Abstract
Sin3 is an evolutionarily conserved corepressor that exists in different complexes with the histone deacetylases HDAC1 and HDAC2. Sin3-HDAC complexes are believed to deacetylate nucleosomes in the vicinity of Sin3-regulated promoters, resulting in a repressed chromatin structure. We have previously found that a human Sin3-HDAC complex includes HDAC1 and HDAC2, the histone-binding proteins RbAp46 and RbAp48, and two novel polypeptides SAP30 and SAP18. SAP30 is a specific component of Sin3 complexes since it is absent in other HDAC1/2-containing complexes such as NuRD. SAP30 mediates interactions with different polypeptides providing specificity to Sin3 complexes. We have identified p33ING1b, a negative growth regulator involved in the p53 pathway, as a SAP30-associated protein. Two distinct Sin3-p33ING1b-containing complexes were isolated, one of which associates with the subunits of the Brg1-based Swi/Snf chromatin remodeling complex. The N terminus of p33ING1b, which is divergent among a family of ING1 polypeptides, associates with the Sin3 complex through direct interaction with SAP30. The N-terminal domain of p33 is present in several uncharacterized human proteins. We show that overexpression of p33ING1b suppresses cell growth in a manner dependent on the intact Sin3-HDAC-interacting domain.
In eukaryotic cells, the DNA is packaged with histones in the form of chromatin (24). The primary unit of chromatin is the nucleosome, composed of 146 bp of DNA wrapped around an octamer of histone proteins (32). The packaging of DNA into chromatin allows for efficient storage of genetic information, although this compaction impedes the access of proteins to DNA. In the nucleus there are machineries that alter chromatin structure and aid proteins in gaining access to their DNA sites (reviewed in references 23 and 51). Types of activities that alter chromatin structure include ATP-dependent nucleosome remodeling and covalent modification of core histones, particularly acetylation-deacetylation of lysine residues and methylation of lysine and arginine residues. These covalent modifications occur primarily within the N-terminal histone tails (46, 61).
Several ATP-dependent nucleosome-remodeling factors have been characterized. These fall into two broad classes: the Swi-Snf and ISWI-related families. Two human complexes related to the yeast Swi-Snf complex. Brg1 and Brm, have been characterized (53, 54). These complexes contain a set of common as well as unique subunits, referred to as Brg1-associated factors (BAFs). Several ISWI-containing complexes have also been isolated from different species (reviewed in reference 50). These chromatin-remodeling complexes possess a DNA-stimulated ATPase activity that resides in the highly conserved Snf2-related subunit.
Different regions of the genome exhibit specific patterns of histone acetylation, which are established by the action of histone acetyltransferases and histone deacetylases (HDACs). Several histone acetyltransferases have been identified. Some of these histone acetylases exist in large complexes and participate in transcriptional activation. HDACs have also been identified. In mammals, there appear to be at least nine different HDACs: HDAC1, HDAC2, and HDAC3 are related to the yeast HDAC Rpd3, whereas HDAC4, HDAC5, HDAC6, HDAC7, HDAC8, and HDAC9 are related to the yeast HDA1 (reviewed in references 6 and 25). Recently, the yeast and mouse Sir2 proteins, which function in the establishment of silencing, were found to possess NAD+-dependent HDAC activity in vitro (21, 29, 45). Some of these HDACs exist in large protein complexes and are directed to specific regions of the genome by interacting, directly or indirectly, with sequence-specific DNA-binding proteins. Yeast Sin3, and the Sin3-associated HDAC Rpd3, have been implicated in promoter-specific repression as well as in silencing (39, 47). Recruitment of the Sin3-Rpd3 complex to the promoter by the DNA-binding repressor Ume6 results in local deacetylation of two nucleosomes in the vicinity of the Ume6-binding site (39). Surprisingly, deletion of Saccharomyces cerevisiae SIN3, RPD3, and HDA1 leads to an increase rather than a loss of silencing (38, 47).
The human Sin3 protein was identified as a corepressor that interacts with the E-box-binding repressor complex Mad-Max (4, 41). Furthermore, interaction with Sin3 was found to be essential for Mad to suppress Myc-induced transformation (15). Subsequent studies revealed that Sin3 also interacts with N-CoR and SMRT (1, 16, 36), corepressors that bind to unliganded nuclear hormone receptors (5, 20).
Two major human HDAC1/2-containing complexes have been characterized biochemically in HeLa cells—Sin3 and NuRD (9, 58–60, 62). Human Sin3 and NuRD complexes share a set of four polypeptides, a presumed catalytic core complex composed of HDAC1, HDAC2, and the histone-binding proteins RbAp46, and RbAp48 (60). In addition to these four polypeptides, a Sin3 complex contains mSIN3A, SAP30, and SAP18 (58). However, several other polypeptides are associated with Sin3-containing complexes, including the CpG-methylated binding protein MeCP2 (37), the Rb-binding protein RBP1 (28), and the corepressors NCoR and SMRT (1, 16, 36). The SAP30 subunit was found to interact with NCoR, SMRT, RBP1, and other polypeptides, suggesting that SAP30 may tether the core HDAC1/2 complex to different polypeptides (26, 62).
The NuRD complex provides an unexpected link between two chromatin-modifying activities. In addition to the HDAC core complex, it contains an ISWI-related polypeptide, CHD4 (Mi2β). CHD4 contains a SNF2-like helicase domain, which enables the NuRD complex to alter the structure of nucleosomes in an ATP-dependent manner. NuRD contains a nucleosome-stimulated ATPase activity. NuRD is able to deacetylate oligonucleosomal histones in vitro in a manner that is stimulated by ATP (49, 52, 55, 59). In light of this finding, it was proposed that access of the HDACs to histone tails in oligonucleosomes requires alteration and/or mobilization of nucleosome structure (49, 59).
In the present studies, we have analyzed polypeptides that associate with the Sin3 complex. We found that the p53-binding protein p33ING1b interacts with the Sin3 complex.
The p53-binding protein ING1 was identified in a genetic selection for genes whose inactivation can cause neoplastic transformation (12). Overexpression of p33ING1 inhibits cell growth in a manner dependent on the wild-type p53, and p33ING1 interacts with p53 in vivo (11). Recently, it was found that the human ING1 gene encodes three alternatively spliced transcripts that give rise to proteins of 47 kDa (ING1a), 33 kDa (ING1b), and 24 kDa (ING1c) (12). All ING1 isoforms share a conserved C terminus containing a plant homeodomain (PHD) Zn finger motif. Curiously, the mouse ING1 gene appears to produce only two alternatively spliced forms, p37 (similar to human ING1b) and p24/31 (similar to the human ING1c) (56). A recent study has identified ING1b as the breast and testis cancer-specific antigen (22).
In this work, we further characterized the human Sin3-HDAC containing ING1. Two different ING1-containing Sin3 complexes were observed in HeLa cells. One of these complexes additionally contains the subunits of the Brg1-based human Swi-Snf chromatin-remodeling complex. We found that the ING1b-Sin3-HDAC-containing complexes possess HDAC and oligonucleosome deacetylase activity and that interaction with the Sin3 complex is important for the growth suppression activity of p33ING1b.
MATERIALS & METHODS
Plasmid construction.
p33ING1b cDNA in the pCI vector (Promega) was a gift of K. Riabowol. For bacterial expression of a His6-p33 fusion protein, p33ING1b cDNA was amplified using primers containing 5′ NdeI and 3′ XhoI adapters (5′-GGGGAATTCCATATGTTGAGTCCTGCCAACGGG-3′ and 5′-CCCGCTCGAGCCTGTTGTAAGCCCTCTCTTTTT-3′) (recognition sequences are underlined), digested with NdeI and XhoI, and subcloned into the pET21a(+) vector (Novagen) linearized with the same enzymes. For mammalian expression of the Gal4-p33 fusion, p33ING1b cDNA was amplified using primers containing 5′ BamHI and 3′ XbaI adapters (5′-GCGCGGATCCGTATGTTGAGTCCTGCCAACGGG-3′ and 5′-CCTAGTCTAGAGCCTGTTGTAAGCCCTCTCTTTTT-3′), digested with BamHI and XbaI, and subcloned into pSG424 vector (40) linearized with the same enzymes.
To generate a glutathione S-transferase (GST)-p33 fusion for bacterial expression, p33 cDNA was amplified using primers containing 5′ BamHI and 3′ XhoI adapters (5′-GCGCGGATCCCGTATGTTGAGTCCTGCCAACGGG-3′ and 5′-CCCGCTCGAGCCTGTTGTAAGCCCTCTCTTTTT-3′), digested with BamHI and XhoI, and subcloned into pGEX-5X-1 vector (Pharmacia) linearized with the same enzymes. To generate the GST-ΔN120 and GST-ΔC59 fusion proteins, p33 cDNA was amplified using the following sets of primers (in each case the direct primer had a BamHI adapter and the reverse primer had a XhoI adapter): 5′-GCGCGGATCCGTCGCGCGCTGATCCGCAGCCAG-3′ and 5′-CCCGCTCGAGCCTGTTGTAAGCCCTCTCTTTTT-3′ (for ΔN120) and 5′-GCGCGGATCCGTATGTTGAGTCCTGCCAACGGG-3′ and 5′-CCCGCTCGAGGATGGGGAGGTCGGCAGGGGGACG-3′ (for ΔC59). The PCR products were digested with BamHI and XhoI and subcloned into the pGEX-5X-1 vector linearized with the same enzymes.
