Abstract
Phosphorylation at a highly conserved serine residue (Ser-10) in the histone H3 tail is considered to be a crucial event for the onset of mitosis. This modification appears early in the G2 phase within pericentromeric heterochromatin and spreads in an ordered fashion coincident with mitotic chromosome condensation. Mutation of Ser-10 is essential in Tetrahymena, since it results in abnormal chromosome segregation and extensive chromosome loss during mitosis and meiosis, establishing a strong link between signaling and chromosome dynamics. Although mitotic H3 phosphorylation has been long recognized, the transduction routes and the identity of the protein kinases involved have been elusive. Here we show that the expression of Aurora-A and Aurora-B, two kinases of the Aurora/AIK family, is tightly coordinated with H3 phosphorylation during the G2/M transition. During the G2 phase, the Aurora-A kinase is coexpressed while the Aurora-B kinase colocalizes with phosphorylated histone H3. At prophase and metaphase, Aurora-A is highly localized in the centrosomic region and in the spindle poles while Aurora-B is present in the centromeric region concurrent with H3 phosphorylation, to then translocate by cytokinesis to the midbody region. Both Aurora-A and Aurora-B proteins physically interact with the H3 tail and efficiently phosphorylate Ser10 both in vitro and in vivo, even if Aurora-A appears to be a better H3 kinase than Aurora-B. Since Aurora-A and Aurora-B are known to be overexpressed in a variety of human cancers, our findings provide an attractive link between cell transformation, chromatin modifications and a specific kinase system.
A critical process in cell division concerns the dynamic chromosomal restructuring that is necessary for the proper transmission of parental genetic information to the two daughter cells (39). Although the molecular steps implicated in chromosome dynamics are essential, the intracellular signaling pathways governing them are only poorly understood. During G2 interphase to M phase, the relaxed interphase chromatin is converted into mitotic condensed chromosomes, a process likely to be essential for the subsequent nuclear division (27). The transition between the condensed and relaxed states of chromatin has been correlated with a number of posttranslational modifications of the histone molecules, including phosphorylation, acetylation, and methylation (7, 25, 29, 53).
Phosphorylation of histones H1 and H3 has been linked with mitotic chromatin condensation. H1 hyperphosphorylation is temporally associated with entry into mitosis and depends on Cdc2 kinase activity (5, 34). Recent studies, however, have revealed that chromatin condensation can occur without H1 hyperphosphorylation (2, 21) or even without H1 itself (40, 41, 52). Therefore, the biological significance of mitotic H1 hyperphosphorylation remains undefined.
Histone H3 is phosphorylated during mitosis on at least two serine residues, Ser-10 (22, 42, 59) and Ser-28 (19). Phosphorylation at Ser-10 begins in early G2 in the pericentromeric heterochromatin of each chromosome (26) and by metaphase has spread throughout all chromosomes, while phosphorylation on Ser-28 starts to be evident only in early mitosis. The different timing of Ser-10 and Ser-28 phosphorylation events suggests that diverse signaling routes are utilized for the two sites. Mitotic H3 phosphorylation at Ser-10 is required for proper execution of mitosis in Tetrahymena (26, 58–60), whereas Saccharomyces cerevisiae strains bearing Ser-to-Ala mutations at positions 10 and/or 28 in the H3 tail have no apparent mitotic defects (28). This may indicate redundancy with modifications on other histones in some specific systems versus others. Moreover, phosphorylation on H3 Ser-10 appears to be involved in the initiation, but not the maintenance, of mammalian chromosome condensation (58). Additional evidence indicates that the N-terminal tail of histone H3 is able to stably bind its mitotic kinase, inhibiting chromatin condensation in an in vitro system (11). In this study an H3 kinase activity could be sequestered in vitro by nucleosome complexes in peptide competition experiments.
Recently, members of the Aurora kinase family have been shown to be involved in mitotic phosphorylation of histone H3 at Ser-10 both in yeast and nematodes (28). While yeast cells contain only one protein of the family, Ipl1 (17), two Aurora-like kinases are present in Drosophila (17, 43) and Caenorhabditis elegans (47, 48) and at least three are present in mammals (3, 18, 31–33, 50, 56). We will refer to the mammalian kinases as Aurora-A, Aurora-B, and Aurora-C, as recently suggested (38). The three mammalian kinases have 67 to 76% homology in their catalytic domain and are expressed in a cell cycle-regulated fashion with a peak of expression at the G2/M phase transition (3, 32, 56). The kinase activity of the Aurora-A subfamily peaks in G2 and in prophase, prior to the full activation of Aurora-B and before the maximal activation of p34cdc2 (3). Importantly, members of this family are overexpressed in a variety of cancers, underscoring the pivotal role that Aurora kinases play in governing cell proliferation (3, 54, 55, 62).
Studies on the intracellular localization of Aurora kinases in mitotic cells have suggested association with mitotic structures (4), in keeping with their distinct subcellular localization: Aurora-A has been found at the centrosomes of interphase cells and at the spindle poles of metaphase cells (3, 18, 31), Aurora-B has been found at the midbody of anaphase cells and at the post-mitotic bridge of telophase cells (3, 56), while Aurora-C appears to be localized to the centrosomes of anaphase cells (32). Increasing evidence points to a potential central role of Aurora-A and Aurora-B in the correct execution of the mitotic program. In particular, the Aurora-A-type enzymes appear to be required to maintain the separation of centrosomes to give normal bipolar spindle structure, as shown genetically in Drosophila (17) and also biochemically in Xenopus, where the corresponding pEg2 kinase is efficiently inhibited by specific antibodies or inactive mutants (16, 44). In contrast, RNA interference (RNAi) experiments with Aurora-A in C. elegans show no prevention of centrosome seperation, although both spindle formation and centrosomal morphology were abnormal (47).
The Aurora-B kinases, on the other hand, appear to be required for cytokinesis, as indicated by ectopic expression of an inactive kinase mutant in cultured mammalian cells (55, 56). An effect on cytokinesis has also been reported by blocking the function for the C. elegans B-type enzyme, Air-2 (30, 48, 51).
The role of Aurora kinases in phosphorylating H3 has also been explored. RNAi experiments with Drosophila and C. elegans indicate that targeting Aurora-B causes a reduction of histone H3 phosphorylation during mitosis (15, 28) whereas, at least in C. elegans, Aurora-A does not appear to be implicated (28). The situation is less clear in Xenopus, where both Aurora-A (49) and Aurora-B (37) were identified as potential histone H3 mitotic kinases. Thus, although a role for Aurora-B in mitotic H3 phosphorylation seems to be conserved throughout metazoans, it is not clear whether Aurora-B directly phosphorylates histone H3. Indeed, Aurora-B could be associated with a distinct mitotic chromosomal kinase or, once localized at the centromere, could regulate the activity of other kinases positioned along the chromosome arms. Another open question is whether Aurora-B acts alone or in concert with other kinases, such as Aurora-A.
We have investigated the spatio-temporal expression of Aurora-A and Aurora-B during the mitotic cell cycle of mammalian cells. The two kinases have a mutually exclusive localization: while Aurora-A is present in the centrosomes, Aurora-B colocalizes with the phosphorylated form of histone H3. Both kinases bind the H3 N-terminal tail, and both phosphorylate Ser-10 in vitro and in vivo. Our results favour a scenario where a dynamic relocalization of Aurora kinases dictates the signaling specificity at distinct mitotic checkpoints.
MATERIALS AND METHODS
Cell culture and synchronization.
