Abstract
In the small, free-living amoeba Acanthamoeba castellanii, rRNA transcription requires, in addition to RNA polymerase I, a single DNA-binding factor, transcription initiation factor IB (TIF-IB). TIF-IB is a multimeric protein that contains TATA-binding protein (TBP) and four TBP-associated factors that are specific for polymerase I transcription. TIF-IB is required for accurate and promoter-specific initiation of rRNA transcription, recruiting and positioning the polymerase on the start site by protein-protein interaction. In A. castellanii, partially purified TIF-IB can form a persistent complex with the ribosomal DNA (rDNA) promoter while homogeneous TIF-IB cannot. An additional factor, TIF-IE, is required along with homogeneous TIF-IB for the formation of a stable complex on the rDNA core promoter. We show that TIF-IE by itself, however, does not bind to the rDNA promoter and thus differs in its mechanism from the upstream binding factor and upstream activating factor, which carry out similar complex-stabilizing functions in vertebrates and yeast, respectively. In addition to its presence in impure TIF-IB, TIF-IE is found in highly purified fractions of polymerase I, with which it associates. Renaturation of polypeptides excised from sodium dodecyl sulfate-polyacrylamide gels showed that a 141-kDa polypeptide possesses all the known activities of TIF-IE.
rRNA transcription initiation in eukaryotic cells is a multistep process (20, 40, 45, 47, 52; reviewed in reference 46). Initially, an unusually stable complex of transcription factors, designated the committed complex, forms on the promoter. This complex is resistant to challenge by a second ribosomal DNA (rDNA) template and persists through multiple rounds of transcription. RNA polymerase I (Pol I), probably in association with other factors, is then recruited to the promoter by protein-protein interactions with the committed complex to form the preinitiation complex (PIC) (31, 37, 48, 57). PIC formation is followed by melting of the double-stranded DNA, formation of the first few phosphodiester bonds, and promoter clearance (4, 27, 35). Following promoter clearance, the committed complex remains promoter bound and functional to recruit additional RNA polymerases for multiple rounds of transcription (45, 47), though a new study suggests there may be differences between species in what components of the committed complex remain bound to the promoter (1).
A single DNA-binding transcription initiation factor is essential and sufficient for the accurate initiation of transcription in vitro (14, 33, 36, 38, 62, 67). This fundamental factor is referred to as transcription initiation factor IB (TIF-IB) in the mouse and Acanthamoeba castellanii, SL1 in the human and the rat, Rib1 in Xenopus spp., factor D or TFID in the rat, and core factor (CF) in the yeast Saccharomyces cerevisiae. In the best-studied committed complex, A. castellanii TIF-IB binds to the minor groove (18) and to the ribosomal initiator element (50) of the core domain of the promoter. In all species, TIF-IB is a multisubunit protein complex composed of TATA-binding protein (TBP) and three or four additional subunits known as Pol I-specific TBP-associated factors (TAFIs), though the tenacity of TBP association with the TAFIs is variable between species (12, 30). The TAFI proteins show sequence similarities approaching 90% between closely related species (22), but between more distantly related species sequence conservation is lost (30).
The presence of TBP, which is a component of the basal transcription machinery used by all three nuclear RNA polymerases (23, 58), puts TIF-IB into the same class of transcription factors as transcription factor IID (TFIID) and TFIIIB in the Pol II and Pol III systems, respectively (47). Just as TIF-IB is responsible for correctly positioning Pol I at the transcription start site (31), TFIID and TFIIIB also serve key roles in recruiting their respective RNA polymerases to the promoter and positioning them (23, 28, 44, 46, 69, 72). Therefore, TIF-IB confers promoter selectivity on Pol I and is the sole DNA-binding factor required for accurate initiation of basal transcription.
The ability of TIF-IB homologues to form stable complexes on the core promoter was initially thought to vary considerably from species to species. The human factor was reported unable to bind the promoter at all, while the rodent and yeast homologues were found to form weak complexes and the A. castellanii TIF-IB fraction completed the spectrum by robustly binding the promoter. However, data suggesting that the human factor can form a promoter complex have been presented recently (42), while we (51) have shown that homogeneous A. castellanii TIF-IB cannot form the robust complex originally described (see below). Reconciling these results, all the homologues form similar stable committed complexes on the promoter when in the presence of accessory transcription factors. In vertebrates, upstream binding factor (UBF) (8, 9,34, 55, 61) serves this accessory role; in S. cerevisiae, upstream activating factor (UAF) in conjunction with TBP facilitates the recruitment of the yeast factor to the rDNA promoter (1, 29, 30, 65); and in A. castellanii, a stabilizing factor, TIF-IE, is involved. Surprisingly, each of these appears to function by a different mechanism (see Discussion).
Partially purified A. castellanii TIF-IB is capable of forming a committed complex with the rDNA promoter that is resistant to template challenge (4–6, 24, 31, 46). However, TIF-IB purified to near-homogeneity loses this ability to commit the template, though it forms a quasistable complex with the promoter (51). Therefore, it appeared that during purification an additional component that helped stabilize the complex of TIF-IB with the promoter had been separated from TIF-IB. This component could not be detected as a side fraction during these last steps in TIF-IB purification, presumably because it was too dilute. However, a novel transcription factor named TIF-IE, capable of conferring on homogeneous TIF-IB the ability to commit the rDNA template, was found associated with Pol I. TIF-IE can be separated partially from the polymerase by rate zonal sedimentation in a glycerol gradient (51). The work presented here establishes that TIF-IE is composed of a single polypeptide and determines that its mechanism of action in conferring commitment ability on TIF-IB is distinct from the mechanism of UBF or UAF, suggesting a previously unrecognized diversity among species in accomplishing this critical task.
MATERIALS AND METHODS
Purification of TIF-IB.
TIF-IB was purified from a crude nuclear extract to apparent homogeneity as described previously (51) with the following modifications: The TIF-IB-containing fraction obtained by ammonium sulfate precipitation of the nuclear extract was dialyzed against 75 mM KCl in HEG10 (50 mM HEPES [pH 7.9], 0.1 mM EDTA, and 10% glycerol). This fraction was loaded onto a DEAE Sepharose Fast Flow column (Pharmacia Biotech) that was later developed with a linear gradient from 100 to 750 mM KCl. TIF-IB was further purified by BioRex 70 (Bio-Rad, Hercules, Calif.) chromatography and two rounds instead of one round of promoter-DNA Sepharose CL-4B affinity chromatography, followed by rate zonal sedimentation in a glycerol gradient.
Purification of RNA Pol I and TIF-IE.
Pol I was purified from a whole-cell extract, and its activity was analyzed by a nonspecific transcription assay, as described previously (63). We have preliminary evidence that some of this Pol I is associated with Acanthamoeba TIF-IA (J. Gogain, unpublished data). TIF-IE was separated from RNA Pol I at the last step of purification, rate zonal sedimentation in a glycerol gradient. At this stage of purification, TIF-IE is not homogeneous (51).
Plasmids and templates.
Plasmid pGG4C and/or plasmid pEBH10 was used for preparation of the DNAs used in electrophoretic mobility shift assays (EMSAs), methidiumpropyl-EDTA · Fe(II) [MPE · Fe(II)] footprinting, and template binding order-of-addition assays. Plasmid pGG4C contains a 114-bp fragment of the A. castellanii core promoter, from −96 to +18 relative to the transcription initiation site (tis) (+1), subcloned into the NotI site of pBluescript II SK(−) (18). Plasmid pEBH10 contains the A. castellanii core promoter from −120 to +80 cloned into the HincII site of pUC8 (5). For EMSAs and MPE · Fe(II) footprinting assays, DNA fragments were prepared from the plasmids by initial digestion with BamHI, which cleaves pGG4C downstream of the +18 end of the insert and cleaves pEBH10 upstream of the −120 end of the insert. For binding competition assays, pGG4C was initially digested upstream of the −96 insert end with SacII. In each case, this was followed by treatment with shrimp alkaline phosphatase to dephosphorylate the 5′ ends. The linear plasmids (2 μg) were then 5′ end labeled with T4 polynucleotide kinase. pGG4C/BamHI and pGG4C/SacII were then digested with SacI and XbaI, respectively. SacI cleaves pGG4C/BamHI upstream of the −96 insert end to generate a 150-bp fragment, while XbaI cleaves pGG4C/SacII downstream of the +18 insert end to generate a fragment of 127 bp. pEBH10/BamHI was digested downstream of the +80 insert end with PstI to generate a 217-bp fragment. Each labeled fragment was separated from the linear vector on a 1.75 to 2% agarose gel, visualized by autoradiography, excised, eluted, and purified with a QIAEX II Gel Extraction Kit (Qiagen) according to the manufacturer’s protocol. After purification, the specific activity of each labeled DNA fragment was estimated by liquid scintillation counting.