For mammalian expression of a FLAG-p33 fusion protein, p33 cDNA was amplified using primers containing 5′-HindIII and 3′-XbaI adapters (5′-CCCAAGCTTATGTTGAGTCCTGCCAACGGGGAG-3′ and 5′-CCTAGTCTAGAGCCTGTTGTAAGCCCTCTCTTTTT-3′), digested with HindIII and XbaI, and subcloned into pCMV-FLAG-2 vector (Sigma) linearized with the same enzymes. For stable selection of FLAG tag fusions of full-length p33 and p33 deletion mutants, p33 cDNA was amplified using the following sets of primers (a direct primer containing FLAG epitope sequences [underlined] and a reverse primer containing two STOP codons [underlined]): 5′-ACCATGGACTACAAAGACGATGACGACAAGATGTTGAGTCCTGCC-3′ and 5′-CTACTACCTGTTGTAAGCCCTCTCTTTTTTGAATTTCTCCAG-3′ (for full-length p33),5′-ACCATGGACTACAAAGACGATGACGACAAGATCCTGAAGGAG-3′and 5′-CTACTACCTGTTGTAAGCCCTCTCTTTTTTGAATTTCTCCAG-3′ (for the N-terminal deletion mutant), and 5′-ACCATGGACTACAAAGACGATGACGACAAGATGTTGAGTCCTGCC-3′ and 5′-CTACTAGGGGAGGTCGGCAGGGGAGCG-3′ (for the C-terminal deletion mutant). These PCR products were directly ligated into the pEF6/V5-His-TOPO vector (Invitrogen) as specified by the manufacturer.
For mammalian expression of green fluorescent protein (GFP) fusion proteins, the FLAG-p33 cDNA was amplified from the pCMV-FLAG-2 vector using the following sets of primers (in each case the direct primer contained the NheI adapter and the reverse primer contained the AgeI adapter): 5′-CTAGCTAGCTAGACCATGGACTACAAA-3′ and 5′-CGACCGGTAGCTCTTGGTATTTCGC-3′ (for N46-GFP), 5′-CTAGCTAGCTAGACCATGGACTACAAA-3′ and 5′-CGACCGGTAGCCGCCGCTTCTGCGC-3′ (for N69-GFP), and 5′-CTAGCTAGCTAGACCATGGACTACAAA-3′ and 5′-CGACCGGTAGGTTGCCCGCTGTGTC-3′ (for N125-GFP). The PCR products were digested with NheI and AgeI and subcloned into the pECFP-Nuc vector (Clontech) linearized with the same enzymes. Note that the pECFP-Nuc vector encodes the enhanced cyan fluorescent protein containing a triple simian virus 40 T-antigen nuclear localization signal which is necessary for nuclear targeting of the p33 truncations (the nuclear localization signal of p33ING1b is located at amino acids 185 to 189).
Immunoaffinity purification of different SIN3-containing complexes and mass spectrometric identification of proteins.
Immunoaffinity purification was based on a previously published procedure (58). Rabbit polyclonal antibodies raised against full-length p33ING1b or SAP30 were affinity purified on an antigen column and cross-linked to protein A-agarose beads (Repligen) with 20 mM dimethylpimelimidate (Pierce) at 2 mg of antibodies per 1 ml (wet volume) of beads as described previously (14). A total of 10 to 20 μl beads was incubated for 12 h at 4°C with 50 to 100 μg of a protein fraction in buffer B (20% glycerol, 50 mM Tris-HCl [pH 7.9] at 4°C, 0.2 mM EDTA, 10 mM 2-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride) containing 100 mM KCl. The beads were washed with buffer B containing 500 mM KCl and 0.05% NP-40, and bound proteins were eluted with 0.1 M glycine (pH 2.6) and resolved on a 5 to 12.5% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (SDS-PAGE) for Western blotting and silver staining.
To identify coimmunoprecipitating proteins, the protein bands were excised from the gels, digested with trypsin (17), and processed for mass spectrometric fingerprinting as described previously (8). Briefly, peptide mixtures were partially fractionated on Poros 50 R2 RP microtips and the resulting peptide pools were analyzed by matrix-assisted laser-desorption/ionization reflectron time-of-flight mass spectrometry (MALDI-reTOF MS) using a Reflex III instrument (Brüker Franzen, Bremen, Germany) and, in most cases, by electrospray ionization (ESI) tandem MS on an API 300 triple quadrupole instrument (PE-SCIEX, Thornhill, Canada), modified with an injection-adaptable fine ionization source (JAFIS) as described previously (13). Selected mass values from the MALDI-reTOF experiments were taken to search the protein nonredundant database (National Center for Biotechnology Information [NCBI], Bethesda, Md.) using the PeptideSearch (33) algorithm. MS-MS spectra from the ESI triple-quadrupole analyses were inspected for y” ion series, and the resultant information was transferred to the SequenceTag (34) and PepFrag (10) programs and used as a search string. Any protein identification was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS-MS data.
Conventional purification of core Sin3 complex and Sin3 complexes I and II.
Approximately 2 g of HeLa nuclear extracts (7) was loaded on a 200-ml phosphocellulose column in buffer B containing 100 mM KCl, and bound proteins were eluted stepwise with buffer B containing 300, 500, and 1,000 mM KCl. The 0.5 M KCl fraction (300 mg) was dialyzed into buffer BC (10% glycerol, 50 mM Tris-HCl [pH 7.9] at 4°C, 0.2 mM EDTA, 10 mM 2-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride) containing 100 mM KCl. The proteins were then loaded onto a 90-ml DE-52 column, and bound proteins were eluted with buffer BC containing 350 mM KCl. The 350-mM KCl eluate from the DE-52 column (155 mg) was dialyzed against buffer BC containing 75 mM ammonium sulfate. The proteins were then loaded onto a 45-ml HPLC DEAE-5PW column (TosoHaas). The column was eluted with a linear gradient of ammonium sulfate from 75 to 400 mM in buffer BC (gradient volume, 750 ml; flow rate, 2.5 ml/min), and 7.5-ml fractions were collected. For conventional purification of Sin3 complexes I and II, the DEAE-5PW fractions corresponding to Sin3 complexes I and II were loaded onto a 1-ml FPLC MonoP column (Pharmacia). Bound proteins were eluted with a linear gradient of KCl from 200 to 1,200 mM in buffer BC (gradient volume, 10 ml; flow rate, 0.2 ml/min), and 200-μl fractions were collected. The Sin3 peak fractions from MonoP were pooled, dialyzed into buffer B containing 100 mM KCl, and loaded onto a 1-ml phosphocellulose column. Bound proteins were step eluted with buffer B containing 500 mM KCl and dialyzed against buffer B containing 100 mM KCl. This concentrated pool was further used for immunoaffinity purification and for nucleosome deacetylation assays. For conventional purification of the core Sin3 complex, the DEAE-5PW flowthrough fraction (∼30 mg) was concentrated on a 5-ml phosphocellulose column as described above. Peak fractions were pooled and saved in two 3-ml aliquots (2 mg/ml). Each 3-ml aliquot was fractionated on a 120-ml Superose 6 XK16/70 column (Pharmacia) in buffer BC containing 500 mM KCl, and 1.3-ml fractions were collected. The Sin3 peak fractions from the two Superose 6 columns were pooled and again concentrated on a 1-ml phosphocellulose column for further use in immunoaffinity purification and deacetylation assays.
Anti-FLAG immunoprecipitation from transiently transfected cells and anti-FLAG immunoaffinity purification from cell lines stably expressing FLAG-tagged Swi-Snf components.
For transient transfections, 293T cells were plated on 10-cm dishes at 106 cells per dish and transfected with 10 μg of pCMV-FLAG-2-SAP30 or pCMV-FLAG-ING1b by calcium phosphate precipitation. Nuclear extracts were prepared from cells harvested 48 h after transfection. For anti-FLAG immunoprecipitations, 300 μg of 293T nuclear extract was incubated overnight at 4°C with 20 μl of anti-FLAG M2 agarose (Sigma) in buffer B containing 100 mM KCl. Bound proteins were washed with buffer B containing 500 mM KCl and 0.05% NP-40 and analyzed by Western blotting.
Establishment of cell lines stably expressing FLAG-tagged INI1 and purification of FLAG-INI1-containing Swi-Snf complexes were described previously (43). Establishment of cell lines stably expressing FLAG-Brg1. FLAG-Brg1 K798R, and FLAG-Brm and purification of FLAG-Brg1- and FLAG-Brm-containing complexes were described previously (42).
HDAC and nucleosome deacetylase assays.
The 3H release assay was performed as described previously (19). Core histones were labeled in vitro with recombinant S. cerevisiae Hat1 in the presence of [3H]acetyl coenzyme A to 3 × 107 dpm/mg as described previously (38). Column fractions or protein complexes bound to antibody beads were incubated for 1 h at 30°C with 0.5 to 2 μg of labeled core histones in a buffer containing 75 mM Tris-HCl (pH 7.0), 150 mM NaCl, 2 mM dithiothreitol, 0.1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. The released [3H]acetic acid was extracted with ethyl acetate and quantified by liquid scintillation counting.
