Abstract
XMAP215 belongs to a family of proteins involved in the regulation of microtubule dynamics. In this study we analyze the function of different parts of XMAP215 in vivo and in Xenopus egg extracts. XMAP215 has been divided into three fragments, FrN, FrM and FrC (for N-terminal, middle and C-terminal, respectively). FrN co-localizes with microtubules in egg extracts but not in cells, FrC co- localizes with microtubules and centrosomes both in egg extracts and in cells, while FrM does not co- localize with either centrosomes or microtubules. In Xenopus egg extracts, FrN stimulates microtubule growth at plus-ends by inhibiting catastrophes, while FrM has no effect, and FrC suppresses microtubule growth by promoting catastrophes. Our results suggest that XMAP215 is targeted to centrosomes and microtubules mainly through its C-terminal domain, while the evolutionarily conserved N-terminal domain contains its microtubule-stabilizing activity.
Keywords: centrosome/MAP/microtubule dynamics/mitotic spindle
Introduction
Microtubules are dynamic polymers that change their distribution during cell cycle and development. In vivo, microtubules are found in two main states, either growing or shrinking. Microtubules transit stochastically between these two states, a behavior known as dynamic instability. Such a transition from growth to shrinkage is called a ‘catastrophe’, while the reverse transition from shortening to extension is called a ‘rescue’ (Walker et al., 1988). Microtubule dynamics has been extensively studied in extracts prepared from Xenopus eggs. Molecules regulating microtubule dynamics have been identified, and their effects on the assembly of purified tubulin into microtubules determined. There are two classes of such molecules. Stabilizing factors, such as the microtubule-associated proteins (MAPs) XMAP215, XMAP230 and XMAP310 (Gard and Kirschner, 1987; Andersen et al., 1994; Andersen and Karsenti, 1997), promote microtubule growth by reducing catastrophes and increasing the growth rate. Destabilizing factors, such as OP18/Stathmin (Belmont and Mitchison, 1996), katanin (McNally et al., 1996) and XKCM1, a member of the Kin I kinesin family (Walczak et al., 1996), increase the catastrophe rate.
The primary MAP that stabilizes microtubules in Xenopus egg extracts is XMAP215 (Gard and Kirschner, 1987; Tournebize et al., 2000). XMAP215 is a 228 kDa protein that belongs to a conserved family of proteins required for the growth of microtubules and mitotic spindle assembly (Charrasse et al., 1998; Cullen et al., 1999; Tournebize et al., 2000). In extracts, XMAP215 exerts its microtubule growth-promoting activity mainly by antagonizing the activity of the catastrophe factor XKCM1 (Tournebize et al., 2000). Ran-GTP-induced microtubule assembly in egg extracts also depends on XMAP215 (Wilde and Zheng, 1999). Previous work on XMAP215-related proteins from various organisms, including yeast (STU2, p93dis1), Dictyostelium discoideum (DdCP224), Caenorhabditis elegans (Zyg-9), Drosophila melanogaster (Msps) and Homo sapiens (ch-TOG), showed that they were all associated with the microtubule network and centrosomes or spindle pole bodies in some or all stages of the cell cycle (Nabeshima et al., 1995; Nakaseko et al., 1996; Wang and Huffaker, 1997; Charrasse et al., 1998; Matthews et al., 1998; Cullen et al., 1999; Gräf et al., 2000). In yeast, C.elegans, Drosophila and Amoeba, mutants show similar phenotypes compatible with defects in microtubule dynamics.
Here we show that, like its orthologs in other organisms, XMAP215 localizes to microtubules and centrosomes. In order to examine in more detail how this MAP affects microtubule dynamics and to address its potential function at the centrosome, we have carried out a functional analysis of several fragments. We have correlated the intracellular distribution of various XMAP215 fragments with their functional properties in egg extracts. Our results indicate that the whole protein is required for proper binding to microtubules in vivo, while its C-terminal part targets XMAP215 to the centrosome. Although both N- and C-terminal parts bind to microtubules in egg extracts, their effects on microtubule growth in mitotic egg extracts are dramatically different. The conserved N-terminal domain suppresses catastrophes both in native extracts and in XMAP215-depleted extracts where it fully complements the loss of function achieved by depletion. The C-terminal fragment destabilizes microtubules in egg extracts in a dominant-negative factor manner.
Results
XMAP215 binds to microtubules and centrosomes throughout the cell cycle
We have previously shown by immunofluorescence that XMAP215 is localized on microtubules (Tournebize et al., 2000). Now we have re-examined its localization in greater detail by both immunofluorescence (see Supplementary figure 1, available at The EMBO Journal Online) and electron microscopy (EM) on fixed cells and centrosomes pre-incubated in Xenopus egg extracts. EM studies revealed the presence of XMAP215 on microtubules and associated with the pericentriolar material (Figure 1). We then examined the intracellular distribution of a green fluorescent protein (GFP)-tagged version of XMAP215 in live cells. A cDNA coding for the full-length XMAP215 protein was fused at either its N- or C-terminus to GFP and the constructs were electroporated into XL177 cells. GFP–XMAP215 localized both to interphase microtubules (Figure 2A, C and D) and the centrosome (Figure 2B and D). In mitotic cells, GFP–XMAP215 localized predominantly to spindle poles and spindle microtubules (Figure 2E). The centrosome staining was very strong, independent of the N- or C-terminal localization of the GFP tag, or the cell cycle stage, and appeared early after transfection. To confirm further the centrosomal localization of XMAP215, we depolymerized microtubules in XL177 cells with nocodazole. Figure 2F shows that XMAP215 remains associated with centrosomes after microtubule depolymerization (the anti-α-tubulin signal reveals centrioles). We conclude that XMAP215 is a microtubule-binding protein which localizes along microtubules and to the centrosome throughout the cell cycle.
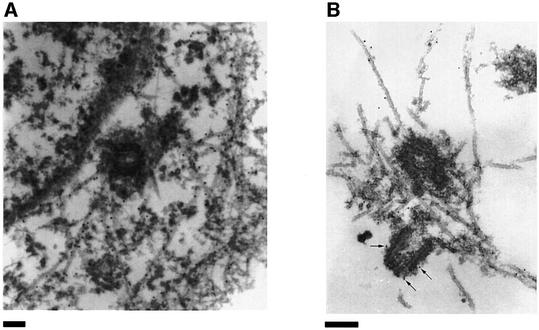
Fig. 1. Immunoelectron microscopic localization of XMAP215. (A) Electron microscopy on XL177 cells. In cultured epithelial cells, anti-N-terminal XMAP215 antibodies stain microtubules and, to a lesser extent, pericentriolar material. (B) In asters, assembled in vitro using mitotic Xenopus egg extracts and human KE37 cell line centrosomes, XMAP215 is localized on microtubules and pericentriolar material. Arrows indicate gold particles associated with pericentriolar material. Scale bar, 200 nm.
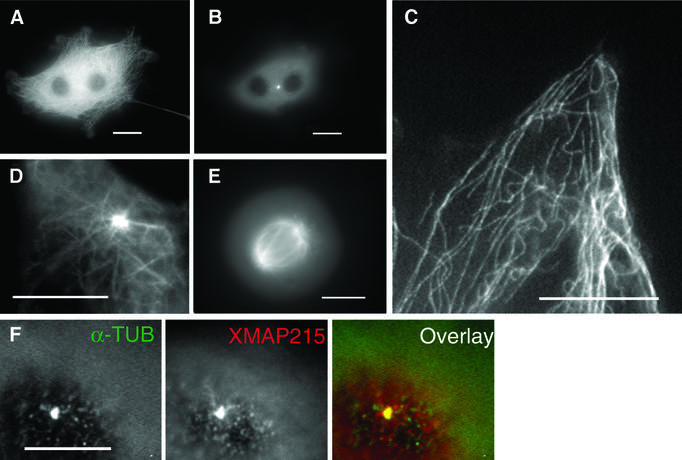
Fig. 2. GFP–XMAP215 associates with microtubules and centrosomes in interphase and mitosis. (A) A live XL177 cell electroporated with GFP–XMAP215. Note that cultured Xenopus cells frequently have more than one nucleus. (B) Same cell as in (A), showing the plane distal from the glass coverslip. The bright fluorescent spot is the centrosome. (C) High-magnification image of a live cell, showing GFP–XMAP215 associated with interphase microtubules at the periphery of the cell. (D) High-magnification image showing GFP–XMAP215 on the centrosome and proximal to centrosome microtubules. (E) In mitosis, GFP–XMAP215 stains the mitotic spindle and spindle poles. (F) Immunolocalization of XMAP215 in fixed XL177 cells after nocodazole-induced depolymerization of microtubules. Scale bars, 10 µm.
