Abstract
Minisatellite DNA is repetitive DNA with a repeat unit length from 15 to 100 bp. While stable during mitosis, it destabilizes during meiosis, altering both in length and in sequence composition. The basis for this instability is unknown. To investigate the factors controlling minisatellite stability, a minisatellite sequence 3′ of the human HRAS1 gene was introduced into the Saccharomyces cerevisiae genome, replacing the wild-type HIS4 promoter. The minisatellite tract exhibited the same phenotypes in yeast that it exhibited in mammalian systems. The insertion stimulated transcription of the HIS4 gene; mRNA production was detected at levels above those seen with the wild-type promoter. The insertion stimulated meiotic recombination and created a hot spot for initiation of double-strand breaks during meiosis in the regions immediately flanking the repetitive DNA. The tract length altered at a high frequency during meiosis, and both expansions and contractions in length were detected. Tract expansion, but not contraction, was controlled by the product of the RAD1 gene. RAD1 is the first gene identified that controls specifically the expansion of minisatellite tracts. A model for tract length alteration based on these results is presented.
Repetitive DNA tracts have been divided into categories based on the size of the individual repeat unit. Microsatellite sequences have repeats ranging from a single base pair up to 14 bp in length (58). The control of microsatellite DNA sequence stability is mediated by the postreplicative mitotic mismatch repair (MMR) system (reviewed in references 16 and 57). Minisatellite sequences have repeat units ranging from 15 bp up to approximately 100 bp in length. Unlike microsatellites, which alter primarily during the mitotic cell cycle, presumably during DNA replication minisatellites (also called variable number tandem repeats [VNTRs]) destabilize upon transit through meiosis, undergoing alterations in both length and repeat sequence (reviewed in references 28 and 29). Minisatellite tracts are useful for mapping and DNA fingerprinting. Some minisatellites (MS1 and MS32, for example) show a very high rate of mutation; these types have been utilized for DNA fingerprinting. Minisatellite alterations are often polar in nature (6, 31). One common hypothesis is that alterations in minisatellite sequences are due to a failure during recombination, rather than replication.
One relatively stable minisatellite that has been characterized in detail is the VNTR associated with the HRAS1 gene in humans. When the DNA sequence of the HRAS1 gene was determined (13) from human lymphocytes and a human bladder carcinoma cell line, an approximately 1.4-kb tract of repetitive DNA was detected 3′ of the gene. The original clone included 29 copies of a 28-bp sequence, although it was determined that the tract had shortened during passage through Escherichia coli. Since that time, a large number of alleles of this minisatellite tract have been identified.
In the HRAS1 minisatellite, there are four main types of 28-bp repeats, differing from one another at two positions (+7 and +15, containing either a G or C [see Fig. 1a]). Shortly after the HRAS1 VNTR was described, it was shown that the human population contains four common variant alleles, designated a1 through a4 (44). These four common alleles have different arrangements of the four repeat types within each tract. A large number of less common alleles of the minisatellite have been identified, many from tumor DNAs. Rare alleles appear to derive from one of the common alleles through expansion and alteration of the internal repeats. Sequencing and minisatellite variant repeat PCR have been used to analyze the structure of many of the common and rare alleles (15, 20).
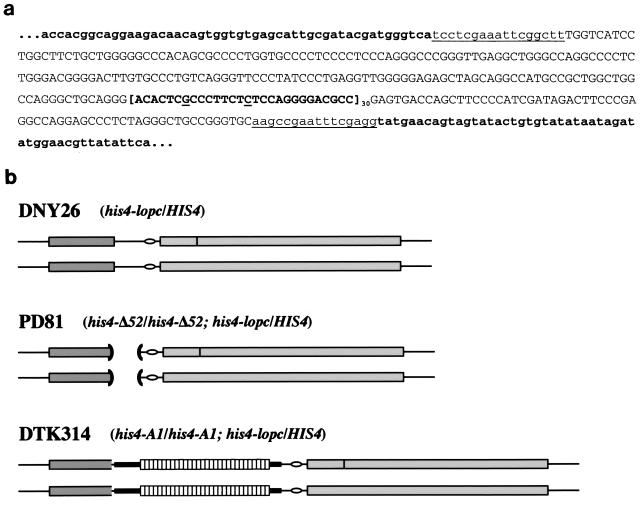
FIG. 1.
The HRAS1 minisatellite sequence and diagram of the HIS4 locus. (a) Sequence of the HRAS1 minisatellite and flanking regions. HIS4 locus sequences are lowercase and in boldface. Lowercase, underlined sequences were inserted during the cloning steps (see Materials and Methods). Capitalized sequences are unique HRAS1 sequences. The capitalized, boldface, bracketed sequence is one HRAS1 minisatellite repeat (type 1)—the underlined letters indicate the two sites of variability between repeats at +7 and +15. As indicated, there are 30 repeats in the human A1a allele utilized for this study. (b) Arrangement of the HIS4 locus in various diploid strains. DNY26 is a diploid strain bearing the wild-type HIS4 promoter sequence and the his4-lopc heterozygous insertion (dark vertical bar) disrupting the HIS4 coding sequence on one chromosome. PD81 is isogenic with DNY26, except that the promoter region has been deleted, as indicated by the open parentheses. DTK314 is also isogenic but contains the HRAS1 minisatellite sequence (white boxes) inserted in place of the region deleted in PD81. Dark gray rectangles are BIK1 coding sequences; light gray rectangles are HIS4 coding sequences; ovals are the TATAA box for HIS4.
Rare alleles of the HRAS1 minisatellite may play a role in the cancer types in which they are found (42, 44). Mutated versions of the HRAS1 gene promote the morphological transformation of cell lines, and inclusion of the minisatellite region increases the efficacy of transformation (14, 60). Gel shift analysis demonstrated that the minisatellite sequence binds a cellular component, the rel/NF-κB transcription factor, specifically the constitutive forms, including p50, p72, and p85 but not p65 (66). Further experiments showed that different Ha-ras VNTR alleles had differing enhancer activities, with some of the rarer alleles having a stronger transcription enhancement than that of the common forms (24), possibly due to enhanced binding of the rel/NF-κB factors. Case studies sought to link the presence of rare alleles with increased risk of various types of cancers, with variable results (see references 9 and 42 to 44).
Alterations in other minisatellite sequences also have been associated with disease phenotypes. For example, progressive myoclonus epilepsy is associated with instability in a minisatellite repeat 5′ of the human cystatin B gene (45, 67). A minisatellite at the IDDM2 locus near the human insulin gene has been connected with insulin-dependent diabetes mellitus (34).
A greater understanding of the mechanisms controlling minisatellite stability and transcription-enhancing activity may provide insight into the etiology of a number of human diseases. However, analysis of the components underlying the meiotic instability and transcription-enhancing activity of the HRAS1 minisatellite is difficult in mammalian systems. To identify factors involved in these processes, we introduced the minisatellite tract into the HIS4 locus of Saccharomyces cerevisiae.
We chose the HIS4 locus because it has been well characterized with respect to both mitotic transcription and meiotic recombination. The HIS4 promoter in our strain background contains the strongest recombination hot spot yet identified in yeast, and characteristics of the HIS4 hot spot and the mechanisms controlling initiation of recombination at HIS4 have been extensively studied (18, 21, 36, 47, 51, 52, 69–72). The HIS4 promoter binds four transcription factors: Rap1p, Bas1p, Bas2p, and Gcn4p (7, 19, 65). Initiation of meiotic recombination requires the binding of three of these factors to the HIS4 promoter: Rap1p, Bas1p, and Bas2p (70, 71). Deletion of any of the gene products or their binding sites at HIS4 eliminates meiotic recombination hot spot activity. However, this dependency on the transcription factors for meiotic recombination is not a requirement for transcription; alterations in the TATAA sequence for HIS4 greatly diminish mRNA production but have no effect on meiotic recombination (69). Therefore, certain transcription factors are capable of stimulating recombination during meiosis independently of their effect on transcription.