Hyperacetylated oligonucleosomes were purified using a published procedure (3). For the nucleosome deacetylation assay, 200 ng of hyperacetylated oligonucleosomes or core histones was incubated for 2 h at 30°C with column fractions in a buffer containing 10 mM HEPES-KOH (pH 7.5), 50 mM KCl, and 5 mM MgCl2 in the presence or absence of 4 mM ATP. Histone deacetylation was analyzed by Western blotting using antibodies specific to H4 acetylated at lysines 5, 8, and 12 and antibodies specific to hyperacetylated H3 (a gift of D. Allis).
GST pull-down assay.
GST-p33, GST-ΔN120, GST-ΔC59, and GST-Dr1 fusion proteins were expressed in bacteria and purified on glutathione-Sepharose (Pharmacia) as specified by the manufacturer. The His6-tagged SAP30, Dr1, and RbAp46 proteins were expressed in bacteria and purified on Ni-nitrilotriacetic acid agarose (Qiagen). The FLAG-tagged HDAC1 was purified from infected Sf9 cells on anti-FLAG M2 agarose (Kodak). The mSin3A protein was obtained by in vitro translation of the pCITE-mSin3A plasmid (a gift of D. Ayer) using the TNT coupled reticulocyte lysate system (Promega).
For a GST pull-down assay, glutathione-Sepharose beads (bed volume, 5 μl) were prebound with 100 ng of a GST fusion protein and incubated for 1 h at 4°C with 100 to 200 ng of SAP30, RbAp46, HDAC1, or mSin3A in buffer B containing 500 mM KCl, 0.1% NP-40, and 0.1 mg of bovine serum albumin (BSA) per ml. The protein complexes were washed three times with the same buffer, eluted with SDS-PAGE loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromphenol blue, 10% glycerol, 0.1 M dithiothreitol), and analyzed by Western blotting.
Luciferase reporter assay.
293T cells were transfected in duplicate on 6-cm plates (2 × 106 cells/plate) by the standard calcium phosphate method. Each transfection mixture included 500 ng of PCH110 plasmid (for expression of β-galactozidase, normalization control [Clontech]), 250 ng of reporter plasmid (Gal4-TK-Luc; a gift of P. Traber), 500 ng of pSG424-p33 plasmid (for expression of Gal4-p33), and variable amounts of pBabe-Brg1 or pBabe-Brg1(K798R) (a gift of R. Kingston). The cells were harvested 48 h after transfection, and β-galactosidase and luciferase reporter assays were performed as specified by the manufacturer (Promega). Luciferase activity was normalized to the β-galactosidase activity.
Colony formation assay.
NIH 3T3 cells were transfected with pEF6 vector (Invitrogen) carrying p33 deletion mutants by using Effectene reagent (Qiagen) as specified by the manufacturer. At 48 h posttransfection, the cells were split into Dulbecco’s modified Eagle’s medium plus 10% fetal calf serum plus 5 μg of blasticidin (Invitrogen) per ml on 10-cm plates (three plates per experiment; 104 cells per plate). After 2 weeks of selection, colonies were stained with Coomassie brilliant blue and counted.
RESULTS AND DISCUSSION
Identification of p33ING1b as a SAP30-associated protein.
We have previously characterized a human Sin3 corepressor complex composed of mSin3A, HDAC1 and HDAC2, histone-binding proteins RbAp46 and RbAp48, and two novel polypeptides, SAP30 and SAP18 (58). To address whether the Sin3-associated polypeptides represent a single complex or are present in distinct complexes and whether these polypeptides are also found in complexes other than Sin3, we performed immunoaffinity purification using antibodies against HDAC1 and SAP30 (58, 62).
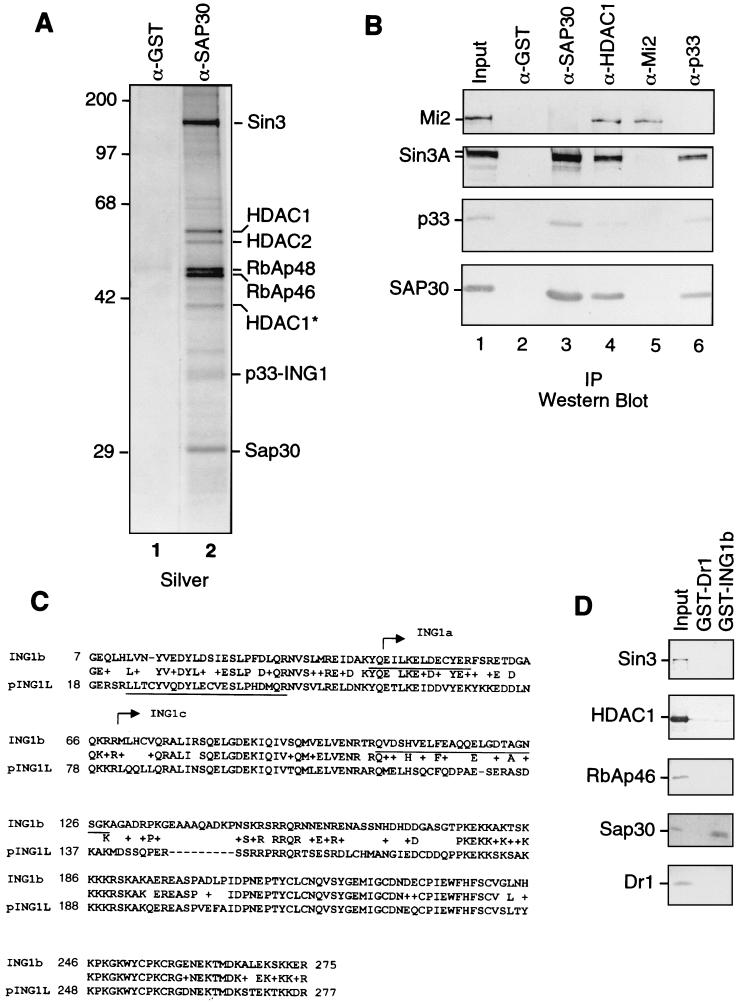
Immunoaffinity purification using anti-HDAC1 antibodies yielded the Sin3 complex but also the novel HDAC1/2-containing complex NuRD (59). Immunoaffinity purification using anti-SAP30 antibodies (62) (Fig. 1) revealed a set of polypeptides that were also retained on an anti-Sin3 column, particularly mSin3a, HDAC1, HDAC2, RbAp46, RbAp48, and SAP30, as well as several polypeptides present in substoichiometric amounts (Fig. 1A). These less abundant polypeptides were consistently observed and were also present in preparations derived from immunopurifications using antibodies against Sin3 and HDAC1 but not in preparations using other antibodies such as MTA2 and Mi2 (data not shown: see reference 59). In light of these findings, we speculated that: (i) the abundant polypeptides correspond to the core HDAC complex (60), composed of HDAC1, HDAC2, RbAp46, and RbAp48, in addition to Sin3 and SAP30; (ii) the less abundant polypeptides represent proteins that associate, directly or indirectly, with the core Sin3-HDAC complex; and (iii) the affinity purification procedure yielded different complexes. Therefore, we identified these polypeptides. The 33-kDa band in the anti-SAP30 immunoprecipitates was subjected to in-gel tryptic digestion and analyzed by MS fingerprinting (8). Two peptides derived from this band (QVDSHVELFEAQQELGDTAGNSGK and YQEILKELDECYER) matched the recently corrected sequence of an isoform of the human growth inhibitor p33ING1b (AF1818501). The human ING1 gene encodes three different transcripts, p47ING1a, p33ING1b, and p24ING1c, all containing a common C-terminal domain but with divergent N-terminal domains (11) (see Fig. 5A). While one peptide obtained from the 33-kDa band matched the sequence of ING1 common to all three isoforms, another peptide was unique for the isoform ING1b (Fig. 1C). Another peptide obtained from the 33-kDa band (LLTCYVQDYLECVESLPHDMQR) did not match p33ING1b but matched exactly the sequence of the protein encoded by the pING1L gene (NP001555), which is ∼50% identical to p33ING1b. This peptide is located at the N terminus of pING1L and thus has homology to the unique region of ING1b.
FIG. 1.
Identification of p33ING1b as a SAP30-associated protein. (A) Silver staining of anti-SAP30 immunoprecipitates. An aliquot of the DEAE-52 bound material (∼100 μg) was immunoprecipitated with anti-SAP30 antibodies, and immunoprecipitated proteins were washed with buffer containing 0.5 M KCl and 0.05% NP-40. Proteins were eluted with 0.1 M glycine (pH 2.6) and separated by SDS-PAGE followed by silver staining. (B) Western blots of anti-SAP30, anti-HDAC1, anti-Mi2, and anti-p33 immunoprecipitates. Immunoprecipitations (IP) were performed as in panel A. Input, 10 μg of the DEAE-52-bound fraction. Lanes 2 to 6 correspond to 1/10 of the total glycine eluate from the corresponding affinity columns. (C) Sequence alignment of p33ING1b and pING1L. Peptides derived from the 33-kDa band in anti-SAP30 immunoprecipitates are underlined. The sequences of p33ING1b shared with other ING1 isoforms (ING1a and ING1c) are indicated by arrows. (D) Direct interaction between p33 and SAP30. GST-p33 or GST-Dr1 proteins (100 ng) were attached to glutathione-Sepharose beads (5 μl) and incubated with 200 ng of recombinant purified HDAC1, RbAp46, SAP30, Dr1, or in vitro-translated Sin3 protein in buffer containing 0.5 M KCl, 0.1% NP-40, and 0.1 mg of BSA per ml. The bound proteins were washed with the same buffer and eluted with SDS loading buffer. A 1/10 aliquot of material bound to the beads was analyzed by Western blotting as indicated in the figure. Input, 20 ng of recombinant protein.