The centrosome and microtubule-binding domains of XMAP215
To determine which domains were required to target XMAP215 to microtubules and centrosomes, we divided the molecule into three fragments, termed FrN, FrM and FrC, for N-terminal, middle and C-terminal, respectively (see Figure 3A). They were chosen according to the regions of homology between XMAP215 and other members of this family of proteins (see Discussion and Figure 9).
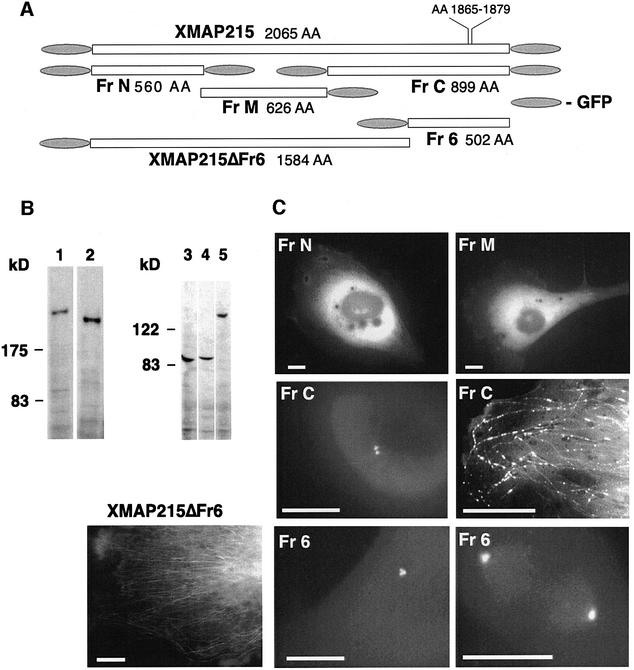
Fig. 3. Intracellular localization of the XMAP215 fragments expressed as GFP fusions. (A) Schematic representation of fragments and GFP fusion constructs of XMAP215 and five XMAP215 fragments. Full-length XMAP215, FrN and FrC were separately expressed as either N- or C-terminal fusions with GFP to exclude artifacts created by hindrance of the GFP tag. Sizes of fragments in amino acids (AA) are shown excluding GFP. (B) Immunoblotting analysis showing protein bands corresponding to the full-length GFP–XMAP215 (1), GFP–XMAP215_ΔFr6 (2), FrN–GFP (3), FrM–GFP (4) and FrC–GFP (5). Lysates from transfected cells were resolved on a polyacrylamide gel, blotted and probed with anti-GFP antibodies. Molecular weight markers are shown on the left-hand side. (C) Expression of the GFP fusions of FrN, FrM, FrC, XMAP215_ΔFr6 and Fr6 in live XL177 cells. Scale bars, 10 µm.
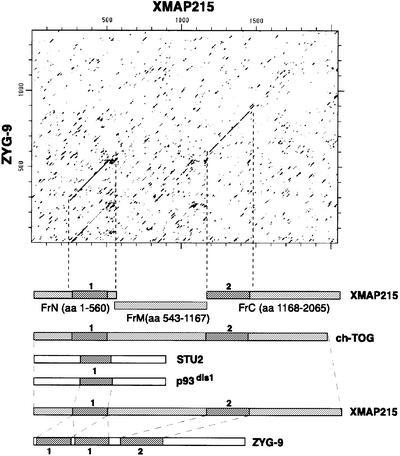
Fig. 9. Domain organization of some members of the XMAP215 family of proteins. On the top is a dot plot of the XMAP215 protein sequence compared with Zyg-9. Two regions of homology were detected: the N-terminal domain (1) and a more distal domain (2). The N-terminal domain is duplicated in Zyg-9. This plot was used as a basis for splitting XMAP215 into three major fragments: FrN, FrM and FrC. Below are some other members of the XMAP215 family with corresponding homology domains. p93dis1 (as well as STU2 protein from S.cerevisiae) is considerably shorter than XMAP215 and contains only domain 1, which is 31% identical to domain 1 in XMAP215. Human ch-TOG (as well as Msps protein from Drosophila and DdCP224 from D.discoideum) contains both domain 1 and 2 and is homologous to XMAP215 over the whole length of the protein. ch-TOG has 76% overall identity with XMAP215. Finally, Zyg-9 is shorter than ch-TOG, msps and DdCP224, and has two copies of domain 1 and one copy of domain 2. The first and the second domain 1 in Zyg-9 show 41 and 45% identity to the corresponding domain 1 in XMAP215.
We first examined the localization of these fragments in vivo, by expressing GFP-tagged proteins in XL177 cells. To exclude the interference of the GFP tag with microtubule- or centrosome-binding properties of these proteins, the N- and C-terminal fragments of XMAP215 were tagged with GFP separately, either at the N- (GFP–Fr) or C-terminus (Fr–GFP). These constructs behaved similarly. Figure 3B shows protein bands corresponding to the major GFP-fusion fragments of XMAP215 used in this study. When expressed in XL177 cells, both FrN–GFP (GFP–FrN) and FrM–GFP showed a diffuse cytoplasmic fluorescence (Figure 3C). In practically all transfected cells, FrC–GFP (or GFP–FrC) was localized on centrosomes throughout the cell cycle (see below). This localization was not affected by nocodazole treatment (data not shown). In addition, in some cells overexpressing FrC–GFP, a less prominent punctate staining was observed along microtubules. Taken together, these results indicate that the C-terminal part of XMAP215 contains a domain that targets XMAP215 to centrosomes and participates in microtubule binding in vivo.
The sequence of the C-terminal part of XMAP215 contains a microtubule-binding domain (consensus: L…KI/VGS-E/DN-K, amino acids 1865–1879 of the XMAP215 sequence) that targets other MAPs to microtubules (Chapin and Bulinski, 1992; Charrasse et al., 1998). In order to define further the centrosome- and microtubule-binding domains of the C-terminal fragment of XMAP215, we created two more deletion mutants: GFP–Fr6 and GFP–XMAP215_ΔFr6 (Figure 3A). The boundary between these two fragments was chosen in order to remove the above-mentioned domain. Thus, GFP–XMAP215_ΔFr6 contains the whole protein minus the microtubule-targeting domain of the C-terminal region. The remaining C-terminal part of XMAP215 was termed Fr6. Similar to the localization of FrC–GFP in transfected cells, GFP–Fr6 intensely stained centrosomes both in interphase and mitosis (Figure 3C; only GFP–Fr6-containing mitotic cell is shown). XMAP215_ΔFr6 did not co-localize with centrosomes, but was weakly associated with microtubules. However, in contrast to the FrC–GFP staining, which appeared both continuous and punctate, GFP–XMAP215_ΔFr6 showed a ‘smooth’ association with microtubules (Figure 3C). Therefore, the C-terminal part of XMAP215 contains the centrosome-targeting domain of XMAP215. The microtubule-binding properties of XMAP215 appear to involve several regions of the molecule, which are necessary for its proper distribution on the microtubule cytoskeleton in vivo.