NIH 3T3 mouse fibroblasts, HeLa human cervical adenocarcinoma cells, and HEK293 human embryonic kidney cells were cultured as recommended by the American Type Culture Collection (Rockville, Md.). HEK293 cells were transfected in 10-cm plates with 10 μg of the indicated plasmid by the calcium phosphate coprecipitation method (20). For synchronization studies, HeLa and NIH 3T3 cells were plated at 80% confluence and after 4 to 6 h were blocked at the G1/S boundary by exposure to 2 mM thymidine (Sigma) for 14 h. The plates were then washed three times with a 1× phosphate-buffered saline solution, and normal growth medium was added. After 11 h, 2 mM thymidine was added for an additional 14 to 16 h. The plates were then washed three times with 1× phosphate-buffered saline, and normal growth medium was added. This time point, corresponding to the G1/S transition, was designated time zero.
Antibodies and plasmids.
cDNAs corresponding to the human Aurora kinases (Aurora-A [accession no. AF008551.1], Aurora-B [accession no. AF0085.5], and Aurora-C [accession no. AF054621.1]) were PCR amplified from a HeLa cells cDNA library and cloned in the expression vectors pCS2myc (45) and pET-28 (Novagen). The mutations in Aurora-A (D274N in the putative catalytic domain), Aurora-B (K106R in the putative ATP-binding site), and Aurora-C (D184N in the putative catalytic domain) were generated using the Quikchange site-directed mutagenesis kit (Stratagene). The antibodies used in the present study were rabbit anti-P.H3 (Upstate Biotechnology), mouse anti-P.H3 (New England Biolabs), anti-myc (E910 mouse monoclonal antibody) (Transduction Laboratories), anti-IAK1 (mouse Aurora-A; referred to in the figures as Aurora-A) (Transduction Laboratories). The polyclonal anti-Aurora-B/Nter anti-Aurora-B/Cter antibodies were generated in our laboratory by immunizing rabbits with keyhole limpet hemocyanin-coupled peptides corresponding to a N-terminal sequence (amino acids [aa] 1 to 17) or to a C-terminal sequence (aa 332 to 344) of the human Aurora-B protein.
In-gel kinase assays.
HeLa cells from different stages of the cell cycle were lysed directly in 0.5 ml of 1× Laemmli buffer (total extract) or in 1 ml of kinase lysis buffer (soluble fraction) (50 mM HEPES [pH 8.0], 600 mM KCl, 0.5% NP-40, 1 mM Na3VO4, 50 mM NaF, 1 mM dithiothreitol [DTT], 1 mM microcystine, protease inhibitor cocktail). The lysate was then centrifuged at 12,000 × g for 10 min at 4°C, and 1 ml of 2× Laemmli sodium dodecyl sulfate (SDS) sample buffer was added to the supernatant. The pellet (insoluble fraction) was then resuspended in 0.5 ml of 1× Laemmli SDS sample buffer.
Kinase activity gel analyses were performed as described previously (36). Protein extracts were loaded on gels containing 0.1 mg of calf thymus H3 (Boehringer) per ml or 0.5 mg of bovine serum albumin (BSA) per ml in the separating portions. After electrophoresis, the SDS was removed by washing the gels with 20% 2-propanol-50 mM Tris-HCl (pH 8.0); proteins were denaturated by treating the gel with 6 M guanidine-HCl-50 mM Tris-HCl (pH 8.0)-5 mM 2-mercaptoethanol and then renatured overnight at 4°C in a buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM 2-mercaptoethanol, 0.04% (vol/vol) Tween 40, 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT. After three washes in the assay buffer (40 mM HEPES [pH 7.4], 2 mM MnCl2, 5 mM MgCl2, 1 mM DTT, 0.2 mM EGTA), the gels were incubated for 4 h at room temperature in 3 ml of assay buffer containing 50 μM ATP and 70 μCi of [γ32-P] ATP (3,000 Ci/mole; NEN). After four washes with 1% tetrasodium pyrophosphate-5% trichloroacetic acid and staining with Coomassie blue R-250, the gels were exposed for autoradiography.
Western analyses.
Protein extracts were resolved by standard SDS-polyacrylamide gel electrophoresis (PAGE). Samples were electroblotted onto Protan nitrocellulose (Schleicher & Schuell). The membranes were incubated for 12 h at 4°C in PBS-5% low-fat milk containing the appropriate antibody. Donkey anti-rabbit- or anti-mouse (Jackson) antibodies conjugated to horseradish peroxidase were used to reveal immunocomplexes by enhanced chemiluminescence (Pierce).
Immunoprecipitations.
At 48 h after transfection, HEK293 cells were lysed in 1 ml of kinase lysis buffer. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatants were precleared at 4°C for 1 h with protein A-Sepharose. The anti-myc antibody was added at a 1:1,000 dilution, and after an incubation of 3 h at 4°C, the immunoprecipitates were collected by adding 20 μl of protein A-Sepharose. The beads were washed three times in lysis buffer and twice in kinase buffer.
In vitro kinase assays.
Assays were performed in 30 μl of kinase buffer (20 mM HEPES [pH 7.4], 150 mM KCl, 5 mM MnCl2, 5 mM NaF, 1 mM DTT, 50 μM ATP, 1 mM microcystine, protease inhibitor cocktail) containing the different substrates (0.5 mg of myelin basic protein (MBP) (Sigma) per ml, 0.5 mg of histone mix (Boehringer) per ml, 0.25 mg of the indicated glutathione S-transferase (GST) fusion per ml, or 0.1 mg of mononucleosomes per ml with or without 20 mM [γ32-P] ATP). The reaction mixtures were incubated at 37°C for 20 min, the reactions were stopped by the addition of 30 μl of 2× Laemmli SDS sample buffer, and the reaction products were denatured for 10 min at 95°C. Proteins were then separated by standard SDS-PAGE and analyzed by autoradiography or by Western blotting.
Mononucleosome preparation.
Nuclei of HeLa cells were prepared as described previously (14). The DNA concentration in the nuclei suspension was determined by measuring the UV absorption at 260 nm in a 2 M NaCl solution. Nuclei were digested with micrococcal nuclease (25 U/200 μg of DNA) at 37°C for 30 min and then incubated in 2 mM EDTA on ice for 10 min. After centrifugation, the supernatant was loaded on a 5 to 20% sucrose gradient (15 mM Tris-HCl [pH 7.4], 0.67 M NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]). After ultracentrifugation (105 × g for 40 h at 4°C), fractions were collected and analyzed by Coomassie staining of an SDS-PAGE (15% polyacrylamide) gel. Positive fractions were dialyzed against 1 mM Tris-HCl (pH 7.4), lyophilized, and dissolved in kinase buffer.
Expression and purification of His-tag Aurora kinases.
pET-28-Aurora constructs were transformed in competent cells of the protease-deficient Escherichia coli strain BL21, and expression was induced by adding isopropyl-β-𝒹-thiogalactopyranoside (IPTG) to 1 mM for 3 h at 30°C. The cells were collected and lysed in a buffer containing 500 mM NaCl, 50 mM sodium phosphate buffer, 20% glycerol, 1% NP-40, 1 mM DTT, 0.5 mM PMSF, and 200 μg of lysozyme per ml. After incubation on ice and three freeze-thaw cycles, 10 μg of DNase I per ml was added and the samples were incubated for a further 30 min on ice. After centrifugation at 9,000 ×g for 30 min, the supernatants were purified using Ni-nitrilotriacetic acid beads as recommend by the manufacturer (Qiagen).
GST fusions.
The GST-H3 tail construct was obtained by cloning a PCR fragment corresponding to aa 1 to 30 of human histone H3 in the pGex-4T1 vector (Pharmacia). Mutation of Ser-10 to alanine was generated using the Quickchange site-directed mutagenesis kit (Stratagene). The GST-H4 plasmid was a gift from A. Imhof. The GST fusions were expressed in E. coli, extracted in 1× PBS by sonication, and purified on glutathione-Sepharose resin (Pharmacia). They were then eluted by adding 10 mM reduced glutathione.
GST pull-down assay.