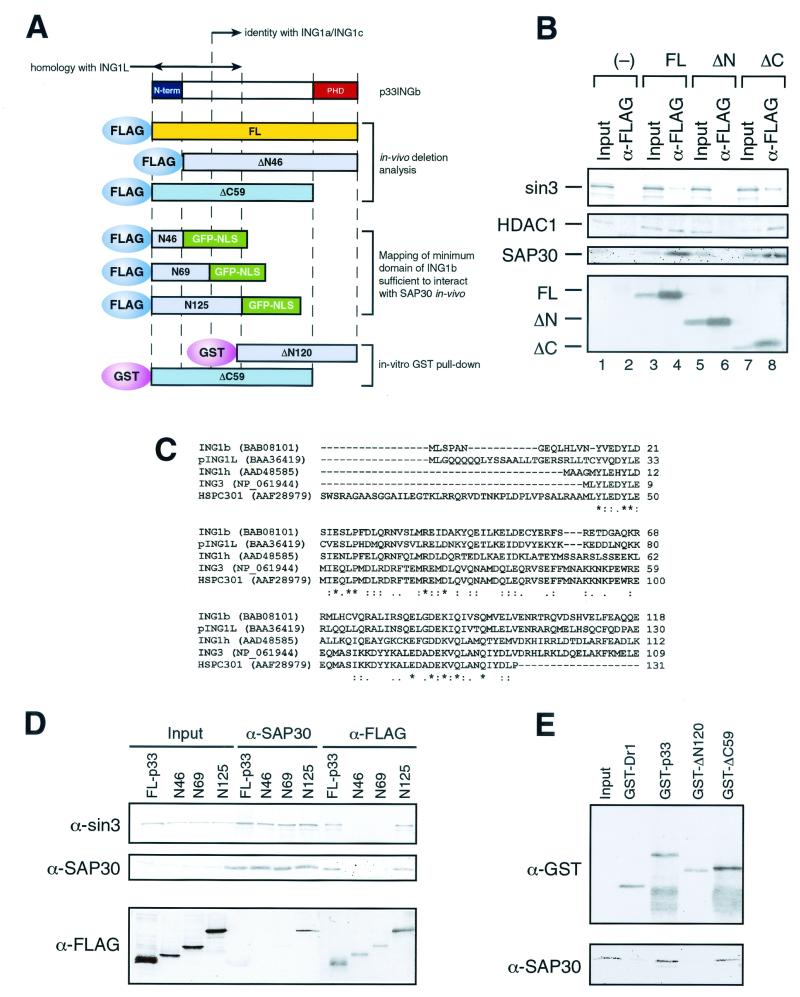
FIG. 5.
The unique N-terminal domain of p33ING1b is required for interaction with the Sin3 complex in vivo. (A) Scheme of ING1 deletion mutants. (B) Western blot analysis of anti-FLAG immunoprecipitates from nuclear extracts of NIH 3T3 cells transfected with FLAG-tagged p33ING1b deletion mutants. Nuclear extracts made from 5 × 107 NIH 3T3 cells transfected with empty vector (−), full-length p33 (FL), and p33 deletion mutants (ΔN and ΔC) were immunoprecipitated with anti-FLAG antibodies. Immunoprecipitated proteins were washed with buffer containing 0.5 M KCl and 0.05% NP-40, eluted with SDS loading buffer, and analyzed by Western blotting using antibodies specific for Sin3,HDAC1, and SAP30. Input corresponds to 1/10 of the nuclear extract used for immunoprecipitation, and α-FLAG corresponds to 1/2 of the eluate from the α-FLAG beads. The bottom panel shows expression of the transfected p33ING1b deletion mutants detected using anti FLAG-antibodies. (C) Sequence alignment of proteins containing the domain similar to the N terminus of p33ING1. Alignment was done using the CLUSTALW program at the European Bioinformatics Institute server (http://www2.ebi.ac.uk/clustalw) (48). Asterisks indicate identical positions, colons indicate conserved substitutions, and dots indicates semiconserved (i.e., conserved in most but not all of the aligned sequences) substitutions. (D) Identification of a minimum domain of p33ING1b sufficient to interact with the Sin3 complex in vivo. 239T cells were transfected with either full-length FLAG-tagged p33 protein (FL-p33) or FLAG-tagged fusion proteins containing the N-terminal 46, 69, or 125 amino acids of the p33 protein fused with GFP (N46, N69, and N125). Nuclear extracts from transfected cells (∼1 mg) were immunoprecipitated either with anti-SAP30 or with anti-FLAG antibody, and aliquots of immunoprecipitated proteins (1/2) were analyzed by Western blotting as indicated on the left side of the panel. Inputs correspond to 5 μg (1/200) of nuclear extracts used for immunoprecipitation. (E) The N terminus of p33 is required for the interaction with SAP30 in vitro. GST pull-down was performed as described in the legend to Fig. 1D, using 100 ng of the indicated GST fusion protein and 100 ng of recombinant SAP30. Aliquots of the reactions (1/2) were analyzed by Western blotting using anti-GST and anti-SAP30 antibodies. Input corresponds to 10 ng of recombinant SAP30.
Next, we analyzed whether p33ING1b is a component of the Sin3-SAP30 complex or interacts with SAP30 independently of Sin3. To address this question, we used immunoaffinity purification with antibodies against components of the Sin3 and NuRD complexes in addition to antibodies against p33ING1b.
As the input material for immunoaffinity purification, we used a crude protein fraction enriched for Sin3, SAP30, and ING1b. After immunoadsorption of the proteins to different immunoaffinity resins, the bound proteins were eluted and analyzed by Western blotting. As expected, antibodies against SAP30 immunoprecipitated the known components of the Sin3 complex. Additionally, the SAP30 antibodies coimmunoprecipitated p33-ING1b (Fig. 1B) but not the NuRD component Mi2β. Consistent with this observation, antibodies against Mi2β failed to immunoprecipitate p33ING1b. In a reciprocal experiment, anti-ING1b antibodies coimmunoprecipitated Sin3 and SAP30 but failed to immunoprecipitate Mi2β. Furthermore, antibodies against HDAC1, along with the components of Sin3 and NuRD complexes, also coimmunoprecipitated p33ING1b (Fig. 1B). These results demonstrate that ING1b is an integral component of a Sin3-HDAC complex.
Given that the Sin3 complex contains several subunits that are shared with another deacetylase complex, NuRD, which does not interact with p33, we wanted to find what determines the specificity of association of p33 with the Sin3 complex. To this end, we analyzed the interactions between p33 and various subunits of the Sin3 complex using a GST pull-down assay with recombinant purified proteins (Fig. 1D). In this assay, we detected a specific and direct interaction between p33 and SAP30. No direct interaction between p33 and Sin3, HDAC1, or RbAp46 was detected. Therefore, it is likely that a direct interaction with SAP30 mediates the association of p33ING1b with the Sin3 complex.
Biochemical separation of two ING1b-Sin3-containing complexes.
Having established that p33ING1b is a component of a Sin3 complex, we next wanted to analyze whether p33ING1b is present in a single or multiple complexes. This was analyzed using conventional chromarography of HeLa cell extracts coupled with affinity purification. We found that ING1b copurifies with Sin3 and SAP30 on various chromatographic resins (data not shown [see below]). The DEAE-5PW cation-exchange resin proved to be particularly informative in resolving different p33ING1b-containing complexes (Fig. 2A). We noticed that a significant fraction of Sin3 and SAP30 was found in the flowthrough of the column. This fraction was relatively devoid of p33ING1b, which bound to the column, and was resolved into two discrete peaks coeluting with Sin3 and SAP30 (Fig. 2B and data not shown). This feature appears specific to Sin3-p33ING1b, since NuRD fractionates as a single peak, displaced from Sin3 (data not shown).
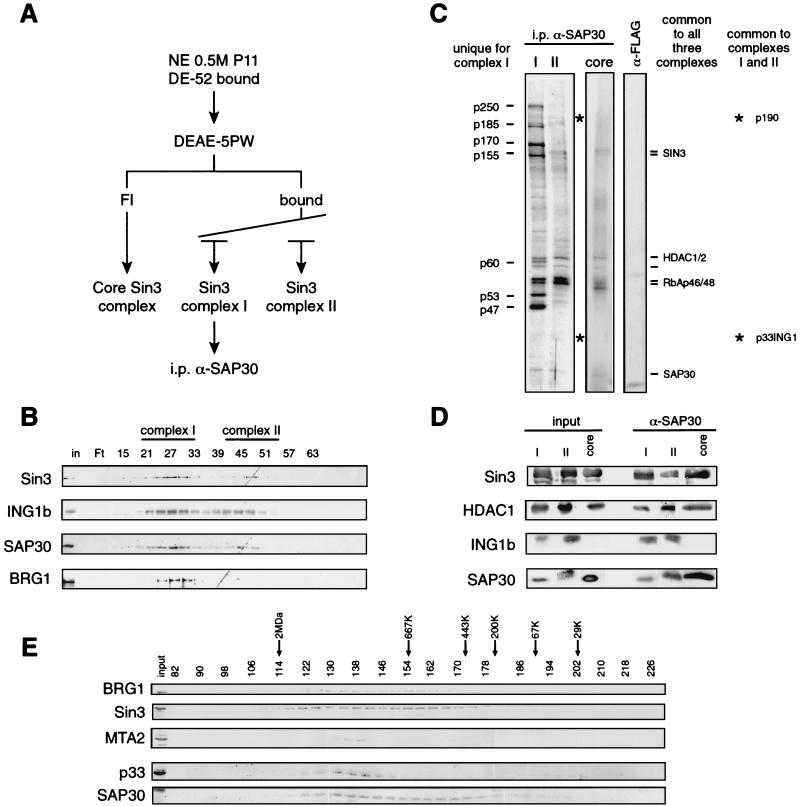
FIG. 2.