The intergenic region between HIS4 and the neighboring gene BIK1 is the site of a meiosis-specific double-strand break (DSB) (21, 46). The DSBs are blunt ended and occur over a short region of approximately 100 bp just 5′ of the binding sites for the HIS4 transcription factors (72). The level of DSBs strongly correlates with the level of hot spot activity as defined genetically by the frequency of aberrant segregation of heterozygous HIS4 mutations (21). Alterations in the wild-type hot spot sequence that reduce the level of aberrant segregation also reduce the level of DSBs, while alterations that increase aberrant segregation also increase DSBs. These results suggest that DSB formation is the initiating step in meiotic recombination.
In this study, we inserted the HRAS1 minisatellite tract in place of the wild-type HIS4 promoter and recombination hot spot. We found that in yeast the minisatellite exhibits many of the phenotypes that it displays in mammalian systems. The insertion stimulates transcription of HIS4, and it acts as a hot spot for meiotic recombination, stimulating the formation of Spo11p-dependent meiosis-specific DSBs and leading to a high level of aberrant segregation of a heterozygous HIS4 alteration. During meiosis, the minisatellite tract increases and decreases in length at high frequency. Furthermore, we find that the meiotic expansions in length, but not the contractions, are under the control of the RAD1 gene. This novel result has direct implications for the mechanism of tract length expansion.
MATERIALS AND METHODS
Media, plasmids, and yeast strains.
Standard media were used (25). Sporulation plates contained 1% potassium acetate, 0.1% yeast extract, 0.05% glucose or galactose, 6 μg of adenine/ml, and 2% agar. Diploids were sporulated at 18°C and dissected onto rich growth medium plates (yeast extract-peptone-dextrose). After colonies formed at 30°C, the plates were replica plated to omission medium plates to determine the segregation patterns of all heterozygous markers. Postmeiotic segregation (PMS) events at HIS4 were detected as sectored His+/His− colonies (18).
The plasmid 37Y8 was used to integrate the HRAS1 minisatellite repeat into the HIS4 locus. To construct the plasmid, PCR was performed on genomic DNA from the EJ bladder carcinoma cell line using the Hras 5′ outer primer and the Hras 3′ outer primer, and the product was ligated into the TA cloning vector pCR2.1. This plasmid was digested with EcoRI, and the ends were filled in using Klenow polymerase. The blunt-ended fragment was inserted into XhoI-digested pPD5 (70) that had also been filled in, creating the plasmid 37Y8. The insert junctions were verified by sequencing. The pPD5 plasmid contains a Sau3AI fragment of HIS4 DNA in which 171 bp of the HIS4 promoter have been deleted and replaced with a XhoI linker (the his4-Δ52 allele [48]). The insertion of the HRAS1 minisatellite into the upstream region of HIS4 created a novel allele of HIS4 that we designated his4-A1.
All strains were derived from the haploid strain AS13 (MATa leu2 ura3 ade6 rme1) or AS4 (MATa trp1 arg4 tyr7 ade6 ura3 spt22) (61). All strains are isogenic except for alterations introduced by transformation. To integrate the HRAS1 minisatellite tract into the HIS4 locus, a two-step integration was performed (56). The plasmid 37Y8 was digested with MfeI and transformed into PD63 (AS4 but his4-Δ52 [18]) and PD57 (AS13 but his4-Δ52 [18]), selecting for Ura+ transformants. Ura− transformants were isolated following growth on 5-fluoroorotic acid medium (10), and strains containing the minisatellite insert were identified by PCR, generating DTK288 and DTK294, respectively. The his4-lopc allele, an insertion of 26 bp into HIS4 that is poorly recognized and repaired during meiosis, was introduced into DTK294 using pDN13 (47), creating DTK305. An isogenic strain containing the wild-type promoter (DNY25 [AS13 but his4-lopc]) has been previously described (47).
DTK288 and DTK305 were transformed with BamHI-digested pDG18 to delete the RAD1 gene, as previously described (37), generating DTK506 and DTK507, respectively. Similarly, DTK288 and DTK305 were transformed with BstXI-digested pJH523 (41) to delete the PMS1 gene, generating DTK514 and DTK516.
For analysis of DSB formation, the rad50S allele was introduced into DTK288 and DTK305 by transformation with BamHI/EcoRI-digested pNKY349 (1), by previously described protocols (21), creating DTK468 and DTK469. Similar procedures were used to generate DTK137 and DTK138, rad50S derivatives of PD63 and PD80 (PD57 but his4-lopc), respectively, containing the his4-Δ52 promoter deletion. DNY107 (rad50S AS4) and HF4 (rad50S DNY25) were constructed previously (21).
The plasmid pNKY58, containing the spo13:HisG-URA3-HisG construct, was used to delete the SPO13 gene (1), in DTK288 and DTK305, creating DTK627 and DTK628, respectively. To delete the SPO11 gene, primers 2771119 and 2771120 were used to amplify the KanMX4 construct on the pFA6-KanMX4 plasmid (68). Transformation with the resulting PCR products removed the complete coding region of the SPO11 gene in DTK627 and DTK628, creating DTK637 and DTK638, respectively, and in DTK468 and DTK469, creating DTK629 and DTK630, respectively.
Isogenic diploid strains were generated by mating the following haploids: DNY26 (AS4 × DNY25), PD81 (PD63 × PD80), DTK314 (DTK288 × DTK305), DTK508 (DTK506 × DTK507), DTK517 (DTK514 × DTK516), DTK472 (DTK468 × DTK469), DTK490 (DTK137 × DTK138), FX3 (DNY107 × HF4), DTK633 (DTK627 × DTK628), DTK634 (DTK629 × DTK630), and DTK639 (DTK637 × DTK638).
PCR primers.
The Ras 5′ primer is CCCTGAGGTTGGGGGAGAGC, and the Ras 3′ primer is GGGCTCCTGGCCTCGGGAAG. These primer sequences correspond to human DNA sequences flanking the repeat units. The Hras 5′ outer primer is TGGTCATCCTGGCTTCTGCT, and the Hras 3′ outer primer is GCACCCGGCAGCCCTAGA. Primer 2771119 is 5′ CTTCACCCTTAAGATTTTACGATTTACTAAGTTCACCTTCTCCGTACGCTGCAGGTCGAC,and primer 2771120 is 5′ CTTGAAAAACATTTTTATAAAGCAACAGCTCCCATTCTTATATCGATGAATTCGAGCTC.
PCR analysis of tract length.
Whole-cell PCR was performed on spore colonies resulting from dissection of diploid strains following meiosis. Spores were allowed to form colonies on yeast extract-peptone-dextrose plates, and then a small amount of cells was placed in 6 μl of H2O in 200-μl microtubes. Tubes were heated at 94°C for 6 min and then placed at −80°C for 5 min. Forty-four microliters of a master mix was added. The mix for 12 samples was 62.5 μl of 10× PCR buffer [450 mM Tris, 160 mM (NH4)2SO4], 45 μl of 25 mM MgCl2, 3 μl each of 40 mM Ras 3′ and 5′ primers, 100 μl of 1.25 mM deoxynucleoside triphosphates, 100 μl of gel loading buffer, 243 μl of H2O, and 22 μl of Taq polymerase. The concentrations of MgCl2 and primers given are only representative—the actual concentration of MgCl2 and primers was determined for each strain and for each batch of primers. PCR was performed in a Hybaid PCRExpress machine using the following parameters: 1 cycle of 94°C for 1 min; 2 cycles of 94°C for 1 min, 65°C for 45 s, and 72°C for 2 min; 9 cycles of 94°C for 1 min, 65 to 56°C for 45 s (temperature decreases 1°C per cycle), and 72°C for 2 min; 30 cycles of 94°C for 1 min, 56°C for 45 s, and 72°C for 2 min; 1 cycle of 72°C for 5 min; and then holding at 4°C.
Southern analysis of DSBs.
To examine DSB formation during meiosis at the HIS4 locus, cells were sporulated in liquid 1% potassium acetate at 25°C (21). DNA was isolated using standard protocols (23), treated with BglII, and separated on a 0.8% agarose gel. Standard Southern analysis was performed using a XbaI-XhoI fragment of pDN42 containing the 5′ sequence of HIS4 (21).
Northern analysis of message levels.
Previously published procedures were used (38). Briefly, total RNA was isolated (59) from cells grown in liquid culture, and standard Northern analysis was performed (54). The RNA-bearing nylon filter was hybridized to a mixture of two probes: the pDN42 HIS4 probe and a 1-kb actin gene probe resulting from a HindIII-XhoI digest of plasmid pGAL-ACT1.
