPO11 homolog in the mouse. Chromosoma, 109, 133–138. [DOI] [PubMed] [Google Scholar]
- Motamayor J.-C., Vezon,D., Bajon,C., Sauvanet,A., Grandjean,O., Marchand,M., Bechtold,N., Pelletier,G. and Horlow,C. (1999) Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sexual Plant Reprod., 12, 209–218. [Google Scholar]
- Nichols M.D., DeAngelis,K., Keck,J.L. and Berger,J.M. (1999) Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J., 18, 6177–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R., Cao,L. and Kleckner,N. (1991) Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell, 66, 1239–1256. [DOI] [PubMed] [Google Scholar]
- Pâques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson B.N., Owen,H.A., Feldmann,K.A. and Makaroff,C.A. (1996) Characterization of three male-sterile mutants of Arabidopsis thaliana exhibiting alterations in meiosis. Sexual Plant Reprod., 9, 1–16. [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J. and Camerini-Otero,R.D. (1999) Cloning, characterization and localization of mouse and human SPO11. Genomics, 61, 156–169. [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz,P. and Jones,G.H. (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res., 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz,P., Armstrong,S.J., Vizir,I., Mulligan,B., Franklin,F.C. and Jones,G.H. (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res., 5, 551–559. [DOI] [PubMed] [Google Scholar]
- Schneitz K., Hülskamp,M. and Pruitt,R.E. (1995) Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J., 7, 731–749. [Google Scholar]
- Shannon M., Richardson,L., Christian,A., Handel,M.A. and Thelen,M.P. (1999) Differential gene expression of mammalian SPO11/TOP6A homologs during meiosis. FEBS Lett., 462, 329–334. [DOI] [PubMed] [Google Scholar]
- Smith K.N. and Nicolas,A. (1998) Recombination at work for meiosis. Curr. Opin. Genet. Dev., 8, 200–211. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco,D., Schultes,N.P. and Szostak,J.W. (1989) Double-strand breaks at an initiation site for meiotic gene conversion. Nature, 338, 87–90. [DOI] [PubMed] [Google Scholar]
- Terasawa M., Shinohara,A., Hotta,Y., Ogawa,H. and Ogawa,T. (1995) Localization of RecA-like recombination proteins on chromosomes of the lily at various meiotic stages. Genes Dev., 9, 925–934. [DOI] [PubMed] [Google Scholar]
- Thorne L.W. and Byers,B. (1993) Stage-specific effects of X-irradiation on yeast meiosis. Genetics, 134, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizir I.Y., Anderson,M.L., Wilson,Z.A. and Mulligan,B.J. (1994) Isolation of deficiencies in the Arabidopsis genome by γ-irradiation of pollen. Genetics, 137, 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner B.M. and Kleckner,N. (1994) Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell, 77, 977–991. [DOI] [PubMed] [Google Scholar]
- Yang M., Hu,Y., Lodhi,M., McCombie,W.R. and Ma,H. (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl Acad. Sci. USA, 96, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D. and Simchen,G. (2000) Meiotic double-strand breaks in Schizosaccharomyces pombe. Curr. Genet., 38, 33–38. [DOI] [PubMed] [Google Scholar]
- Zickler D. and Kleckner,N. (1999) Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet., 33, 603–754. [DOI] [PubMed] [Google Scholar]