p33ING1b is the component of a subset of Sin3-HDAC complexes. (A) Scheme of the immunoaffinity purification of the different SAP30-containing complexes, i.p., immunoprecipitation. (B) Western blot analysis of the DEAE-5PW column fractions. The column was developed as indicated in Materials and Methods. Aliquots of the fractions (5 μl) were separated by SDS-PAGE, and the fractionation of the indicated polypeptides was analyzed by Western blotting using antibodies against Sin3, SAP30, and p33ING1b. Pools used for subsequent anti-SAP30 and anti-p33 immunoprecipitations are indicated (complex I and complex II). (C) Silver staining of anti-SAP30 immunoprecipitates from DEAE-5PW fractions corresponding to the three Sin3 complexes. Equal SAP30 Western blot amounts of DEAE-5PW fractions corresponding to the three Sin3 complexes (I, II, and core; ∼100 μg) were immunoprecipitated with anti-SAP30 antibodies. Bound proteins were washed with buffer containing 0.5 M KCl and 0.05% NP-40 and eluted with 0.1 M glycine (pH 2.6). A 1/10 aliquot of eluted proteins was separated by SDS-PAGE and visualized by silver staining. Polypeptides unique to complex I are indicated on the left side of the panel. Other polypeptides in the three pools are indicated on the right side of the panel. (D) Western blot analysis of anti-SAP30 immunoprecipitates from the DEAE-5PW fractions corresponding to the three Sin3 complexes (I, II, and core). Anti-SAP30 immunoprecipitation was performed as in panel C, and bound proteins were analyzed by Western blotting using antibodies indicated in the figure. Inputs correspond to 1/10 (∼10 μg) of the material used for immunoprecipitation, and the α-SAP30 lanes correspond to 1/10 of the corresponding glycine eluates. (E) Western blot analysis of nuclear extract Sephacryl-400 fractions. Unfractionated HeLa nuclear extract (approximately 10 mg of total protein) was fractionated on a 120-ml Sephacryl-400 column in buffer BC (see Materials and Methods) containing 500 mM KCl, 0.1% NP-40, and 40 μg of ethidium bromide per ml. Aliquots (20 μl) of of each fraction (0.5 ml) were analyzed by Western blotting as indicated.
For simplicity, we will refer to the DEAE-5PW flowthrough fraction as the core Sin3 complex (see below) and to the two DEAE-5PW-bound complexes as Sin3 complex I (early peak) and Sin3 complex II (late peak).
Having established the existence of three biochemically distinct Sin3-containing complexes, we next wanted to characterize their polypeptide composition. To this end, we used immunoaffinity purification with antibodies against SAP30 (Fig. 2A). DEAE-5PW fractions corresponding to the different Sin3 complexes were pooled, concentrated, and loaded onto anti-SAP30 immunoaffinity columns. Bound proteins were eluted and analyzed by Western blotting and silver staining. As seen in Fig. 2D, Sin3, SAP30, and HDAC1 were present in all three complexes whereas ING1b was present in complexes I and II but was absent from the core Sin3 complex.
Silver-staining analysis revealed that the different Sin3 complexes have different polypeptide compositions (Fig. 2C). The Sin3 core complex contains HDAC1/2 and RbAp46/48, together with Sin3 and SAP30. Sin3 complex II, in addition to the polypeptides present in the core complex, includes p33ING1b and a 190-kDa polypeptide, which was identified as the retinoblastoma-binding protein 1 (RBP1) (Fig. 2C and data not shown). Recent studies have shown that RBP1 can recruit the HDAC1/2-containing complexes to the pocket of Rb through an interaction with SAP30 (27). Sin3 complex I contains all the polypeptides present in complex II as well as several additional polypeptides. The same set of polypeptides were recovered in complexes I and II after immunoprecipitation using antibodies against p33ING1b (see Fig. 4A).
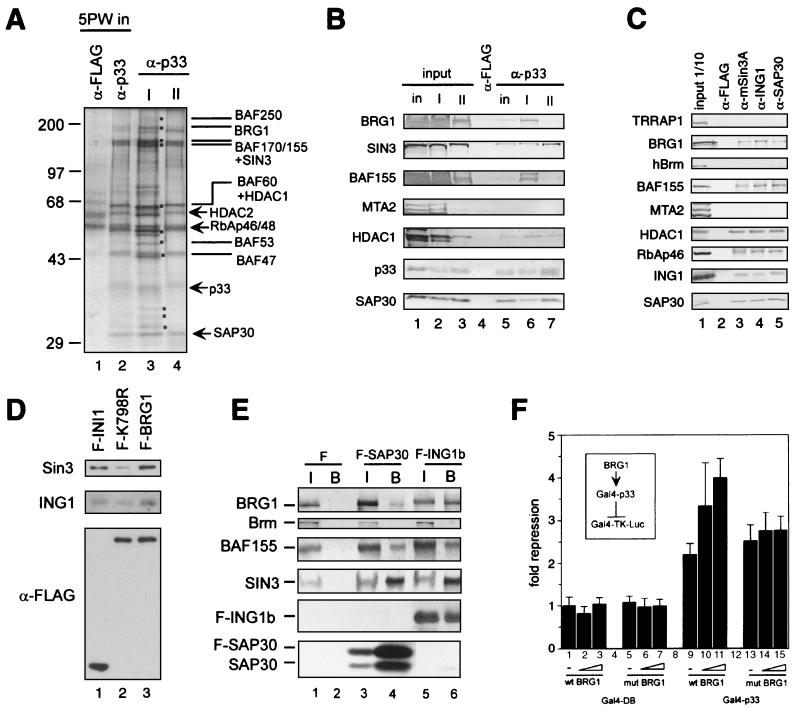
FIG. 4.
Interaction between the Sin3 complex I and the Brg1-based Swi-Snf complex. (A) Silver staining of anti-p33 immunoprecipitates derived from the DEAE-5PW fractions corresponding to Sin3 complexes I and II. Equal p33 Western blot units of DEAE-5PW fractions corresponding to Sin3 complexes I and II or DEAE-5PW input (∼100 μg) were immunoprecipitated with anti-p33 or anti-FLAG antibodies, washed with buffer containing 0.5 M KCl and 0.05% NP-40, and eluted with 0.1 M glycine (pH 2.6). Immunoprecipitated proteins (one-quarter of the total glycine eluate) were resolved by SDS-PAGE and visualized by silver staining. Polypeptides specific for complex I are marked by dots. The subunits of the Sin3 and Swi-Snf complexes are indicated. (B) Western blot of anti-p33 immunoprecipitates from DEAE-5PW input and DEAE-5PW fractions corresponding to Sin3 complexes I and II. Anti-p33 immunoprecipitation from DEAE-5PW input (in) and DEAE-5PW fractions corresponding to Sin3 complexes I (I) and II (II) was performed as described in the legend to panel A, followed by Western blot analysis using different antibodies described on the left side of the panel. Input lanes correspond to 10% of the material used for immunoprecipitation. Lanes corresponding to α-p33 immunoprecipitates contain one-quarter of the total glycine eluates. As a negative control, anti-FLAG immunoprecipitation was performed from the DEAE-5PW input material (column labeled α-FLAG). (C) Western blots of anti-Sin3, anti-ING1b, and SAP30 immunoprecipitates from HeLa nuclear extract. Approximately 300 μg of nuclear extracts was immunoprecipitated with antibodies, as indicated at the top of the panel. Samples were washed and proteins were eluted as described in the legend to panel A. Half of the total glycine eluate was separated by SDS-PAGE and analyzed by Western blotting using the antibodies indicated on the left side of the panel. (D) Sin3 and p33ING1b are associated with the Brg1-based Swi-Snf complex. Nuclear extracts from HeLa cells stably expressing FLAG-tagged wild-type Brg1 (F-BRG1) and ATPase-deficient mutant (K798R) (F-K798R) and FLAG-tagged INI1 (F-INI1) were fractionated on an anti-FLAG agarose column, and bound proteins were eluted with excess FLAG peptide and analyzed by Western blotting as described previously (42). To ensure equal efficiency of immunoprecipitation of FLAG-tagged INI1, Brg1, and Brg1(K798R), the same immunoprecipitates were analyzed by Western blotting with anti-FLAG antibodies (bottom panel). Cell lines have been described previously (42, 43). (E) Endogenous Brg1 and BAF155 but not Brm coimmunoprecipitate with transiently overexpressed FLAG-tagged SAP30 and p33ING1b. Nuclear extracts were prepared from 5 × 107 293T cells transfected with empty vector (F), FLAG-SAP30 (F-SAP30), or FLAG-p33ING1b (F-ING1b) expression vectors. These extracts (300 μg) were immunoprecipitated with anti-FLAG antibodies, and bound proteins were washed with buffer containing 0.5 M KCl and 0.05% NP-40 and eluted with the SDS loading buffer followed by Western blot analysis using antibodies described on the left side of the panel. Input (I) corresponds to 1/10 of the nuclear extract used for immunoprecipitation, whereas the α-FLAG bead-bound fraction (B) corresponds to 1/2 of the material eluted from the beads. (F) Brg1 enhances repression by Gal4-p33ING1b. 293T cells (106 cells per 6-cm plate) were transfected with a luciferase reporter driven by a promoter containing five copies of the Gal4 DNA binding site (Gal-TK-Luc; 250 ng), together with a Gal4-p33 fusion protein (Gal4-p33; 500 ng) or Gal4 DNA-binding domain alone (Gal4-DB; 500 ng) in the presence or absence of the Brg1 expression vector (wtBRG1, or mutBRG1; 250, 500, or 1,000 ng). Luciferase activity was measured as described in Materials and Methods. Fold repression was normalized to reporter activity in the absence of Gal4-p33.