Statistical analysis.
Comparisons were done with Instat 1.12 (GraphPad) for Macintosh, using either a chi-square or Fisher’s exact variant test. Results are considered statistically significant if P was 0.05 or less.
RESULTS
We constructed an integrating plasmid to introduce the HRAS1 minisatellite tract into the yeast genome. The complete sequence of the A1a allele (13, 15), including some flanking nonrepetitive DNA and 30 tandem copies of the 28-bp repeat unit, from an EJ bladder carcinoma cell line, was cloned into the yeast vector pPD5 (70) (Fig. 1a). pPD5 contains DNA from the HIS4 locus on chromosome III of S. cerevisiae in which 171 bp of the promoter for HIS4 have been deleted and replaced with an XhoI linker (the his4-Δ52 allele [48]). The minisatellite tract was inserted into the plasmid at this XhoI site, replacing the HIS4 promoter region with the minisatellite tract (Fig. 1b). This novel allele of HIS4 is designated his4-A1.
The HRAS1 VNTR stimulates transcription in yeast.
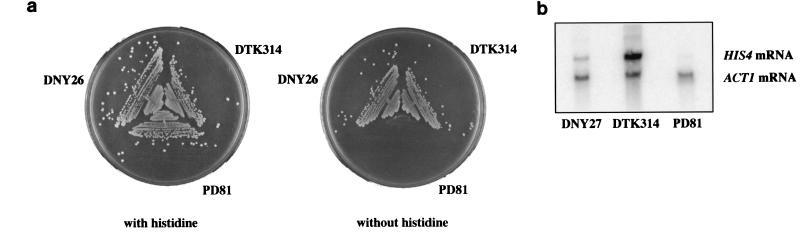
The HRAS1 minisatellite acts as an enhancer of transcription in mammalian cells (66). To determine its effect on transcription of the HIS4 locus, we examined the ability of strains containing the minisatellite to grow on medium lacking histidine. A yeast strain containing the his4-Δ52 allele (the deletion of the HIS4 promoter) cannot grow without exogenous histidine, while cells with a wild-type promoter region do not require histidine in the medium. Upon integration of the minisatellite tract, the resulting transformants became His+. Diploid strains homozygous for the wild-type promoter sequence (DNY26) or the minisatellite insertion (DTK314) gave rise to colonies on medium lacking histidine with equal efficiency, while a strain with a homozygous deletion of the promoter (PD81) failed to generate colonies (Fig. 2a). Northern analysis of the level of HIS4 mRNA in these strains demonstrated that the minisatellite tract stimulated mRNA production at a higher level than did the wild-type promoter (Fig. 2b).
FIG. 2.
The HRAS1 minisatellite affects HIS4 transcription. (a) Strains containing the minisatellite insertion grow on medium lacking histidine. DNY26 is wild type for the HIS4 promoter. PD81 has a deletion of the promoter region, while DTK314 contains the minisatellite insertion into the site of the promoter deletion at HIS4. Strains were grown on solid media with or without histidine. (b) HIS4 mRNA production is stimulated by the minisatellite insertion. Total RNA was isolated from DNY26, PD81, and DTK314. Northern analysis was performed using probes to HIS4 and ACT1. ACT1 was used as a control to normalize for RNA levels in each lane.
The HRAS1 VNTR stimulates meiotic recombination.
The level of recombination at a particular locus in the yeast genome can be determined using genetic or physical methods. A high frequency of non-Mendelian segregation of a heterozygous alteration at the locus of interest indicates that the alteration is located near a recombination hot spot. The nomenclature utilized for segregation of eight-spored fungi is routinely used to describe aberrant segregation tetrads. By this nomenclature, standard Mendelian segregation is 4:4, and gene conversion events are either 6:2 (3 wild-type spore colonies:1 mutant spore colony) or 2:6 (1 wild-type spore colony:3 mutant spore colonies). A tetrad with a spore colony that is sectored for the wild-type and mutant alleles is known as a PMS tetrad and will be described below as a 5:3 (2 wild-type spore colonies:1 mutant spore colony:1 sectored colony) or 3:5 (1 wild-type spore colony:2 mutant spore colonies:1 sectored colony) segregation.
The level of aberrant segregation of a heterozygous alteration in HIS4 (his4-lopc) has been analyzed in detail previously in strains containing a wild-type HIS4 promoter (DNY26) and the his4-Δ52 promoter deletion (PD81). The his4-lopc allele is an insertion of 26 bp into HIS4, disrupting the coding sequence. We compared the level of aberrant segregation of his4-lopc in those strains to the level observed in DTK314, containing the minisatellite tract in place of the HIS4 promoter (Table 1). As another test for the level of recombination in the vicinity of the minisatellite insertion, we monitored the level of intergenic recombination between HIS4 and LEU2. Diploid strains were sporulated, and the four meiotic spores from a single diploid cell were isolated. After the spores had formed colonies, the segregation pattern of the his4-lopc marker was determined. As shown in Table 1, a strain with a wild-type promoter has a very high level of aberrant segregation (51%)—the wild-type promoter acts as a hot spot for meiotic recombination initiation. The hot spot activity is significantly reduced when the HIS4 promoter is no longer present (21%). Introduction of the minisatellite tract (the his4-A1 allele) creates a meiotic recombination hot spot at HIS4 once again; aberrant segregation of his4-lopc is restored nearly to wild-type levels (44%). Similarly, the genetic distance between HIS4 and LEU2 is greatly reduced in PD81 but is identical in DNY26 and DTK314 (31 centimorgans [cM] [Table 1]).
TABLE 1.
Comparison of recombination in strains containing promoter alterations
| Strainc | HIS4 promoter | Relevant mutations | No. of tetrads with various segregation patternsa
|
% Ab. Seg. | cMb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total tetrads | 4:4 | 6:2 | 2:6 | 5:3 | 3:5 | Ab 4:4 | Other Ab. Seg.d | |||||
| DNY26 | HIS4/HIS4 | Wild type | 493 | 244 | 29 | 17 | 88 | 67 | 15 | 33 | 51 | 31 |
| PD81 | his4-Δ52/his4-Δ52 | Wild type | 398 | 313 | 14 | 6 | 26 | 32 | 5 | 2 | 21 | 19 |
| DTK314 | his4-A1/his4-A1 | Wild type | 558 | 310 | 29 | 16 | 75 | 85 | 25 | 18 | 44 | 31 |
| DTK508 | his4-A1/his4-A1 | Δrad1/Δrad1 | 487 | 279 | 28 | 24 | 56 | 66 | 22 | 12 | 43 | 25 |
| DTK517 | his4-A1/his4-A1 | Δpms1/Δpms1 | 238 | 108 | 13 | 10 | 43 | 33 | 20 | 12 | 55 | 26 |
For all segregation patterns, the first number represents the wild-type allele and the second represents the mutant his4-lopc allele. The segregation patterns include 4:4 (normal Mendelian segregation), 6:2 and 2:6 (gene conversion), 5:3 and 3:5 (tetrads with a single PMS event), and Ab 4:4 (aberrant 4:4; one wild-type, one mutant, and two sectored colonies). The Other Ab. Seg. class includes aberrant 6:2 and 2:6 tetrads as well as tetrads with three or four PMS or gene conversion events.
Genetic map distance between LEU2 and HIS4.
DNY26 data are from reference 21; PD81 data are from reference 18.
Ab. Seg., aberrant segregation.
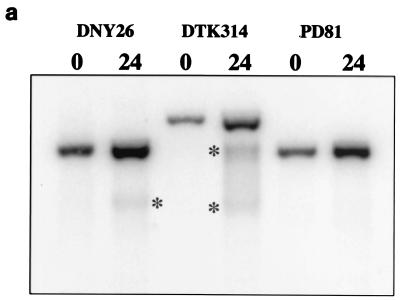
Recombination hot spots also can be identified through physical analysis of the meiotic genome. Meiotic recombination in S. cerevisiae is initiated by a DSB, which can be detected by Southern analysis (62). At the wild-type HIS4 hot spot, these breaks occur in a single well-defined region 5′ of the HIS4 gene (21, 46, 72). To determine if the hot spot activity of the minisatellite tract insertion is associated with DSBs, we created a rad50S derivative of DTK314. The rad50S mutation prevents the processing of DSBs during meiosis, enhancing the ability to detect the lesions (2).
