Abstract
Nucleotide excision repair (NER) is a major DNA repair mechanism that recognizes a broad range of DNA damages. In Escherichia coli, damage recognition in NER is accomplished by the UvrA and UvrB proteins. We have analysed the structural properties of the different protein–DNA complexes formed by UvrA, UvrB and (damaged) DNA using atomic force microscopy. Analysis of the UvrA2B complex in search of damage revealed the DNA to be wrapped around the UvrB protein, comprising a region of about seven helical turns. In the UvrB–DNA pre-incision complex the DNA is wrapped in a similar way and this DNA configuration is dependent on ATP binding. Based on these results, a role for DNA wrapping in damage recognition is proposed. Evidence is presented that DNA wrapping in the pre-incision complex also stimulates the rate of incision by UvrC.
Keywords: atomic force microscopy/damage recognition/Escherichia coli/nucleotide excision repair/UvrABC
Introduction
Nucleotide excision repair (NER) is a multi-step process that removes a wide variety of lesions from the DNA. In Escherichia coli, the UvrA, UvrB and UvrC proteins initiate the repair reaction. The joint action of these proteins results in incisions on both sides of the lesion (for reviews see Van Houten, 1990; Sancar, 1996; Goosen et al., 1998). In solution, UvrA forms a dimer that interacts with UvrB to form the UvrA2B complex. After this complex has bound to a damaged site, UvrB is loaded onto the DNA to produce a stable UvrB–DNA pre-incision complex. Upon binding of UvrC to the pre-incision complex, two incisions are made. The first incision is made at the 4th or 5th phosphodiester bond 3′ to the lesion, immediately followed by incision at the 8th phosphodiester bond at the 5′ side of the lesion. The catalytic sites for 3′ and 5′ incisions are located in different domains of UvrC (Lin and Sancar, 1992; Verhoeven et al., 2000).
One of the most fascinating aspects of the UvrABC system is its ability to repair a vast array of structurally unrelated DNA lesions. In general, it has been proposed that a change in base-pair stacking caused by the damage is the determinant for damage recognition (Van Houten, 1990), but as yet little is known on the mechanism of recognition of such altered stacking interactions. It is generally believed that the UvrA subunit of the UvrA2B complex initially recognizes a damage, as the UvrA protein on its own binds preferentially to damaged DNA (Mazur and Grossman, 1991). After damage recognition, the UvrA2B complex brings the UvrB protein close to the damage site. This is most likely achieved by the opening up of the DNA helix. UvrB can specifically recognize a damage in the absence of UvrA, when this damage is located close to the end of a double-stranded fragment (Moolenaar et al., 2000a) or in the unpaired region of a bubble substrate (Zou and Van Houten, 1999). Next, the UvrB protein probes the DNA again for the presence of structural aberrations, possibly by checking the base-pair stacking interactions. When a lesion is detected, the UvrB protein hydrolyses its bound ATP molecule, leading to formation of the pro-pre-incision complex (Moolenaar et al., 2000b). Finally, an active UvrB–DNA pre-incision complex is formed by the binding of a new ATP molecule, and after binding of UvrC to this complex the two incisions are made (Moolenaar et al., 2000b).
The structural properties of the UvrA2B–DNA and UvrB–DNA complexes will help in understanding the mechanism of damage recognition. Using atomic force microscopy (AFM) we could distinguish and analyse the protein–DNA complexes at a defined damaged site and those on undamaged DNA. We show that the DNA is wrapped around UvrB in both the UvrB–DNA pre-incision complex and in the UvrA2B–DNA complexes on non-damaged DNA. The initial presence of UvrA imposes asymmetry, relative to the damaged site, on the DNA wrapped around UvrB.
Results
Binding of UvrAB to a 1020 bp substrate with a defined cholesterol lesion
To study UvrAB-mediated protein–DNA complexes with AFM, we have constructed a 1020 bp DNA fragment containing a cholesterol moiety attached to a propanediol backbone at a defined position as described in Materials and methods (Figure 1A). It has been shown that this lesion is a substrate for UvrABC, resulting in incisions at the 5th phosphodiester bond 3′ and the 8th phosphodiester bond 5′ of the damage (Moolenaar et al., 1998).
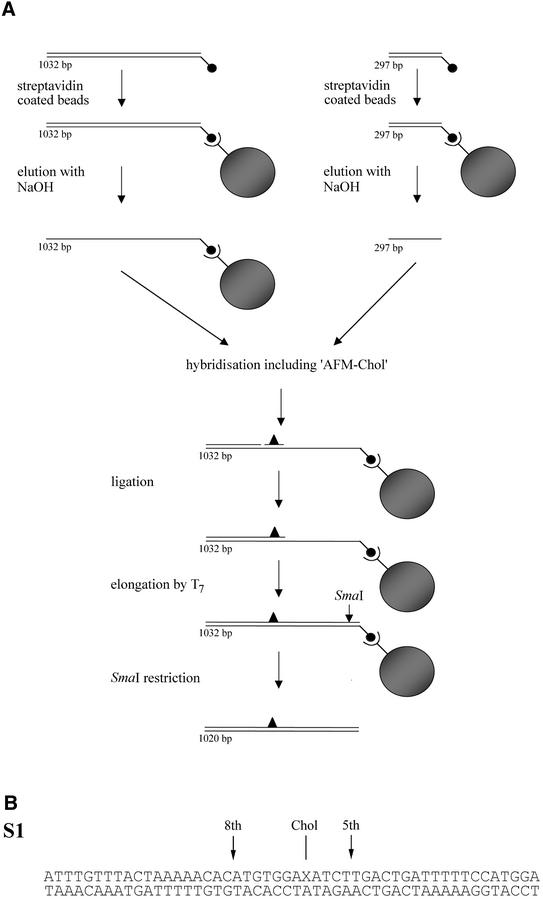
Fig. 1. (A) Schematic representation of the construction of the double-stranded AFM substrate. The different DNA strands of the 5′ biotinylated PCR fragments (indicated by small filled circles) are isolated using streptavidin-coated magnetic beads (indicated by large filled circles). The 297 nt top strand is added to the immobilized 1032 nt bottom strand together with the 50 nt phosphorylated ‘AFM-chol’ oligo containing the cholesterol adduct. After hybridization and ligation, the incomplete top strand is extended by T7 polymerase to produce a completely double-stranded fragment. Digestion with SmaI yields a 1020 bp DNA fragment containing a single cholesterol adduct at position 324 in the top strand. (B) DNA sequence of the 50 bp S1 substrate. The cholesterol adduct (Chol) is introduced at position 27 (X) in the top strand. The incision positions are indicated with arrows.