Therefore, the three biochemically distinct human Sin3 complexes have different polypeptide compositions. The DEAE-5PW flowthrough Sin3 complex contains mSin3A, HDAC1/2, RbAp46/48, and SAP30 and probably corresponds to the previously reported core Sin3 complex (58). Sin3 complex II contains ING1b and p190, in addition to the polypeptides present in the core complex. Sin3 complex I additionally contains seven polypeptides, p250, p185, p170, p155, p60, p53, and p47.
To address whether the different complexes described above represent breakdown products of a larger complex, size fractionation analysis of the unfractionated HeLa nuclear extract was performed on a Sephacryl-400 column (Fig. 2E). This analysis demonstrates that Sin3 elutes broadly, probably in at least two peaks. SAP30 also elutes in a broad peak and is contained within most of the Sin3-containing fractions. Importantly, p33 coelutes only with the early Sin3 peak and is absent from the later fractions where both Sin3 and SAP30 coeluted. Therefore, the p33-devoid core Sin3 complex is present in the unfractionated HeLa nuclear extract and is probably not a result of breakdown but is a reflection of other Sin3-containing complexes. In this analysis it is difficult to distinguish Sin3-p33 complexes I and II; however, the elution profile of p33 is much broader than the peak of MTA2, a subunit of the ∼1.5-MDa NuRD complex. Therefore, it is likely that the p33-containing fractions include both p33/Sin3-containing complexes. Importantly, the majority of the p33 protein present in HeLa nuclear extract is contained in the high-molecular-weight Sin3 complexes, whereas only a small part of p33 was detected in fractions that correspond to the monomeric p33 protein.
HDAC and nucleosome deacetylase activity of the three Sin3 complexes.
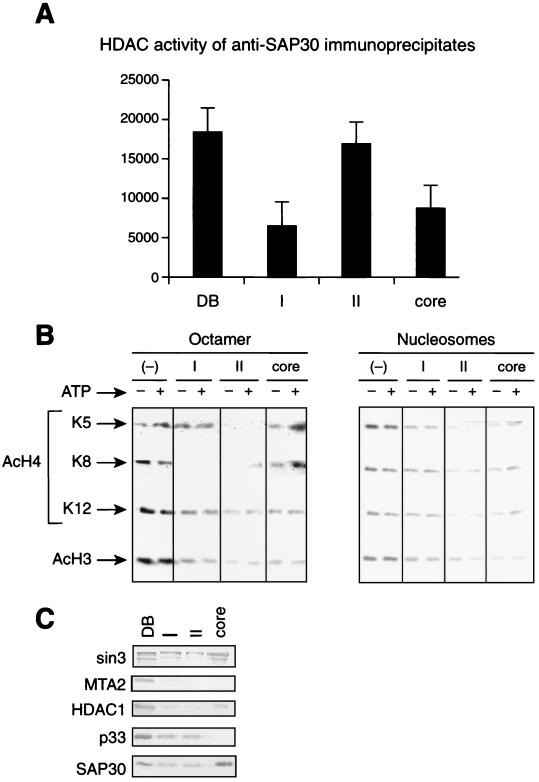
To analyze whether the three Sin3 complexes represent functional HDAC enzymes, we assayed their activity using core histones and oligonucleosomes as substrate (Fig. 3). First, we analyzed the HDAC activity of the different Sin3 complexes adsorbed onto anti-SAP30 beads by measuring the 3H release from labeled core histones (Fig. 3A). Equal Western blot units (anti-HDAC1 and anti-SAP30, Fig. 3C) of the different complexes were used. The results demonstrate that all three complexes possess comparable HDAC activity, with Sin3 complex II being somewhat more active than the other two complexes. Next, we analyzed the nucleosome deacetylation activity of the three Sin3 complexes and their specificity to particular acetylation sites by using antibodies that recognize specific acetylated lysines (Fig. 3B). Since nucleosome deacetylation activity is inefficient when complexes are attached to beads (62), we used the native Sin3 complexes. The core Sin3 complex was moderately active on H3 and H4 lysine 12, whereas it was almost inactive on H4 lysines 5 and 8, in both the octamer and in oligonucleosomes. Sin3 complex II possessed significant deacetylase activity on all H4 and H3 sites tested in both the octamer and oligonucleosome, with a particular preference for H4 lysines 5 and 8. Sin3 complex I was active only in deacetylation of H4 lysine 8, and the activity was blocked in oligonucleosomes.
FIG. 3.
HDAC and nucleosome deacetylase activity of the different Sin3 complexes. (A) 3H release assay using core histones. Equal SAP30 Western blot units of each of the Sin3 complexes immunoprecipitated from the DEAE-5PW column fractions using anti-SAP30 antibodies as described in the legend to Fig. 2 (I, II, and core) were incubated with [3H]acetyl-coenzyme A-labeled core histones (0.5 μg, 3 × 107 dpm/μg). Deacetylation activity was measured by quantification of the amount of [3H]acetate released from histones. Anti-SAP30 immunoprecipitate from the DE-52 bound fraction (DB) was used as a positive control. (B) Assay of oligonucleosome deacetylase activity of the native Sin3 complexes I, II, and core, using antibodies specific to particular acetylated lysines. Equal SAP30 Western blot units of partially purified Sin3 complexes were incubated with hyperacetylated core histones or oligonucleosomes (average, five nucleosomes) in the presence or absence of 4 mM ATP, and histone deacetylation was analyzed by Western blotting with antibodies that recognize particular acetylated lysine residues. As a negative control, the same amount of substrate was incubated with a crude fraction devoid of SAP30-HDAC1 (−). (C) Western blot analysis of the partially purified Sin3 complexes (I, II, and core) used for the HDAC assays in panel B. DB corresponds to 5 μg of the DEAE-5PW input. Each lane corresponds to the amount of fraction used for HDAC assays in panel B.
Sin3 complex I contains components of the Brg1-based Swi-Snf complex.
Both anti-SAP30 and anti-p33 immunioprecipitates from Sin3 complex I displayed a subset of polypeptides that were absent from complex II or core complex; these are p250, p185, p170, p155, p60, p53, and p47 (Fig. 2C and 4A). To identify these polypeptides, we performed MS fingerprinting. Of 20 peptides derived from p185, 18 matched perfectly with peptides found in the human Brg1 protein (NCBI 4507073). Interestingly, no peptides corresponding to Brm were detected. Eleven peptides derived from the p170 band matched BAF170 (NCBI 4507081), whereas 9 of 24 peptides obtained from p155 matched mSin3A and 7 matched BAF155 (NCBI 4507081). Therefore, approximately stoichiometric amounts of mSin3A- and BAF155-derived peptides were obtained from the p155 band. This result suggests that the Swi-Snf components are associated with Sin3 complex I. The p60, p55, and p47 polypeptides present in complex I were identified as subunits of the Swi-Snf complex by Western blot analysis (data not shown). The MS identification was confirmed by Western blot analysis. Both Brg1 and BAF155 were readily detected in anti-p33 and anti-SAP30 immunoprecipitates derived from Sin3 complex I but not from Sin3 complex II or the core Sin3 complex (Fig. 4B and data now shown). To analyze whether SAP30 and p33 are present in a subset of Swi-Snf complexes independently of Sin3, we performed immunoprecipitation from HeLa nuclear extracts using antibodies against Sin3, p33, and SAP30 and detected BRG1 and BAF155 in each immunoprecipitate (Fig. 4C). In this analysis we also analyzed for the presence of TRRAP1, the human homologue of the Tra1 subunit of the yeast NuA4 complex, which has been suggested to interact with the yeast homologue of p33ING1b (30). Our immunoprecipitation analysis failed to detect TRRAP1 (Fig. 4C).
Next we wanted to analyze whether the subunits of the Sin3 complex could be detected in purified human Swi-Snf complexes. For this purpose, we used the HeLa-derived cell lines stably expressing FLAG-tagged Swi-Snf components INI1, Brg1, and an ATPase-deficient Brg1 mutant (43). As seen from Fig. 4D, Swi-Snf immunopurified through a tag on INI1 or Brg1 also contained Sin3 and p33ING1b. Interestingly, Sin3 and p33ING1b were partially depleted from the Swi-Snf complex immunopurified from a cell line carrying a FLAG tag on the ATPase-deficient mutant of Brg1 (Fig. 4D). The decreased levels of Sin3 and p33ING1b in the mutant Brg1-based Swi-Snf complex were not due to instability of the mutant protein or to decreased levels of Brg1 expression (Fig. 4D).