We performed a Southern analysis of the HIS4 region in rad50S derivatives of DNY26 (wild-type strain), PD81 (promoter deletion strain), and DTK314 (minisatellite insertion strain), using DNA from cells prior to meiosis induction and 24 h after induction (Fig. 3a). Consistent with previous reports (21), the HIS4-specific probe detected no DSBs at HIS4 in the promoter deletion strain and a single meiosis-specific break in the wild-type strain. The strain containing the minisatellite insertion had two distinct DSBs. The sizes of the fragments indicate that the breaks flank the minisatellite insertion but do not occur within the repetitive DNA (Fig. 3b). These results indicate that the minisatellite tract insertion stimulates DSB formation during meiosis, which presumably leads to the high frequency of aberrant segregation of the his4-lopc allele.
FIG. 3.
DSB formation during meiosis at HIS4. (a) The minisatellite insertion stimulates DSB formation during meiosis. DNA from rad50S derivatives of DNY26 (wild-type promoter [FX3]), DTK314 (minisatellite insertion [DTK472]), and PD81 (promoter deletion [DTK490]) was isolated from strains prior to meiosis and 24 h after meiotic induction. Southern analysis was performed using a HIS4-specific probe. Asterisks mark the bands resulting from meiosis-specific DSBs. (b) Processing events following DSB formation during meiosis. A diagram of the HIS4 locus in DTK314 is shown. The asterisks mark the approximate locations of the meiosis-specific DSBs. The arrows (labeled A, B, C, and D) underneath the diagram represent the direction of processing events subsequent to DSB formation.
Spo11p is the endonuclease that creates the meiosis-specific DSBs during meiosis in S. cerevisiae (33). We analyzed the physical and genetic consequences of deleting SPO11 for DSB formation and recombination at HIS4 to verify that these processes are initiated by Spo11p. SPO11 was deleted in the rad50S derivative of DTK314, creating DTK634, and the formation of DSBs during meiosis was determined by Southern analysis as described above. Neither of the meiotic DSBs present in the minisatellite strain (Fig. 3b) was detected in DTK634 (data not shown). To determine the effect of the SPO11 deletion on genetic recombination at HIS4, two strains were created. DTK633 is a derivative of DTK314 in which the SPO13 gene has been eliminated, while DTK639 is a derivative in which both SPO13 and SPO11 have been deleted. Strains lacking SPO11 arrest during meiosis, but the spo13 mutation allows a spo11 strain to proceed through meiosis, as mutation of SPO13 causes a bypass of the first meiotic division, leading to formation of two diploid meiotic products rather than four haploid products (39). While each diploid will be heterozygous for his4-lopc, the diploids will be phenotypically His+ unless a recombination event leads to a gene conversion event or PMS event involving the wild-type HIS4 allele. Following meiosis and dissection of dyads in DTK633 (spo13), 41% of the diploid colonies are His− or His+/− (Table 2). In DTK639 (spo13 spo11), the percentage of colonies with a His− or His+/− phenotype is 4%. Other heterozygous loci in these strains (LEU2, ARG4, TRP1, and TYR7) exhibit a similar reduction. These data indicate that Spo11p is responsible for the generation of meiotic DSBs and genetic recombination at HIS4 in strains containing the minisatellite tract.
TABLE 2.
His+ segregation in spo13 and spo11 spo13 backgrounds
| Strain | Relevant mutations | HIS4 allele | Total dyads | No. of dyads with His+ segregationa
|
% Ab. Seg.b | ||||
|---|---|---|---|---|---|---|---|---|---|
| 4:0 | 3:1 | 2:2 | 1:3 | 0:4 | |||||
| DTK633 | spo13/spo13 | his4-lopc/HIS4 | 214 | 127 | 23 | 61 | 2 | 1 | 41 |
| DTK639 | spo11/spo11 | his4-lopc/HIS4 | 216 | 207 | 0 | 9 | 0 | 0 | 4 |
| spo13/spo13 | |||||||||
SPO13 mutations eliminate meiosis I divisions, leading to two diploid products rather than four haploid products. Each diploid should be heterozygous at HIS4 unless a recombination event occurs during meiosis. 4:0 segregation, both colonies are His+; 3:1, one His+, one His+/−; 2:2, one His+, one His−; 1:3, one His−, one His+/−; 0:4, both His−.
Ab. Seg., aberrant segregation.
Minisatellite tracts alter in length during meiosis.
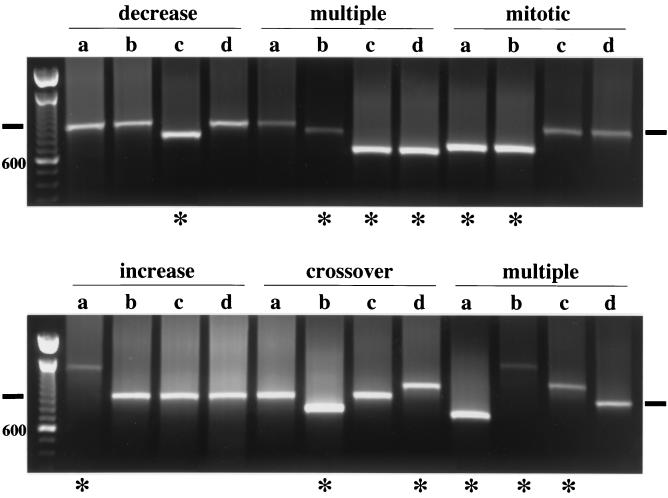
In mammalian systems in which minisatellite stability has been studied, the minisatellite tracts often alter in length following passage through meiosis. We evaluated the stability of the his4-A1 minisatellite allele during meiosis using PCR to determine the length of the tract. Whole-cell PCR was performed (see Materials and Methods) on spore colonies following meiosis. In DTK314, the minisatellite tract frequently altered in length during meiosis (Table 3); 45% of the tetrads examined (96 of 213) had at least one spore colony with an altered minisatellite tract. This frequency of alteration is equivalent to the level of aberrant segregation of the his4-lopc allele in this same strain. Five classes of tetrads were observed: tetrads that had increases or decreases in the minisatellite length in a single spore, tetrads with two spore alterations, tetrads with three altered spores, and tetrads with four altered spores (Table 3). Representative PCRs from these classes are shown in Fig. 4. Sixty-three percent (60 of 96) of the altered tetrads had one spore colony that exhibited an alteration. One-third of these were increases, while two-thirds were decreases. In the class with two spores that exhibited altered length, 10 tetrads exhibited alteration patterns suggestive of a mitotic rearrangement prior to meiosis—both altered minisatellite tracts were identical in length, while three tetrads had a pattern characteristic of an unequal crossover event, i.e., one spore had an increase in tract length while another had a concomitant decrease (Fig. 4). Finally, a number of tetrads exhibited complex rearrangements, with three or four spores having tracts with altered lengths. In Fig. 4, one multiple tetrad had two tracts of the same decreased length (resembling a mitotic rearrangement) and a third with a smaller contraction in length. Another tetrad had two spores exhibiting increases, of differing lengths, and one with a decrease—this tetrad could have arisen from an unequal crossover event and an independent length expansion. A few colonies had both a wild-type and an altered-tract-length PCR product (data not shown), appearing to have undergone a PMS event indicative of heteroduplex formation during meiosis.
TABLE 3.
Distribution of tract length alterations in various strain backgrounds
| Strain | Relevant mutations | Total tetrads examined | No. (%) of tetrads with change:
|
|||||
|---|---|---|---|---|---|---|---|---|
| No alteration | Single spore decrease | Single spore increase | Two spore alterations | Three spore alterations | Four spore alterations | |||
| DTK314 | Wild type | 213 | 117 (55) | 41 (19) | 19 (9) | 22 (10) | 13 (6) | 1 (0.5) |
| DTK508 | Δrad1/Δrad1 | 275 | 142 (52) | 67 (24) | 9 (3) | 44 (16) | 11 (4) | 2 (0.7) |
| DTK517 | Δpms1/Δpms1 | 109 | 46 (42) | 26 (24) | 9 (8) | 22 (20) | 6 (5) | 0 |
FIG. 4.