First we analysed complex formation of the 1020 bp double-stranded AFM substrate with UvrA and UvrB in a bandshift assay, using the standard UvrAB binding conditions (i.e. 50 mM Tris–HCl and 85 mM KCl). Two protein–DNA complexes were observed on a native agarose gel (Figure 2, lane 1). The upper complex shifts to the slot in the presence of antibodies raised against UvrA, while both complexes shift to the slot in the presence of antibodies raised against UvrB, indicating the formation of the UvrA2B–DNA and UvrB–DNA complexes, respectively (Figure 2, lanes 2 and 3).
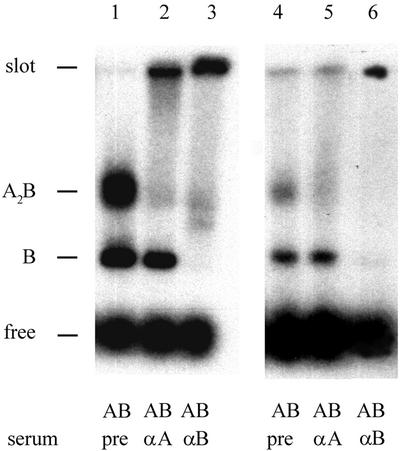
Fig. 2. Binding of UvrA and UvrB to the 1020 bp double-stranded AFM substrate. UvrA (5 nM) and UvrB (100 nM) were incubated with the double-stranded AFM substrate for 10 min at 37°C in Uvr-endo buffer containing 50 mM Tris–HCl, 85 mM KCl (lanes 1–3) or 15 mM Tris-HCl, 40 mM KCl (lanes 4–6). After incubation, pre-serum, UvrA antiserum or UvrB-antiserum was added to the samples as indicated. The complexes were analysed on a native 0.7% agarose gel as described in Materials and methods.
Deposition of protein–DNA complexes on mica for AFM analysis is hampered by monovalent cations. We therefore also tested UvrAB binding in a buffer more suitable for AFM (i.e. 15 mM Tris–HCl and 40 mM KCl). Complex formation was qualitatively similar when compared with standard binding conditions, although the total amount of complexes was somewhat lower (Figure 2B, lanes 4–6). This shows that the 1020 bp cholesterol-containing DNA substrate is suitable for AFM analysis of protein–DNA complexes formed by UvrA and UvrB.
The DNA in the UvrB–DNA pre-incision complex is wrapped around the UvrB protein
As is shown in Figure 2, incubation of UvrA and UvrB with damaged DNA results in two protein–DNA complexes, the UvrA2B–DNA and the UvrB–DNA complex. To be able to discriminate between the two complexes in the AFM images, we first specifically analysed the UvrB–DNA pre-incision complex by purifying it on streptavidin-coated beads as described previously (Moolenaar et al., 2000b). This method has been shown to remove all UvrA, from solution as well as from the DNA. In short, the immobilized AFM substrate was incubated with UvrA and UvrB to form the UvrB–DNA complex. The free proteins and the non-specifically bound UvrA2B complexes were washed away with a buffer containing high salt and the remaining UvrB–DNA complexes were cut from the beads with SmaI. The resulting mixture containing the isolated UvrB–DNA complexes was diluted in deposition buffer and deposited onto mica.
We first analysed complexes that were isolated by washing in the presence of ATP, to prevent loss of the ATP molecule in the UvrB–DNA pre-incision complex. The AFM images show complexes in which a protein is bound to the DNA at about one third of the DNA length (Figure 3A and B). This position corresponds to the location of the cholesterol adduct, indicating that UvrB is specifically bound to the damaged site. In many of the complexes observed, the DNA seems to be kinked at the position where the protein is bound, since the two flanking DNA arms are leaving the complex at a relatively sharp angle. From electron microscopic and flow linear dichroism studies, the introduction of a kink in the DNA upon formation of the pre-incision complex had been proposed before and it was postulated that the DNA might be wrapped around the UvrB protein (Shi et al., 1992; Takahashi et al., 1992). To analyse this, we measured the amount of DNA contained within the UvrB–DNA complex, by determining the contour length of the DNA from one end through the complex to the other end. The contour length of naked DNA fragments in the same depositions was compared with that of protein-bound DNA fragments. Images were collected from different depositions, using the same conditions and always having the same scan size. The data are summarized in Figure 4A. For the UvrB-complexed DNA a clear shift in the contour length distribution of ∼23 nm with respect to the naked DNA is observed (Table I). From the ratio of the contour length of the naked DNA and the number of base pairs in the fragment the DNA rise per base pair was determined at 0.32 nm/bp. This means that the length of the DNA fragment is shortened by ∼72 bp (6.7 helix turns) as a result of UvrB binding. Such a DNA shortening is not expected to be caused by DNA compaction within the protein, but is most likely to be the result of wrapping of the DNA around UvrB.
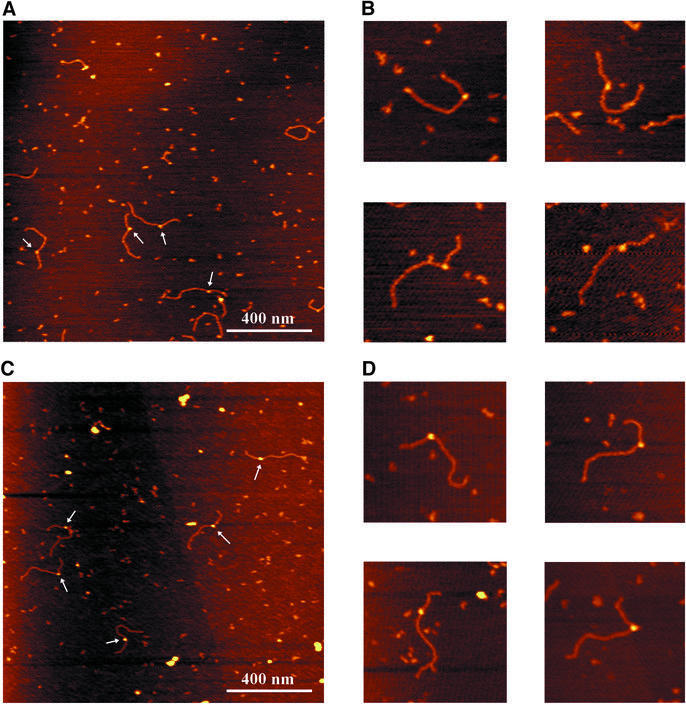
Fig. 3. Atomic force images of UvrB–DNA complexes formed on the immobilized AFM substrate. The colour scale ranges from 0.0 to 3.0 nm (from dark to bright). (A) Purified UvrB–DNA complexes washed in the presence of ATP. (B) Zoomed images of ATP-containing UvrB–DNA complexes. Image size, 400 nm. (C) Purified UvrB–DNA complexes after removal of ATP. (D) Zoomed images of UvrB–DNA complexes without ATP. Image size, 400 nm.