The GST fusions were expressed in E. coli, extracted in BC0 (20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 20% glycerol, 1 mM DTT, 0.5 mM PMSF) containing 500 mM KCl and 1% NP-40, sonicated, and purified on glutathione-Sepharose resin (Pharmacia). Portions (500 ng) of different GST, GST-H3, GST-H3S10A, and GST-H4 fusions were incubated overnight at 4°C with 1 mg of HeLa cell protein extract, prepared by lysing the cells in EBC buffer (50 mM Tris-HCl [pH 8.0], 170 mM NaCl, 0.5% NP-40, 1 mM PMSF, 50 mM NaF, 1 mM microcystine, protease inhibitor cocktail). After three washes in EBC buffer, the beads were used for in vitro kinase assays or Western blot analyses.
Immunofluorescence.
Cells were grown on 35-mm plates, fixed with 4% paraformaldehyde in 1× PBS, and permeabilized with 0.2% Triton-1× PBS. After blocking for 1 h in 5% BSA in 1× PBS-0.05% Tween 20, the plates were incubated with the indicated antibody overnight at 4°C (rabbit anti-P.H3 antibody, 1:2,000; mouse anti-P.H3 antibody, 1:200; anti-Aurora-A antibody, 1:250; anti-Aurora-B/Nter antibody, 1:500). The plates were then washed with 1× PBS and incubated with a secondary antibody (Cy3-conjugated anti-mouse serum and a fluorescein isothiocyanate-conjugated anti-rabbit serum at 1:100 and 1:200, respectively [Jackson]). Immunostained samples were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Boehringer) before being subjected to microscopy. Image collection was done on a DMLB Leica microscope with an HBO 100-W lamp.
RESULTS
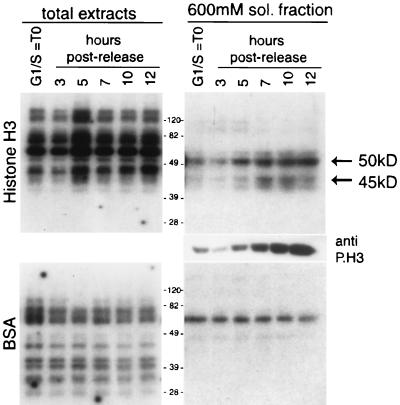
The activity of two H3 kinases, of 50 and 45 kDa, correlates with histone H3 mitotic phosphorylation.
The in-gel kinase assay has already been successfully applied to identify histone H3 Ser-10 mitogen-induced kinases from cell lysates (46). We used this method to determine changes in histone H3 protein kinase activity during the cell cycle. The cell synchronization protocol used in this study, the double thymidine block and release, yields about 60 to 70% mitotic cells at 10 h for HeLa cells and 6 h for NIH 3T3 cells, as analyzed by flow cytometry (data not shown). Protein extracts from synchronized HeLa cells were collected at different times after release from the thymidine block during the G1/S-to-M-phase transition. Equivalent amounts of protein lysates were analyzed by an in-gel kinase activity assay using purified histone H3 as substrate (Fig. 1). Several protein species were detected in total-cell extracts (Fig. 1, left panel), but none displayed activity kinetics paralleling changes in H3 phosphorylation (as detected with an anti-P.H3 antibody [26]). In contrast, the soluble fraction contained two dominant protein species of about 50 and 45 kDa, whose activity tightly correlated with histone H3 phosphorylation on Ser-10 (see Western analysis of total extracts, right middle panel). Activity is not due to kinase autophosphorylation, as demonstrated when identical samples were analyzed with BSA (Fig. 1, bottom panel). Thus, at least two H3 kinases are regulated during the cell cycle in parallel with the specific Ser-10 phosphorylation event. The molecular size, the timing of enzymatic activation postrelease, and the identity of the recently identified yeast H3 mitotic kinase (28) suggested that the catalytically active enzymes might be Aurora-A and Aurora-B.
FIG. 1.
The activity of two H3 kinases of 50 and 45 kDa correlates with histone H3 mitotic phosphorylation. HeLa cells were synchronized by a double thymidine block and harvested at 0, 3, 5, 7, 10, and 12 h after being released from the block. Progression through the cell cycle and the consequent accumulation of phosphorylated histone H3 was verified by Western blotting using the anti-phosphorylated histone H3 antibody (anti-P.H3). Protein extracts (10 μg) (total extract and 600 mM soluble fractions), prepared at the indicated times, were analyzed by an in-gel kinase assay, using histone H3 (top) or BSA (bottom) as substrates. Equal loading was checked by Coomassie staining.
Kinetics of Aurora-A and Aurora-B levels in the HeLa and NIH 3T3 cell cycle.
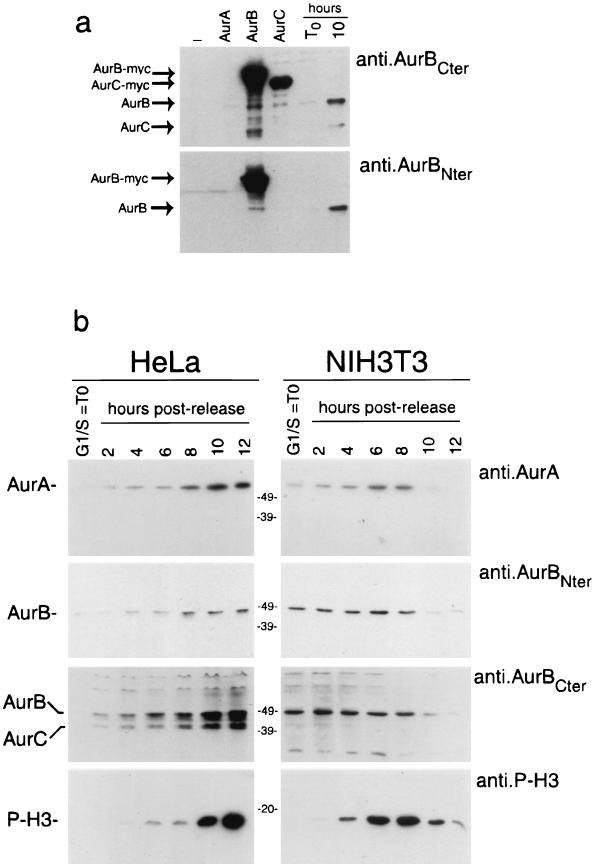
To verify the identity of the mitotically regulated H3 kinases, we set out to generate Aurora-specific antibodies. While reliable anti-Aurora-A antibodies are commercially available (18), we needed to produce antibodies specific for Aurora-B. To do so, two peptides, corresponding to the N-terminal and C-terminal amino acid sequences at aa 1 to 17 and aa 332 to 344, respectively, were synthesized. Polyclonal antibodies were generated and are referred to as anti-AurB-Nter and anti-AurB-Cter. To test the specificity of these antibodies, we performed Western blot analyses on HEK293 cells transfected with the three Myc-tagged Aurora kinases. The anti-AurB-Nter antibody is highly specific since it recognizes only Aurora-B and no other member of the family, even if overexpressed; the anti-AurB-Cter antibody recognizes both Aurora-B and Aurora-C but not Aurora-A (Fig. 2a). When these antibodies were tested for the detection of endogenous proteins, using extracts from HeLa cells arrested in G1/S or 10 h postrelease, the original Aurora-B and Aurora-C proteins were detected, paralleling the same specificity as obtained using the Myc-tagged proteins (Fig. 2a).
FIG. 2.
Coordinated accumulation of Aurora kinases and H3 phosphorylation during the G2/M transition. (a) Western blot analysis using anti-Aurora-B-Cter and anti-Aurora-B-Nter on 10 μg of total-protein extracts prepared from HeLa cells nontransfected, transfected with myc-tagged Aurora-A, Aurora-B, or Aurora-C, and arrested in G1/S or 10 h after release. (b) Total-protein extracts (10 μg) prepared from synchronized HeLa and NIH 3T3 cells harvested at the indicated times were analyzed by Western blot analysis using anti-Aurora-A, anti-Aurora-B-Cter, anti-Aurora-B-Nter, and anti-P.H3.