Tract length of the minisatellite alters during meiosis at high frequency. Representative whole-cell PCRs across the minisatellite tract in spore colonies (a, b, c, and d) derived from individual diploid cells are shown. The horizontal black bars mark the band size of the unaltered minisatellite tract. Asterisks indicate the spore colonies exhibiting altered-length alleles. The size standards are 100-bp ladders; as indicated, the brighter lower band is the 600-bp band.
In the 60 tetrads with a single altered-length spore colony, 6 tetrads had the same colony exhibit both an altered minisatellite tract and a PMS event for his4-lopc, while 14 tetrads had different spore colonies demonstrate his4-lopc PMS and a tract length alteration. Seven tetrads underwent gene conversion of his4-lopc, making it impossible to determine if the same spore colony was involved. The remaining 33 tetrads did not exhibit aberrant segregation of his4-lopc.
The number of crossovers between HIS4 and LEU2 in tetrads containing a single length alteration event was determined and compared to the overall level of intergenic recombination. Of the 60 tetrads exhibiting an alteration in tract length in one spore (Table 3), 31 had a parental configuration, 2 had a nonparental configuration, and 19 were tetratype, giving a distance of 29.8 cM between HIS4 and LEU2. The HIS4-LEU2 distance for all DTK314 tetrads was 31 cM (243 parental ditype, 14 nonparental ditype, and 198 tetratype). We conclude that events which alter the tract length do not occur by a mechanism that results in preferentially crossover or noncrossover configurations (3). This result is similar to results from a study of minisatellite tract alterations during mitotic recombination (50).
Identification of RAD1 as a gene controlling tract length alteration.
One model (see Discussion and Fig. 5) for the generation of altered-length alleles of the minisatellite tract during meiosis involves a modification of the DSB model for meiotic recombination initiation (64). Briefly, during strand invasion following DSB formation, misalignment of the donor and recipient strands can allow the formation of loops consisting of an integral number of repeat units. Repair of these mismatches potentially can lead to increases or decreases in tract length. A meiosis-specific pathway involved in large loop repair (LLR) has been described previously (37). One component of this repair pathway is the protein encoded by the RAD1 gene. A second repair pathway specific for the repair of base-base and other small mismatches involves the PMS1 gene product. To determine the impact of these repair pathways on recombination and minisatellite length alteration, we investigated meiotic recombination and tract length alterations in strain backgrounds lacking either the RAD1 or the PMS1 gene.
FIG. 5.
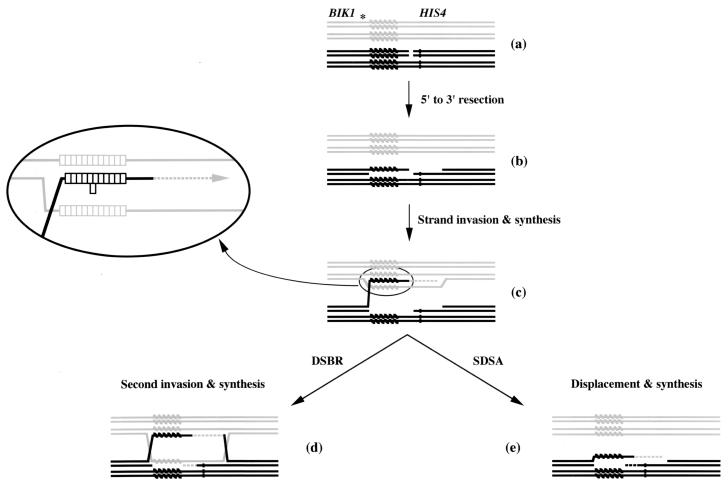
A model for tract length alteration during meiosis. Individual single-stranded DNA molecules are depicted, with one homolog in black and the other in gray. The his4-lopc allele is shown as a dark bar, while the minisatellite tract is shown as a series of diagonal bars. In panel a, a DSB is shown between the minisatellite tract and his4-lopc. The asterisk marks the relative site of the second DSB observed (Fig. 3). In panel b, the ends of the break have been resected. Strand invasion, heteroduplex formation, and the beginning of repair synthesis are depicted in panel c. The enlarged portion shows the potential misalignment of minisatellite repeats following strand invasion. One repeat unit has formed a loop on the invading strand; other configurations are possible. The DSBR model (64) predicts invasion of the second end followed by repair synthesis and Holliday junction resolution (d). The SDSA model (reviewed in reference 49) predicts a strand displacement event followed by repair synthesis (e).
The deletion of RAD1 did not affect the level of recombination observed at the his4-lopc allele. However, there was a decrease in the level of intergenic recombination between HIS4 and LEU2 (Table 1); this decrease was statistically significant (P = 0.05). A decrease in intergenic recombination upon deletion of RAD1 was not seen previously in this strain background (32, 37). The RAD1 deletion had a strong effect on the types of alterations detected in the minisatellite tract. Tetrads in which the minisatellite tract had undergone an expansion were significantly reduced (Table 3). Nine single-spore-increase tetrads were detected; the decrease in this class of altered tetrads represented a significant (P = 0.01) change relative to the distribution of classes in the wild-type strain. Based on these results, we conclude that the LLR pathway has a direct role in the meiotic expansion of the minisatellite tract. A model for this activity is presented in the Discussion.
Unlike RAD1, deletion of the PMS1 gene increased the level of aberrant segregation of the his4-lopc allele (P = 0.01) but had no significant effect on the level of intergenic recombination between HIS4 and LEU2 (Table 1). The PMS1 deletion also did not have any significant effect on the types of size alterations detected in the minisatellite tract (Table 3). From these data, we conclude that the meiotic alteration in tract length of the minisatellite does not involve the PMS1-dependent mismatch repair pathway. However, the number of tetrads exhibiting two altered spores was significantly increased in the pms1 background (P = 0.025).
Minisatellite tracts are significantly more stable during mitotic growth.
To determine the frequency of minisatellite tract alteration during normal mitotic growth, DTK314 (wild type), DTK508 (Δrad1), and DTK517 (Δpms1) were grown in liquid culture. Serial dilutions of each culture were plated on solid medium, and colonies were allowed to form for 3 days at 30°C. These growth conditions are approximately equivalent to the sporulation and dissection protocol, but without the exposure to meiosis-inducing conditions. Whole-cell PCR was performed on approximately 100 colonies from each strain. The PCR analysis of the minisatellite tract revealed that the tract was stable during mitotic growth. DTK314 had only one colony that exhibited a tract length alteration (1 of 101), DTK508 had three alterations (3 of 102), and DTK517 had none (0 of 101). All of the alterations were deletions (data not shown). From these data, we conclude that the repetitive DNA in the minisatellite tract is much more stable during mitosis than during meiosis, even in repair-deficient strain backgrounds.
DISCUSSION
Activities of the HRAS1 minisatellite in yeast.
The HRAS1 minisatellite tract exhibits three distinct phenotypes in mammalian systems: it enhances transcription of the HRAS1 gene, it destabilizes during meiosis but not during mitosis, and it alters in length and sequence composition. To determine the genetic components capable of modulating these phenotypes, we inserted the minisatellite tract into the S. cerevisiae genome at the HIS4 locus on chromosome III in place of a well-characterized deletion of the HIS4 promoter sequence (Fig. 1b). We found that the minisatellite tract in yeast recapitulates the phenotypes observed in mammalian systems.
The minisatellite tract stimulated mRNA production at HIS4 through an unknown mechanism. It is possible that the tract is binding a yeast transcription factor; the minisatellite tract in mammalian cells binds a subset of the rel/NF-κB transcription factors (66). Alternatively, the repetitive nature of the minisatellite tract could be stimulating transcription independent of a transcription factor. Poly(A) and poly(G) tracts in yeast can act as promoter elements, for example (27), possibly by creating an open chromatin configuration that favors transcription initiation.
Insertion of the HRAS1 minisatellite creates a meiotic recombination hot spot by a number of criteria: meiosis-specific DSB formation was stimulated at HIS4 (Fig. 3a), aberrant segregation of a nearby heterozygous marker (his4-lopc) increased (Table 1), and intergenic recombination between HIS4 and LEU2 was elevated (Table 1).