Further support of a physical association between the p33ING1-Sin3 and Swi-Snf complexes was obtained from experiments utilizing transient overexpression of the components of these complexes. Nuclear extracts were prepared from 293T cells transiently transfected with expression vectors carrying FLAG-tagged SAP30 or FLAG-tagged ING1b followed by immunoprecipitation using anti-FLAG antibodies. Western blot analysis revealed that the endogenous Brg1 and BAF155 subunits, but not Brm, are specifically coimmunoprecipitated with overexpressed FLAG-SAP30 or FLAG-p33ING1b (Fig 4E). These studies collectively establish that the p33ING1b-Sin3 complex interacts with the Brg1-based Swi-Snf complex in vivo.
What is the physiological significance of the interaction between the Sin3-ING1b and the Swi-Snf complexes? There is increasing evidence of the role of Swi-Snf complexes in transcriptional repression both in yeast and in mammals. Particularly, the human Swi-Snf complex facilitates repression of the c-fos promoter (35) and cooperates with HDACs in repression of certain Rb-regulated promoters (57). Moreover, recent results have established that Sin3 is associated with Swi-Snf subunits in vivo (42). Therefore, we asked if overexpression of Brg1 could affect the repression of a reporter gene by a tethered Sin3-HDAC complex. To this end, we transfected 293T cells with a luciferase reporter driven by a promoter containing five copies of the Gal4 DNA-binding site together with a Gal4-p33 fusion protein in the presence or absence of the Brg1 expression vector. As seen in Fig. 4F, expression of Gal4-p33 resulted in modest repression of reporter activity (ca. twofold), which has been shown to be due to the recruitment of the Sin3-HDAC complex (44). This repression could be further stimulated by overexpression of Brg1, whereas overexpression of Brg1 in the absence of Gal4-p33 had no effect on the reporter expression. Furthermore, Brg1 containing a mutation in the ATP-binding domain was unable to stimulate repression by Gal4-p33. Therefore, Brg1 was able to stimulate repression by a tethered p33-Sin3 complex in a manner dependent on its ATPase activity.
The unique N-terminal domain of p33ING1b is required for interaction with SAP30.
Human p33ING1 was identified as a negative growth regulator (11). Therefore, we wanted to analyze whether the interaction of p33 with the Sin3-HDAC complex was important for growth suppression activity. To this end, we first identified the domain of p33ING1b necessary for association with the Sin3 complex in vivo.
Full-length p33ING1b and deletion mutants in which the PHD (ΔC) or the unique N terminus (ΔN) were deleted were cloned into the pEF6 mammalian expression vector in frame with the FLAG epitope (Fig. 5A). These constructs were transfected into NIH 3T3 cells, and nuclear extracts from transfected cells were prepared and used for immunoprecipitation with anti-FLAG antibodies. The ability of p33ING1b deletion mutants to be incorporated into Sin3 complexes was monitored by coimmunoprecipitation of endogenous Sin3, HDAC1, and SAP30. As seen in Fig. 5B, overexpressed full-length p33ING1b was coimmunoprecipitated with endogenous Sin3, HDAC1, and SAP30 whereas the N-terminal deletion mutant was not, despite equal levels of expression. The deletion of the conserved C terminus, containing the PHD Zn finger motif, did not impair the ability of p33ING1b to be incorporated into Sin3 complexes.
The result that the unique N terminus, rather than the conserved C terminus of p33 which contains the PHD zinc finger, was responsible for interaction with the Sin3-SAP30 complex prompted us to analyze the database for the presence of proteins that contain sequence similarity to the N terminus of p33ING1b.
To this end, we used the 46 N-terminal amino acids of p33ING1b to search the nonredundant protein database (NCBI) using the BLAST algorithm (2). This search yielded a set of proteins from different species showing at least 45 to 50% identity to p33ING1b through the entire length of the N terminus. In particular, a total of five human proteins share this motif: p33ING1b, pING1L, and three uncharacterized proteins, AAD48585 (named ING1 homologue, or ING1h). NP061944 (named ING1 family member 3, or ING3), and HSPC301 (Fig. 5C).
Interestingly, with the exception of the small protein HSPC301, all the other human proteins detected also have the PHD Zn finger motif at their C termini, and in the case of NP061944, a serine-rich spacer separates these two domains (data not shown). Similarly, an uncharacterized Drosophila protein, CG6632, which is related to NP061944, contains a long histidine-rich spacer between its N terminus and the PHD finger (data not shown). The presence of a variable, nonconserved spacer supports the notion that the N terminus of p33ING1b contains a defined protein motif, which we tentatively called SAID (for SAP30-interacting domain).
Whereas p33ING1b, pING1L, ING1h, and ING3 contain both the N-terminal motif and a PHD finger, other human ING1 isoforms (ING1a and ING1c) contain only the PHD finger (Fig. 5A). In contrast, HSPC301 protein contains only the N-terminal domain and lacks the PHD finger. This protein probably represents an alternatively spliced form of NP061944. These observations suggest that these proteins define a family containing two functionally important motifs: the N-terminal SAID motif and the PHD finger, which might be involved in combinatorial regulation of chromatin structure.
While the deletion of the N-terminal 46 amino acids, corresponding to ca. one-third of SAID, abrogates the interaction of ING1b with the Sin3 complex, this does not demonstrate that this domain is responsible for the interaction with Sin3 complex. We therefore investigated the minimum portion of p33ING1b that is sufficient to interact with the Sin3 complex when fused to a heterologous protein. We used p33 truncations containing the N-terminal 46, 69, and 125 amino acids (which correspond to unique part of ING1b [N46], part of ING1b different from ING1c [N69], and the complete SAID domain [N125], respectively). These protein fragments were fused to a GFP containing three copies of simian virus 40 T antigen nuclear localization signal and an N-terminal FLAG epitope. DNA constructs expressing these recombinant proteins were transfected into 293T cells, and their association with endogenous Sin3 complexes was tested using immunoprecipitation with anti-FLAG or anti-SAP30 antibodies. As seen in Fig. 5D, the N46 or N69 fusion proteins were unable to interact with the Sin3 complex; however, the N125-GFP fusion was coimmunoprecipitated with Sin3 and SAP30 as efficiently as the full-length p33ING1b protein was. Therefore, the full-length SAID is necessary and sufficient for the interaction with the Sin3 complex in vivo.
Since we have established that SAP30 is the only subunit of the Sin3 complex that makes a direct contact with p33, we investigated whether deletion of SAID abrogates the interaction between p33ING1b and SAP30 in vitro. In agreement with the results presented above, the deletion of SAID abrogated the interaction with recombinant SAP30, as demonstrated using a GST pull-down assay (Fig. 5E). Importantly, and in full agreement with the in vivo results presented above (Fig. 5B), deletion of the p33ING1b C-terminal PHD finger did not affect the interaction with SAP30.
Therefore, the N terminus of p33ING1b contains a putative protein-protein interaction domain which is necessary and sufficient to interact with the Sin3 complex in vivo, and this interaction is mediated by a direct contact with SAP30.
Interaction of p33ING1b with the Sin3-HDAC complex is important for growth suppression.
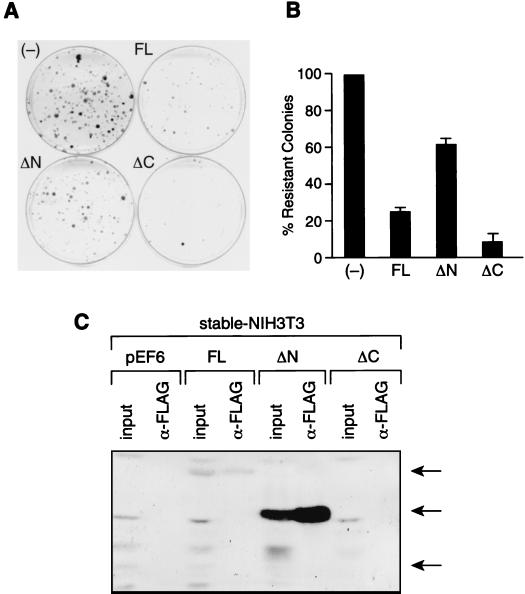
To address whether the interaction between the Sin3-SAP30 complex and p33ING1b is important in regulating cell growth, we used the colony formation assay. NIH 3T3 cells were transfected with the pEF6 vector carrying various deletion mutants of p33ING1b, and colonies were selected on blasticidin for 2 weeks. Expression of full-length p33ING1b results in a ca. fourfold reduction in colony formation (Fig. 6A and B). Therefore, in NIH 3T3 cells, p33ING1b acts as a growth suppressor. Importantly, the deletion of the N-terminal domain of p33ING1b, which is required for the interaction with the Sin3 complex, partially (ca. threefold) reduced the growth suppression effect of p33ING1b. Interestingly, deletion of the PHD finger, which is dispensable for the interaction with the Sin3 complex, did not reduce but, rather, enhanced the growth suppression effect of p33ING1b. To confirm that high levels of expression of p33ING1b are incompatible with cell survival, we analyzed the expression of the transfected deletion mutants in nuclear extracts prepared from equal numbers of cells that have survived blasticidin selection. In clones stably transfected with full-length p33, extremely low levels of FLAG-p33 were detected, whereas no expression was detected in the few clones surviving stable transfection with the C-terminal deletion mutant (Fig. 6C). In contrast, the N-terminal deletion mutant was expressed at high levels in the blasticidin-resistant clones, confirming that deletion of the N terminus significantly impairs the growth suppression properties of p33ING1b.