We then set out to study the comparative kinetics of Aurora kinase accumulation during mitosis in HeLa and NIH 3T3 cells. Protein extracts from synchronized cells were collected at different times after release from the block during the S-to-M-phase transition. These were analyzed for the expression of Aurora kinases in correlation to H3 phosphorylation (Fig. 2b).
HeLa and NIH 3T3 cells show remarkable differences in the kinetics of accumulation of the Aurora kinases: in HeLa cells all three isoforms are present during the G2/M transition and their accumulation parallels the phosphorylation of the histone H3 at Ser-10; in contrast, NIH 3T3 cells arrested at the G1/S boundary already display high levels of Aurora-A and Aurora-B, which only doubles during progression in mitosis. Finally, the anti-AurB-Cter reveals an extra band, undetectable with the anti-AurB-Nter antibody, which corresponds in the molecular weight to Aurora-C in HeLa cells extracts.
Temporal and spatial correlation between Aurora kinase expression and the mitotic phosphorylation of histone H3.
Studies of the intracellular localization of Aurora kinases in mitotic cells have shown that these are associated mainly with mitotic structures (4). In particular, Aurora-A has been found in the centrosomes of interphase cells and the spindle poles of metaphase cells (3, 18, 31), Aurora-B has been found at the midbody of anaphase cells and at the postmitotic bridge of telophase cells (3, 56), while Aurora-C appears to be localized to centrosomes of anaphase cells (32).
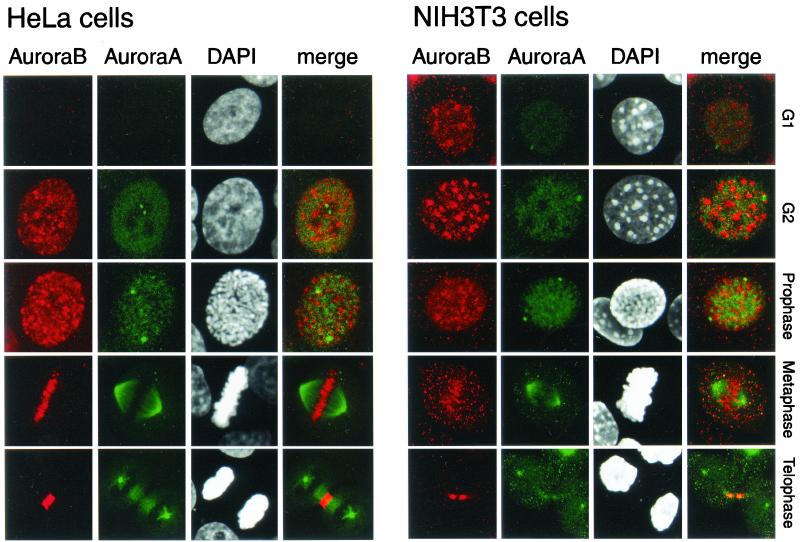
We wanted to study in detail the intracellular localization of Aurora-A and Aurora-B in conjunction with the mitotic signal of H3 phosphorylation. Immunofluorescence analyses were performed for both HeLa and NIH 3T3 cells arrested at different phases of the cell cycle (Fig. 3). Confirming the Western analysis (Fig. 2b), there was a drastic difference between HeLa and NIH 3T3 cells at G1/S. Indeed, while both Aurora-A and Aurora-B were expressed in NIH 3T3 cells, they were both absent in HeLa cells. This is the only notable difference between the two cell types, since in the other phases of the cell cycle the intracellular distributions of the two kinases were basically identical. In G2, Aurora-A was both nuclear and centrosomic while Aurora-B had a punctuate distribution throughout all regions of condensing chromosomes, from prophase throughout metaphase. At prophase and metaphase the concentration of Aurora-A in the centrosomic region and in the spindle poles was strongly increased. Aurora-B was present mostly in the centromeric region and relocated by cytokinesis to the midbody region is telophase (Fig. 3). These results clearly show an almost mutually exclusive distribution of the two kinases indicating, independently from the similarity in their primary sequence, a specificity of substrate.
FIG. 3.
Localization of Aurora-A and Aurora-B in HeLa (left panels) and NIH 3T3 (right panels) cells in G1/S, G2, prophase, metaphase, and telophase. HeLa and NIH 3T3 cells at various stages of the cell cycle were stained for Aurora-B (using anti-Aurora-B-Nter), Aurora-A (using anti-IAK-1), and DNA (using DAPI). Double labeling is indicated as a merge. The cell cycle phases were identified by the DAPI staining.
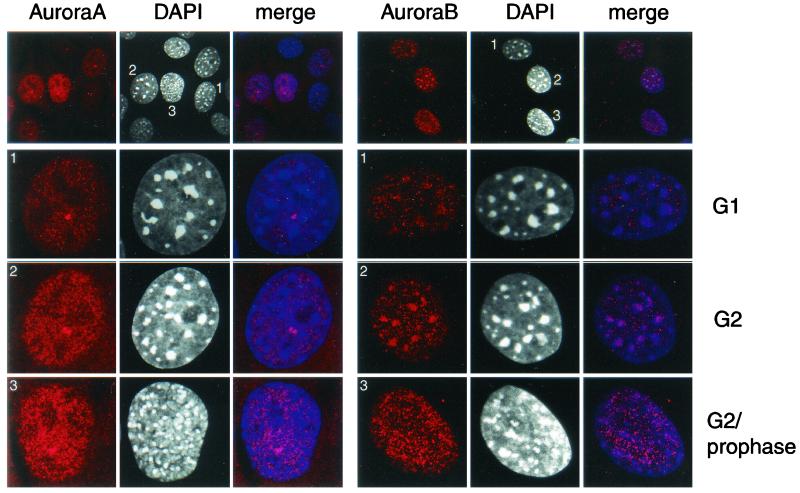
To better investigate the distribution of Aurora-A and Aurora-B during the G1 and G2 phases, we performed confocal analysis on NIH 3T3 cells. Several fields were studied, and a representative analysis is presented in Fig. 4. This field contains cells at different stages of the G1-S-G2 transition. Confocal imaging was performed with a 40× objective, as shown in the top panel, and at higher resolution (×100 with a 4× zoom) as shown in the other panels. There is a clear increase in the intensity of the signal corresponding to both kinases during the G1-S-G2 transition. However, while the localization of Aurora-A does not change during time, that of Aurora-B does. During interphase, Aurora-B is partially localized at the G1 heterochromatin sites, but by the G2 phase, staining clearly correlates with condensed regions of DNA observed by DAPI staining.
FIG. 4.
Localization of Aurora-A (left panels) and Aurora-B (right panels) in NIH 3T3 cells in G1/S, G2, and G2/prophase. NIH 3T3 cells at various stages of the cell cycle were stained for Aurora-B (using anti-Aurora-B-Nter), Aurora-A (using anti-IAK-1), and DNA (using DAPI). Double labeling is indicated as a merge. The cell cycle phases were identified by the DAPI staining.