We next characterized the centrosome-binding domain contained within the C-terminal part of XMAP215. A series of deletion mutants was made within the C-terminal fragments of XMAP215 (Figure 4A). The two smallest fragments, Fr9 and Fr10, were tagged with GFP separately, either on the N- or C-terminus, to exclude interference of the GFP tag with centrosome binding. For the same reason, the ΔC6 deletion was made on FrC rather than Fr6. The constructs were electroporated into frog cells and the intensity of the centrosomal staining in live cells was compared with the intensity of the cytoplasmic GFP fluorescence (Figure 4B). GFP–Fr6 was found tightly bound to centrosomes, while deletion of 112, 184 and 201 amino acids (constructs ΔC3–ΔC5) from the C-terminus gradually decreased the affinity of the constructs for the centrosome. Deletion of 282 amino acids from GFP–FrC (construct FrC_ΔC6) led to the loss of binding to centrosomes. The results summarized in Figure 4C (and also presented schematically in Figure 4A) suggest that the region between amino acids 1784 (C-terminus of FrC_ΔC6) and the C-terminus of XMAP215 is necessary for the centrosomal localization of XMAP215. This region includes the classical microtubule-binding domain of Tau2, MAP4, MAP2b, with a KXGS phosphorylation site (aa 1871–1874 of the XMAP215 sequence) (Drewes et al., 1997). Phosphorylation of the KXGS motif by MARK kinase reduces the affinity of some MAPs for the microtubule lattice (Ebneth et al., 1999). To determine whether the putative microtubule-binding domain played a role in the centrosomal localization of XMAP215, we performed a site-specific mutagenesis on the FrC–GFP-expressing construct, replacing the KIGS site and two flanking basic amino acids (KKIGSK) by six irrelevant (NGYLAQ, mutation ‘M6’) amino acids. The centrosome-binding properties of this mutant (FrC_M6–GFP) were compared with those of the wild-type construct after transfecting them into XL177 cells (Figure 4D). A quantification of fluorescence in transfected cells shown in Figure 4E indicates that the intensity of the centrosomal staining for the M6 mutant was consistently ∼50% lower than that observed in cells transfected with wild-type FrC. Therefore, this motif is clearly involved in targeting of XMAP215 to the centrosome.
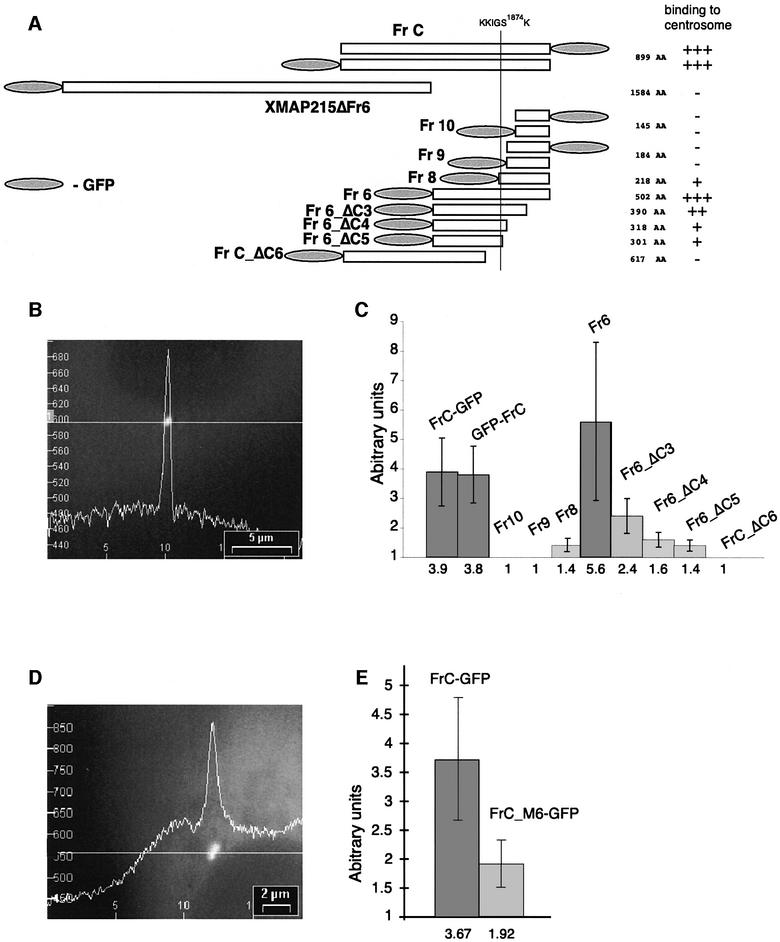
Fig. 4. The centrosome-binding site is localized at the C-terminal part of XMAP215. (A) Schematic representation of the GFP fusion constructs of XMAP215 designed to map the centrosome-binding domain. FrC, Fr9 and Fr10 were separately expressed as either N- or C-terminal fusions with GFP to exclude artifacts created by hindrance of the GFP tag. KKIGS1874K is a six amino acid sequence which was mutated in FrC–GFP to assess its role in centrosome binding. A vertical line through the C-terminus of XMAP215 shows the position of the KKIGSK sequence in different constructs. Relative intensity of centrosome staining indicated on the right is based on the results shown in (C). (B) Intensity profile of GFP fluorescence in a cell expressing FrC–GFP. (C) Ratios of intensities of centrosome/cytoplasm in live XL177 cells expressing different C-terminal fragments (see A). To calculate the ratios, the camera background was first subtracted from cytoplasm and centrosome fluorescence. The scale of the y-axis begins at 1, representing a cell in which no difference in fluorescence intensity between centrosome and cytoplasm exists, for example, in cells transfected with GFP fusions of Fr9, Fr10 and FrC_ΔC6. For each bar, 20 cells were assessed in a blind assay. Values of intensity ratios are indicated under the columns. GFP–FrC and FrC–GFP represent fusion proteins with N- or C-terminal positions of GFP. (D) Intensity profile of GFP fluorescence in a live XL177 cell expressing FrC_M6–GFP, where M6 stands for the mutation introduced into FrC with amino acids 1870–1875 (KKIGSK) replaced by six irrelevant amino acids (NGYLAQ). Note that the level of GFP fluorescence in the cytoplasm is higher than in (B). (E) Ratios of intensities of centrosome/cytoplasm in live XL177 cells, electroporated with FrC–GFP and FrC_M6–GFP. For each bar, 40 cells were measured. Note that the centrosome/cytoplasm fluorescence ratio of FrC–GFP in this experiment is very close to the data in (C). The scale of the y-axis begins at 1 (as in C).
Altogether, this analysis shows that XMAP215 is targeted to the centrosome by a domain localized between amino acids 1784 and 2065.
Microtubule-binding properties of FrN, FrM and FrC
The results reported in the preceding section indicate how each of the three fragments of XMAP215 binds to microtubules in vivo. Since several parts of the molecule seemed to be required to provide the staining pattern of the full-length XMAP215, we decided to compare the microtubule-binding properties of these fragments in vitro. First, we used bacterially expressed fusions of FrN, FrM and FrC with GFP. These proteins were incubated with taxol-stabilized microtubules and sedimented through a glycerol cushion without fixation. Results shown in Figure 5A indicate that all three fragments could bind to microtubules while GFP alone did not. To assess this binding quantitatively, we used the same three XMAP215 fragments, which were tagged with a c-myc peptide instead of GFP. Various amounts of these fragments were incubated with 1 µM taxol-stabilized microtubules. Figure 5B and C shows that microtubule-binding capacity for fragments N and M was exceeded in a 1:4 ratio, whereas FrC bound to microtubules in a 1:1 ratio. More than half of FrC co-pelleted with microtubules at 0.5 and 0.25 µM, whereas <30% of FrN and FrM were found in the microtubule pellet at these concentrations. These results indicate that FrN and FrM have a very low affinity for microtubules on their own, and bind to microtubules at a low stoichiometry relative to tubulin dimers. In contrast, FrC can bind to microtubules proportionally to the concentration of the polymerized tubulin.