Plasmids pAr6 and pEBH10 were used for preparation of the templates used in the template commitment assays. pAr6 contains the A. castellanii rRNA promoter from −683 to +219 cloned into the SmaI site of pUC8 (51). Restriction digestion of pAr6 with HindIII and of pEBH10 with NdeI produce 240- and 309-nucleotide runoff RNAs, respectively.
EMSAs.
The 5′-end-labeled 150-bp BamHI/SacI fragment of pGG4C and the 217-bp BamHI/PstI fragment of pEBH10 were used in EMSAs. Binding conditions were the same as those described by Geiss et al. (18) except that bovine serum albumin was used at 0.5 mg/ml instead of 0.05 mg/ml. DNA was incubated with proteins for 20 min at 25°C. Reactions were stopped on ice, and reaction products were loaded immediately on a low-cross-linking, nondenaturing polyacrylamide gel (5% acrylamide; 80:1 [wt/wt] ratio of acrylamide to N,N′-methylene bisacrylamide) as described by Gong et al. (19). Gels were run at 200 V for 1.5 h, dried, and exposed to phosphor storage screens. Data were analyzed on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and quantified by using ImageQuant software (version 5.1).
Template commitment assay.
Template commitment assay conditions were the same as those described by Radebaugh et al. (51). The minimum amount of template required for binding all the available TIF-IB in the reaction was determined for pEBH10/NdeI (DNA A) and pAr6/HindIII (DNA B) and used in the following protocol. The first template(s) (DNA A and/or DNA B) was preincubated either with TIF-IB alone or with TIF-IB plus TIF-IE for 10 min at 25°C in the presence of nucleoside triphosphates. The second template (DNA B) or buffer was then added, and preincubation continued for another 10 min. RNA synthesis was initiated by addition of Pol I and proceeded for another 30 min. Runoff RNAs were analyzed as described by Radebaugh et al. (51).
MPE · Fe(II) footprinting.
The 5′-end-labeled 150-bp BamHI/SacI fragment of pGG4C was used as the template strand of the rRNA promoter. Binding conditions for footprinting were the same as those for EMSAs. DNA and proteins, in a final reaction volume of 20 μl, were incubated for 20 min at 25°C. One microliter of a 70 μM MPE and 50 μM (NH4)2Fe(SO4)2 solution and 1 μl of 0.5 M dithiothreitol were added to the reaction mixtures, and incubation continued for another 15 min at 25°C. MPE · Fe(II) reactions were stopped, and reaction products were processed, as described by Geiss et al. (18). DNA was analyzed on denaturing (7 M urea) 10% sequencing gels with 1× TBE run buffer (53). Gels were run at 20 mA for 2 to 3 h; then they were dried and exposed to phosphor storage screens overnight. Data were analyzed by use of a PhosphorImager and ImageQuant software as described above.
SDS-polyacrylamide gel electrophoresis (PAGE) and staining of proteins.
TIF-IE fractions were precipitated with chloroform-methanol as described by Wessel and Flugge (68). Protein pellets were resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer and electrophoresed through an SDS-polyacrylamide gel by standard methods (17). Gels were stained with Coomassie brilliant blue R-250 as described previously (53) or with silver stain (11).
Renaturation of proteins from SDS-polyacrylamide gels.
Renaturation was performed essentially as described by Hager and Burgess (21) but with some of the modifications reported by Kretzschmar et al. (32) and Briggs et al. (13). To precipitate proteins, 5 volumes of cold acetone (at −20°C) were added to the TIF-IE sample. The sample was allowed to precipitate for 30 min. in a dry-ice-ethanol bath and then centrifuged for 30 min at 10,000 × g at 4°C. The acetone supernatant was removed, and the protein pellet was dried under a vacuum for 2 min. The protein pellet was resuspended in 20 to 30 μl of 1× SDS loading buffer, heated for 5 min at 95°C, and electrophoresed on an SDS-PAGE gel (6 or 10% polyacrylamide). To visualize and excise the protein bands, a Zinc Stain & Destain Kit for Electrophoresis (Bio-Rad) was used, and staining and destaining were carried out according to the manufacturer’s protocol. Individual gel slices were then excised, put into siliconized microcentrifuge tubes, and soaked in two changes of 1 ml of 1 mM dithiothreitol for 15 min. The 1 mM dithiothreitol solution was decanted and discarded. A 0.5- to 1-ml volume (depending on the size of gel slice) of elution buffer was added to each gel slice, and the gel was crushed with several strokes of a small Teflon pestle that fit tightly inside the 1.7-ml microcentrifuge tube. The elution buffer contained 50 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 0.1% SDS, 5 mM dithiothreitol, and 150 mM NaCl. Proteins were allowed to elute for 21 h at room temperature with continuous agitation. Residual polyacrylamide was removed by centrifugation, and eluates were transferred to 15-ml siliconized Corex tubes. To remove SDS, eluates were then precipitated with 5 volumes of cold acetone as described above and centrifuged at 10,000 × g for 30 min at 4°C. The acetone supernatants were poured off, and precipitates were rinsed once gently with 1 ml of ice-cold 80% acetone and 20% dialysis buffer (100 mM KCl, 0.2% Nonidet P-40, 50 mM HEPES [pH 7.9], 20% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) to remove the last traces of residual SDS. This was followed by centrifugation at 10,000 × g for 10 min at 4°C. Supernatants were poured off, and protein pellets were allowed to dry at room temperature. The resulting protein pellets were resuspended in 50 μl of a buffer containing 6 M guanidine hydrochloride, 100 mM KCl, 0.2% Nonidet P-40, 50 mM HEPES [pH 7.9], 20% glycerol, 0.1 mM EDTA, and 1 mM dithiothreitol. Pellets were dissolved thoroughly and allowed to stand at room temperature for 30 to 40 min. The solution was then dialyzed against 500 ml of dialysis buffer (composition described above) with 0.1 mM phenylmethanesulfonyl fluoride and 1 mM benzamidine for 21 h at 4°C to remove the guanidine hydrochloride and permit the proteins to renature.
Template binding order-of-addition assay.
The 127-bp SacII/XbaI fragment of pGG4C (DNA A) and 217-bp BamHI/PstI fragment of pEBH10 (DNA B) that had been 5′ end labeled with 32P were used in this assay. Binding conditions were the same as those in EMSAs. The labeled fragment(s) was preincubated with either TIF-IB alone, TIF-IB plus TIF-IE, or TIF-IE alone for 15 min at 25°C. For the formation of a complex with TIF-IB on each template, TIF-IE amounts were limiting. The second template and either TIF-IB or TIF-IE were then added in the secondary 15-min incubation period. Reactions were stopped on ice, and reaction products were loaded on 5% nondenaturing polyacrylamide gels as described by Gong et al. (19). Gels were run at 200 V for 3.5 h, dried, and exposed to phosphor storage screens. Data were analyzed by phosphorimaging and quantified by using ImageQuant software.
RESULTS
TIF-IE is required along with glycerol gradient-purified TIF-IB to form a stable (committed) complex on the A. castellanii rRNA promoter.
We reported previously (51) that while TIF-IB purified through the first round of promoter-DNA affinity chromatography could form a committed complex with the rRNA core promoter in a template commitment assay, glycerol gradient-purified TIF-IB could not. A novel transcription factor, TIF-IE, that is required along with TIF-IB for the formation of this stable complex was discovered. TIF-IE is found associated with Pol I but can be separated from it (see below).
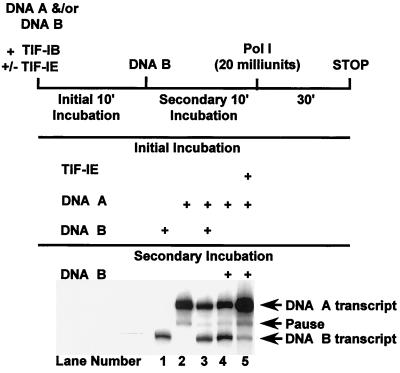
We confirmed that glycerol gradient-purified TIF-IB cannot form a stable complex with the rDNA promoter in a template commitment assay. In this assay, during an initial incubation period, sufficient DNA template is added to sequester all the DNA-binding factor(s). This complex is then challenged with a second promoter DNA during a subsequent incubation. Formation of a committed complex on the first DNA precludes transcription of the second. When only DNA A was preincubated with glycerol gradient-purified TIF-IB in the initial period (Fig. 1, lane 4), transcription levels from both templates were nearly identical to those obtained when both DNAs were present during the first incubation period (compare lanes 3 and 4), verifying that glycerol gradient-purified TIF-IB was unable to form a stable complex on the first DNA. However, upon addition of TIF-IE to TIF-IB in the initial incubation period, the second template failed to be transcribed efficiently (lane 5). TIF-IE also stimulated transcription about threefold (compare lanes 2 and 5). Stimulation occurred even in the absence of a second template and thus could not be attributed to the additional DNA sequestering an inhibitor. We conclude that TIF-IE is required along with glycerol gradient-purified TIF-IB for the formation of a stable (committed) complex on the rDNA core promoter.