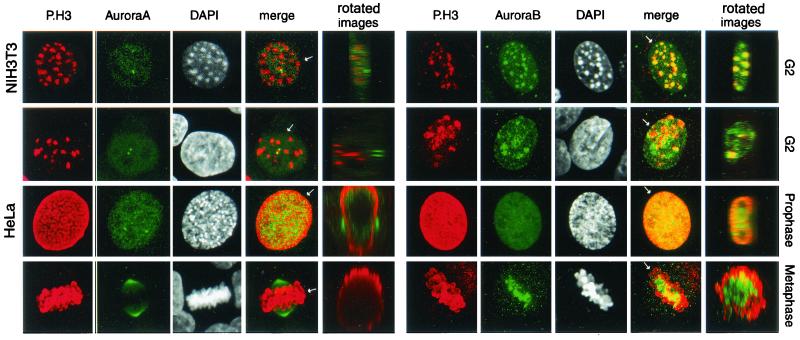
To investigate the possible coupling between the localization of either Aurora-A or Aurora-B and the phosphorylation of histone H3, confocal analysis was performed on HeLa and NIH 3T3 cells arrested in G2, prophase, and metaphase. Aurora-B colocalized with the condensing chromosome, a location where H3 began to be phosphorylated in both NIH 3T3 (Fig. 5, upper left panel) and HeLa (Fig. 5, second panel on the left) cells. In contrast, although Aurora-A was nuclear in G2, no colocalization with phosphorylated H3 was observed. During prophase and later in metaphase, Aurora-A distribution appeared independent from phosphorylated H3 while Aurora-B was in the centromeric region and partially colocalized with phosphorylated H3. Optical section were taken from the apical to basal side of the cells. The whole stack of images was subsequently rotated by 90° around the axis to produce a Z-section (panel “rotated images” in Fig. 5), indicating that only Aurora-B colocalizes with the phosphorylated histone H3 from the G2 phase to metaphase. The subsequent relocalization in the midbody region during telophase indicates that Aurora-B appears to behave as a so so-called passenger protein, maybe through the interaction with INCENP (inner centromere protein) as shown in Drosophila (1, 30).
FIG. 5.
Localization of Aurora-A and Aurora-B in relation to that of P.H3 in HeLa and NIH 3T3 cells during the G2, phase, prophase, and metaphase. NIH 3T3 cells in G2 (top row) and HeLa cells in G2, prophase, and metaphase (bottom three rows) were stained with anti-P.H3 in combination either with anti-IAK-1, to identify Aurora-A (left panels), or with anti-Aurora-BNter, to identify Aurora-B (right panels). The cell cycle phases were identified by the DAPI staining. Double labeling is indicated as a merge. The whole stack of images for each individual panel was subsequentely rotated by 90° on the axis (see arrow) to produce a Z-section (panel “rotated images”).
Both Aurora-A and Aurora-B physically interact with the H3 tail.
The observations concerning the intracellular localization of Aurora proteins suggested that these may be directly involved in phosphorylating H3. The recent description that the amino-terminal tail of histone H3 is able to stably bind its mitotic kinase (11) prompted us to question whether Aurora kinases may be able to interact with the H3 tail. Indeed, an excess of a peptide corresponding to the H3 N-terminal tail was shown to prevent DNA condensation in Xenopus egg extracts (11), probably because it was able to interfere with H3 kinase activity. To test whether Aurora-A and Aurora-B could physically associate with the H3 N-terminal tail, we performed GST pull-down experiments (Fig. 6).
FIG. 6.
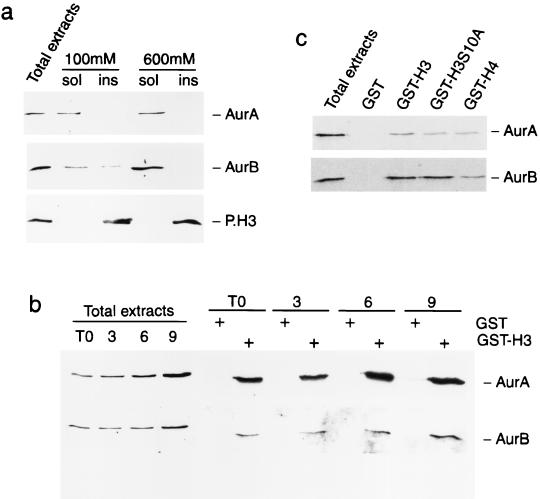
Interaction between the N-terminal tail of histone H3 and Aurora-A and Aurora-B. (a) Different susceptibilities of Aurora-A, Aurora-B, and P.H3 to increasing concentrations of salt. Protein extracts were prepared from HeLa cells using buffers with 100 and 600 mM salt concentration and analyzed by Western blotting with anti-IAK-1 to identify Aurora-A (top panel) or with anti-Aurora-B-Nter to identify Aurora-B (bottom panel). (b) Aurora-A and Aurora-B interact with the histone H3 tail. Total-protein extracts from synchronous HeLa cells were incubated with GST alone or GST-H3, and the pulled-down products were tested by Western blotting with anti-Aurora-A (top panel) and anti-Aurora-B-Nter (bottom panel) antibodies. (c) Aurora-A and Aurora-B interact with the histone H4 tail less efficiently than with the H3 tail. Equal amounts of GST fusions (not shown) were verified by Coomassie staining.
To optimize the extraction protocol of the Aurora kinases, exponentially growing HeLa cells were lysed directly in Laemmli buffer (total-cell extracts) or in lysis buffer (soluble fraction; sol in Fig. 6), containing either 100 or 600 mM NaCl. The lysate was then centrifuged, and the pellet (insoluble fraction; ins in Fig. 6) was resuspended in Laemmli SDS sample buffer. Aurora-A (Fig. 6a, top panel) was readily soluble at 100 mM NaCl. In contrast Aurora-B (Fig. 6a, middle panel) was fully solubilized only at higher salt concentrations, indicating its tight association with chromatin structures. As expected, the phosphorylated H3 was insoluble at both salt concentrations (Fig. 6a, bottom panel). Interestingly, the salt concentration used in this experiment to reveal differential solubility of the two Aurora kinases also allowed the detection of H3 kinase activities in the in-gel kinase assay (Fig. 1).
Protein extracts prepared by lysis in 600 mM salt from HeLa cells arrested in G1/S or at 3, 6, or 9 h postrelease were incubated with a bacterially generated H3 tail fused to GST or with GST alone as a control. Both Aurora-A and Aurora-B kinases specifically interact with the GST-H3 fusion but not with GST alone (Fig. 6b). A moderate but reproducible increase in the amount of protein bound to GST-H3 was observed, in accordance with the increased amount of Aurora-A and Aurora-B during the G1/S-to-M transition. Control unrelated kinases were tested for specificity (data not shown).
The capacity of Aurora-A and Aurora-B to associate with an H3 tail where the Ser-10 is mutated into Ala was also analyzed. Both kinases still show binding to the mutated tail, although with a slightly decreased efficacy, especially for Aurora-A (Fig. 6c). This result is important because it stresses that the site of interaction of the Aurora kinases on the H3 tail involves additional residues as well as Ser-10. We have also used the tail of histone H4. Interestingly, the H4 tail can also bind the Aurora kinases (Fig. 6c), although with a weaker efficacy than the H3 tail for both kinases. While it is unclear whether this result is physiologically meaningful, it may indicate that Aurora kinases could have a number of possible chromatinic substrates. At any rate, our results show that both Aurora kinases efficiently interact with the H3 tail.
Histone H3 kinase activity of Aurora kinases.
The in-gel kinase assays we performed (Fig. 1) were suggestive of H3 being a direct substrate of Aurora kinases. Several lines of evidence indicate that Aurora-B, more than Aurora-A, may induce Ser-10 phosphorylation of histone H3 during mitosis and thereby play a role in chromatin condensation (1, 28). However, it is yet unclear whether Aurora kinases directly phosphorylate Ser-10 or if they activate a yet unidentified additional H3 kinase.