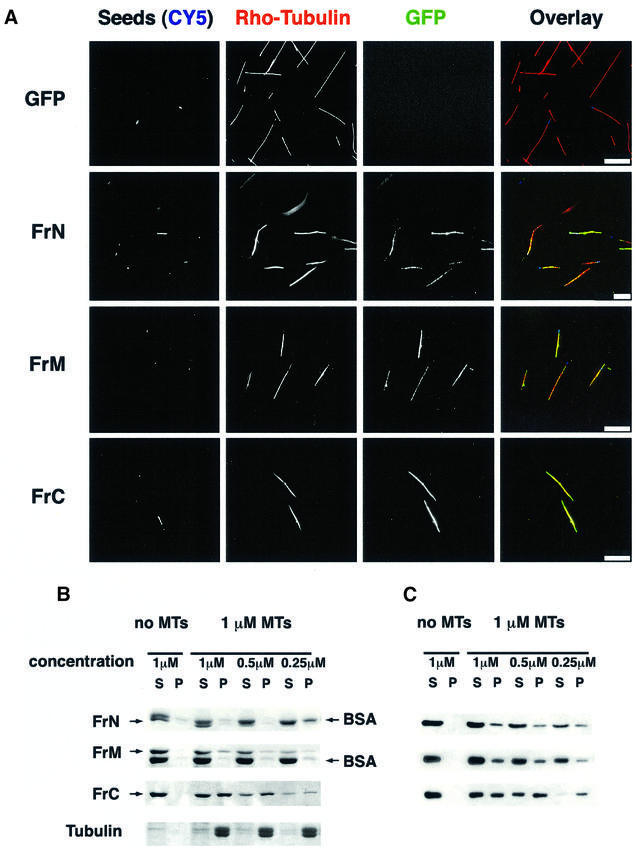
Fig. 5. The C-terminal fragment of XMAP215 has the highest affinity for pure tubulin microtubules. (A) GFP fusion proteins of FrN, FrM and FrC were incubated with pure tubulin microtubules and sedimented without fixation. CY5-labeled microtubule seeds are shown in a blue pseudocolor. Note that all three of the fragments studied are associated with microtubules whilst GFP alone does not bind to microtubules. (B) Microtubule spin-down assay. c-myc-tagged FrN, FrM and FrC were incubated with 1 µM pure tubulin microtubules. After sedimentation, pellets and supernatants were analyzed by SDS–electrophoresis followed by Coomassie Blue staining. BSA was present in reactions to prevent precipitation and/or absorption of XMAP215 fragments on the walls of test tubes. Note that the FrN band (62 kDa) is just under the BSA band. (C) Immunoblotting results of the same experiment shown in (B).
Functional effects of XMAP215 fragments on microtubule dynamics in egg extracts
We then examined the effect of the XMAP215 fragments on microtubule dynamics in egg extracts. We first examined the localization properties of the fragments in extracts. When added at a low concentration of no more than 0.1 µM, both GFP-tagged N- and C-terminal fragments bound to the microtubule asters growing from centrosomes (Figure 6A), whereas fragment M did not. These results suggested that FrN and FrC were more likely to have an effect on microtubule dynamics in extracts.
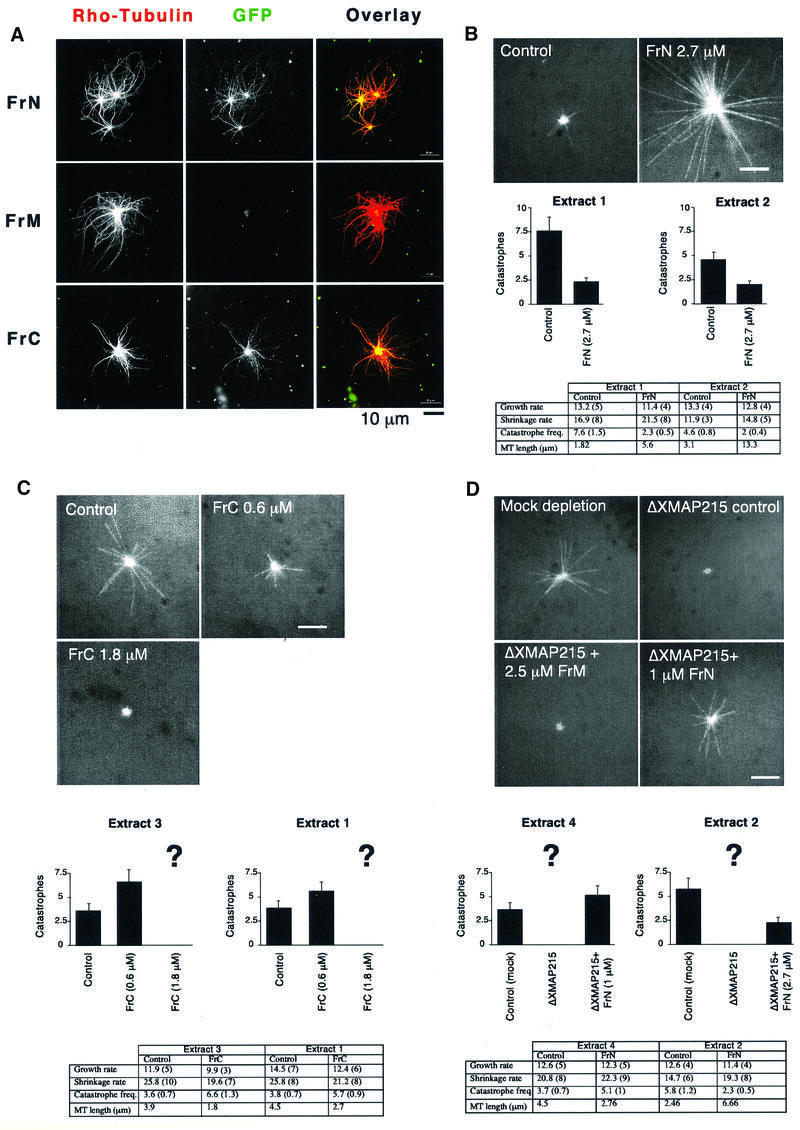
Fig. 6. N- and C-terminal parts of XMAP215 bind to centrosomal asters in egg extracts and have opposite effects on microtubule dynamics. (A) Only GFP–FrN and GFP–FrC bind to the microtubule asters assembled in a mitotic Xenopus egg extract. Asters were assembled in the presence of rhodamine–tubulin and 0.1 µM recombinant XMAP215 fragments fused to GFP. Scale bar, 10 µm. (B) Addition of FrN to mitotic egg extract reduces the number of catastrophes. At the top are the images of microtubule asters as observed in mitotic Xenopus egg extracts during recording by fluorescence video microscopy. Results in the table summarize two separate experiments performed in two different extracts. Control reactions show microtubule dynamics after the addition of the buffer. The final concentration of FrN in the reaction was 2.7 µM. (C) FrC augments the catastrophe rate in mitotic egg extracts. Note that at 1.8 µM FrC practically no microtubules are visible (corresponding catastrophe rate is shown in the chart as ?). Therefore, effects of FrC on microtubule dynamics were recorded at a final concentration of 0.6 µM. Results from two different extracts are presented. (D) The phenotype of the XMAP215 depletion can be reversed by addition of FrN. Results from two different extracts are presented. Note that at 1 µM, FrN partially restores microtubule dynamics to the level of the mock-depleted extract, but at 2.7 µM the number of catastrophes was even lower than before the depletion of XMAP215. Scale bar, 5 µm. Note that the number of rescues is not indicated in the tables of (B), (C) and (D) because in most cases the number of events observed was too low.
To investigate this issue, we added His6-tagged FrN, FrM and FrC to mitotic egg extracts and measured aster size and dynamic parameters of microtubule growth. The N-terminal fragment markedly increased the aster size (Figure 6B), while FrM did not have any effect on microtubule length (not shown). At 2.7 µM, FrN increased average microtubule length 3- to 4-fold and reduced the catastrophe rate 2- to 3-fold. This addition of FrN represents an ∼10-fold molar excess over the amounts of endogenous XMAP215 (0.3 µM, calculated from the results of Gard and Kirschner, 1987). FrN addition had no significant effect on the growth or shrinkage rates. We next investigated the effect of FrC on microtubule dynamics. At 1.8 µM, the C-terminal fragment destabilized microtubules, yielding centrosomes practically devoid of microtubules (Figure 6C). To determine which parameters of microtubule growth were affected by FrC, we titrated down the final concentration of FrC 3-fold to 0.6 µM and measured the dynamics of individual microtubules. Under these conditions, FrC decreased microtubule length 1.6- to 2-fold depending on different extracts. The main parameter affected was the catastrophe frequency. It almost doubled relative to the control (50 and 83% increase in two different experiments). To see whether this effect of FrC was specific to egg extracts, we added FrC to preformed microtubule asters growing from centrosomes in the presence of pure tubulin. Figure 7 shows that not only did FrC not induce microtubule depolymerization under these conditions, but it also brought about microtubule bundling. The total mass of polymerized tubulin in the asters increased 2.8-fold in the presence of 2 µM FrC compared with the control reaction. Therefore, the catastrophe-inducing activity of FrC is specific to egg extracts.