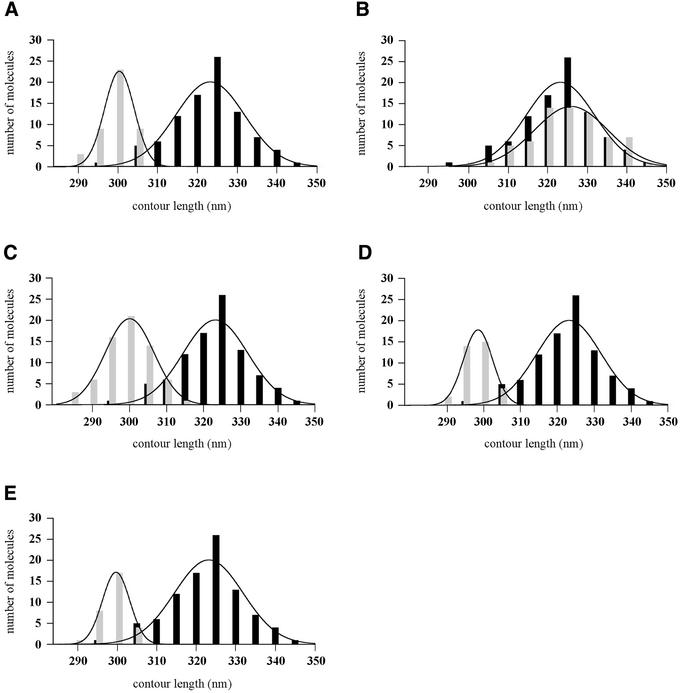
Fig. 4. Contour length distributions of free DNA molecules and UvrB–DNA complexes. The lines represent the Gaussian fitting of the distribution and the values obtained are presented in Table I. In all cases the free DNA is represented with black bars and the protein–DNA complexes with grey bars. (A) Free DNA molecules and purified UvrB–DNA complexes washed in the presence of ATP. (B) Free DNA molecules and purified UvrB–DNA complexes washed in the absence of ATP. (C) Free DNA molecules and purified UvrB–DNA complexes after reintroduction of ATP. (D) Free DNA molecules and purified UvrB–DNA complexes after reintroduction of ATPγS. (E) Free DNA molecules and UvrB–DNA complexes observed in direct depositions of the UvrAB incubation mixture.
Table I. Contour length of free DNA molecules, UvrB–DNA and UvrA2B–DNA complexes.
| Contour length (nm) (Gaussian fit) | Difference from free DNA (nm) | No. of molecules | |
|---|---|---|---|
| UvrB–DNA complexes | |||
| free DNA with damage | 323 ± 12 | 92 | |
| UvrB–DNA washed in presence of ATP | 300 ± 5 | 23 | 44 |
| UvrB–DNA without ATP | 325 ± 13 | –2 | 69 |
| UvrB–DNA after reintroduction of ATP | 300 ± 9 | 23 | 66 |
| UvrB–DNA after introduction of ATPγS | 298 ± 6 | 25 | 36 |
| UvrB–DNA, not washed | 299 ± 5 | 24 | 30 |
| UvrA2B–DNA complexes | |||
| free DNA without damage | 323 ± 12 | 122 | |
| one UvrA2B complex | 298 ± 9 | 25 | 93 |
| two UvrA2B complexes | 271 ± 6 | 52 | 44 |
| UvrA–DNA complexes | |||
| free DNA with damage | 322 ± 12 | 102 | |
| UvrA–DNA complex on damaged site | 322 ± 12 | 0 | 67 |
| UvrA–DNA complex on non-damaged site | 323 ± 11 | –1 | 32 |
The DNA contour length values are the mean of the Gaussian fit of the distributions reported in Figures 9, and .
Since the lesion is located asymmetrically within the DNA template, it is possible to determine the fraction of each flanking DNA arm that participates in the wrapping. The arm ratio for the naked AFM substrate is 0.47 (324/696 bp). If the DNA wrapped around UvrB were provided exclusively by the arm on either the 5′ or the 3′ side of the lesion, the expected ratios would be 0.36 (249/696 bp) and 0.52 (324/621 bp), respectively. If both arms participated in the wrapping, the measured ratio would be in-between these values. The ratio of the measured contour lengths of the arms obtained from AFM images is 0.51 ± 0.02. Figure 5A shows the ratio of arm lengths for each individual molecule, demonstrating that in all complexes the DNA exclusively on the 3′ side of the lesion is participating in the DNA wrapping.
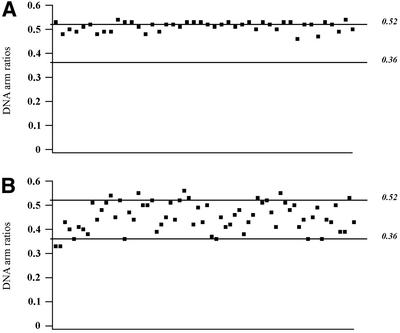
Fig. 5. (A) DNA arm ratios of UvrB–DNA complexes washed in the presence of ATP. (B) DNA arm ratios of UvrB–DNA complexes after reintroduction of ATP. Each dot represents an individual protein–DNA complex. The expected ratios when the DNA wrapped around UvrB is provided exclusively by the arm on the 5′ or the 3′ side of the damage are 0.36 (249/696 bp) and 0.52 (324/621 bp), respectively, as indicated.
DNA wrapping is dependent on ATP binding
In the pre-incision complex, UvrB is stably bound to the damaged site and contains ATP. Removal of the cofactor results in the pro-pre-incision complex, which is thought to be an intermediate complex formed after ATP hydrolysis by UvrB and before binding of a new ATP molecule (Moolenaar et al., 2000b). To analyse whether the DNA in the pro-pre-incision complex is also wrapped around UvrB, we removed the nucleotide cofactor from the pre-incision complexes by washing the immobilized UvrB–DNA complexes in the absence of ATP. The AFM images of the pro-pre-incision complexes appear similar to those washed in the presence of ATP (Figure 3C and D). However, length measurements of these complexes indicate that the DNA is no longer wrapped around UvrB (Figure 4B). Apparently, the presence of ATP in the UvrB–DNA complex is required for DNA wrapping.
Next, we reintroduced ATP or ATPγS by incubating the washed UvrB–DNA complexes in the presence of 1 mM ATP or ATPγS for 5 min at room temperature prior to deposition. The reintroduction of both ATP and ATPγS restored the wrapping (Figure 4C and D; Table I), showing that ATP binding and not hydrolysis is required for this particular DNA configuration. Surprisingly, although the total amount of DNA that is wrapped around UvrB is the same, the ratio of the measured contour lengths of the protruding arms is different for each individual molecule, ranging from 0.36 to 0.52 (Figure 5B). This means that the part of the DNA participating in wrapping is no longer restricted to the region 3′ to the adduct. Some molecules have only the 3′ or the 5′ arm wrapped around UvrB and in some molecules both arms participate in the wrapping. The total amount of DNA that contacts the surface of UvrB is the same in all complexes, which implies that the same contact points on the surface of the protein have the potential to interact with either of the two arms. This random type of wrapping is clearly different from the unique wrapping of the 3′ arm observed in the UvrB–DNA complexes that were purified in the presence of ATP. Apparently, wrapping is unidirectional when it occurs immediately after the loading of UvrB, i.e. in the presence of UvrA, but is random when it occurs in a later stage starting from an unwrapped UvrB–DNA complex.