FIG. 1.
TIF-IE is required along with glycerol gradient-purified TIF-IB to form a committed complex on the rDNA promoter. Template commitment assays of glycerol gradient-purified TIF-IB alone (lane 4) or with the addition of TIF-IE (lane 5) were performed as described in Materials and Methods. DNA A (pEBH10/NdeI) and DNA B (pAr6/HindIII) produced 309- and 240-nucleotide runoff RNAs, respectively (lanes 1, 2, and 3). The RNA transcripts synthesized from DNA A and DNA B in lanes 3, 4, and 5 were quantified, and their ratios (A/B) in each of these lanes are 1.1, 1.4, and 20.4, respectively. Components used for the initial or secondary incubation are indicated.
Direct measurement of TIF-IB binding activity.
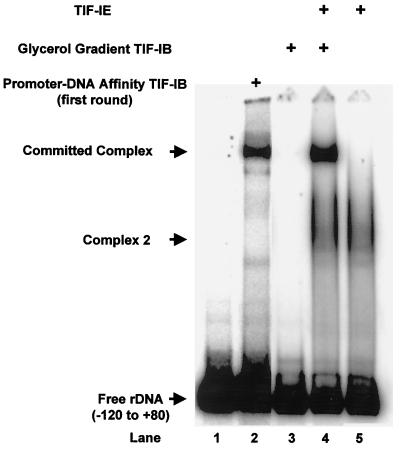
TIF-IE could formally be stimulating a later step in the transcription process, e.g., Pol I recruitment, DNA melting, or promoter clearance, as has been reported for mouse TIF-IA or TIF-IC (55). To more directly test the ability of TIF-IE to stabilize the binding of TIF-IB to the rRNA core promoter, EMSAs were used to determine the binding of TIF-IB alone, TIF-IB plus TIF-IE, and TIF-IE alone to the rDNA promoter. TIF-IB purified through one round of promoter-DNA affinity chromatography was capable of binding stably to the promoter (Fig. 2, lane 2). However, further purification of TIF-IB through a second round of promoter-DNA affinity chromatography, followed by rate zonal sedimentation in a glycerol gradient, caused TIF-IB to lose its ability to bind stably to the rDNA core promoter (Fig. 2, lane 3). In agreement with the template commitment assays, this shows directly that glycerol gradient-purified TIF-IB cannot form a committed complex on the promoter.
FIG. 2.
TIF-IE is required along with glycerol gradient-purified TIF-IB for the formation of a stable complex on the rDNA promoter. EMSAs on the rRNA promoter of TIF-IB purified through one round of promoter-DNA affinity chromatography (lane 2), of glycerol gradient-purified TIF-IB alone (lane 3), of TIF-IB in the presence of TIF-IE (lane 4), and of TIF-IE alone (lane 5) were performed as described in Materials and Methods. Lane 1, probe DNA alone.
As in the template commitment assays, addition of TIF-IE to glycerol gradient-purified TIF-IB allowed the formation of a stable complex with the core promoter (Fig. 2, lane 4) with an electrophoretic mobility identical to that of the complex formed with promoter-DNA affinity-purified TIF-IB (Fig. 2, lane 2). The identical electrophoretic mobilities of the two complexes suggest that they contain identical protein components, i.e., the TIF-IE purified from Pol I is likely the component lost during rate zonal sedimentation of TIF-IB. TIF-IB and TIF-IE are both required for the formation of a stable complex on the rDNA core promoter and for template commitment. The TIF-IE sample alone produced a different complex on the template (Fig. 2, lanes 4 and 5, complex 2). Since at this stage of purification TIF-IE was not homogeneous, it was not clear whether this DNA-binding activity observed was due to TIF-IE or to a contaminant; further experiments demonstrated that this complex 2 was due to a contaminant (see below).
TIF-IE associates with Pol I but is partially separated from it by rate zonal sedimentation.
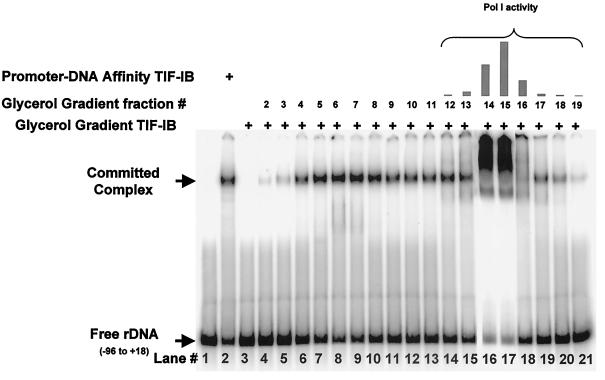
We have not been able to recover significant amounts of TIF-IE either from the DNA-affinity column washes or from the glycerol gradient fractions of TIF-IB. We can barely detect TIF-IE activity in the glycerol gradient fractions of TIF-IB, presumably because TIF-IE is highly diluted in these fractions. However, incubation of homogeneous TIF-IB with Pol I in the initial incubation of a template commitment assay led to stable complex formation (data not shown). Radebaugh et al. (51) found that TIF-IE was present in significant amounts in the Pol I heparin-Sepharose fraction (63) and could be partially separated from Pol I during the last step of purification, rate zonal sedimentation in a glycerol gradient. Glycerol gradient fractions were assayed for TIF-IE activity in the EMSA stimulation assay (Fig. 3). As shown above, glycerol gradient-purified TIF-IB cannot form a stable complex on the ribosomal promoter (Fig. 3; compare lane 2, impure TIF-IB, with lane 3, pure TIF-IB), but addition of fractions from rate zonal sedimentation of Pol I (Fig. 3, lanes 4 to 21) showed that TIF-IE is present in the fractions, with the peak activity sedimenting in fractions 6 and 7 (lanes 8 and 9). Pol I activity was detected by a nonspecific transcription assay in fractions 12 to 19, with the peak activity present in fractions 14 and 15 (Fig. 3, bar graph). This explains the huge complexes detected in lanes 16 and 17 of the EMSA. In these assays, TIF-IB, TIF-IE, and Pol I should form a complex on the promoter DNA, and this is supported by the promoter-dependent transcriptional activity of these fractions (data not shown). Alternatively, the polymerase may nonspecifically bind the DNA ends to produce these shifts. TIF-IE is spread over a number of fractions from the gradient, suggesting that either it forms multimeric complexes or the factor “bleeds” off of the polymerase as it sediments. The TIF-IE used in all subsequent experiments was obtained from the heparin-Sepharose-purified Pol I by rate zonal sedimentation. Peak fractions between 5 and 7 were used for most experiments, except that of Fig. 10.
FIG. 3.
Rate zonal sedimentation of TIF-IE in a glycerol gradient. Shown are results of EMSAs on the rRNA promoter of promoter-DNA affinity-purified TIF-IB (lane 2) and of glycerol gradient-purified TIF-IB alone (lane 3) or with 0.5 μl of fractions 2 to 19 from a glycerol gradient sedimentation of Pol I (lanes 4 to 21). The bar graph above lanes 14 to 21 shows nonspecific Pol I activity in the fractions.
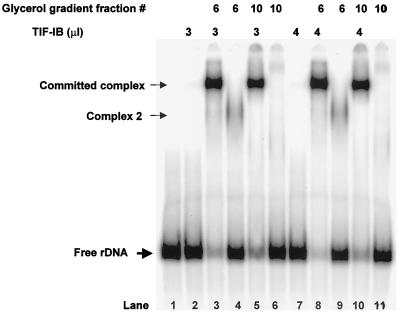
FIG. 10.
TIF-IE activity does not correlate with the DNA-binding activity (complex 2) found in impure fractions. Shown are EMSAs of glycerol gradient-purified TIF-IB alone (lanes 2 and 7) or with 0.5 or 0.7 μl of glycerol gradient fractions 6 and 10 (Fig. 1), respectively (lanes 3, 5, 8, and 10). EMSAs of 0.5 and 0.7 μl of glycerol gradient fractions 6 and 10 in the absence of TIF-IB are shown in lanes 4, 6, 9, and 11. Complex 2 is a protein-rRNA promoter DNA complex formed by a contaminating protein in fraction 6.