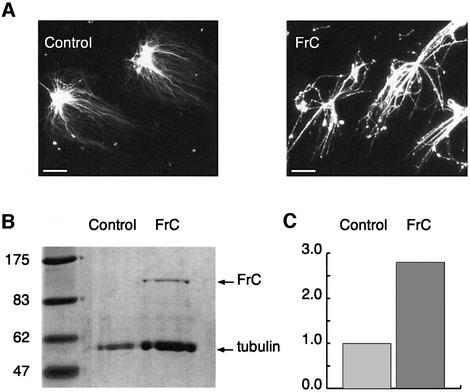
Fig. 7. FrC does not have an intrinsic microtubule-depolymerizing activity. (A) FrC was added to preformed microtubule asters growing from centrosomes in the presence of pure tubulin. Note the extensive bundling of microtubules in the presence of FrC. (B) Coomassie-stained gel with samples from the experiment shown in (A). (C) Quantification of the tubulin protein bands shown in (B). The mass of polymerized tubulin in the asters increased 2.8-fold in the presence of 2 µM FrC compared with the control reaction.
Since the addition of FrN to extracts diminished the number of catastrophes, we wondered whether it could rescue the phenotype of an XMAP215-depleted extract. Therefore, we added FrN to an XMAP215-depleted extract (see Figure 8A for an example of the XMAP215 depletion). If >90% of XMAP215 is depleted, centrosomes nucleate microtubules that are so short they are barely visible (Tournebize et al., 2000; see also Figure 6D) and no microtubule dynamics parameters can be measured. FrN reversed this effect. Microtubule length, recorded after addition of 1 µM FrN, was 61% of the length of microtubules in the mock-depleted extract, with a catastrophe frequency of 5.1 events/min (compared with that of 3.7 events/min in the control; Figure 6D). In contrast, FrM even at 2.5 µM did not have any effect on microtubule growth. When FrN was added at 2.7 µM, the resulting microtubules were 2.7 times longer (6.6 µm compared with 2.46 µm) than in the control reaction, reflecting a >2-fold drop in the catastrophe frequency.
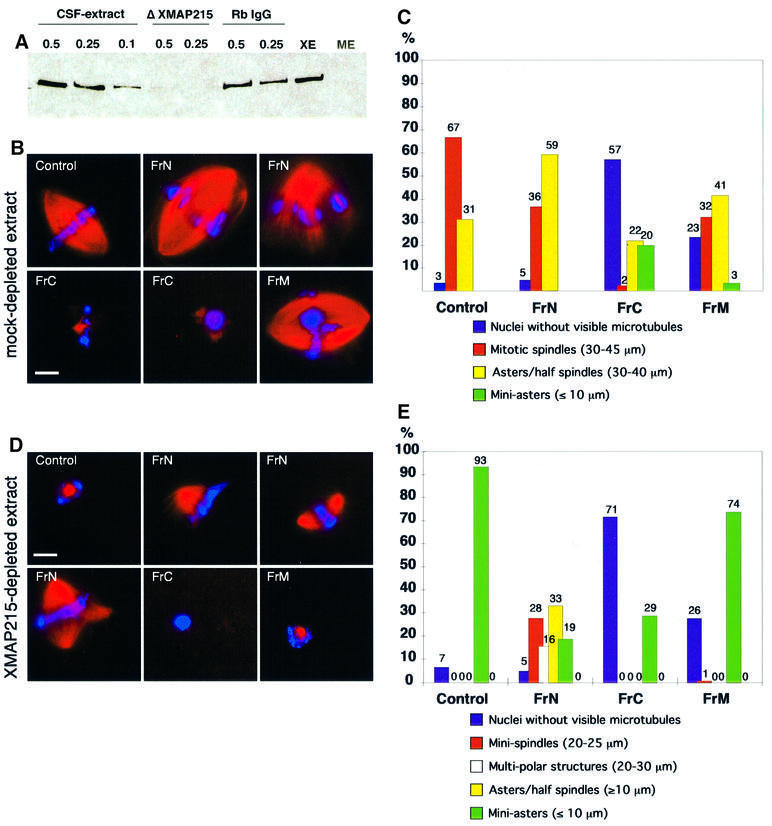
Fig. 8. The N-terminal part of XMAP215 partially rescues mitotic spindle formation in XMAP215-depleted extracts. The C-terminal part of XMAP215 inhibits microtubule growth around sperm nuclei. (A) Immunoblot with anti-XMAP215 antibodies showing the efficiency of XMAP215 depletion from Xenopus egg mitotic extracts. Lanes 1–3 represent 0.5, 0.25 and 0.1 µl of the extract resolved on a 6% SDS–gel. Lanes 4–5 show 0.5 and 0.25 µl of an XMAP215-depleted extract (ΔXMAP215). Lanes 6–7 show 0.5 and 0.25 µl of a mock-depleted extract (Rb IgG, non-immune rabbit serum IgG). XE, eluate from Dynabeads loaded with anti-XMAP215 antibodies after depletion. ME, eluate from Dynabeads loaded with Rb IgG after mock depletion. (B) Formation of mitotic spindles in the cycled extracts after the addition of recombinant XMAP215 fragments. Most frequent phenotypes are shown. (C) Quantification of the phenotypes shown in (B). More than one hundred nuclei per reaction were observed and the microtubule structures associated with these nuclei were classified. (D) Formation of spindles and other structures in the cycled XMAP215-depleted extracts in the presence of recombinant XMAP215 fragments. (E) Quantification of the phenotypes shown in (D). More than one hundred nuclei per reaction were estimated. Scale bar, 10 µm.
We conclude that the N- and C-terminal parts of XMAP215 have opposite activities on the microtubule dynamics in egg extracts: the N-terminal fragment suppresses catastrophes, while the C-terminal fragment promotes them.
The N-terminal fragment rescues spindle assembly in XMAP215-depleted extracts whereas the C-terminal fragment blocks spindle assembly
We have recently shown that immunodepletion of XMAP215 from egg extracts abolished spindle formation (Tournebize et al., 2000). We reproduced this experiment by adding XMAP215 fragments to depleted extracts. After removal of >90% of XMAP215 from the extracts (Figure 8A), no spindles were observed (Figure 8D and E). Adding back FrN (1 µM) partially rescued the depletion phenotype, resulting in the formation of minispindles or multipolar spindles around chromosomes in 50–60% of nuclei-associated structures. Addition of FrC at 1.8 µM resulted in a dramatic inhibition of microtubule growth. In control mock-depleted extracts, addition of FrN (1 µM) and FrM (2.5 µM) had only a minor effect on the percentage of structures formed around sperm nuclei, while FrC markedly inhibited microtubule growth (Figure 8B and C).
In conclusion, we have found that FrN rescues spindle formation in XMAP215-depleted egg extracts, whereas FrC prevents microtubule polymerization both in depleted and control extracts.
Discussion
Localization of XMAP215 in relation to its orthologs
In all species examined so far, XMAP215 orthologs have been found on microtubules and centrosome/spindle pole bodies, although some variability exists among the published data (Table I). Our results indicate that XMAP215 interacts both with microtubules and centrosomes in all stages of the cell cycle (Figures 1–3 and Supplementary figure 1). The EM data indicate that in the centrosome, XMAP215 is mostly associated with the pericentriolar material. This is also supported by the observation that a D.discoideum analog of XMAP215 (DdCP224; Gräf et al., 2000) is a resident centrosomal protein. Since D.discoideum possesses a very compact centrosome lacking centrioles (Moens, 1976), this finding suggests that DdCP224 and other members of the XMAP215 family bind to the pericentriolar material, rather than directly to centrioles. The microtubule localization of XMAP215 seems different from that reported for the closest ortholog studied, ch-TOG (Charrasse et al., 1998). The reason for this difference is unclear but could be due to the fact that this group used antibodies directed against the C-terminal domain of ch-TOG. In our hands, using cells in interphase, antibodies raised against the C-terminal domain of XMAP215 did not stain either microtubules or centrosomes (not shown). However, anti-N-terminal antibodies used on fixed human HeLa cells show a pattern of staining identical to that of XMAP215 both in interphase and mitosis (see Supplementary figure 2).