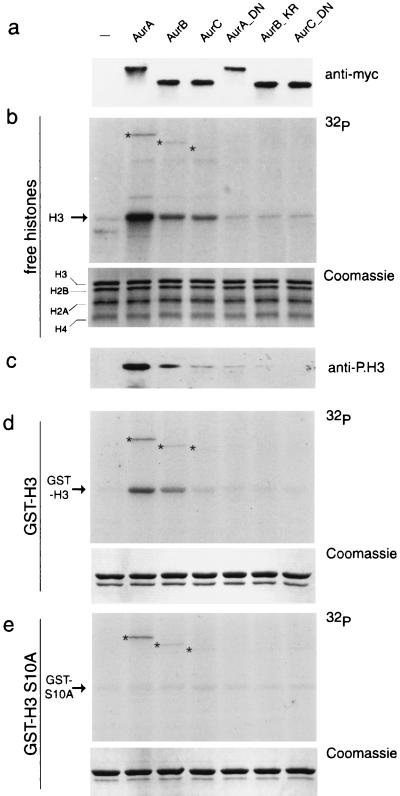
We assessed the ability of all three Aurora kinases to phosphorylate histone H3. To this aim, Myc-tagged Aurora kinases were ectopically expressed in HEK293 cells and then tested in kinase assays after immunoprecipitation (Fig. 7a). Although in this assay a mixture of histones was used, both Aurora-A and Aurora-B efficiently and specifically phosphorylate histone H3 (Fig. 7b). Phosphorylation occurred predominantly on Ser-10, as indicated by the use of the anti-phospho-H3 antibody on the products of the kinase reaction (26) (Fig. 7c). To validate this observation, we performed an analogous experiment using as substrates GST-H3 and its mutated form GSTH3-S10A, where Ser-10 is mutated into Ala. All three Aurora kinases readily phosphorylated GST-H3, although with different efficacy (Fig. 7d), while the Ser-to-Ala mutation at position 10 abolished phosphorylation (Fig. 7e). It is noteworthy that in all three kinase assays in Fig. 7, the levels of autophosphorylation of each Aurora kinase were unchanged. This experiment also demonstrates that Ser-28 is a poor substrate for Aurora kinases. The different ability of the three Aurora kinases to phosphorylate H3 is not due to different levels of expression (Fig. 7a, top panel). Finally, no H3 phosphorylation was seen with catalytically inactive Aurora proteins (Aur-A_D274N, Aur-B_K106R, and Aur-C_D184N; [Fig. 7]), indicating that H3 phosphorylation is not due to Aurora-associated kinases copurified in the immunoprecipitation.
FIG. 7.
Preferential phosphorylation of H3 at Ser-10 by Aurora kinases. (a) HEK293 cells were transfected with expression plasmids encoding Myc-tagged Aurora-A, Aurora-B, and Aurora-C kinases and the mutated isoforms Aur-A_D274N, Aur-B_K106R, and Aur-C_D184N. Protein extracts prepared from transfected cells were subjected to immunoprecipitation using the anti-Myc antibody. Immunoprecipitated complexes were analyzed by Western blotting to test the expression level of each kinase. (b and c) In vitro kinase assay performed on immunoprecipitated complexes using a histone mix as the substrate. Autoradiography (b, top panel) and Western blotting with anti-P.H3 antibody (c, bottom panel) are shown. The equal amounts of histones in each reaction were verified by Coomassie staining (middle panel). (d and e) In vitro kinase assay performed using the same samples described in panel b and GST-H3 and GSTH3-S10A as substrates; autoradiography (top panel) and Coomassie staining (bottom panel) are shown. The asterisks indicate the bands corresponding to the autophosphorylated Aurora kinases.
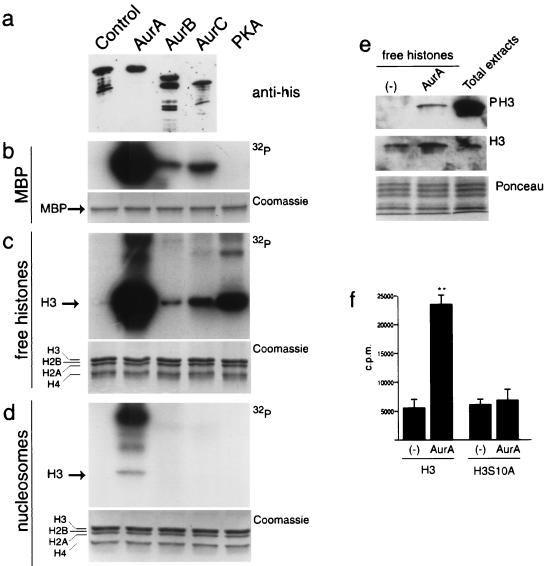
We extended our analysis by testing the ability of recombinant Aurora-A, Aurora-B, and Aurora-C to phosphorylate H3. His-tagged recombinants were produced in E. coli, and their kinase activity was tested on different substrates. Although with remarkable differences in efficacy, all three Aurora kinases phosphorylated MBP (Fig. 8b) and H3 in a free mixture of histones (Fig. 8c). Aurora-A displayd a kinase activity significantly greater than the other two members of the family. This higher activity possibly explains why, when the three kinases were tested on mononucleosome preparations, only Aurora-A appeared to phosphorylate histone H3 (Fig. 8d). The different abilities of the three Aurora kinases to phosphorylate H3 and MBP are not due to different levels of expression (Fig. 8a, top panel) but are likely to be directly related to their different intrinsic activities.
FIG. 8.
Kinase activity of the recombinant bacterial Aurora kinases. (a) Western blot analysis performed on the recombinant bacterial Aurora kinases using the anti-His antibody. (b to d) In vitro kinase assay performed with recombinant bacterial Aurora-A, Aurora-B, and Aurora-C and protein kinase A on MBP (b), free histones (c), and nucleosomes (d). The presence of equal amounts of histones in each reaction was verified by Coomassie staining. (e) In vitro kinase assay performed with recombinant bacterial Aurora-A analyzed by Western blotting. (f) In vitro kinase assay performed with recombinant bacterial Aurora-A on wild-type and mutated (S10A) histone H3 peptides (aa 1 to 30).
Western blot analyses using the anti-phospho-H3 antibody confirm that Aurora-A (Fig. 8e) and Aurora-B (data not shown) phosphorylate H3 on Ser-10. Finally, we performed in vitro kinase assays with Aurora-A on a N-terminal H3 peptide (aa 1 to 30) and on an equivalent peptide carrying the Ser-10-to-Ala mutation. The results reveal that the mutation of the Ser-10 site impairs phosphorylation by Aurora-A (Fig. 8f).
DISCUSSION
Mitotic H3 phosphorylation is a modification conserved in evolution that is thought to be essential for chromatin condensation during cell division (24). It first appears in the G2/M phase, initiating at pericentromeric heterochromatin, and then progresses along the chromosomal arms until it spreads to the whole chromosome (15, 23, 26). While it is still unclear how H3 phosphorylation may elicit the condensation process, a causal link was established by the finding that a Ser-to-Ala mutation at the conserved Ser-10 residue in Tetrahymena results in abnormal chromosomal segregation and massive chromosome loss (60). The critical role of the single Ser-10 residue is not confirmed in other systems (28), where it is possible that combined modifications at other sites may be needed. Indeed, phosphorylation at Ser-28 on the same H3 tail has also been correlated with chromosome condensation during mitosis (19). A likely and fascinating scenario may involve concerted modifications of other histone tails in conjunction with H3 Ser-10 phosphorylation, so that a specific “condensation code” or “program” may need to be activated to achieve proper chromosomal condensation. Although we know only very little about this possible program—and the temporal cascade of events involved—it is evident that deciphering of the signaling pathways governing it will be crucial. Here we have reported on the kinase involved in the mitotic phosphorylation of histone H3 Ser-10.