Even after one round of rate zonal sedimentation, Pol I was still associated with TIF-IE. Small additional amounts of TIF-IE could be resolved from Pol I by a second round of rate zonal sedimentation, but the majority of the TIF-IE activity was still coincident with the nonspecific polymerase activity (data not shown). This strongly suggests an interaction between Pol I and TIF-IE so that some, but not all, TIF-IE is released from the enzyme during rate zonal sedimentation (see Discussion). We have not succeeded in obtaining Pol I that is completely free of TIF-IE. We were initially concerned that TIF-IE could be a loosely associated Pol I subunit, but as shown below, the molecular weight of TIF-IE does not match that of any known A. castellanii RNA Pol I subunit (15, 63).
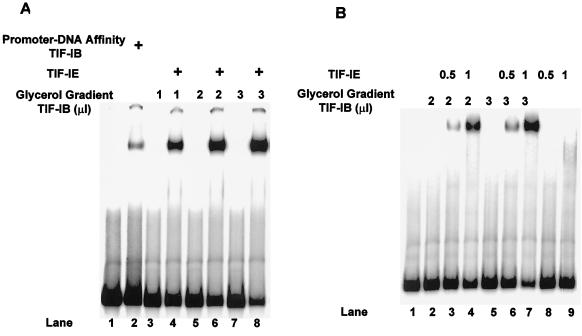
Stimulation of TIF-IB binding to the rDNA core promoter by TIF-IE is dose dependent.
Because the transcription stimulation reaction is time dependent (see below), we were concerned that TIF-IE might act catalytically rather than stoichiometrically. Addition of increasing amounts of glycerol gradient-purified TIF-IB alone to the promoter DNA did not result in a detectable complex in EMSAs (Fig. 4A, lanes 3, 5, and 7, and 4B, lanes 2 and 5). However, in the presence of excess TIF-IE, titration of TIF-IB resulted in the formation of stable complexes in amounts that were dependent on the dose of TIF-IB (Fig. 4A, lanes 4, 6, and 8). More important, the amount of complex was equally dependent on the amount of TIF-IE added (Fig. 4B). In this experiment, neither TIF-IB nor DNA amounts were limiting, and titration of TIF-IE (Fig. 4B; compare lane 3 with lane 4 and lane 6 with lane 7) resulted in a parallel increase in the amounts of complex formed. TIF-IE alone did not form a complex on this DNA (Fig. 4B, lanes 8 and 9).
FIG. 4.
Stimulation of TIF-IB binding in an EMSA by TIF-IE is dose dependent. (A) Lane 1 is the probe DNA alone; lane 2 has added promoter-DNA affinity column-purified TIF-IB. Lanes 3, 5, and 7 have increasing amounts of glycerol gradient-purified TIF-IB alone; lanes 4, 6, and 8 are the same with 0.5 μl of TIF-IE added. (B) EMSAs of glycerol gradient-purified TIF-IB alone (lanes 2 and 5) or with increasing amounts of TIF-IE, as indicated (lanes 3, 4, 6, and 7). Lanes 8 and 9 contain the indicated amounts of TIF-IE alone.
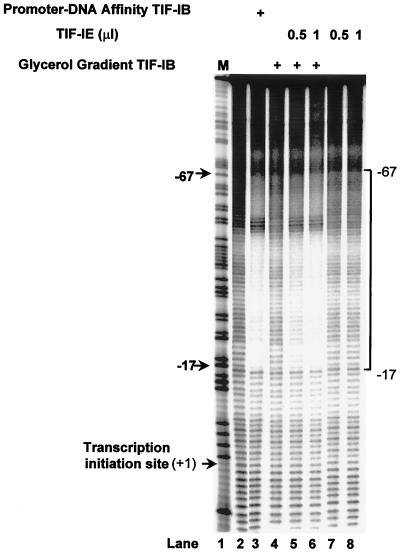
The TIF-IE dose dependence of complex formation was confirmed by MPE · Fe(II) footprinting. Promoter-DNA affinity-purified TIF-IB alone protected the rRNA promoter from MPE cleavage in a region extending from −67 to −17 with respect to the tis (+1) (Fig. 5, lane 3), while glycerol gradient-purified TIF-IB did not (lane 4). However, addition of increasing amounts of TIF-IE to glycerol gradient-purified TIF-IB (Fig. 5, lanes 5 and 6) stimulated the formation of a complex on the rRNA promoter that produced the same pattern of protection from MPE cleavage as promoter-DNA affinity-purified TIF-IB (Fig. 5, lane 3). The extent of protection from MPE cleavage was also dependent on the dose of TIF-IE (Fig. 5; compare lanes 5 and 6). TIF-IE alone did not protect the promoter from MPE cleavage in the promoter region (Fig. 5, lanes 7 and 8). Therefore, the region of the rDNA promoter extending from −67 to −17 was protected from MPE cleavage only by the cooperative binding of both TIF-IB and TIF-IE. The dose dependency observed suggests that TIF-IE enters into the complex stoichiometrically rather than catalytically modifying TIF-IB. Additionally, the EMSA and MPE · Fe(II) footprinting reactions are carried out in the absence of nucleoside triphosphates, acetyl coenzyme A, and other small-molecule donors of common modification reactions, and it is extremely unlikely that the highly purified protein fractions used would contain any residual small molecules of this type. Thus, we conclude that TIF-IE most likely carries out its function by participation in the formation of the complex directly.
FIG. 5.
TIF-IE is required along with glycerol gradient-purified TIF-IB for the formation of a footprint on the rDNA, and its effect is dose dependent. Shown are MPE · Fe(II) footprints of the template strand of the rRNA promoter. DNA was preincubated either with promoter-DNA affinity-purified TIF-IB (lane 3), with glycerol gradient-purified TIF-IB alone (lane 4), with glycerol gradient-purified TIF-IB and increasing amounts of TIF-IE (lanes 5 and 6), or with TIF-IE alone (lanes 7 and 8) before MPE · Fe(II) treatment. The previously determined footprint (−17 to −67) of the committed complex is indicated. M, marker lane; lane 2, probe DNA alone, treated with MPE · Fe(II) as described in Materials and Methods.
Subunit composition of TIF-IE.
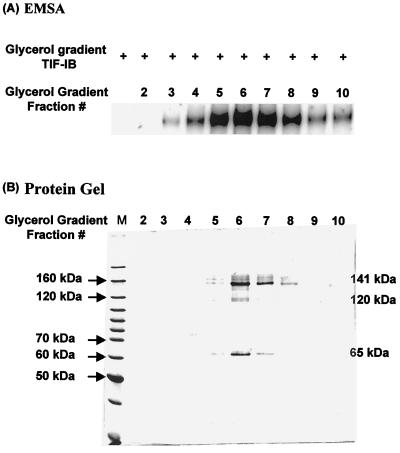
TIF-IE sediments near the top of a glycerol gradient of Pol I, in fractions 3 to 10, with the peak activity in fraction 6 (as shown in an EMSA of TIF-IB plus TIF-IE [Fig. 6A]), at a sedimentation velocity between those of bovine serum albumin (66 kDa) and aldolase (158 kDa). In order to determine the subunit makeup of TIF-IE, these fractions were analyzed by SDS-10% PAGE and stained with Coomassie blue (Fig. 6B). Three major polypeptides were consistently found to sediment with TIF-IE activity. These had relative molecular weights of 65,000, 120,000, and 141,000 (Fig. 6, fractions 5 to 8).
FIG. 6.
Three major polypeptides with relative molecular sizes of 65, 120, and 141 kDa consistently sedimented with TIF-IE activity. (A) EMSA showing complexes formed between glycerol gradient-purified TIF-IB and 0.5 μl of glycerol gradient fractions of Pol I (fractions 2 to 10) on an rDNA promoter fragment (−120 to +80). (B) SDS-10% polyacrylamide gel of 40 μl of the glycerol gradient fractions assayed in panel A, stained with Coomassie blue. Molecular masses of markers are shown on the left, and estimated molecular masses of the three prominent polypeptides are shown on the right.