The DNA is already wrapped around UvrB in the UvrA2B complex
We have also tried to analyse UvrA2B complexes bound to damaged DNA by performing a direct deposition experiment. In this experiment, the AFM substrate was cut from the beads prior to incubation with UvrA and UvrB, and the incubation mixture was directly diluted and deposited onto mica. As expected, two different types of protein–DNA complex were observed. We observed complexes similar to the purified UvrB–DNA complexes, with protein bound at one third of the DNA length. Contour length measurements showed that also in these complexes, which are most likely to be UvrB–DNA complexes, the DNA is wrapped around the protein (Figure 4E; Table I) and the ratio of the arm lengths (0.50 ± 0.02) indicates that DNA on the 3′ side of the damage is participating in this wrapping exclusively. In addition we observed much larger protein–DNA complexes, which are most likely to be formed by UvrA2B. In contrast to the UvrB–DNA complexes, however, these larger UvrA2B–DNA complexes were found at random positions on the DNA (Figure 6), indicating that UvrA2B binding to non-damaged sites on the 1020 bp substrate is stable enough to be visualized by AFM. Because of this random binding, damage-bound UvrA2B–DNA complexes could not be identified with certainty. Possibly these complexes are immediately converted into UvrB–DNA complexes.
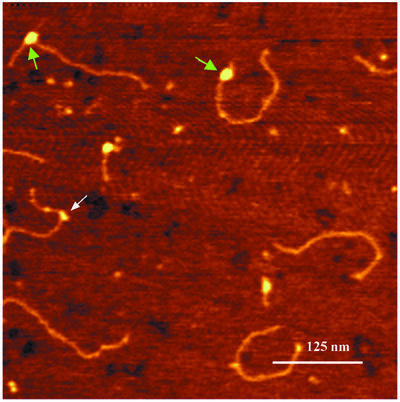
Fig. 6. Atomic force images of UvrA2B–DNA and UvrB–DNA complexes on damaged DNA. The white arrow indicates a UvrB–DNA pre-incision complex and green arrows indicate the UvrA2B–DNA complexes. The colour scale ranges from 0.0 to 3.0 nm (from dark to bright).
The UvrA2B complexes bound to non-damaged DNA were analysed in more detail. For that purpose we incubated UvrA and UvrB with the same 1020 bp substrate without the cholesterol adduct. Again randomly distributed UvrA2B complexes were observed that were clearly larger than the UvrB–DNA complexes on damaged DNA (Figure 7A and B). The majority of the DNA molecules contained one UvrA2B complex, but a substantial number contained two or more protein complexes. We measured the contour lengths of the DNA molecules containing one or two UvrA2B complexes (Figure 8; Table I). For the DNA molecules with one UvrA2B complex, a shift in the length distribution is observed similar to the shift for the ATP-containing UvrB–DNA complexes on damaged DNA. Interestingly, when two UvrA2B complexes were bound to the DNA, the contour length reduction is 52 nm, which is approximately twice that observed with one UvrA2B complex (Figure 8; Table I). Since the contour length reduction is identical to the reduction observed for pre-incision complexes, it is likely that the DNA in the UvrA2B–DNA complex is wrapped around the UvrB protein in a similar way to that in the UvrB–DNA pre-incision complex. To test this assumption, we incubated the damaged AFM substrate with UvrA alone in a direct deposition experiment (Figure 9A). It has been shown in filter binding assays that UvrA alone preferentially binds to damaged DNA (Mazur and Grossman, 1991). Indeed, damage-specific binding by UvrA was observed in the AFM images, since 65% of the protein–DNA complexes contained the UvrA protein at the site of the damage (Table I). Binding of UvrA was considered to be damage specific when the measured arm ratio was within one standard deviation from the calculated value. More importantly, contour length analysis shows that UvrA does not wrap the DNA, irrespective of whether UvrA is bound to the damage or not (Table I; Figure 9B), confirming the hypothesis that in the UvrA2B–DNA complex the DNA is wrapped around UvrB. The observation that the DNA is already wrapped around UvrB prior to damage recognition strongly suggests that DNA wrapping plays a role in damage recognition by the UvrA2B complex.
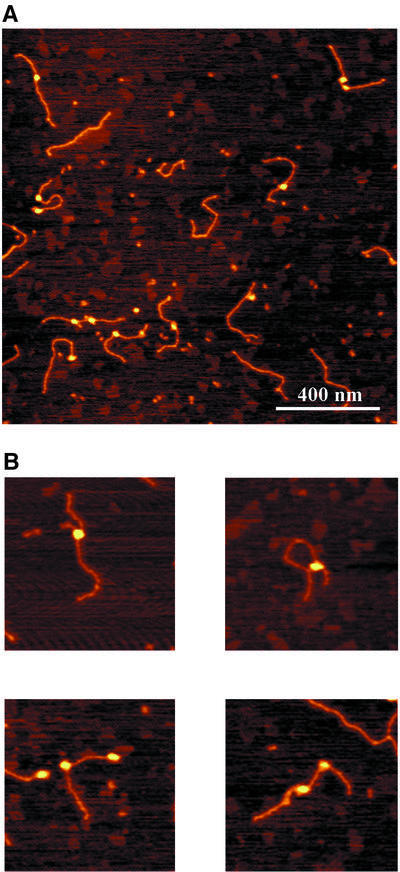
Fig. 7. Atomic force images of UvrA2B complexes formed on the non-damaged DNA substrate. The colour scale ranges from 0.0 to 3.0 nm (from dark to bright). (A) Image of UvrA2B–DNA complexes. (B) Zoomed images of DNA molecules with one or two UvrA2B complexes. Image size, 400 nm.
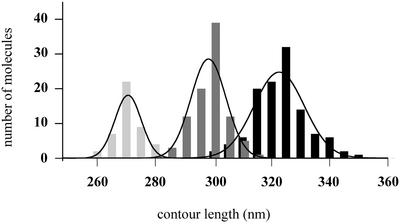
Fig. 8. Contour length distributions of free DNA molecules (black bars), DNA molecules with one UvrA2B complex (dark grey bars) and DNA molecules with two UvrA2B complexes (light grey bars). The lines represent the Gaussian fitting of the distribution and the values obtained are presented in Table I.