The Ser-10 residue in the histone H3 tail appears to be the target of a number of signaling kinases (8, 35, 46, 57). In response to mitogenic or hormonal stimuli, the Ser-10 site is phosphorylated with early-response kinetics which parallel the specific transcriptional activation of selected genes (9, 10, 35, 46, 57). In these settings, phosphorylation concerns only a selected subset of H3 molecules, possibly corresponding to loci of chromatin decondensation and promoter opening (6, 8, 46). Mitotic phosphorylation, on the other hand, correlates with condensation and concerns most if not all H3 molecules (26, 58, 59). This striking difference between the two “personalities” of H3 phosphorylation begs the question of how signaling specificity is achieved.
Mitotic H3 phosphorylation in yeasts and nematodes is controlled by the Ipl1/AIR-2 kinase (28), while in Aspergillus nidulans the NIMA kinase has been implicated (12). Although these are divergent kinases, their regulation during the cell cycle and genetic loss-of-function experiments strongly support their involvement. However, the question of how these kinases are regulated is still open. Both Ipl1 and NIMA are phosphoproteins and are likely to be placed downstream of cell cycle-regulated signaling pathways. Indeed, NIMA is direcly phosphorylated by NIMX, the Aspergillus homologue of p34cdc2 (61), while the upstream kinase for Ipl1 has not been identified yet.
The data presented here indicate that the mammalian mitotic H3 kinase belongs to the Aurora family, a group of proteins evolutionarily analogous to the yeast Ipl1. In mammals, however, the picture is more complex, since three members of the family are present. Our results show that the expression of Aurora-A, Aurora-B, and Aurora-C is tightly coordinated with H3 phosphorylation during the G2/M transition in HeLa cells, while in NIH 3T3 cells the accumulation of the Aurora kinases precedes the phosphorylation of histone H3. Taking advantage of a highly specific antibody for Aurora-B developed in our laboratory, we describe for the first time the intracellular colocalization of Aurora-B and phosphorylated histone H3 in the G2 phase, in both HeLa and NIH 3T3 cells.
Importantly, our in-gel kinase assay revealed two major kinases, which, because of their timing of activation and molecular weight, most probably correspond to Aurora-A and Aurora-B. Even if this assay limits the detection of activities to single subunits, it was useful to reveal multiple kinase activities during the cell cycle. Indeed, within a specific time point, the simultaneous measurement of both Aurora-A and Aurora-B shows that they can phosphorylate H3 with distinct kinetics. That both enzymes are bona fide H3 kinases is also suggested by our GST pulldown experiments demonstrating that both Aurora-A and Aurora-B physically interact with the H3 tail, independent of the reduced solubility of Aurora-B. These data are of particular interest considering the finding that the amino-terminal tail of histone H3 is able to stably bind its mitotic kinase from Xenopus egg extracts (11).
Evidence that Aurora-A and Aurora-B may act as H3 kinases is provided by our in vivo and in vitro studies. Both Aurora-A and Aurora-B efficiently phosphorylate histone H3 on Ser-10, whether the kinases are derived from immunoprecipitates of transfected HEK293 cells (Fig. 6) or from bacteria (Fig. 7). In all our assays, Aurora-A appeared to be a better H3 kinase than Aurora-B. This difference in kinase activity could be due to different signaling requirements of Aurora-B. Indeed, while the recombinant Aurora-A was efficiently autophosphorylated, Aurora-B was much less active. This may indicate that the activity of Aurora-B could be more strictly dependent on a yet undefined upstream kinase. This lack of activity of the recombinant Aurora-B probably also explains its inability to phosphorylate H3 within a nucleosomal context. The search for upstream signaling pathways leading to the cell cycle-regulated activity of Aurora-B would be of great importance. The intracellular colocalization of the Aurora-B kinase with the phosphorylated histone H3 observed in our confocal analysis is very suggestive and is strengthened by the result obtained by RNAi showing that decreased levels of Aurora-B, but not Aurora-A, reduce mitotic H3 phosphorylation in C. elegans and Drosophila (15, 28). Thus, the biochemical data obtained with Xenopus (49) and mammals (this study) indicating that Aurora-A is a more efficient histone H3 kinase would probably be challenged by future investigations aimed at identifying the kinase lying upstream of Aurora-B.
Since kinases of the Aurora family have been found to be amplified in a variety of human cancers (3, 50, 54, 62) and aberrant regulation of their function has been associated with chromosomal missegregation, aneuploidy, and tumor progression (13, 55, 56), our studies have wide implications. Additional work aimed at the elucidation of the signaling routes governing Aurora kinases during the cell division would provide important further insights. Our results stress the multiplicity of signaling pathways converging on the same chromatin modification leading to alternative remodeling of the chromatin architecture.
Acknowledgments
C. Crosio, G. M. Fimia, and R. Loury contributed equally to this work.
We thank P. Cheung, J. L. Vonesch, E. Nigg, M. Boeglin, A. Imhof, G. D. Plowman, E. Argentini, and E. Heitz for help, discussions, and generous gifts of reagents, and we thank E. Heitz and M. Rastegar for valuable technical assistance. We thank all members of the Sassone-Corsi laboratory for discussions and help.
C.C. is supported by a postdoctoral fellowship from the European Community and from the Fondation pour la Recherche Medicale; G.M.F. was supported by a postdoctoral fellowship of the Schering Foundation (Berlin). This work was supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Fondation de la Recherche Médicale, Association Française contre les Myopathies, Université Louis Pasteur, Human Frontier Science Program, Organon (Akzo/Nobel), and Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Adams, R. R., H. Maiato, W. C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajiro, K., K. Yoda, K. Utsumi, and Y. Nishikawa. 1996. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 271:13197–13201. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, J. R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, C. S. Chan, M. Novotny, D. J. Slamon, and G. D. Plowman. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, J. R., and G. D. Plowman. 1999. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454–459. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury, E. M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays 14:9–16. [DOI] [PubMed] [Google Scholar]
- 6.Chadee, D. N., M. J. Hendzel, C. P. Tylipski, C. D. Allis, D. P. Bazett-Jones, J. A. Wright, and J. R. Davie. 1999. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 274:24914–24920. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103:263–271. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905–915. [DOI] [PubMed] [Google Scholar]
- 9.Clayton, A. L., S. Rose, M. J. Barratt, and L. C. Mahadevan. 2000. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19:3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosio, C., N. Cermakian, C. D. Allis, and P. Sassone-Corsi. 2000. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 3:1241–1247. [DOI] [PubMed] [Google Scholar]
- 11.de la Barre, A. E., V. Gerson, S. Gout, M. Creaven, C. D. Allis, and S. Dimitrov. 2000. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 19:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Souza, C. P., A. H. Osmani, L. P. Wu, J. L. Spotts, and S. A. Osmani. 2000. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102:293–302. [DOI] [PubMed] [Google Scholar]
- 13.Farruggio, D. C., F. M. Townsley, and J. V. Ruderman. 1999. Cdc20 associates with the kinase aurora2/Aik. Proc. Natl. Acad. Sci. USA 96:7306–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritton, H. P., A. E. Sippel, and T. Igo-Kemenes. 1983. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 11:3467–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giet, R., and D. M. Glover. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giet, R., and C. Prigent. 2000. The Xenopus laevis aurora/Ipl1p-related kinase pEg2 participates in the stability of the bipolar mitotic spindle. Exp. Cell Res. 258:145–151. [DOI] [PubMed] [Google Scholar]