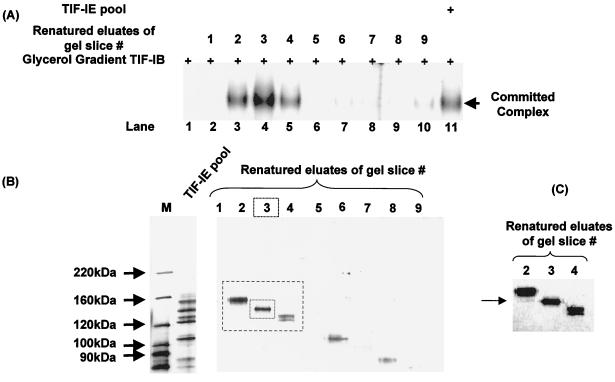
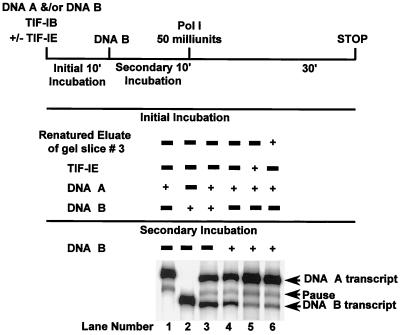
To identify which of the three polypeptides is associated with TIF-IE activity, polypeptide bands were excised from SDS-polyacrylamide gels and renatured. A TIF-IE sample was initially fractionated on an SDS-10% PAGE gel, and proteins were subsequently eluted and renatured from seven different gel slices. The renatured eluates were assayed for TIF-IE activity in an EMSA stimulation assay. The renatured eluate exhibiting activity contained several polypeptides with relative molecular weights between 100,000 and 160,000 (data not shown). These results eliminate the 65,000-molecular-weight polypeptide as a required TIF-IE subunit. To pin down which 100- to 160-kDa polypeptide(s) has TIF-IE activity, we subjected another TIF-IE sample to SDS-6% PAGE in order to better resolve these polypeptides. A negative (zinc) staining kit (Bio-Rad) was used to visualize the polypeptide bands before their excision from the gel. This staining showed clear protein bands on an opaque white background and allowed the excision of nine gel slices containing only one or two polypeptides each. The proteins contained in each gel slice were eluted, renatured, and assayed for TIF-IE activity in an EMSA (Fig. 7A). TIF-IE activity was detected in renatured eluates of gel slices 2 to 4 (Fig. 7A, lanes 3 to 5), with a strong peak in slice 3 (lane 4). Each renatured eluate was resolved by SDS-PAGE and silver stained (Fig. 7B). The eluate of gel slice 3 contained only a 141-kDa polypeptide. TIF-IE activity detected in the gel slices flanking gel slice 3 (Fig. 7A, lanes 3 and 5) was due to the presence of small amounts of the 141-kDa polypeptide in these slices, probably because of diffusion during processing (Fig. 7C shows a darker rendition of the peak lanes). To provide conclusive evidence that the 141-kDa renatured protein possessed all the activities attributed to TIF-IE, we examined it in a template commitment assay (Fig. 8). Indeed, the 141-kDa renatured protein exhibited template commitment activity comparable to that of the starting material (Fig. 8; compare lanes 5 and 6). Therefore, we conclude that TIF-IE is made up of a single polypeptide with a relative molecular weight of 141,000. The sedimentation rate of TIF-IE is consistent with its being predominantly a monomer.
FIG. 7.
TIF-IE has an apparent molecular mass of 141 kDa. (A) EMSAs showing complexes formed between the rRNA promoter and glycerol gradient-purified TIF-IB either alone (lane 1), with the renatured eluates of gel slices 1 to 9 (lanes 2 to 10), or with the TIF-IE applied to the SDS gel (lane 11). (B) SDS-6% polyacrylamide gel of the renatured eluates of gel slices 1 to 9 assayed in panel A, stained with silver. (C) Higher-sensitivity image of the region of the SDS-polyacrylamide gel boxed in panel B, showing polypeptides in eluates of gel slices 2 to 4. Arrow indicates the presence of a small amount of the 141-kDa polypeptide in gel slice 2.
FIG. 8.
The 141-kDa renatured polypeptide is able to confer commitment on glycerol gradient-purified TIF-IB. A template commitment assay was run as described for Fig. 1. Glycerol gradient-purified TIF-IB either alone (lane 4), with 1 μl of TIF-IE (lane 5), or with 5 μl of the 141-kDa renatured eluate of gel slice 3 (lane 6) was used in the assay. The RNA transcripts synthesized from DNA A and DNA B in lanes 3, 4, 5, and 6 were quantified, and their ratios (A/B) in each of these lanes are 1.7, 2.3, 11.96, and 8.3, respectively.
It is noteworthy that in the course of the renaturation experiments, we determined that TIF-IE is extremely stable to heating. We could heat the glycerol gradient-purified sample to 95°C for 10 min, chill it on ice, and recover close to 90% of the activity in an EMSA (data not shown).
Mechanism of action of TIF-IE.
In some vertebrates, formation of the committed complex requires an accessory transcription factor, UBF, in addition to TIF-IB. UBF binds stably to the upstream promoter element (UPE) on its own and has been reported to help recruit TIF-IB to the template (9, 34, 36), both stabilizing the TIF-IB-promoter complex and stimulating transcription. Similarly, in yeast, an additional transcription factor, UAF, binds first to the UPE and, in concert with TBP, helps recruit CF to the promoter, resulting in the formation of a committed complex and stimulating transcription (29, 30). TIF-IE is similar to these two factors in that it is required for the formation of the committed complex in A. castellanii and stimulates transcription.
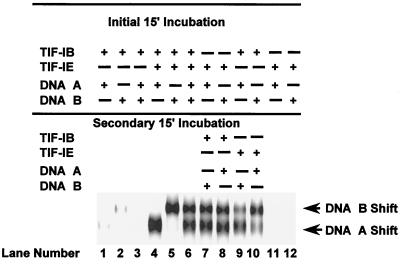
In each of these cases, the stimulatory factor binds the template first. Therefore, it is important to elucidate whether or not initial binding of TIF-IE to the rRNA promoter is required for recruiting glycerol gradient-purified TIF-IB and committing the rDNA template. To test for initial TIF-IE binding, we developed a template binding order-of-addition assay in which two 5′-end-labeled rRNA promoter templates of different lengths were used. The nearly twofold difference in template lengths allowed separation of the different sized committed complexes in an EMSA (Fig. 9, lanes 4 and 5). Glycerol gradient-purified TIF-IB did not bind the DNAs significantly without TIF-IE (Fig. 9, lanes 1 and 2). When both DNA templates were present with TIF-IE and TIF-IB in the initial incubation period, equal amounts of complexes were formed on the two templates (lane 6). To determine whether TIF-IE binds stably to the rRNA promoter before the recruitment of TIF-IB, TIF-IE was incubated with only one of the templates in an initial 15-min incubation period, followed by addition of the second template along with TIF-IB in a secondary incubation period (lanes 7 and 8). The ratio of the complexes formed was the same as when both templates were present simultaneously in the initial incubation period (compare lanes 6, 7, and 8). Therefore, in the initial incubation, TIF-IE did not form a stable complex with the template that resulted in preferred committed complex formation on that DNA. In contrast, TIF-IB formed a weak complex in the initial incubation (lanes 1 and 2) that was later stabilized by the addition of TIF-IE in the secondary incubation period (compare lanes 9 and 10 with lanes 7 and 8). This weak complex of TIF-IB with the template is not readily discernible in template commitment assays (see, e.g., Fig. 1) or in a simple EMSA (Fig. 2) but can be detected with this template binding order-of-addition assay. Addition of TIF-IE alone to the DNA templates did not result in formation of any stable complex in this portion of the gel (lanes 11 and 12). We conclude that, in contrast to UBF and UAF, the initial binding of TIF-IE to the promoter is not required for the recruitment of TIF-IB, and it appears that TIF-IB may bind weakly to the promoter before TIF-IE joins the complex.
FIG. 9.
The initial binding of TIF-IE to the rRNA promoter is not required for the recruitment of homogeneous TIF-IB. EMSAs show complexes formed during the initial incubation period between DNA A and TIF-IB either alone (lane 1) or with TIF-IE (lane 4); between DNA B and TIF-IB either alone (lane 2) or with TIF-IE (lane 5); or between both DNA templates and TIF-IB either alone (lane 3) or with TIF-IE (lane 6). EMSAs in lanes 7 to 10 show the effects of order of addition of the different factors and DNA templates on complex formation, as indicated above the lanes. EMSAs of TIF-IE alone with DNA A and DNA B are shown in lanes 11 and 12, respectively. The complexes formed in lanes 6, 7, 8, 9, and 10 on both templates were quantified, and their ratios (B/A) in each of these lanes are 0.89, 0.84, 0.76, 0.5, and 1.9, respectively.
Glycerol gradient-purified TIF-IE contains a component that binds DNA in an EMSA, forming a complex with greater electrophoretic mobility than the committed complex (Fig. 2, complex 2). However, consistent with our conclusion above, renatured TIF-IE did not form this complex in an EMSA, suggesting that complex 2 was due to a contaminating protein in the glycerol gradient fraction.