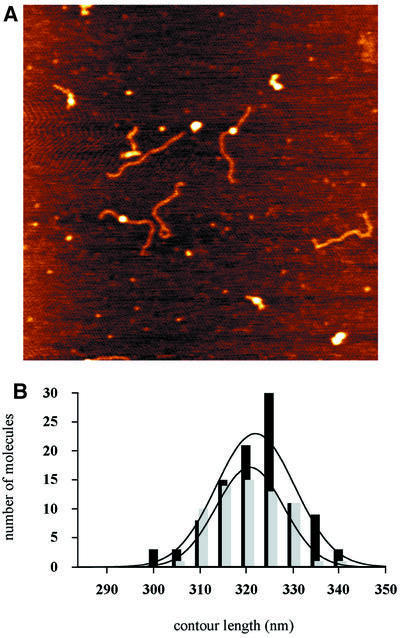
Fig. 9. (A) Image of UvrA–DNA complexes. (B) Contour length distributions of free DNA molecules (black bars) and UvrA–DNA complexes on the damaged site (grey bars).
Wrapping DNA around UvrB enhances the rate of incision
It has been shown that binding of ATP or ATPγS to the pro-pre-incision complex is required for formation of an active pre-incision complex and subsequently allows incision of the substrate after binding of UvrC (Moolenaar et al., 2000b). Here we have shown that binding of ATP or ATPγS also causes the DNA to wrap around UvrB, indicating that DNA wrapping might also play a role in the actual incision step. To study this we constructed a double-stranded 50 bp fragment by hybridizing the complementary strand to the ‘AFM-chol’ oligo used in the synthesis of the 1020 bp AFM substrate (S1, Figure 1B). The resulting S1 substrate contains the same damage in the same sequence context as in the 1020 bp AFM substrate. Whereas the DNA of the 1020 bp fragment will be wrapped around UvrB, substrate S1, which only contains 25 bp at the 3′ side of the adduct, will be too short to support complete wrapping. We have analysed whether this difference in DNA length would influence the incision efficiency.
To study the incision rates independent of the efficiency of protein–DNA complex formation, first the DNA substrates were incubated with UvrA and UvrB to allow UvrB–DNA complex formation. Next, UvrC was incubated with the UvrB–DNA pre-incision complex on ice for 5 min, allowing UvrBC complex formation but not incision (Moolenaar et al., 2000b). After UvrC binding, the reaction mixtures were transferred to 37°C and at different time points samples were analysed on a denaturing gel to determine the incision frequencies (Figure 10). After 2 min, 51% of the 1020 bp AFM substrate is incised compared with 13% for substrate S1. In time, the difference in incision efficiency decreases and eventually both substrates reach the same level of incision. This indicates that DNA wrapping is not essential for the incision itself, but it does stimulate the initial incision rate.
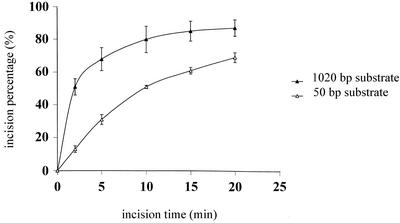
Fig. 10. Kinetics of the incision of the 50 bp S1 substrate (open triangles) and the 1020 bp AFM substrate (filled triangles). The data are the average of three independent experiments.
Discussion
Using AFM we have shown that in both the UvrA2B complexes on undamaged DNA and the UvrB–DNA pre-incision complex the DNA is wrapped around the UvrB protein. A comparable amount of DNA, approximately seven helical turns, participates in wrapping in both complexes, suggesting that in the UvrA2B complex the DNA is wrapped around the surface of UvrB in a similar way to in the pre-incision complex.
UvrA2B complexes bound to undamaged DNA are likely to reflect UvrA2B complexes in search of damage, and the DNA wrapping in these complexes might therefore play an important role in this process. Damage recognition during NER is a multi-step process in which damage is initially recognized by the UvrA subunit of the UvrA2B complex, followed by loading of the UvrB subunit onto the damaged site. It has been shown that in the absence of UvrA, UvrB is capable of damage-specific binding when the damage is located close to the 5′ end of a double-stranded substrate (Moolenaar et al., 2000a) or in the unpaired region of a bubble substrate (Zou and Van Houten, 1999). Based on these observations it was proposed that the function of the UvrA2B complex is to transiently unwind the DNA helix to allow UvrB access to the damaged site. Recently the crystal structure of UvrB has shown it to be structurally related to helicases (Machius et al., 1999; Nakagawa et al., 1999; Theis et al., 1999). In addition there is a β-hairpin with a high content of hydrophobic residues, which has been proposed to be involved in the damage-specific binding of UvrB. In one model (Theis et al., 1999), the β-hairpin inserts between the two strands of the DNA helix. Combining our results with this UvrB–DNA binding model we propose the following model for damage recognition and subsequent formation of the pre-incision complex. In a UvrA2B complex in search of damage, the DNA is wrapped around the UvrB protein, thereby conformationally constraining the DNA on the surface. The stress imposed on the DNA wrapped around UvrB may cause local melting of the DNA, which might occur more readily at or near a damaged site. This local unwinding will allow insertion of the β-hairpin into the DNA. The β-hairpin then probes the DNA for damage and upon encountering a lesion the ATPase activity of UvrB will be triggered to form the pro-pre-incision complex. Binding of a new ATP molecule will finally lead to the UvrB–DNA pre-incision complex (Moolenaar et al., 2000b).
A role for DNA wrapping in DNA strand separation has also been proposed for the initiation of transcription by RNA polymerases (Coulombe and Burton, 1999). AFM analysis of open promoter complexes formed by the E.coli RNA polymerase showed the DNA to be wrapped around the surface of the enzyme (Rivetti et al., 1999). Topo logical experiments of open complexes revealed unwinding of the DNA in the complexes (Amouyal and Buc, 1987). Evidence for DNA wrapping around eukaryotic RNA polymerases, assisted by transcription factors, has also been presented (Coulombe and Burton, 1999) and many prokaryotic transcription factors have been shown to bend the DNA in the upstream promoter region, thereby promoting DNA wrapping of the upstream DNA around RNA polymerase (Crothers and Steitz, 1992; Finkel and Johnson, 1992; Goosen and van de Putte, 1995).
Interestingly, it has been shown that T7 RNA polymerase also contains a β-hairpin that is inserted in the DNA helix in an initiation complex (Cheetham et al., 1999). This insertion of the β-hairpin facilitates melting of the promoter duplex. Recently, it was found that binding of T7 RNA polymerase induces a bend in the DNA, which is proposed to play a role in promoter melting (Ujvari and Martin, 2000). In analogy with our proposed model for damage recognition by UvrB, DNA bending at the promoter by T7 RNA polymerase might melt the DNA helix just enough to allow insertion of the β-hairpin.