- 17.Glover, D. M., M. H. Leibowitz, D. A. McLean, and H. Parry. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81:95–105. [DOI] [PubMed] [Google Scholar]
- 18.Gopalan, G., C. S. Chan, and P. J. Donovan. 1997. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J. Cell Biol. 138:643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto, H., Y. Tomono, K. Ajiro, H. Kosako, M. Fujita, M. Sakurai, K. Okawa, A. Iwamatsu, T. Okigaki, T. Takahashi, and M. Inagaki. 1999. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 274:25543–25549. [DOI] [PubMed] [Google Scholar]
- 20.Graham, F. L., and A. J. van der Eb. 1973. Transformation of rat cells by DNA of human adenovirus 5. Virology 54:536–539. [DOI] [PubMed] [Google Scholar]
- 21.Guo, X. W., J. P. Th’ng, R. A. Swank, H. J. Anderson, C. Tudan, E. M. Bradbury, and M. Roberge. 1995. Chromosome condensation induced by fostriecin does not require p34cdc2 kinase activity and histone H1 hyperphosphorylation, but is associated with enhanced histone H2A and H3 phosphorylation. EMBO J. 14:976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurley, L. R., J. A. D’Anna, S. S. Barham, L. L. Deaven, and R. A. Tobey. 1978. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84:1–15. [DOI] [PubMed] [Google Scholar]
- 23.Gurley, L. R., R. A. Walters, and R. A. Tobey. 1974. Cell cycle-specific changes in histone phosphorylation associated with cell proliferation and chromosome condensation. J. Cell Biol. 60:356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hans, F., and S. Dimitrov. 2001. Histone H3 phosphorylation and cell division. Oncogene 20:3021–3027. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, J. C., C. Tse, and A. P. Wolffe. 1998. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17637–17641. [DOI] [PubMed] [Google Scholar]
- 26.Hendzel, M. J., Y. Wei, M. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348–360. [DOI] [PubMed] [Google Scholar]
- 27.Hirano, T. 2000. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69:115–144. [DOI] [PubMed] [Google Scholar]
- 28.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, R. Lin, M. Mitchell Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291. [DOI] [PubMed] [Google Scholar]
- 29.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266–273. [DOI] [PubMed] [Google Scholar]
- 30.Kaitna, S., M. Mendoza, V. Jantsch-Plunger, and M. Glotzer. 2000. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10:1172–1181. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, M., S. Kotani, T. Hattori, N. Sumi, T. Yoshioka, K. Todokoro, and Y. Okano. 1997. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J. Biol. Chem. 272:13766–13771. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, M., Y. Matsuda, T. Yoshioka, and Y. Okano. 1999. Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J. Biol. Chem. 274:7334–7340. [DOI] [PubMed] [Google Scholar]
- 33.Kimura, M., Y. Matsuda, T. Yoshioka, N. Sumi, and Y. Okano. 1998. Identification and characterization of STK12/Aik2: a human gene related to aurora of Drosophila and yeast IPL1. Cytogenet. Cell Genet. 82:147–152. [DOI] [PubMed] [Google Scholar]
- 34.Langan, T. A., J. Gautier, M. Lohka, R. Hollingsworth, S. Moreno, P. Nurse, J. Maller, and R. A. Sclafani. 1989. Mammalian growth-associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol. Cell. Biol. 9:3860–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahadevan, L. C., A. C. Willis, and M. J. Barratt. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65:775–783. [DOI] [PubMed] [Google Scholar]
- 36.Monteiro, M. J., and T. I. Mical. 1996. Resolution of kinase activities during the HeLa cell cycle: identification of kinases with cyclic activities. Exp. Cell Res. 223:443–451. [DOI] [PubMed] [Google Scholar]
- 37.Murnion, M. E., R. R. Adams, D. M. Callister, C. D. Allis, W. C. Earnshaw, and J. R. Swedlow. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656–26665. [DOI] [PubMed] [Google Scholar]
- 38.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nature (Rev. Mol. Cell Biol.) 2:21–32. [DOI] [PubMed] [Google Scholar]
- 39.Nurse, P. 2000. A long twentieth century of the cell cycle and beyond. Cell 100:71–78. [DOI] [PubMed] [Google Scholar]
- 40.Ohsumi, K., C. Katagiri, and T. Kishimoto. 1993. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science 262:2033–2035. [DOI] [PubMed] [Google Scholar]
- 41.Patterton, H. G., C. C. Landel, D. Landsman, C. L. Peterson, and R. T. Simpson. 1998. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 273:7268–7276. [DOI] [PubMed] [Google Scholar]
- 42.Paulson, J. R., and S. S. Taylor. 1982. Phosphorylation of histones 1 and 3 and nonhistone high mobility group 14 by an endogenous kinase in HeLa metaphase chromosomes. J. Biol. Chem. 257:6064–6072. [PubMed] [Google Scholar]
- 43.Reich, A., A. Yanai, S. Mesilaty-Gross, A. Chen-Moses, R. Wides, and B. Motro. 1999. Cloning, mapping, and expression of ial, a novel Drosophila member of the Ipl1/aurora mitotic control kinase family. DNA Cell Biol. 18:593–603. [DOI] [PubMed] [Google Scholar]
- 44.Roghi, C., R. Giet, R. Uzbekov, N. Morin, I. Chartrain, R. Le Guellec, A. Couturier, M. Doree, M. Philippe, and C. Prigent. 1998. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 111:557–572. [DOI] [PubMed] [Google Scholar]
- 45.Roth, M. B., A. M. Zahler, and J. A. Stolk. 1991. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol. 115:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sassone-Corsi, P., C. A. Mizzen, P. Cheung, C. Crosio, L. Monaco, S. Jacquot, A. Hanauer, and C. D. Allis. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285:886–891. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher, J. M., N. Ashcroft, P. J. Donovan, and A. Golden. 1998. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development 125:4391–4402. [DOI] [PubMed] [Google Scholar]
- 48.Schumacher, J. M., A. Golden, and P. J. Donovan. 1998. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scrittori, L., F. Hans, D. Angelov, M. Charra, C. Prigent, and S. Dimitrov. 2001. pEg2 Aurora-A kinase, histone H3 phosphorylation and chromosome assembly in Xenopus egg extract. J. Biol. Chem. 276:30002–30010. [DOI] [PubMed] [Google Scholar]
- 50.Sen, S., H. Zhou, and R. A. White. 1997. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene 14:2195–2200. [DOI] [PubMed] [Google Scholar]
- 51.Severson, A. F., D. R. Hamill, J. C. Carter, J. Schumacher, and B. Bowerman. 2000. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10:1162–1171. [DOI] [PubMed] [Google Scholar]
- 52.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 82:47–56. [DOI] [PubMed] [Google Scholar]
- 53.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature. 403:41–45. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, T., M. Kimura, K. Matsunaga, D. Fukada, H. Mori, and Y. Okano. 1999. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 59:2041–2044. [PubMed] [Google Scholar]
- 55.Tatsuka, M., H. Katayama, T. Ota, T. Tanaka, S. Odashima, F. Suzuki, and Y. Terada. 1998. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58:4811–4816. [PubMed] [Google Scholar]
- 56.Terada, Y., M. Tatsuka, F. Suzuki, Y. Yasuda, S. Fujita, and M. Otsu. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17:667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson, S., A. L. Clayton, C. A. Hazzalin, S. Rose, M. J. Barratt, and L. C. Mahadevan. 1999. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 18:4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Hooser, A., D. W. Goodrich, C. D. Allis, B. R. Brinkley, and M. A. Mancini. 1998. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. 111:3497–3506. [DOI] [PubMed] [Google Scholar]
- 59.Wei, Y., C. A. Mizzen, R. G. Cook, M. A. Gorovsky, and C. D. Allis. 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. USA 95:7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei, Y., L. Yu, J. Bowen, M. A. Gorovsky, and C. D. Allis. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99–109. [DOI] [PubMed] [Google Scholar]
- 61.Ye, X. S., S. L. Lee, T. D. Wolkow, S. L. McGuire, J. E. Hamer, G. C. Wood, and S. A. Osmani. 1999. Interaction between developmental and cell cycle regulators is required for morphogenesis in Aspergillus nidulans. EMBO J. 18:6994–7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, H., J. Kuang, L. Zhong, W. L. Kuo, J. W. Gray, A. Sahin, B. R. Brinkley, and S. Sen. 1998. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20:189–193. [DOI] [PubMed] [Google Scholar]