We also noted that the DNA binding activity that produced complex 2 sedimented similarly, but not identically, to TIF-IE in the glycerol gradient of Pol I (data not shown). To test this further, two fractions from the glycerol gradient, one that exhibited complex 2 formation (fraction 6) and one that did not (fraction 10), were used to stimulate TIF-IB binding in an EMSA. The TIF-IE activities of the two fractions were normalized and tested side by side (Fig 10). Clearly, both fractions could stimulate TIF-IB binding equally (Fig. 10; compare lane 3 with lane 5 and lane 8 with lane 10), but only fraction 6 formed complex 2. In these experiments, TIF-IB was not limiting because the same amounts of committed complex were formed when an additional increment of TIF-IB was added (compare lane 3 with lane 8 and lane 5 with lane 10). Therefore, the two TIF-IE activities were equalized. We conclude from all of the above that TIF-IE does not form a complex with the DNA template in the absence of TIF-IB.
Furthermore, in transcription experiments with short (2-min) RNA synthesis phases, stimulation occurred only when TIF-IE was preincubated with both TIF-IB and the DNA for several minutes before transcription was initiated, suggesting a time-dependent reaction which we believe is the formation of the committed complex (Fig. 11). In this experiment, transcription was first performed for 2 min without any preincubation, either in the absence or in the presence of additional TIF-IE (lanes 1 and 2). Under these conditions, TIF-IE stimulated transcription only modestly (1.6-fold), presumably because even during the short 2-min incubation, TIF-IE could stabilize the complex and multiple rounds of transcription initiation were initiated. This fold stimulation is also limited because there is a modest amount of TIF-IE in the RNA Pol I fraction, which increases “basal” transcription (lane 1). However, when all the components were preincubated for 5 min before the reaction was started by addition of nucleotides, an additional 1.5-fold stimulation resulted (compare lanes 2 and 6). Significantly, this extra stimulation did not occur if DNA (lane 3) or TIF-IB (lane 5) was omitted from the preincubation, again showing the need for formation of a ternary complex of TIF-IB, TIF-IE, and promoter DNA. Unfortunately, because we cannot separate TIF-IE completely from polymerase, we cannot completely omit TIF-IE from the reaction. Nevertheless, these results are in agreement with the hypothesis that TIF-IE can join and stabilize a complex with TIF-IB on the DNA. It is this stabilization of the committed complex that leads to the stimulation of multiple rounds of transcription observed in these reactions. In contrast, the results are less in tune with the notion that TIF-IE associates first with TIF-IB in solution, changing its conformation to stimulate the kinetics of binding of TIF-IB. If this were the mechanism, preincubation of TIF-IB and TIF-IE in the absence of DNA would lead to stimulation, which is not the case (Fig. 11, lane 3). It is also noteworthy that stimulation does not require the presence of nucleoside triphosphates during preincubation, which, as we mentioned above, suggests that TIF-IE does not covalently modify TIF-IB, for example, by phosphorylation, during preincubation.
FIG. 11.
Transcriptional stimulation by TIF-IE requires preincubation with TIF-IB and DNA and does not require nucleoside triphosphates. Various combinations of transcriptional components were preincubated (preinc) (lanes 3 to 6) or not preincubated (lanes 1 and 2) for 5 min prior to initiation of transcription by addition of the missing component(s) plus nucleoside triphosphates. Transcription proceeded for 2 min before the reaction was stopped and the RNA products were processed as described in Materials and Methods.
DISCUSSION
When we purified the fundamental transcription initiation factor from A. castellanii, TIF-IB, to apparent homogeneity (51) by an additional round of promoter-DNA affinity chromatography, this preparation could neither commit the template nor form a complex stable to electrophoresis under native conditions. We separated an additional transcription factor, TIF-IE, associated with Pol I, that is needed to confer commitment on TIF-IB. We have purified, characterized, and determined the mechanism of action of this novel transcription factor.
Complex stabilization by TIF-IE is dose dependent; both EMSA and MPE · Fe(II) footprinting assays were used to test this, suggesting that TIF-IE acts stoichiometrically rather than catalytically. Furthermore, its abilities to stabilize the complex of TIF-IB on the promoter (Fig. 5, 9, and 10) and to stimulate transcription (Fig. 11) are not dependent on the presence of nucleoside triphosphates.
Renaturation of TIF-IE after SDS-PAGE revealed that it is a 141-kDa polypeptide, and the sedimentation rate suggests that it is predominantly a monomer of this subunit. TIF-IE does not form a stable complex with promoter DNA on its own (Fig. 10). TIF-IE also stimulates transcription, but the extent of this stimulation cannot be quantitatively evaluated because Pol I cannot be entirely separated from the factor. We conclude that the committed complex is made up of promoter DNA bound by a combination of TIF-IB and TIF-IE. In contrast to our earlier conclusions, A. castellanii TIF-IB alone can form only a quasistable complex on the promoter. This TIF-IB-DNA complex is stabilized by protein-protein interaction with TIF-IE, probably by TIF-IE eliciting a conformational change of TIF-IB rather than by an interaction of TIF-IE with the DNA, though interaction of TIF-IE with the DNA when in the complex cannot be entirely ruled out from our results. The time course of transcriptional stimulation suggests that TIF-IE interacts with TIF-IB only once it is bound to the promoter; preincubation of TIF-IB and TIF-IE in the absence of DNA does not lead to the same stimulation as when DNA is present (Fig. 11).
TIF-IE can only be partially separated from Pol I by rate zonal sedimentation. Pol I purified through two rounds of glycerol gradient sedimentation is still associated with TIF-IE, even though these gradients contain the nonionic detergent NP-40 at a concentration that weakens protein-protein interactions. This explains why glycerol gradient-purified Pol I and TIF-IB can mediate efficient specific transcription without TIF-IE, even though this TIF-IB does not bind tightly to the rDNA promoter on its own. We do not know the fate of the TIF-IB following promoter clearance by Pol I under these circumstances; in the presence of TIF-IE, TIF-IB remains bound to the promoter through multiple rounds of initiation, as evidenced by the persistence of its footprint and lack of switching to competing promoter-DNA-containing templates (45).
Relationship to other Pol I-specific transcription factors.
We have ruled out the formal possibility that TIF-IE is a subunit of Acanthamoeba Pol I. The three nuclear RNA polymerases from A. castellanii have been purified to homogeneity and their subunits compared (15, 16, 63, 64). A pattern of subunits similar to those found in the yeast S. cerevisiae and subsequently in other eukaryotes was found (43). There are only two subunits with molecular weights greater than 100,000 in all RNA polymerases (59, 66). In A. castellanii, the two large subunits have relative molecular weights of 185,000 and 133,000, clearly distinct from the molecular weight of TIF-IE, 141,000. The fact that a subunit of this size is not detected in stained gels tells us that the amount of TIF-IE residually associated with Pol I is quite small. This low stoichiometry is reminiscent of the substoichiometric amounts of the required transcription factor TIF-IA/Rrn3p, which also is found associated with Pol I in mammals and yeast (56, 70). We have found immuno-cross-reactivity between a polyclonal antibody raised against yeast Rrn3p and a substoichiometric polypeptide in A. castellanii Pol I that we suspect is the TIF-IA homologue (J. Gogain, unpublished data). This anti-Rrn3p antibody does not cross-react with purified TIF-IE, and the sizes of TIF-IE and the putative TIF-IA are quite different, suggesting that TIF-IE is unrelated to TIF-IA/Rrn3p.
TIF-IE appears to be more closely related to another transcription factor identified in Acanthamoeba and designated EBF, for enhancer binding factor (71). This factor binds to Acanthamoeba enhancers and mediates their stimulatory activity by affecting the binding of TIF-IB to the core promoter. Thus, EBF has activity similar to that of TIF-IE. However, it differs in several respects from the factor described here: First and foremost, EBF does not confer commitment ability on TIF-IB (unpublished data). Second, TIF-IE does not form the same complex on enhancers as EBF in an EMSA (data not shown). Third, EBF does not copurify with RNA Pol I, nor with a 141-kDa protein. Finally, EBF does not exhibit the extreme heat stability that is characteristic of TIF-IE. Thus, while we cannot completely eliminate the possibility that TIF-IE contaminated the impure EBF fraction that was previously characterized, we would have to conclude that the impure preparation contained activities that altered TIF-IE’s template commitment ability, DNA binding, and heat stability. This is extremely unlikely.