In S. cerevisiae, meiotic recombination initiates when Spo11p generates a DSB (33). At the HIS4 locus containing the HRAS1 minisatellite insertion, we detected DSBs on each side of the insertion (Fig. 3). This result contrasts with the DSB generated at HIS4 alleles with a wild-type promoter region, where only one break is detected (21). Based on the sizes of the meiotic DSB fragments, the DSBs occur in localized regions adjacent to the repeats rather than in the repetitive portion of the minisatellite insertion. The relative intensities of the two bands indicate that the regions on each side of the minisatellite are equivalent with respect to the frequency of DSB formation.
Following DSB formation, meiotic breaks are processed to generate a long single-stranded 3′ tail (63). If the minisatellite insertion initiates DSB formation in the same manner as with the wild-type promoter, the breaks flanking the minisatellite tract should be processed equally toward BIK1 (A and C in Fig. 3b) or HIS4 (B and D in Fig. 3b). Of those events in which the single-stranded tail encompasses the minisatellite repeats (B and C), only half (B) can include the his4-lopc marker. We examined those tetrads in which the minisatellite tract had altered in length in one spore, presumably indicating a single recombination initiation event. Of the 60 single events in DTK314 (Table 3), 45% (27 of 60) had an aberrant segregation of the his4-lopc allele (data not shown). This value is the expected value if the DSB and subsequent processing occur as diagrammed in Fig. 3b and recombination events initiate from either DSB with equal frequency.
While the minisatellite insertion stimulated aberrant segregation of his4-lopc during meiosis, the level of aberrant segregation was not quite as high as that seen with the wild-type promoter region. This result could be due to the two DSBs flanking the minisatellite. While the proximal DSB is approximately the same distance from the his4-lopc marker as the wild-type promoter DSB, the distal DSB is over a kilobase away (Fig. 3b). This difference means that recombination events initiated from the distal DSB must undergo a much longer processing event to include the his4-lopc marker in heteroduplex DNA. In support of this interpretation, the level of intergenic recombination is identical in the wild-type and minisatellite hot spot strains (Table 1). As intergenic recombination does not require the marker alleles to be included in the heteroduplex, it should be much less sensitive to distance variations. Also, PCR across the minisatellite tract in spore colonies (Table 3) from DTK314 revealed that the minisatellite tract was altered in length in 45% of the 213 tetrads examined. This level of alteration is identical to the level of aberrant segregation of his4-lopc and suggests that the frequency of alteration and aberrant segregation are related. Finally, on a per-spore (or gamete) level, this represents an alteration rate of 17.3% (147 events among 852 examined)—human minisatellite tracts have been altered in 1 to 10% of gametes examined (30).
A model for tract length alteration during meiosis.
Two models for meiotic recombination are the DSB repair (DSBR) model (64) and the synthesis-dependent strand annealing (SDSA) model (reviewed in reference 49). Both models initiate recombination via a DSB, followed by resection to generate a 3′ single-stranded tail, which then invades the homolog, creating a region of heteroduplex DNA (Fig. 5). In the DSBR model, the second end invades and double Holliday junctions are created, as are regions of heteroduplex DNA on both homologs. Resolution of these junctions leads to crossover and noncrossover molecules. In the SDSA model, only one strand invades, to one side of the initiating DSB. Following synthesis, this strand is displaced to anneal with the other DSB end and no crossover product is formed. In a strain with a wild-type hot spot, the initial strand invasion occurs in unique DNA sequences. In contrast, a DSB adjacent to the minisatellite tract, followed by degradation of one strand into the repetitive sequence, will set up a situation in which the single-stranded invasion occurs in a region of repetitive DNA. The repetitive invading strand can misalign, causing a loop to form (Fig. 5). This loop will be composed of an integral number of repeat units in length (28 bases in the case of the HRAS1 minisatellite). Figure 5 shows a one-repeat loop formed on the invading strand as the result of a misalignment during invasion. As there are four different types of 28-bp repeats, varying at either position 7 or position 15, a series of base-base mismatches will occur in the misaligned tract adjacent to the loop.
S. cerevisiae possesses at least three distinct meiotic mismatch repair pathways (reviewed in reference 35). One pathway acts on base-base mismatches and is akin to the well-characterized postreplicative MMR pathway, consisting of the MSH2, MSH3, MSH6, PMS1, and MLH1 gene products. Another pathway is involved in LLR during meiosis (32, 37) and involves the RAD1, RAD10, MSH2, and MSH3 gene products. Components of the third pathway have not been identified, but genetic data indicate that it functions in LLR, as some repair of large loops is detected in a rad1 strain (37). The loop extruded during heteroduplex DNA formation could be a substrate for the LLR pathway, while the subsequent base-base mismatches could be substrates for the MMR pathway. This model predicts that elimination of the LLR pathway would affect the generation of altered-length alleles, while elimination of the MMR pathway would not affect length alteration. To test this prediction, we deleted the RAD1 or PMS1 genes and assayed the effect of the loss on minisatellite tract length during meiosis. Rad1p has no role in MMR but a major role in LLR, while Pms1p has a major role in MMR but no role in LLR. Msh2p and Msh3p were not analyzed, as they play roles in both MMR and LLR (32, 37).
We found that deletion of RAD1 specifically affected the class of tetrads exhibiting an increase in the length of the minisatellite tract during meiosis (Table 3). Genetic evidence from previous studies (32, 37) indicates that during meiotic LLR Rad1p acts to cleave the strand opposite the extruded loop, rather than cleaving off the loop itself. If, following the cleavage of the opposing strand, the loop is used as a template for repair synthesis, the cleaved strand will increase in length by the same number of repeat units as the loop originally contained. Removal of the Rad1p function would lead to loss of these events, consistent with the data reported here. The mechanism leading to contraction in the tract length is unknown. It is possible that the second, uncharacterized LLR pathway described above may function in this type of alteration. The components of meiotic LLR (Rad1/10p, Msh2p, and Msh3p) also function during mitotic recombination to remove regions of nonhomology (reviewed in reference 49). However, while the same complex of proteins is involved, the meiotic and mitotic actions differ, as sequence is lost during the mitotic action and gained during meiosis. The basis of this difference is not known.
The initial steps in both DSBR and SDSA are identical, leading to formation of a region of heteroduplex DNA (Fig. 5c). In the DSBR model, this region remains and a second is formed following the invasion of the other DNA strand (Fig. 5d). Both heteroduplexes may contain mismatches; in Fig. 5, a DSB between the minisatellite tract and his4-lopc will lead to both being incorporated into the heteroduplex if the break is processed in both directions as indicated. Depending on which strand in the heteroduplex is repaired and the direction of Holliday junction resolution, the mismatches may remain on the same chromosome or segregate to different chromosomes. If the break occurs to the HIS4-distal side of the tract (as indicated by the asterisk in Fig. 5a), a single heteroduplex tract can form that contains both the minisatellite tract and his4-lopc. However, in the SDSA model, the initial heteroduplex region is removed, and a second heteroduplex forms on the chromosome that initiated the recombination event (Fig. 5e). Unless a mismatch that occurs in the initial strand invasion step (Fig. 5c) is repaired prior to the strand displacement step, the minisatellite and his4-lopc alterations will occur on the same chromosome. In DTK314 tetrads containing a single spore exhibiting alteration in tract length and an aberrant segregation of his4-lopc, only 30% (6 of 20) of the events were detected in the same spore. These data are more consistent with the DSBR model, although more complicated variations of the SDSA model, in which multiple rounds of strand invasion can occur (17, 49), are also possible.
We saw a statistically significant increase in the pms1 strain for the class of tetrads with two altered spores (Table 3). This increase is surprising, considering that the pms1 deletion had no significant effect on the frequency of single-spore alterations. Pms1p is known to have a role in limiting recombination between diverged sequences (homologous recombination [11]). Perhaps this function of Pms1p normally acts to limit the number of chromosomes involved in heteroduplex formation during a single meiosis. However, the frequency of two spore alterations (10%) in DTK314 (Table 3) is not significantly different from the expected value calculated from the single-spore alteration frequencies (28% × 28% = 7.8%).
Mitotic rearrangement of the minisatellite tract.