Table I. Localization of the proteins of the XMAP215 family.
| Found on | p93dis1a | STU2b | Mspsc | Zyg-9d | ch-TOGe | ch-TOG (this work)f | XMAP215 (this work)g | DdCP224h |
|---|---|---|---|---|---|---|---|---|
| Microtubules in interphase | + GFP | ± GFP | – IF | – IF | – IF | + IF | + IF, GFP | + IF, GFP |
| MTOC in interphase | –? GFP | + GFP | – IF | – IF | – IF | – IF | – IF, + GFP | + IF, GFP |
| Microtubules in mitotic spindle | ± GFP | ± GFP | + IF | + IF | + IF | + IF | + IF, GFP | + IF, GFP |
| Spindle poles | + GFP | + GFP | + IF | + IF | + IF | + IF | + IF, GFP | + IF, GFP |
IF and GFP represent localization results obtained either by immunofluorescence on fixed cells or with GFP mutants in live cells, respectively. +, – and ? stand for a positive signal, lack of signal and unclear result, respectively.
fSee Supplementary figure 2.
Our conclusion is that, as with other proteins of this family, XMAP215 interacts both with the centrosomes and microtubules.
Functional and structural homologies between members of the XMAP215 family
Sequence analyses reveal that, although all relatives of XMAP215 in species ranging from yeast to human do have regions of considerable homology, their size and domain composition vary. This is shown in Figure 9, which compares schematically the primary structure of the XMAP215 orthologs. On this scheme, ch-TOG represents proteins like the Msps protein (product of the mini spindles gene), DdCP224 and a putative protein from Arabidopsis thaliana (DDBJ/EMBL/GenBank No. 4263790), which shares homology with XMAP215 over the whole protein length. Yeast proteins like p93dis1, STU2 and an 809 amino acid putative protein from Schizosaccharomyces pombe (DDBJ/EMBL/GenBank No. 4164426) are only homologous to the N-terminus of XMAP215. The structure of Zyg-9 is unusual because it contains a duplicated domain 1 in the N-terminus, which is highly conserved in all the proteins discussed. As illustrated in Figure 9, in all species examined, the N-terminal part contains the conserved domain 1 (see also Cullen et al., 1999; Gräf et al., 2000).
We have shown that FrN is responsible for the catastrophe-suppressing activity of XMAP215. Moreover, additional experiments (data not shown) indicate that a recombinant protein much shorter than FrN, containing only domain 1, is sufficient to stabilize microtubule growth in egg extracts. This observation is important because it defines a new domain that stabilizes microtubules most likely by opposing the ‘catastrophic’ kinesins. FrN binds to asters in egg extracts, but does not seem to be associated with microtubules in live cells. However, this can be explained by the low affinity of FrN for microtubules; a high amount of GFP–FrN in the cytoplasm of living cells would prevent observation of individual microtubules by reducing the contrast.
Deletion/mutation analysis shows that the centrosome-targeting domain of XMAP215 includes a classical microtubule-binding motif, ‘L-----KI/VGS-E/DN-K’. This domain is conserved in XMAP215, ch-TOG, DdCP224 and to a lesser extent in the putative protein from A.thaliana (DDBJ/EMBL/GenBank No. 4263790) and the Msps protein from D.melanogaster. However, a centrosomal (or spindle pole) localization was also reported for STU2, p93dis1 and Zyg-9, in which this domain is absent, and the whole C-terminal part of the molecule has no homology to the XMAP215/ch-TOG/DdCP224/Msps proteins. Mutational analysis of p93dis1 (Nakaseko et al., 1996) is interesting because this group used a deletion/expression of GFP-tagged proteins approach similar to ours, and the deletion of the C-terminal region of p93dis1 resulted in a lack of spindle pole body localization. This suggests that while p93dis1, XMAP215 and related proteins share a catastrophe-suppressing domain in the N-terminal region (domain 1 in Figure 9), they are targeted to the MTOC through another sequence in the C-terminal domain. Taken together, this suggests the existence of a functional homology between these two groups of proteins (XMAP215/ch-TOG/DdCP224/Msps and STU2/p93dis1/Zyg-9) even in the absence of sequence similarity in the C-terminal region.
The function of the microtubule-targeting motif found in XMAP215 and other proteins is intriguing. In XMAP215, this site seems to be part of the centrosomal-targeting domain. Indeed, mutation of the KIGS motif, which was shown to be a target site for the MARK kinase, clearly reduced the amount of FrC–GFP bound to the centrosomes. However, the actual centrosome localization domain goes beyond the mutated KKIGSK sequence, and we do not know yet whether the phosphorylation of Ser1874 in the KIGS motif affects the affinity of XMAP215 for centrosomes or microtubules. Also, although FrC of XMAP215 clearly binds to microtubules, the C-terminal part of ch-TOG was not found to bind microtubules in vitro (Spittle et al., 2000). It was instead reported to interact with tubulin dimers. On the other hand, FrC of XMAP215 is not associated with α/β-tubulin dimers in Xenopus egg extracts when analyzed by centrifugation in sucrose gradients (data not shown). These differences could be explained either by a functional difference between the ch-TOG and XMAP215 molecules, or by the fact that Spittle et al. (2000) used a C-terminal fragment of ch-TOG much shorter than FrC of XMAP215 (743 aa of TOGΔ14–1129 versus 899 amino acids of FrC). Another interesting possibility is that FrC of XMAP215 may target in live cells/extracts not only α/β-tubulin dimers but also other centrosome-specific tubulins such as γ-tubulin and/or ε/δ-tubulins (Chang and Stearns, 2000; Smrzka et al., 2000).
How does XMAP215 regulate microtubule dynamics?
Mutational analysis of XMAP215 made it possible for us to draw several conclusions. We have identified the microtubule-stabilizing and centrosome-binding domains in the N- and C-terminal parts of the protein, respectively. We have found that the whole protein is required for proper binding to microtubules in vivo and, most interestingly, we found that the C-terminal fragment has an activity in egg extracts opposite to that of full-length XMAP215: it strongly destabilizes microtubules and blocks spindle assembly. However, it is also the fragment that binds most avidly to microtubules.
The stabilizing effect of fragment N could be explained in the following ways. It could bind directly to microtubule ends. Indeed, the N-terminal end of ch-TOG binds preferentially to short microtubules, rather than to long ones (Spittle et al., 2000) and we do see a binding of the N-terminal fragment of XMAP215 to microtubules in vitro. However, we have not seen GFP–XMAP215 or FrN–GFP exclusively co-localized with microtubule ends either in extracts or in live cells (although its activity there cannot be excluded). The second possibility is that FrN does not need to bind to microtubules to oppose catastrophes in vivo, but rather interacts with Kin I kinesins near growing microtubule ends (see model in Figure 10). More work will be required to understand how the conserved domain contained within FrN stabilizes microtubules in egg extracts.
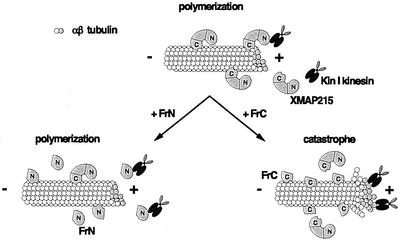
Fig. 10. Model explaining the activity of the XMAP215 fragments in egg extracts. XMAP215 binds to microtubules and prevents the induction of microtubule catastrophes by Kin I kinesins. The N-terminal fragment of XMAP215 (FrN) opposes the Kin I kinesin activity. The C-terminal fragment of XMAP215 (FrC) displaces the endogenous XMAP215, and thus exposes the plus-ends of microtubules to the destabilizing activity of the catastrophic kinesins.
The destabilizing effect of fragment C could be explained by a dominant-negative effect: it binds to microtubules and prevents the endogenous XMAP215 from carrying out its function (Figure 10). Another possibility is that the C-terminal fragment directly increases the catastrophe frequency by binding to microtubules. However, this does not seem to be the case because it does not affect the length of microtubules assembled from pure tubulin and actually increases the mass of polymerized tubulin (Figure 7).