After damage recognition and formation of the pre-incision complex, the DNA remains wrapped around UvrB. The asymmetric location of the cholesterol adduct in the pre-incision complex enabled us to determine that the 3′ side of the DNA with respect to the lesion participates in the wrapping exclusively. Footprint experiments showed that binding of UvrB to a damaged site conferred protection from DNase I digestion to a region of only 19–24 bp (Van Houten et al., 1987; Bertrand-Burggraf et al., 1991; Visse et al., 1991; Zou et al., 1995). This is much shorter than the 70–75 bp that presumably contact UvrB as a result of the DNA wrapping. Since the substrates used in these studies might be too short to support complete wrapping, we subjected the purified pre-incision complex on the 1020 bp AFM substrate to DNase I treatment. Also on this substrate, however, we could not detect an extended protection as a result of wrapping (data not shown). Apparently the contacts involved in DNA wrapping are too weak to interfere with the DNase I enzyme. When ATP is bound to the pre-incision complex, a DNase I hypersensitive site is observed that is absent when the ATP is washed out of the complex (Moolenaar et al., 2000b). This means that the conformational changes in UvrB induced upon ATP binding result in a distortion in the region where UvrB has strong interactions with the DNA. The same ATP binding is also required for the DNA wrapping, suggesting that the ATP-induced distortion of the DNA is essential to allow the DNA to wrap around UvrB.
When the cofactor is washed out of the pre-incision complex, resulting in the pro-pre-incision complex, the DNA is no longer wrapped around UvrB. Apparently, the interactions between the DNA and the surface of UvrB cannot resist the conformational changes in the complex resulting from the removal of the cofactor. In the absence of ATP, however, the UvrB protein remains stably bound to the DNA, probably via the strong interactions that are responsible for the observed protection in the footprint experiments.
From electron microscopy (Shi et al., 1992) and flow linear dichroism studies (Takahashi et al., 1992) using randomly damaged DNA substrates, it was already suggested that binding of Uvr(A)B to a damage induces bending of the DNA and from these results DNA wrapping was postulated. Also, in the AFM images of the pre-incision complexes, kinks can be observed, which might be the result of the DNA wrapping. There is, however, a large variation in the bend angles of the DNA in the UvrB–DNA complexes (Figure 3A and data not shown). Most likely the angles are largely determined by the way the DNA ‘drops’ on the mica surface during the deposition. Therefore, no accurate measurements on these angles could be done.
When ATP or ATPγS is reintroduced in the pro-pre-incision complexes, DNA wrapping is restored, showing that the conformational changes induced by ATP binding are sufficient to allow contacts between the surface of UvrB and the flanking DNA. These additional contacts, however, are now made in a random fashion, indicating that the same residues on the surface of UvrB can interact with either of the two DNA arms. Apparently the conformation of UvrB determines whether the DNA becomes wrapped around the protein, but it does not govern with which side of the DNA the contacts are made.
In UvrB–DNA complexes from direct depositions or UvrB–DNA complexes isolated in the presence of ATP, exclusively the 3′ arm is wrapped around UvrB. The difference between these complexes and the ones obtained after reintroduction of ATP or ATPγS must be ascribed to the presence or absence of the UvrA protein during the wrapping process. The isolation procedure of UvrB–DNA complexes via the magnetic beads removes all UvrA protein from the sample. Therefore, wrapping of the DNA by adding ATP to purified pro-pre-incision occurs in the total absence of UvrA. Apparently under these conditions both DNA arms can contact the surface of UvrB in a random fashion. In the direct depositions, UvrA is still present in the incubation mixture, and as a result only the 3′ arm becomes wrapped around UvrB, indicating that it is the binding of UvrA to UvrB that obstructs wrapping of the 5′ arm, resulting in surface contacts with the 3′ arm only.
It has been shown that formation of a pre-incision complex requires first hydrolysis of ATP, followed by exchange of the resulting ADP with a new ATP molecule (Moolenaar et al., 2000b). This means that during this process the UvrB molecule goes through a stage where it has no cofactor bound. The unidirectional wrapping that is observed in the isolated pre-incision complexes can be explained in two ways. (i) Wrapping is maintained throughout the different steps of the formation of the pre-incision complex, because of a very fast exchange of the cofactor, thereby preventing unwrapping after the ATP hydrolysis step. (ii) After the ATP hydrolysis the DNA becomes unwrapped, but UvrA remains bound to the complex, thereby directing the wrapping after binding of the new ATP molecule.
Although the DNA in the pre-incision complex is wrapped, this conformation seems not to be required for a productive complex, since substrate S1, which is too short for complete wrapping, is efficiently incised. The kinetic experiment, however, shows that DNA wrapping does stimulate the rate of incision, suggesting that in addition to its proposed role in damage recognition, it also stimulates the 3′ incision step. Zou and Van Houten (1999) have shown that with bubble substrates in which five bases around a BPDE lesion are unpaired, the incision efficiency is increased in comparison with normal double-stranded substrates. From these results it was postulated that in the UvrBC–DNA incision complex the DNA around the lesion is unwound, thereby positioning the catalytic site for incision. DNA wrapping could stimulate incision by promoting this local strand separation.
Finally, DNA wrapping might also play a role in the post-incision events. UvrC efficiently induces incisions when ATPγS is bound to the UvrB–DNA complex (Moolenaar et al., 2000b), indicating that during the incision step no ATP hydrolysis is required. This could mean that after the incisions, the UvrB–DNA complex is still in the ATP-bound conformation and as a result the DNA would still be wrapped. It has been shown that UvrB remains stably bound to the DNA even after removal of the damaged oligo (Orren et al., 1992). Removal of the damaged oligo will probably result in a reduction of protein–DNA contacts in the UvrB–DNA complex, making the interactions with the flanking DNA (i.e. DNA wrapping) more important for the stability of the post-incision complex.
In conclusion, we have shown that the UvrA2B complex in search of a damage wraps the DNA around the UvrB protein. We propose an important role for DNA wrapping in damage recognition by favouring an opening of the DNA helix thereby allowing insertion of the β-hairpin of UvrB. After damage recognition, the DNA in the pre-incision complex remains wrapped and this wrapping stimulates the rate of incision. Finally, DNA wrapping might also play a role in the stability of the UvrB–DNA complex after incision, suggesting that DNA wrapping is important throughout the NER reaction.
Materials and methods
Uvr proteins and DNA substrates
The UvrA, UvrB and UvrC proteins were purified as described (Visse et al., 1992).
Substrate S1. The 50mer oligonucleotide ‘AFM-chol’ was provided by Eurogentec (Seraing, Belgium). The oligonucleotide contains a cholesterol moiety, which is attached to a propanediol backbone instead of a nucleoside at position 27. The sequence (Figure 1B) corresponds to nucleotides 1163–1212 of the URA3 gene of Saccharomyces cerevisiae, and the cholesterol adduct replaces a T residue at position 1189. The 5′ terminally labelled substrate S1 was constructed by hybridization of the damaged oligo with the complementary bottom strand as described previously (Moolenaar et al., 1998).