In some vertebrates, formation of the committed complex requires an accessory transcription factor, UBF, in addition to TIF-IB. UBF interacts with the UPE and the core promoter and helps recruit TIF-IB to the template (9, 25, 26, 34, 36), possibly by altering the architecture of the DNA (7, 39). Similarly, in yeast, an additional transcription factor, UAF, is necessary for the formation of the committed complex. Like UBF, UAF binds to the UPE and, in conjunction with TBP, facilitates the recruitment of CF to the rDNA promoter and enables the factors to form a transcriptionally active complex (29, 30). Acanthamoeba TIF-IE is functionally similar to these two factors in that it is required for the formation of the committed complex. Unlike UAF, TIF-IE alone is not sufficient for template commitment. In fact, we have no evidence that TIF-IE binds DNA, in distinct contrast to UBF and UAF, and no UPE has been identified in Acanthamoeba. Indeed, in A. castellanii the template can be deleted to −55 (approximately 12 bp inside the upstream border of the TIF-IB footprint) without affecting the requirement for TIF-IE (data not shown), and the promoter can be deleted to −48 without compromising template commitment (24). TIF-IE also differs from UBF and UAF in structure. UBF purifies as a homodimer of 80- to 100-kDa subunits, depending on the species (2, 3, 41, 49). UAF bears no resemblance to UBF, consisting of six dissimilar subunits: three Pol I-specific subunits, Rrn5p, Rrn9p, and Rrn10p, with apparent molecular masses of 58, 50, and 17 kDa; histones H3 and H4, with relative molecular masses of 18 and 15 kDa, respectively; and a recently characterized 30-kDa protein, Uaf30p (29, 60). Comparison of cloned genes for these yeast components and mammalian UBF reveals no sequence similarities, nor do genome sequence comparisons reveal likely homologues. On the other hand, TIF-IE is a single 141-kDa polypeptide. Thus, TIF-IE appears distinct from both UBF and UAF.
In other eukaryotic systems, no factor similar to TIF-IE has been reported yet. In these other species, the core promoter-binding factors form either weak (9, 10, 30, 61) or strong (54, 55) complexes with the promoter, depending on the species. In Acanthamoeba, our initial studies suggested that TIF-IB could form extremely robust complexes in the absence of additional components. However, as demonstrated by Radebaugh et al. (51) and in the present study, we have found that an additional component is needed to form the committed complex when TIF-IB has been purified further. Curiously, when the subunits of human or mouse TIF-IB are cloned, it is difficult or impossible to reassemble active factor (22). Could this be because the TIF-IE homologue is missing in the cloned components but contaminates the purified factor? We suggest that it is possible that the TIF-IB purified from other species is contaminated with various amounts of TIF-IE homologues, allowing either strong or limited complex formation. Indeed, such differences could explain the disparity between recent (42) and older (8, 9, 34) studies of the human Pol I transcription system with regard to the binding of SL1 to the core promoter in the absence of UBF. Alternatively, it appears that different species could have evolved a variety of mechanisms to strengthen the committed complex; some use UPEs and their associated factors, and possibly Acanthamoeba has largely done away with the need for a separate binding site for the strengthening factor and resorted to a factor that has an activation domain without the DNA-binding domain.
Acknowledgments
This work was supported by Public Health Service grant GM22580 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Aprikian, P., B. Moorefield, and R. H. Reeder. 2001. New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol. 21:4847–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachvarov, D., and T. Moss. 1991. The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res. 19:2331–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachvarov, D., M. Normandeau, and T. Moss. 1991. Heterogeneity in the Xenopus ribosomal transcription factor xUBF has a molecular basis distinct from that in mammals. FEBS Lett. 288:55–59. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, E., L. Hoffman, C. Iida, W. Kubaska, P. Kownin, P. Risi, M. Zwick, and M. R. Paule. 1989. Eukaryotic RNA polymerase I binds to promoters by protein-protein interactions and melts DNA in a separate step following binding, p.259–269. In T. Cech and J. Gralla (ed.), Molecular biology of RNA- and DNA-protein interactions in transcription. Alan R. Liss, Inc., New York, N.Y.
- 5.Bateman, E., C. T. Iida, P. Kownin, and M. R. Paule. 1985. Footprinting of ribosomal RNA genes by transcription initiation factor and RNA polymerase I. Proc. Natl. Acad. Sci. USA 82:8004–8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman, E., and M. R. Paule. 1988. Events during eukaryotic rRNA transcription initiation and elongation: conversion from the closed to the open promoter complex requires nucleotide substrates. Mol. Cell. Biol. 8:1940–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazett-Jones, D. P., B. Leblanc, M. Herfort, and T. Moss. 1994. Short-range looping by the Xenopus HMG-box transcription factor, xUBF. Science 264:1134–1137. [DOI] [PubMed] [Google Scholar]
- 8.Bell, S. P., H.-M. Jantzen, and R. Tjian. 1990. Assembly of alternative multiprotein complexes directs rRNA promoter selectivity. Genes Dev. 4:943–954. [DOI] [PubMed] [Google Scholar]
- 9.Bell, S. P., R. M. Learned, H.-M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241:1192–1197. [DOI] [PubMed] [Google Scholar]
- 10.Bell, S. P., C. S. Pikaard, R. H. Reeder, and R. Tjian. 1989. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell 59:489–497. [DOI] [PubMed] [Google Scholar]
- 11.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99. [Google Scholar]
- 12.Bodeker, M., C. Cairns, and B. McStay. 1996. Upstream binding factor stabilizes Rib 1, the TATA-binding-protein-containing Xenopus laevis RNA polymerase I transcription factor, by multiple protein interactions in a DNA-independent manner. Mol. Cell. Biol. 16:5572–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briggs, M. R., J. T. Kadonaga, S. P. Bell, and R. Tjian. 1986. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234:47–52. [DOI] [PubMed] [Google Scholar]
- 14.Clos, J., D. Buttgereit, and I. Grummt. 1986. A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc. Natl. Acad. Sci. USA 83:604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Alessio, J. M., P. J. Perna, and M. R. Paule. 1979. DNA-dependent RNA polymerases from Acanthamoeba castellanii: comparative subunit structure of the homogeneous enzymes. J. Biol. Chem. 254:11282–11287. [PubMed] [Google Scholar]
- 16.D’Alessio, J. M., S. R. Spindler, and M. R. Paule. 1979. DNA-dependent RNA polymerase II from Acanthamoeba castellanii: purification and subunit structure of the homogeneous enzyme. J. Biol. Chem. 254:4085–4091. [PubMed] [Google Scholar]
- 17.Garfin, D. E. 1990. One-dimensional gel electrophoresis. Methods Enzymol. 182:425–441. [DOI] [PubMed] [Google Scholar]
- 18.Geiss, G. K., C. A. Radebaugh, and M. R. Paule. 1997. The fundamental ribosomal RNA transcription factor, TIF-IB (SL1, factor D), binds to the rRNA core promoter primarily by minor groove contacts. J. Biol. Chem. 272:29243–29254. [DOI] [PubMed] [Google Scholar]
- 19.Gong, X., C. A. Radebaugh, G. K. Geiss, M. N. Simon, and M. R. Paule. 1995. Site-directed photo-cross-linking of ribosomal RNA initiation complexes. Mol. Cell. Biol. 15:4956–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grummt, I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62:109–154. [DOI] [PubMed] [Google Scholar]
- 21.Hager, D. A., and R. R. Burgess. 1980. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results from sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal. Biochem. 109:76–86. [DOI] [PubMed] [Google Scholar]
- 22.Heix, J., J. C. B. M. Zomerdijk, A. Ravanpay, R. Tjian, and I. Grummt. 1997. Cloning of murine RNA polymerase I-specific TAF factors: conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc. Natl. Acad. Sci. USA 94:1733–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor. Genes Dev. 7:1291–1308. [DOI] [PubMed] [Google Scholar]
- 24.Iida, C. T., P. Kownin, and M. R. Paule. 1985. Ribosomal RNA transcription: proteins and DNA sequences involved in preinitiation complex formation. Proc. Natl. Acad. Sci. USA 82:1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jantzen, H.-M., A. Admon, S. P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA binding motif with homology to HMG proteins. Nature 344:830–836. [DOI] [PubMed] [Google Scholar]
- 26.Jantzen, H.-M., A. M. Chow, D. S. King, and R. Tjian. 1992. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6:1950–1963. [DOI] [PubMed] [Google Scholar]
- 27.Kahl, B., H. Li, and M. R. Paule. 2000. DNA melting and promoter clearance by eukaryotic RNA polymerase I. J. Mol. Biol. 299:75–89. [DOI] [PubMed] [Google Scholar]
- 28.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235–245. [DOI] [PubMed] [Google Scholar]
- 29.Keys, D. A., B. S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10:887–903. [DOI] [PubMed] [Google Scholar]
- 30.Keys, D. A., L. Vu, J. S. Steffan, J. A. Dodd, R. T. Yamamoto, Y. Nogi, and M. Nomura. 1994. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 8:2349–2362. [DOI] [PubMed] [Google Scholar]
- 31.Kownin, P., E. Bateman, and M. R. Paule. 1987. Eukaryotic RNA polymerase I promoter binding is directed by protein contacts with transcription initiation factor and is DNA sequence-independent. Cell 50:693–699. [DOI] [PubMed] [Google Scholar]
- 32.Kretzschmar, M., M. Meisterernst, C. Scheidereit, G. Li, and R. G. Roeder. 1992. Transcriptional regulation of the HIV-1 promoter by NF-κB in vitro. Genes Dev. 6:761–774. [DOI] [PubMed] [Google Scholar]
- 33.Learned, R. M., S. Cordes, and R. Tjian. 1985. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol. Cell. Biol. 5:1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Learned, R. M., T. K. Learned, M. M. Haltiner, and R. T. Tjian. 1986. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell 45:847–857. [DOI] [PubMed] [Google Scholar]
- 35.Lofquist, A. K., H. Li, M. A. Imboden, and M. R. Paule. 1993. Promoter opening (melting) and transcription initiation by RNA polymerase I requires neither nucleotide beta-gamma hydrolysis nor protein phosphorylation. Nucleic Acids Res. 21:3233–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McStay, B., C. H. Hu, C. S. Pikaard, and R. H. Reeder. 1991. xUBF and Rib1 are both required for formation of a stable polymerase I promoter complex in Xenopus laevis. EMBO J. 10:2297–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. B. M. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishima, Y., I. Financsek, R. Kominami, and M. Muramatsu. 1982. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 10:6659–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moss, T., V. Y. Stefanovsky, and G. Pelletier. 1998. The structural and architectural role of upstream binding factor, UBF, p.75–94. In M. R. Paule (ed.), Transcription of eukaryotic ribosomal RNA genes by RNA polymerase I. Springer-Verlag New York Inc., New York, N.Y.