In summary, we have identified the microtubule-stabilizing domain of the XMAP215 family of proteins. This N-terminal domain has a powerful microtubule stabilization effect that seems to be achieved by opposing destabilizing factors such as Kin I kinesins. We have shown that the whole protein length is required for its proper distribution on microtubules in live cells. We have also identified the centrosome-binding domain of XMAP215 at the C-terminal part of the protein, although the function of XMAP215 and its orthologs at the centrosome is much less clear. By being concentrated on MTOCs, these molecules could locally increase the growth of microtubules and, therefore, stabilize microtubules nucleated from the γ-tubulin ring complexes (Wiese and Zheng, 2000).
Materials and methods
Antibody production
The first 560 amino acid N-terminal fragment of XMAP215 (FrN; see Figure 3A) was expressed as a His6-tagged recombinant protein, purified, and used to obtain affinity-purified rabbit polyclonal antibodies according to standard protocols (Harlow and Lane, 1988; Tournebize et al., 2000). C-terminal antibodies used for XMAP215 depletion from egg extracts were raised against a peptide containing the last 15 amino acids of XMAP215, and affinity purified against this peptide by Sigma-Genosys, UK.
Cell culture, electroporation and live cell microscopy
XL177 cells (Miller and Daniel, 1977) were grown in 70% Leibowitz-15 medium with 10% fetal calf serum. For electroporation, cells from one confluent 90 mm Petri dish were collected, washed in 70% phosphate-buffered saline (PBS), resuspended in 200 µl of 70% PBS with 20 µg of supercoiled plasmid DNA and electroporated in 0.2 cm cuvettes at 220 V, 200 Ω and 250 µF. Following electroporation, cells were plated on 15 mm poly-l-lysine-coated glass coverslips, and the phenotype was scored 24–96 h later. For immunofluorescence visualization of XMAP215 on centrosomes after microtubule depolymerization, the cells were incubated with 20 µM nocodazole at 4°C for 4 h before fixing. For live cell imaging the coverslips were mounted in 70% serum-free Dulbecco’s modified Eagle’s medium in slide chambers fitted for 15 mm round coverslips, and the images were acquired using a Zeiss Axioscope2 and an F-View CCD 12-bit black and white camera (Soft Imaging Systems). After mounting the coverslips with live cells, another person changed labels on them, so that a blind assay could be assured.
To quantify the amounts of GFP–XMAP215 fusion constructs bound to centrosomes, we took pictures of randomly observed green cells after transient transfection. For measuring the centrosome/cytoplasm ratios, we used images of cells expressing low amounts of the GFP–XMAP215 fragments, defined as those showing peak intensity of the staining at the centrosome <800 U (after subtracting the background of the camera). This threshold was accepted as it was found that cells showing intensities of GFP fluorescence above this level did not differ significantly in their centrosome/cytoplasm GFP ratio. Before calculating the ratio of intensities of centrosome/cytoplasm, the camera background in units was subtracted from the brightness of the centrosome and cytoplasm. These ratios were found to be reproducible between experiments. For each bar, at least 20 cells were assessed in a blind assay.
Immunofluorescence and immunoblotting
For indirect immunofluorescence we used the same conditions as described in Tournebize et al. (2000). Pictures were acquired with an LSM 510 confocal microscope (Zeiss) or Zeiss Axioscope2.
For immunoblotting analysis, cells were electroporated with the respective GFP-fusion expression constructs, lysed 24–96 h later in the SDS–electrophoresis loading buffer and resolved on a 6 or 8% polyacrylamide gel. Proteins were blotted onto nitrocellulose membranes and probed with anti-GFP antibodies (Roche, Catalog No. 1 814 460), followed by anti-mouse IgG peroxidase conjugate and the ECL detection system (Amersham).
EM localization of XMAP215 in XL177 cells and on centrosomal asters
For the EM localization of XMAP215 in XL177 cells and microtubule asters assembled in egg extracts, we used the same conditions as described in Le Bot et al. (1998) and Wittmann et al. (1998). Cells and asters were stained with anti-N-terminal XMAP215 antibody at 10 µg/ml (see Supplementary data).
Recombinant fragments of XMAP215
Cloning of the full-length XMAP215 was described elsewhere (Tournebize et al., 2000). In this study we used the following XMAP215 fragments: FrN (aa 1–560), FrC (aa 1168–2065), FrM (aa 543–1167), Fr6 (aa 1565–2065), XMAP215_ΔFr6 (aa 1–1584), Fr8 (aa 1847–2065), Fr9 (aa 1882–2065), Fr10 (aa 1922–2065), Fr6_ΔC3 (aa 1565–1982), Fr6_ΔC4 (aa 1565–1881), Fr6_ΔC5 (aa 1565–1864) and FrC_ΔC6 (aa 1168–1784). Information on the cloning, expression and purification of these fragments is included in the Supplementary data.
Microtubule sedimentation assays
Information on microtubule sedimentation assays is presented in the Supplementary data.
Xenopus egg extracts, centrosomal asters, spindle assembly and XMAP215 immunodepletion
Xenopus egg extracts and spindle assembly were performed as previously described (Murray, 1991; Sawin and Mitchison, 1991). Depletion of XMAP215 was performed using 30 µg of anti-C-terminal peptide (the last 15 amino acids of XMAP215) antibodies and 10 µg of anti-N-terminal XMAP215 antibodies coupled to 40 µl of protein A Dynabeads (Dynal) per 130 µl of CSF-arrested Xenopus egg extract (CSF-ME). For control ‘depletion’ (‘mock-depleted’ extracts), we used the same amount (40 µg) of rabbit serum IgG (Dianova). After binding the antibodies, beads were washed twice with PBST and then three times with XB-PI buffer (XB: 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose and 10 mM K–HEPES pH 7.7; PI: protease inhibitors, final concentration of 10 µg/ml each of leupeptin, pepstatin and aprotinin). After removing as much buffer as possible, 130 µl of egg extract were added to the beads and incubated on ice for 1.5 h with occasional gentle pipetting.
Depleted and mock-depleted extracts were then used to assay spindle assembly. The efficiency of XMAP215 depletion was estimated by immunoblotting the same amounts of XMAP215- and mock-depleted extracts. Both antibodies used for depletion recognized the same ∼230 kDa band in egg extracts, which was also eluted from the antibody-coated Dynabeads after depletion (see Figure 8A). Spindles were assembled around sperm nuclei. These 15 µl of XMAP215- or mock-depleted extract were mixed with 1 µl of Xenopus sperm nuclei and 0.12 µl of rhodamine–tubulin at 7 mg/ml. The extract was driven into the interphase by addition of 2 µl of 4 mM CaCl2 for 90 min. At this point another 15 µl of the XMAP215- or mock-depleted extract were added together with a protein fragment of XMAP215 in the CSF-XB at one-tenth of the total extract volume. FrN and FrM were added at 1 µM final concentration, while FrC was added at 2 µM final concentration. In a control reaction, the same volume of the CSF-XB buffer with 0.1 mg/ml bovine serum albumin (BSA) was added. The proteins were added to the extract on ice and left for 10 more minutes on ice before transferring the tubes to 20°C for a 30–60 min incubation to allow the extract to cycle into mitosis and form mitotic spindles around sperm DNA. To score the results, 2 µl of extract were fixed in the spindle fix (Heald et al., 1998) and images were collected using Axiophot 2.
To study the localization of the GFP-labeled FrN, FrM and FrC on microtubule asters growing from centrosomes, frog egg extract was prepared as described above and supplemented with rhodamine–tubulin at 140 µg/ml. Human KE37 cell line centrosomes at a concentration of 2 × 108 per ml (prepared according to Moudjou and Bornens, 1998) were added in a 2 µl volume to 40 µl of extracts, followed by 1 µl of the GFP-labeled proteins at a final concentration of 0.1 µM. As a control we used GFP alone, which did not stain centrosome asters (data not shown). Extracts were incubated at 20°C for 10 min and fixed by the addition of 1 ml of fixing solution (BRB80, 30% glycerol, 0.5% Triton X-100 and 0.25% glutaraldehyde). Fixed asters were immediately centrifuged through a 40% glycerol cushion in BRB80 in an HB6 rotor in a Sorvall centrifuge at 12 000 r.p.m., 16°C for 15 min onto coverslips as described (Sawin and Mitchison, 1991). Glutaraldehyde was quenched by incubation in 1 mg/ml NaBH4 in PBS and the coverslips were mounted in Mowiol. Images were obtained using Axiophot 2 and an F-View CCD 12-bit black and white camera (Soft Imaging Systems).