AFM substrate. Construction of the 1020 bp double-stranded AFM fragment containing a defined cholesterol lesion is shown schematically in Figure 1A. Using the URA3 gene from S.cerevisiae as a template, PCR with primers U3 (GAAGGAAGAACGAAGGAAGGAGC) and UH4NB, which is 5′ biotinylated (TTTCCCGGGGGGCCCGGGTAATAACTGATATAATT), produced a fragment of 1032 nucleotides. The PCR product was purified with a GFX™ column (Amersham). The purified biotinylated fragment was incubated with 1 mg of streptavidin-coated magnetic beads (Dynal M-280) for 2 h at 37°C in 200 µl of BW1 buffer (5 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 3 M LiCl). Using a magnetic particle concentrator (Dynal), the immobilized DNA was washed three times with 100 µl of 2× BW1 buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 6 M LiCl). Subsequently, the DNA was washed twice with 100 µl of TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA). After washing, the top strand of this fragment was eluted using denaturing conditions (3 min in 10 µl of 0.1 M NaOH), yielding an immobilized single-stranded bottom strand. A second PCR using primers U3 and Bio1078B, which is 5′ biotinylated (TTTGGGACCTAATGCTTCAAC), produced a fragment of 297 nucleotides, corresponding to the first 297 nucleotides of the PCR product mentioned above. Following purification with a GFX™ column, this fragment was incubated with 1 mg of streptavidin-coated magnetic beads (Dynal M-280) for 2 h at 37°C in 200 µl of BW2 buffer (5 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 1 M NaCl). After washing this fragment three times with 100 µl of 2× BW2 buffer and twice with 100 µl of TE, the top strand of the 297 bp fragment was isolated by 0.1 M NaOH treatment. The isolated single-stranded top strand was added to the beads containing the 1032 nt immobilized bottom strand together with 40 pmol of phosphorylated ‘AFM-chol’. After hybridization the top oligos were ligated in 100 µl of buffer containing 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM dithiothreitol (DTT), 1 mM ATP and T4 ligase. Following ligation the beads were washed twice with 100 µl of 2× BW2 and once with 100 µl of TE. The immobilized fragment was incubated at 37°C for 10 min with 0.2 U of Sequenase™ DNA polymerase Version 2.0 (Amersham) in 100 µl containing 40 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM DTT and 200 µM dNTPs to produce a completely double-stranded fragment. The presence of the cholesterol adduct, which is located in an EcoRV recognition site, was confirmed by restriction analysis (data not shown).
The non-damaged DNA fragment was obtained by incubating the product of PCR with primers U3 and UH4NB on the beads, extensive washing and cutting it from the beads with SmaI in 100 µl of SmaI buffer (10 mM Tris–acetate pH 7.5, 10 mM magnesium acetate and 50 mM potassium acetate).
Bandshift assay
To construct a 5′ terminally labelled AFM substrate, the immobilized 297 bp PCR fragment was 5′ terminally labelled using T4 polynucleotide kinase (10 U) in 50 µl of kinase buffer (70 mM Tris–HCl pH 7.6, 10 mM MgCl2, 5 mM DTT) and 5 pmol of [γ-32P]ATP (7000 Ci/mmol). After removal of the free nucleotides by G50 gel filtration, the labelled top strand was eluted and used to construct the AFM substrate as described above.
The labelled AFM substrate (40 fmol) was incubated with 5 nM UvrA and 100 nM UvrB in 10 µl of Uvr-endo 1 buffer. After 10 min at 37°C, 1 µl of pre- or anti-serum (Visse et al., 1992) was added as indicated and the samples were loaded on a 0.7% agarose gel in 0.5× TBE including 10 mM MgCl2 and 1 mM ATP. The gel was run at room temperature at 85 V while circulating the buffer (0.5× TBE, 10 mM MgCl2 and 1 mM ATP). The gel was dried and the protein–DNA complexes were visualized using autoradiography.
To study complex formation in a buffer suitable for AFM (reduced salt concentration), the substrates were incubated in 10 µl of Uvr-endo 2 buffer (15 mM Tris–HCl pH 7.5, 10 mM MgCl2, 40 mM KCl and 1 mM ATP).
Incision assay
To study the incision rate, the terminally labelled S1 and AFM substrates were incubated with 2.5 nM UvrA and 100 nM UvrB in 20 µl of Uvr-endo 1 buffer [50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM KCl, 0.1 µg bovine serum albumin (BSA)/µl and 1 mM ATP]. After UvrB–DNA complex formation, 20 nM UvrC was added and the mixture was incubated on ice for 5 min to allow UvrC binding. After UvrBC–DNA complex formation, the reaction mixture was transferred to 37°C and the incision reaction was stopped at different times by addition of 2 µl of 0.5 M EDTA. To substrate S1, 2 µl of glycogen (2 µg/ml) were added, followed by ethanol precipitation, and the samples were run on a 15% acrylamide gel containing 7 M urea.
The incision reaction with the AFM substrate was done with the DNA immobilized on the magnetic beads and the incision products were isolated by eluting the top strand with 10 µl of 0.1 M NaOH. To the single-stranded top strand 4 µl of formamide were added. The samples were incubated at 95°C for 3 min and run on a 3.5% denaturing acrylamide gel. The incision products were visualized using autoradiography.
Atomic force microscopy
The immobilized AFM substrate (250 fmol) was incubated with 20 nM UvrA and 400 nM UvrB in 20 µl of Uvr-endo 1 buffer. After 15 min at 37°C, the mixture was transferred to the magnetic particle concentrator and the formed complexes were purified. For this purification the complexes were first washed three times with 50 µl of wash buffer 1 (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.5 M KCl, 0.1 µg/µl BSA) with or without 1 mM ATP, and then twice with 50 µl of wash buffer 2 (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 M KCl, 0.1 µg/µl BSA) with or without 1 mM ATP. Finally, the UvrB–DNA complexes were incubated with SmaI in 10 µl of SmaI buffer with or without ATP to cut the DNA from the beads. For reintroduction of the cofactor, ATP or ATPγS was added to the purified UvrB–DNA complexes to a final concentration of 1 mM and the mixture was incubated for 5 min at room temperature. The final mixture was diluted 10 times in deposition buffer (5 mM HEPES–KOH pH 7.8, 5 mM MgCl2) and deposited onto freshly cleaved mica. After ∼30 s the mica was washed with 4 ml of glass distilled water and dried in a stream of air.
In the direct deposition experiment, the AFM substrate was cut from the beads using SmaI, and 150 fmol of this DNA were incubated with 20 nM UvrA and 400 nM UvrB in 10 µl of Uvr-endo 2 buffer. After 20 min at 37°C, 2 µl of the binding mixture were diluted into 20 µl of deposition buffer (5 mM HEPES–KOH pH 7.8, 5 mM MgCl2), deposited onto mica and further treated as described above. The non-damaged substrate (200 fmol) was incubated with 50 nM UvrA and 200 nM UvrB in 10 µl of Uvr-endo 2 buffer and treated further as described above. UvrA–DNA complexes were formed by incubating 50 nM UvrA with 200 fmol of AFM substrate in the same buffer.