- 40.Nomura, M. 1998. Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation, p.155–172. In M. R. Paule (ed.), Transcription of eukaryotic ribosomal RNA genes by RNA polymerase I. Springer-Verlag New York Inc., New York, N.Y.
- 41.O’Mahony, D. J., and L. I. Rothblum. 1991. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc. Natl. Acad. Sci. USA 88:3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panov, K. I., J. K. Friedrich, and J. C. B. M. Zomerdijk. 2001. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol. Cell. Biol. 21:2641–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paule, M. R. 1981. Comparative subunit composition of eukaryotic nuclear RNA polymerases. Trends Biochem. Sci. 6:128–131. [Google Scholar]
- 44.Paule, M. R. 1990. Transcription initiation: in search of the single factor. Nature 344:819–820. [DOI] [PubMed] [Google Scholar]
- 45.Paule, M. R. 1998. Function of the initiation complex on the core promoter, p.107–120. In M. R. Paule (ed.), Transcription of eukaryotic ribosomal RNA genes by RNA polymerase I. Springer-Verlag New York Inc., New York, N.Y.
- 46.Paule, M. R. 1998. Structure of the fundamental transcription initiation factor from a simple eukaryote, Acanthamoeba castellanii, p.59–74. In M. R. Paule (ed.), Transcription of eukaryotic ribosomal RNA genes by RNA polymerase I. Springer-Verlag New York Inc., New York, N.Y.
- 47.Paule, M. R., and R. J. White. 2000. Transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyroche, G., P. Milkereit, N. Bischler, H. Tschochner, P. Schultz, A. Sentenac, C. Carles, and M. Riva. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19:5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pikaard, C. S., B. McStay, M. C. Schultz, S. P. Bell, and R. H. Reeder. 1989. The Xenopus ribosomal gene enhancers bind an essential polymerase I transcription factor, xUBF. Genes Dev. 3:1779–1788. [DOI] [PubMed] [Google Scholar]
- 50.Radebaugh, C. A., X. Gong, B. Bartholomew, and M. R. Paule. 1997. Identification of previously unrecognized common elements in eukaryotic promoters: a ribosomal RNA gene initiator element for RNA polymerase I. J. Biol. Chem. 272:3141–3144. [DOI] [PubMed] [Google Scholar]
- 51.Radebaugh, C. A., W. M. Kubaska, L. H. Hoffman, K. Stiffler, and M. R. Paule. 1998. A novel transcription initiation factor (TIF), TIF-IE, is required for homogeneous Acanthamoeba castellanii TIF-IB (SL1) to form a committed complex. J. Biol. Chem. 273:27708–27715. [DOI] [PubMed] [Google Scholar]
- 52.Reeder, R. H. 1999. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 62:293–327. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Schnapp, A., J. Clos, W. Hädelt, R. Schreck, A. Cvekl, and I. Grummt. 1990. Isolation and functional characterization of TIF-IB, a factor that confers promoter specificity to mouse RNA polymerase I. Nucleic Acids Res. 18:1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnapp, A., and I. Grummt. 1991. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J. Biol. Chem. 266:24588–24595. [PubMed] [Google Scholar]
- 56.Schnapp, A., C. Pfleiderer, H. Rosenbauer, and I. Grummt. 1990. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 9:2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnapp, A., G. Schnapp, B. Erny, and I. Grummt. 1993. Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol. 13:6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultz, M. C., R. H. Reeder, and S. Hahn. 1992. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell 69:697–702. [DOI] [PubMed] [Google Scholar]
- 59.Sentenac, A., M. Riva, P. Thuriaux, J.-M. Buhler, I. Treich, C. Carles, M. Werner, A. Ruet, J. Huet, C. Mann, N. Chiannilkulchai, S. Stettler, and S. Mariotte. 1992. Yeast RNA polymerase subunits and genes, p.27–54. In S. L. McKnight and K. R. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Siddiqi, I., J. Dodd, L. Vu, K. Eliasson, M. Oakes, J. Keener, R. Mooney, M. K. Young, and M. Nomura. 2001. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J. 20:4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, S. D., D. J. O’Mahony, B. T. Kinsella, and L. I. Rothblum. 1993. Transcription from the rat 45S ribosomal DNA promoter does not require the factor UBF. Gene Expr. 3:229–236. [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, S. D., E. Oriahi, D. Lowe, H.-F. Yang-Yen, D. J. O’Mahony, K. Rose, K. Chen, and L. I. Rothblum. 1990. Characterization of factors that direct transcription of rat ribosomal DNA. Mol. Cell. Biol. 10:3105–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spindler, S., G. L. Duester, J. M. D’Alessio, and M. R. Paule. 1978. A rapid and facile procedure for the preparation of RNA polymerase I from Acanthamoeba castellanii: purification and subunit structure. J. Biol. Chem. 253:4669–4675. [PubMed] [Google Scholar]
- 64.Spindler, S. R., G. L. Duester, J. M. D’Alessio, and M. R. Paule. 1978. DNA-dependent RNA polymerase III from Acanthamoeba castellanii: a rapid procedure for the large scale preparation of homogeneous enzyme. J. Biol. Chem. 253:6242–6248. [PubMed] [Google Scholar]
- 65.Steffan, J. S., D. A. Keys, J. A. Dodd, and M. Nomura. 1996. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 10:2551–2563. [DOI] [PubMed] [Google Scholar]
- 66.Thuriaux, P., and A. Sentenac. 1992. Yeast nuclear RNA polymerases, p.1–48. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), Molecular and cellular biology of the yeast Saccharomyces: gene expression, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 67.Tower, J., V. C. Culotta, and B. Sollner-Webb. 1986. Factors and nucleotide sequences that direct ribosomal DNA transcription and their relationship to the stable transcription complex. Mol. Cell. Biol. 6:3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141–143. [DOI] [PubMed] [Google Scholar]
- 69.White, R. J., B. C. E. Khoo, J. A. Inostroza, D. Reinberg, and S. P. Jackson. 1994. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science 266:448–450. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto, R. T., Y. Nogi, J. A. Dodd, and M. Nomura. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 15:3964–3973. [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, Q., C. A. Radebaugh, W. Kubaska, G. K. Geiss, and M. R. Paule. 1995. Acanthamoeba castellanii contains a ribosomal RNA enhancer binding protein which stimulates TIF-IB binding and transcription under stringent conditions. Nucleic Acids Res. 23:4345–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zawel, L., and D. Reinberg. 1995. Common themes in assembly and function of eukaryotic transcription complexes. Annu. Rev. Biochem. 64:533–561. [DOI] [PubMed] [Google Scholar]