Recording of microtubule dynamics
For microtubule dynamics, single microtubules growing from centrosomes were recorded as described (Tournebize et al., 1997), using a Zeiss Axioskop, a 100× Apochromate lens (NA 1.4) and a rhodamine filter cube. Images were recorded every 2 s on an 8-bit black and white camera (Sony SSC-M370CE) with an image processor (model Argus 10; Hamamatsu) and stored on a Macintosh using NIH Image software. Data analysis was performed using a Microsoft Excel macro. For each condition (control experiment and in the presence of XMAP215 fragments), ∼25 microtubules were analyzed for no longer than 10 min. In control reactions, only the buffer, into which individual fragments were dialysed, was added. We found that addition of the PIPES buffer (80 mM PIPES–HCl pH 6.8, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.1 mg/ml BSA) led to a higher number of catastrophes in control reactions than that of CSF-XB supplemented with 0.1 mg/ml BSA. Differences between experiments, with and without a specific XMAP215 fragment, were evaluated using a Student’s t-test with a confidence level of 95%. The average length of microtubules (‘steady-state’ length) was calculated from the dynamic parameters as described previously using the following equation (Verde et al., 1992):
<L> = (VgFres – VsFcat)/(Fcat + Fres)
where Vg, Vs, Fcat and Fres are growth rate, shrinkage rate, catastrophe frequency and rescue frequency, respectively. In some experiments, only a few rescues were observed, therefore the calculated average microtubule length should be considered as an approximation.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Fabrice Senger for cell culture, Torsten Wittmann for much advice, Vladimir Gelfand for advice on Xenopus cell electroporation, Alena Bogdanova for help with cloning, and Torsten Wittmann and Celia Antonio for numerous egg extract donations. We would like to thank Torsten Wittmann, Heidi McBride and Wofgang Zachariae for critical reading and suggestions for the manuscript. A.V.P. and K.K. were both supported by HFSPO long-term fellowships.
REFERENCES
- Andersen S. and Karsenti,E. (1997) XMAP310: a Xenopus rescue-promoting factor localized to the mitotic spindle. J. Cell Biol., 139, 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S.S., Buendia,B., Dominguez,J.E., Sawyer,A. and Karsenti,E. (1994) Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopus laevis eggs and dividing cells [published erratum appears in J. Cell Biol., 1995, 128, following 988]. J. Cell Biol., 127, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L. and Mitchison,T.J. (1996) Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell, 84, 623–631. [DOI] [PubMed] [Google Scholar]
- Chang P. and Stearns,T. (2000) δ-tubulin and ε-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nature Cell Biol., 2, 30–35. [DOI] [PubMed] [Google Scholar]
- Chapin S.J. and Bulinski,J.C. (1992) Microtubule stabilization by assembly-promoting microtuble-associated proteins: a repeat per formance. Cell Motil. Cytoskeleton, 23, 236–243. [DOI] [PubMed] [Google Scholar]
- Charrasse S., Schroeder,M., Gauthier-Rouviere,C., Ango,F., Cassimeris,L., Gard,D.L. and Larroque,C. (1998) The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci., 111, 1371–1383. [DOI] [PubMed] [Google Scholar]
- Cullen C.F., Deak,P., Glover,D.M. and Ohkura,H. (1999) mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol., 146, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G., Ebneth,A., Preuss,U., Mandelkow,E.M. and Mandelkow,E. (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell, 89, 297–308. [DOI] [PubMed] [Google Scholar]
- Ebneth A., Drewes,G. and Mandelkow,E. (1999) Phosphorylation of MAP2c and MAP4 by MARK kinases leads to the destabilization of microtubules in cells. Cell Motil. Cytoskeleton, 44, 209–224. [DOI] [PubMed] [Google Scholar]
- Gard D. and Kirschner,M. (1987) A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus end. J. Cell Biol., 105, 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf R., Daunderer,C. and Schliwa,M. (2000) Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J. Cell Sci., 113, 1747–1758. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Heald R., Tournebize,R., Vernos,I., Murray,A., Hyman,A. and Karsenti,E. (1998) In vitro assays for mitotic spindle assembly and function. In Celis,J.E. (ed.), Cell Biology: A Laboratory Handbook, 2nd edn. Academic Press, San Diego, CA, pp. 326–335.
- Le Bot N., Antony,C., White,J., Karsenti,E. and Vernos,I. (1998) Role of xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the Golgi apparatus. J. Cell Biol., 143, 1559–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L.R., Carter,P., Thierry-Mieg,D. and Kemphues,K. (1998) ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol., 141, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J., Okawa,K., Iwamatsu,A. and Vale,R.D. (1996) Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J. Cell Sci., 109, 561–567. [DOI] [PubMed] [Google Scholar]
- Miller L. and Daniel,J.C. (1977) Comparison of in vivo and in vitro ribosomal RNA synthesis in nucleolar mutants of Xenopus laevis. In Vitro, 13, 557–563. [DOI] [PubMed] [Google Scholar]
- Moens P.B. (1976) Spindle and kinetochore morphology of Dictyostelium discoideum. J. Cell Biol., 68, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M. and Bornens,M. (1998) Method of centrosome isolation from cultured animal cells. In Celis,J.E. (ed.), Cell Biology: A Laboratory Handbook, 2nd edn. Academic Press, San Diego, CA, pp. 111–119.
- Murray A. (1991) In Kay,B.K. and Peng,H.B. (eds), Cell Cycle Extracts. Vol. 36. Academic Press, San Diego, CA, pp. 581–605.
- Nabeshima K., Kurooka,H., Takeuchi,M., Kinoshita,K., Nakaseko,Y. and Yanagida,M. (1995) p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev., 9, 1572–1585. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y., Nabeshima,K., Kinoshita,K. and Yanagida,M. (1996) Dissection of fission yeast microtubule associating protein p93Dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells, 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Sawin K.E. and Mitchison,T.J. (1991) Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol., 112, 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrzka O.W., Delgehyr,N. and Bornens,M. (2000) Tissue-specific expression and subcellular localisation of mammalian δ-tubulin. Curr. Biol., 10, 413–416. [DOI] [PubMed] [Google Scholar]
- Spittle C., Charrasse,S., Larroque,C. and Cassimeris,L. (2000) The interaction of TOGp with microtubules and tubulin. J. Biol. Chem., 275, 20748–20753. [DOI] [PubMed] [Google Scholar]
- Tournebize R., Andersen,S.S., Verde,F., Doree,M., Karsenti,E. and Hyman,A.A. (1997) Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J., 16, 5537–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R. et al. (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nature Cell Biol., 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Verde F., Dogterom,M., Stelzer,E., Karsenti,E. and Leibler,S. (1992) Control of microtubule dynamics and length by cyclin A and cyclin B dependent kinases in Xenopus egg extracts. J. Biol. Chem., 118, 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C., Mitchison,T.J. and Desai,A.B. (1996) XKCM1: A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell, 84, 37–47. [DOI] [PubMed] [Google Scholar]
- Wang P.J. and Huffaker,T.C. (1997) Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol., 139, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R.A., O’Brien,E.T., Pryer,N.K., Sobeiro,M.F., Voter,W.A., Erickson,H.P. and Salmon,E.D. (1988) Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol., 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C. and Zheng,Y. (2000) A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nature Cell Biol., 2, 358–364. [DOI] [PubMed] [Google Scholar]
- Wilde A. and Zheng,Y. (1999) Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science, 284, 1359–1362. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Boleti,H., Antony,C., Karsenti,E. and Vernos,I. (1998) Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein and dynein. J. Cell Biol., 143, 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]