FIG. 6.
Interaction of p33ING1b with the Sin3-HDAC complex is important for growth suppression. (A) Colony formation assay with NIH 3T3 cells stably transfected with p33ING1b deletion mutants. NIH 3T3 cells were transfected with empty pEF6 vector (−), pEF6 vector containing full-length p33 (FL), or pEF6 vector containing p33 deletion mutants ΔN and ΔC. Cells expressing pEF6 constructs were selected on blasticidin for 2 weeks, and resistant colonies were stained with Coomassie brilliant blue. (B) Quantification of data in panel A. Blasticidin-resistant colonies were counted manually. Error was determined based on three independent experiments. (C) Analysis of the expression of p33 deletion mutants in transfected cells that survived selection on blasticidin. Nuclear extracts were prepared from equal numbers of blasticidin-resistant cells (mass cultures) that have been stably transfected with empty vector (pEF6) or with pEF6 vector carrying different p33 deletion mutants (FL, ΔN, and ΔC). Aliquots of these extracts (30 μg) were analyzed by Western blotting using anti-FLAG antibodies (columns labeled input). Also, aliquots of these extracts (300 μg) were immunoprecipitated with anti-FLAG antibodies followed by Western blot analysis (columns labeled α-FLAG).
These results suggest that p33ING1b is a negative growth regulator and that its ability to inhibit cell growth depends on its interaction with the Sin3-HDAC complex.
Concluding remarks.
In these studies we demonstrate that the candidate tumor suppressor p33ING1b, which interacts in vivo with p53, is a subunit of a Sin3-HDAC complex and directly interacts with SAP30. Our findings are in agreement with the studies described by Skowyra et al. (44) and Loewith et al. (31), which demonstrated that p33ING1b, and the yeast homologue Pho23, respectively, are subunits of a Sin3-HDAC complex. However, our studies have been extended to demonstrate that p33 is present in a subset of cellular Sin3 complexes and that the p33-containing Sin3 complex interacts with the Brg1-based Swi-Snf nucleosome-remodeling complex. This latter finding is in agreement with the results of Sif et al. (42). Importantly, however, our studies demonstrate that the interaction with the Brg1 complex is specific for the p33-containing Sin3 complex. We also demonstrate that the N terminus of p33ING1b is both necessary and sufficient for interaction with the Sin3-HDAC complex in vivo and is required for direct interaction with SAP30 in vitro. Importantly, the N-terminal domain of p33 appears to be present in a family of proteins. We suggest that the N terminus of p33ING1b constitutes a domain that mediates interaction with the Sin3-HDAC complex through SAP30. We have termed this domain SAID (for SAP30-interacting domain).
Acknowledgments
We thank R. Kingston and S. Sif for providing us with the data presented in Fig. 4d and for providing mammalian expression vectors encoding wild-type and mutant Brg1 proteins, D. Allis for providing antibodies specific to acetylated histones, K. Riabowol and A. Gudkov for providing p33 cDNA, and L. Lacomis for helping with mass spectrometric analysis.
This work was supported by grants from Howard Hughes Medical Institute and NIH (grant GM-48518) to D. Reinberg and NCI Cancer Center (grant P30 CA08748) to P. Tempst.
REFERENCES
- 1.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePlnho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49–55. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausio, J., and K. E. van Holde. 1986. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry 25:1421–1428. [DOI] [PubMed] [Google Scholar]
- 4.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by temary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767–776. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454–457. [DOI] [PubMed] [Google Scholar]
- 6.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1–16. [DOI] [PubMed] [Google Scholar]
- 7.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582–598. [DOI] [PubMed] [Google Scholar]
- 8.Erdjument-Bromage, H., M. Lui, L. Lacomis, A. Grewal, R. S. Annan, D. E. McNulty, S. A. Carr, and P. Tempst. 1998. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. Ser. A 826:167–181. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Q., and Y. Zhang. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenyo, D., J. Qin, and B. T. Chait. 1998. Protein identification using mass spectrometric information. Electrophoresis 19:998–1005. [DOI] [PubMed] [Google Scholar]
- 11.Garkavtsev, I., I. A. Grlgorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov, and A. V. Gudkov. 1998. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391:295–298. [DOI] [PubMed] [Google Scholar]
- 12.Garkavtsev, I., A. Kazarov, A. Gudkov, and K. Riabowol. 1996. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 14:415–420. [DOI] [PubMed] [Google Scholar]
- 13.Geromanos, S., G. Freckleton, and P. Tempst. 2000. Tuning of an electrospray ionization source for maximum peptide-ion transmission into a mass spectrometer. Anal. Chem. 72:777–790. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Harper, S. E., Y. Qiu, and P. A. Sharp. 1996. Sin3 corepressor function in Myc-induced transcription and transformation. Proc. Natl. Acad. Sci. USA 93:8536–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43–48. [DOI] [PubMed] [Google Scholar]
- 17.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451–455. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendzel, M. J., G. P. Delcuve, and J. R. Davie. 1991. Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem. 266:21936–21942. [PubMed] [Google Scholar]
- 20.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397–404. [DOI] [PubMed] [Google Scholar]
- 21.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD- dependent histone deacetylase. Nature 403:795–800. [DOI] [PubMed] [Google Scholar]
- 22.Jager, D., E. Stockert, M. J. Scanlan, A. O. Gure, E. Jager, A. Knuth, L. J. Old, and Y. T. Chen. 1999. Cancer-testis antigens and ING1 tumor suppressor gene product are breast cancer antigens: characterization of tissue-specific ING1 transcripts and a homologue gene. Cancer Res. 59:6197–6204 [PubMed] [Google Scholar]
- 23.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339–2352. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294. [DOI] [PubMed] [Google Scholar]
- 25.Kuzmichev, A., and D. Reinberg. 2001. Role of histone deacetylase complexes in the regulation of chromatin metabolism. Curr. Top. Microbiol. Immunol. 254:35–58. [DOI] [PubMed] [Google Scholar]
- 26.Laherty, C. D., A. N. Billin, R. M. Lavinsky, G. S. Yochum, A. C. Bush, J. M. Sun, T. M. Mullen, J. R. Davie, D. W. Rose, C. K. Glass, M. G. Rosenfeld, D. E. Ayer, and R. N. Elsenman. 1998. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 2:33–42. [DOI] [PubMed] [Google Scholar]
- 27.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSin3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21:2918–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, A., J. M. Lee, W. M. Yang, J. A. DeCaprio, W. G. Kaelin, Jr., E. Seto, and P. E. Branton. 1999. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol. 19:6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landry, J., J. T. Slama, and R. Sternglanz. 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278:685–690. [DOI] [PubMed] [Google Scholar]
- 30.Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol, and D. Young. 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20:3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman, J. D. Boeke, and D. Young. 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 29:24068–24074. [DOI] [PubMed] [Google Scholar]
- 32.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260. [DOI] [PubMed] [Google Scholar]
- 33.Mann, M., P. Hojrup, and P. Roepstorff. 1993. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol. Mass Spectrom. 22:338–345. [DOI] [PubMed] [Google Scholar]
- 34.Mann, M., and M. Wilm. 1994. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66:4390–4399. [DOI] [PubMed] [Google Scholar]
- 35.Murphy, D. J., S. Hardy, and D. A. Engel. 1999. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 19:2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373–380. [DOI] [PubMed] [Google Scholar]
- 37.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Elsenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389. [DOI] [PubMed] [Google Scholar]
- 38.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831–835. [DOI] [PubMed] [Google Scholar]
- 40.Sadowski, I., and M. Ptashne. 1989. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 17:7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber-Agus, N., L. Chin, K. Chen, R. Torres, G. Rao, P. Guida, A. I. Skoultchi, and R. A. DePinho. 1995. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80:777–786. [DOI] [PubMed] [Google Scholar]
- 42.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sif, S., P. T. Stukenberg, M. W. Kirschner, and R. E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skowyra, D., M. Zeremski, N. Neznanov, M. Li, Y. Choi, M. Uesugi, C. A. Hauser, W. Gu, A. V. Gudkov, and J. Qin. 2001. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 276:8734–8739. [DOI] [PubMed] [Google Scholar]
- 45.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41–45. [DOI] [PubMed] [Google Scholar]
- 47.Sun, Z. W., and M. Hampsey. 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152:921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917–921. [DOI] [PubMed] [Google Scholar]
- 50.Tyler, J. K., and J. T. Kadonaga. 1999. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell 99:443–446. [DOI] [PubMed] [Google Scholar]
- 51.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843–846. [DOI] [PubMed] [Google Scholar]
- 53.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117–2130. [DOI] [PubMed] [Google Scholar]
- 55.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 2:851–861. [DOI] [PubMed] [Google Scholar]
- 56.Zeremski, M., J. E. Hill, S. S. Kwek, I. A. Grigorian, K. V. Gurova, I. V. Garkavtsev, L. Diatchenko, E. V. Koonin, and A. V. Gudkov. 1999. Structure and regulation of the mouse ing1 gene. Three alternative transcripts encode two phd finger proteins that have opposite effects on p53 function. J. Biol. Chem. 274:32172–32181. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79–89. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357–364. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279–289. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343–2360. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1:1021–1031. [DOI] [PubMed] [Google Scholar]