Analysis of the stability of the HRAS1 minisatellite tract during meiosis revealed that it was quite unstable. However, a number of events were detected that corresponded to the expected pattern for a mitotic rearrangement in the tract (Fig. 4 shows an example). We analyzed the mitotic stability of the tract in the wild-type, rad1, and pms1 backgrounds. The tract is much more stable during mitotic growth than during passage through meiosis; only 1.3% of the diploid colonies examined had a rearrangement during mitosis. This level of mitotic alteration is still very high relative to other mitotic events such as mutation or recombination and appears to be diploid specific. Tract length alteration at this frequency during vegetative growth should be detectable in the PCRs on spore colony tetrads (Fig. 4), leading to multiple bands in the reaction, with the relative intensities of the bands dependent on the time at which the rearrangement occurred during colony formation. No such band pattern has been detected in any of the haploid spore colonies that were analyzed (more than 1,300 colonies were examined). It is possible that generation of the mitotic alterations could require either homologous chromosome pairs or a diploid-specific protein. We note, however, that during our meiotic tetrad analysis the level of alterations that we detected that appear to be mitotic events originating in the diploid cell prior to meiosis (10 of 213 in DTK314 [4.7%] [data not shown]) is higher than the rate that we determined for mitotic rearrangements. It is possible that some of these events are due to another pathway that is meiosis specific.
Comparison to activities of other minisatellite tracts in yeast.
Three other research groups have investigated the stability of minisatellite sequences in the yeast genome during meiosis, each using a different minisatellite sequence. First, the human minisatellite MS32 was integrated near the LEU2 recombination hot spot (12) and examined by random spore analysis (1,700 spore colonies [5]) or tetrad analysis (approximately 100 tetrads [4]). MS32 had a high meiotic, but not mitotic, mutation frequency. A mechanism proposed to explain these recombination events could explain the subset of events (described above) detected in our study with the HRAS1 minisatellite that appear to be mitotic gene conversion events but may be meiosis specific. Second, the human CEB1 locus was integrated into the ARG4 locus of yeast and analyzed genetically (17). CEB1 insertions also showed a high degree of meiotic destabilization, with alterations that were indicative of heteroduplex formation followed by gene conversion. Furthermore, meiosis-specific DSBs were detected and shown to be required for the destabilization of the CEB1 tracts. Third, a naturally occurring yeast minisatellite sequence normally associated with subtelomeric regions was introduced into the LEU2 locus. Tetrad analysis demonstrated that the minisatellite tract underwent gene conversions at high frequency (8); either the DSBR model (64) or a type of SDSA (26) was implicated in these events.
At one level, these data are all consistent with our results. Minisatellite tracts introduced into the yeast genome rearrange at a high frequency, and the mechanism for this rearrangement is recombinational. However, a closer comparison reveals intriguing differences. For example, the stability of the CEB1 minisatellite was affected by deletion of PMS1 (17), but no effect of PMS1 deletion was seen in our study or the study of the subtelomeric minisatellite (8). Also, the types of rearrangements and their frequency differed among the studies. The most likely explanation for many of these differences lies in the primary sequence of the minisatellites themselves, as each group utilized tracts with different primary sequences and lengths of repeats. Furthermore, the different studies utilized minisatellite alleles of two different lengths in the same diploid. By the model given above, invoking misalignment during heteroduplex formation as a mechanism for length alteration, in these diploids every recombination event would give rise to an extruded loop. Therefore, every recombination event would require a loop repair event, a situation that may not reflect the actual conditions during meiosis. Clearly, minisatellite stability during meiosis is a complex phenomenon affected by many factors.
Implications for mammalian systems.
Our finding that the expansion of the HRAS1 minisatellite tract during meiosis in S. cerevisiae requires RAD1 suggests the possibility that the human homolog of RAD1 might have a similar effect on minisatellite stability during gametogenesis. The role of XPF/ERCC4, the human homolog of the RAD1 gene, in LLR during meiosis has not been explored. Because of the potential correlation of increased tract length and oncogenesis (see references 9 and 42 to 44), limiting the meiotic expansion of the HRAS1 minisatellite is desirable. In addition to its role in LLR, the yeast RAD1 gene is essential for nucleotide excision repair, and XPF/ERCC4 also functions in this type of repair. Unfortunately, the loss of XPF/ERCC4 function leads to a type of skin cancer known as xeroderma pigmentosum (reviewed in references 22, 40, 53, and 55). If XPF/ERCC4 does contribute to minisatellite tract expansion in humans, a method of inactivating the protein selectively in the germ line would need to be developed for the results of this study to have a clinical application.
Acknowledgments
This work was supported by a Special Fellowship from the Society for Leukemia and Lymphoma (D.T.K.) and by a Basil O’Connor Starter Scholarship (research grant no. 5-FY00-573) from the March of Dimes Birth Defects Foundation to D.T.K.
We are grateful to J. Berman, R. Herman, C. Kirkpatrick, P. Magee, T. Petes, and A. Rougvie for advice and suggestions during the course of this work and to Eric Alani for the pNKY58 plasmid.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419–436. [DOI] [PubMed] [Google Scholar]
- 3.Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47–57. [DOI] [PubMed] [Google Scholar]
- 4.Applegren, H., H. Cederberg, and U. Rannug. 1999. Meiotic interallelic conversion at the human minisatellite MS32 in yeast triggers recombination in several chromatids. Gene 239:29–38. [DOI] [PubMed] [Google Scholar]
- 5.Applegren, H., H. Cederberg, and U. Rannug. 1997. Mutations at the human minisatellite MS32 integrated in yeast occur with high frequency in meiosis and involve complex recombination events. Mol. Gen. Genet. 256:7–17. [DOI] [PubMed] [Google Scholar]
- 6.Armour, J. A. L., P. C. Harris, and A. J. Jeffreys. 1993. Allelic diversity at minisatellite MS205 (D16S309): evidence for polarized variability. Hum. Mol. Genet. 2:1137–1145. [DOI] [PubMed] [Google Scholar]
- 7.Arndt, K. T., C. A. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874–880. [DOI] [PubMed] [Google Scholar]
- 8.Bishop, A. J., E. J. Louis, and R. H. Borts. 2000. Minisatellite variants generated in yeast meiosis involve DNA removal during gene conversion. Genetics 156:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm, T. L. J., H.-P. Hirth, B. Kornhuber, and D. Drahovsky. 1987. Oncogene amplifications, rearrangements, and restriction fragment length polymorphisms in human leukemia. Eur. J. Clin. Oncol. 23:623–629. [DOI] [PubMed] [Google Scholar]
- 10.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346. [DOI] [PubMed] [Google Scholar]
- 11.Borts, R. H., W.-Y. Leung, W. Kramer, B. Kramer, M. Williamson, S. Fogel, and J. E. Haber. 1990. Mismatch repair-induced meiotic recombination requires the PMS1 gene product. Genetics 124:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089–1101. [DOI] [PubMed] [Google Scholar]
- 13.Capon, D. J., E. Y. Chen, A. D. Levinson, P. H. Seeburg, and D. V. Goeddel. 1983. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature 302:33–37. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, J. B., M. V. Walter, and A. D. Levinson. 1987. A repetitive sequence element 3′ of the human c-Ha-ras1 gene has enhancer activity. J. Cell. Physiol. 5(Suppl.):75–81. [DOI] [PubMed] [Google Scholar]
- 15.Conway, K., S. N. Edmiston, B. S. Hulka, P. A. Garrett, and E. T. Liu. 1996. Internal sequence variations in the Ha-ras variable tandem number repeat—rare and common alleles identified by minisatellite variant repeat polymerase chain reaction. Cancer Res. 56:4773–4777. [PubMed] [Google Scholar]
- 16.Crouse, G. F. 1996. Mismatch repair systems in Saccharomyces cerevisiae, p.411–448. In M. F. Hoekstra and J. A. Nickoloff (ed.), DNA damage and repair—biochemistry, genetics, and cell biology. Humana Press, Totowa, N.J.