The complexes were imaged using a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) in the tapping mode. DNA lengths were measured using the Image SXM (v. 1.62) software (an NIH Image version modified for use with SPM images by Dr Steve Barrett, Surface Science Research Centre, University of Liverpool, UK). Brightness and contrast of the images were optimized for the purpose of clarity.
Acknowledgments
Acknowledgements
We thank Marcel Tijsterman for kindly providing primers U3 and UH4NB, and Rob Visse and Remus Dame for fruitful discussions. This work was supported by the J.A.Cohen Institute for Radiopathology and Radiation Protection (IRS), the Centre for Biomedical Genetics and the Division of Chemical Sciences of the Dutch Organisation for Science (NWO).
References
- Amouyal M. and Buc,H. (1987) Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J. Mol. Biol., 195, 795–808. [DOI] [PubMed] [Google Scholar]
- Bertrand-Burggraf E., Selby,C.P., Hearst,J.E. and Sancar,A. (1991) Identification of the different intermediates in the interaction of (A)BC excinuclease with its substrates by DNase I footprinting on two uniquely modified oligonucleotides. J. Mol. Biol., 219, 27–36. [DOI] [PubMed] [Google Scholar]
- Cheetham G.M., Jeruzalmi,D. and Steitz,T.A. (1999) Structural basis for initiation of transcription from an RNA polymerase–promoter complex. Nature, 399, 80–83. [DOI] [PubMed] [Google Scholar]
- Coulombe B. and Burton,Z.F. (1999) DNA bending and wrapping around RNA polymerase: a ‘revolutionary’ model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev., 63, 457–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D.M. and Steitz,T.A. (1992) Transcriptional activation by E.coli CAP protein. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 501–534.
- Finkel S.E. and Johnson,R.C. (1992) The Fis protein: it’s not just for DNA inversion anymore. Mol. Microbiol., 6, 3257–3265. [DOI] [PubMed] [Google Scholar]
- Goosen N. and van de Putte,P. (1995) The regulation of transcription initiation by integration host factor. Mol. Microbiol., 16, 1–7. [DOI] [PubMed] [Google Scholar]
- Goosen N., Moolenaar,G.F., Visse,R. and van de Putte,P. (1998) Functional domains of the Escherichia coli UvrABC proteins in nucleotide excision repair. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology: DNA Repair. Springer Verlag, Berlin, Germany, pp. 101–123.
- Lin J.J. and Sancar,A. (1992) Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J. Biol. Chem., 267, 17688–17692. [PubMed] [Google Scholar]
- Machius M., Henry,L., Palnitkar,M. and Deisenhofer,J. (1999) Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl Acad. Sci. USA, 96, 11717–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur S.J. and Grossman,L. (1991) Dimerization of Escherichia coli UvrA and its binding to undamaged and ultraviolet light damaged DNA. Biochemistry, 30, 4432–4443. [DOI] [PubMed] [Google Scholar]
- Moolenaar G.F., Bazuine,M., van Knippenberg,I.C., Visse,R. and Goosen,N. (1998) Characterization of the Escherichia coli damage-independent UvrBC endonuclease activity. J. Biol. Chem., 273, 34896–34903. [DOI] [PubMed] [Google Scholar]
- Moolenaar G.F., Monaco,V., van Der Marel,G.A., van Boom,J.H., Visse,R. and Goosen,N. (2000a) The effect of the DNA flanking the lesion on formation of the UvrB–DNA preincision complex. Mechanism for the UvrA-mediated loading of UvrB onto the damage. J. Biol. Chem., 275, 8038–8043. [DOI] [PubMed] [Google Scholar]
- Moolenaar G.F., Herron,M.F., Monaco,V., Van der Marel,G.A., van Boom,J.H., Visse,R. and Goosen,N. (2000b) The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J. Biol. Chem., 275, 8044–8050. [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Sugahara,M., Masui,R., Kato,R., Fukuyama,K. and Kuramitsu,S. (1999) Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem., 126, 986–990. [DOI] [PubMed] [Google Scholar]
- Orren D.K., Selby,C.P., Hearst,J.E. and Sancar,A. (1992) Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC–DNA complex by helicase II and DNA polymerase I. J. Biol. Chem., 267, 780–788. [PubMed] [Google Scholar]
- Rivetti C., Guthold,M. and Bustamante,C. (1999) Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J., 18, 4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- Shi Q., Thresher,R., Sancar,A. and Griffith,J. (1992) Electron microscopic study of (A)BC excinuclease. DNA is sharply bent in the UvrB–DNA complex. J. Mol. Biol., 226, 425–432. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Bertrand-Burggraf,E., Fuchs,R.P. and Norden,B. (1992) Structure of UvrABC excinuclease-UV-damaged DNA complexes studied by flow linear dichroism. DNA curved by UvrB and UvrC. FEBS Lett., 314, 10–12. [DOI] [PubMed] [Google Scholar]
- Theis K., Chen,P.J., Skorvaga,M., Van Houten,B. and Kisker,C. (1999) Crystal structure of UvrB, a helicase adapted for nucleotide excision repair. EMBO J., 18, 6899–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A. and Martin,C.T. (2000) Evidence for DNA bending at the T7 RNA polymerase promoter. J. Mol. Biol., 295, 1173–1184. [DOI] [PubMed] [Google Scholar]
- Van Houten B. (1990) Nucleotide excision repair in Escherichia coli. Microbiol. Rev., 54, 18–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B., Gamper,H., Sancar,A. and Hearst,J.E. (1987) DNase I footprint of ABC excinuclease. J. Biol. Chem., 262, 13180–13187. [PubMed] [Google Scholar]
- Verhoeven E.E.A., van Kesteren,M., Moolenaar,G.F., Visse,R. and Goosen,N. (2000) Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem., 275, 5120–5123. [DOI] [PubMed] [Google Scholar]
- Visse R., de Ruijter,M., Brouwer,J., Brandsma,J.A. and van de Putte,P. (1991) Uvr excision repair protein complex of Escherichia coli binds to the convex side of a cisplatin-induced kink in the DNA. J. Biol. Chem., 266, 7609–7617. [PubMed] [Google Scholar]
- Visse R., de Ruijter,M., Moolenaar,G.F. and van de Putte,P. (1992) Analysis of UvrABC endonuclease reaction intermediates on cisplatin-damaged DNA using mobility shift gel electrophoresis. J. Biol. Chem., 267, 6736–6742. [PubMed] [Google Scholar]
- Zou Y. and Van Houten,B. (1999) Strand opening by the UvrA2B complex allows dynamic recognition of DNA damage. EMBO J., 18, 4889–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Liu,T.M., Geacintov,N.E. and Van Houten,B. (1995) Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(–)-anti-BPDE. Biochemistry, 34, 13582–13593. [DOI] [PubMed] [Google Scholar]