- 17.Debrauwere, H., J. Buard, J. Tessier, D. Aubert, G. Vergnaud, and A. Nicolas. 1999. Meiotic instability of human minisatellite CEB1 in yeast requires DNA double-strand breaks. Nat. Genet. 23:367–371. [DOI] [PubMed] [Google Scholar]
- 18.Detloff, P., M. A. White, and T. D. Petes. 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devlin, C., K. Tice-Baldwin, D. Shore, and K. T. Arndt. 1991. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol. Cell. Biol. 11:3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding, S., G. P. Larson, K. Foldenauer, G. Zhang, and T. G. Krontiris. 1999. Distinct mutation patterns of breast cancer-associated alleles of the HRAS1 minisatellite locus. Hum. Mol. Genet. 8:515–521. [DOI] [PubMed] [Google Scholar]
- 21.Fan, Q., F. Xu, and T. D. Petes. 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15:1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedberg, E. C., and R. D. Wood. 1996. DNA excision repair pathways, p.249–269. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 23.Goyon, C., and M. Lichten. 1993. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol. Cell. Biol. 13:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green, M., and T. G. Krontiris. 1993. Allelic variation of reporter gene activation by the HRAS1 minisatellite. Genomics 17:429–434. [DOI] [PubMed] [Google Scholar]
- 25.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., San Diego, Calif.
- 26.Hastings, P. J. 1988. Recombination in the eukaryotic nucleus. Bioessays 9:61–64. [DOI] [PubMed] [Google Scholar]
- 27.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14:2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarman, A. P., and R. A. Wells. 1989. Hypervariable minisatellites: recombinators or innocent bystanders? Trends Genet. 5:367–371. [DOI] [PubMed] [Google Scholar]
- 29.Jeffreys, A. J., D. L. Neil, and R. Neumann. 1998. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 17:4147–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffreys, A. J., N. J. Royle, V. Wilson, and Z. Wong. 1988. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332:278–281. [DOI] [PubMed] [Google Scholar]
- 31.Jeffreys, A. J., K. Tamaki, A. MacLeod, D. G. Monckton, D. L. Neil, and J. A. L. Armour. 1994. Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet. 6:136–145. [DOI] [PubMed] [Google Scholar]
- 32.Kearney, H. M., D. T. Kirkpatrick, J. L. Gerton, and T. D. Petes. 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158:1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375–384. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy, G. C., M. S. German, and W. J. Rutter. 1995. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 9:293–298. [DOI] [PubMed] [Google Scholar]
- 35.Kirkpatrick, D. T. 1999. Roles of the DNA mismatch repair and nucleotide excision repair proteins during meiosis. Cell. Mol. Life Sci. 55:437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkpatrick, D. T., Q.-Q. Fan, and T. D. Petes. 1999. Maximal stimulation of meiotic recombination by a yeast transcription factor requires the transcription activation domain and a DNA-binding domain. Genetics 152:101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkpatrick, D. T., and T. D. Petes. 1997. Repair of DNA loops involves DNA mismatch and nucleotide excision repair proteins. Nature 387:929–931. [DOI] [PubMed] [Google Scholar]
- 38.Kirkpatrick, D. T., Y.-H. Wang, M. Dominska, J. D. Griffith, and T. D. Petes. 1999. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol. Cell. Biol. 19:7661–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klapholz, S., and R. E. Esposito. 1980. Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics 96:589–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraemer, K. H., M. M. Lee, and J. Scotto. 1987. Xeroderma pigmentosum: cutaneous, ocular and neurologic abnormalities in 830 published cases. Arch. Dermatol. 123:241–250. [DOI] [PubMed] [Google Scholar]
- 41.Kramer, W., B. Kramer, M. S. Williamson, and S. Fogel. 1989. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J. Bacteriol. 171:5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krontiris, T. G. 1995. Minisatellites and human disease. Science 269:1682–1683. [DOI] [PubMed] [Google Scholar]
- 43.Krontiris, T. G., B. Devlin, D. D. Karp, N. J. Robert, and N. Risch. 1993. An association between the risk of cancer and mutations in the HRas1 minisatellite locus. N. Engl. J. Med. 329:517–523. [DOI] [PubMed] [Google Scholar]
- 44.Krontiris, T. G., N. A. DiMartino, M. Colb, and D. R. Parkinson. 1985. Unique allelic restriction fragments of the human Ha-ras locus in leukocyte and tumour DNAs of cancer patients. Nature 313:369–374. [DOI] [PubMed] [Google Scholar]
- 45.Lafreniere, R. G., D. L. Rochefort, N. Chretien, J. M. Rommens, J. I. Cochius, R. Kalvaiainen, U. Nousiainen, G. Patry, K. Farrell, B. Soderfeldt, A. Federico, B. R. Hale, O. H. Cossio, T. Sorensen, M. A. Pouliot, T. Kmiec, P. Uldall, J. Janszky, M. R. Prantzatelli, F. Andermann, E. Andermann, and G. A. Rouleau. 1997. Unstable insertion in the 5′ flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat. Genet. 15:298–302. [DOI] [PubMed] [Google Scholar]
- 46.Nag, D. K., and T. D. Petes. 1993. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nag, D. K., M. A. White, and T. D. Petes. 1989. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340:318–320. [DOI] [PubMed] [Google Scholar]
- 48.Nagawa, F., and G. R. Fink. 1985. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 82:8557–8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paques, F., G.-F. Richard, and J. E. Haber. 2001. Expansions and contractions in 36-bp minisatellites by gene conversion in yeast. Genetics 158:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petes, T. D., R. E. Malone, and L. S. Symington. 1991. Recombination in yeast, p.407–521. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Porter, S. E., M. A. White, and T. D. Petes. 1993. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics 134:5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prakash, S., P. Sung, and L. Prakash. 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27:33–70. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43–81. [DOI] [PubMed] [Google Scholar]
- 56.Sherman, F., G. R. Fink, and J. B. Hicks. 1982. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Sia, E. A., S. Jinks-Robertson, and T. D. Petes. 1997. Genetic control of microsatellite stability. Mutat. Res. 383:61–70. [DOI] [PubMed] [Google Scholar]
- 58.Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17:2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solomon, F., L. Connell, D. Kirkpatrick, V. Praitis, and B. Weinstein. 1992. Methods for studying the cytoskeleton in yeast, p.197–221. In K. L. Carraway and C. A. C. Carraway (ed.), The cytoskeleton: a practical approach. IRL Press, New York, N.Y.
- 60.Spandidos, D. A., and L. Holmes. 1987. Transcriptional enhancer activity in the variable tandem repeat DNA sequence downstream of the human Ha-ras1 gene. FEBS Lett. 218:41–46. [DOI] [PubMed] [Google Scholar]
- 61.Stapleton, A., and T. D. Petes. 1991. The Tn3 B-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics 127:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338:87–90. [DOI] [PubMed] [Google Scholar]
- 63.Sun, H., D. Treco, and J. W. Szostak. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64:1155–1161. [DOI] [PubMed] [Google Scholar]
- 64.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand break model for recombination. Cell 33:25–35. [DOI] [PubMed] [Google Scholar]
- 65.Tice-Baldwin, K., G. R. Fink, and K. T. Arndt. 1989. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246:931–935. [DOI] [PubMed] [Google Scholar]
- 66.Trepicchio, W. L., and T. G. Krontiris. 1992. Members of the rel/NF-kB family of transcriptional regulatory proteins bind the HRAS1 minisatellite DNA sequence. Nucleic Acids Res. 20:2427–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Virtaneva, K., E. D’Amato, J. Miao, M. Koskiniemi, R. Norio, G. Avanzini, S. Franceschetti, R. Michelucci, C. A. Tassinari, S. Omer, L. A. Pennacchio, R. M. Myers, J. L. Dieguez-Lucena, R. Krahe, A. de la Chapelle, and A.-E. Lehesjoki. 1997. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat. Genet. 15:393–396. [DOI] [PubMed] [Google Scholar]
- 68.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808. [DOI] [PubMed] [Google Scholar]
- 69.White, M. A., P. Detloff, M. Strand, and T. D. Petes. 1992. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr. Genet. 21:109–116. [DOI] [PubMed] [Google Scholar]
- 70.White, M. A., M. Dominska, and T. D. Petes. 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White, M. A., M. Wierdl, P. Detloff, and T. D. Petes. 1991. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc. Natl. Acad. Sci. USA 88:9755–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu, F., and T. D. Petes. 1996. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics 143:1115–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]