Abstract
CpG methylation is involved in a wide range of biological processes in vertebrates as well as in plants and fungi. To date, three enzymes, Dnmt1, Dnmt3a, and Dnmt3b, are known to have DNA methyltransferase activity in mouse and human. It has been proposed that de novo methylation observed in early embryos is predominantly carried out by the Dnmt3a and Dnmt3b methyltransferases, while Dntm1 is believed to be responsible for maintaining the established methylation patterns upon replication. Analysis of the sites methylated in vivo using the bisulfite genomic sequencing method confirms the previous finding that some regions of the plasmid are much more methylated by Dnmt3a than other regions on the same plasmid. However, the preferred targets of the enzyme cannot be determined due to the presence of other methylases, DNA binding proteins, and chromatin structure. To discern the DNA targets of Dnmt3a without these compounding factors, sites methylated by Dnmt3a in vitro were analyzed. These analyses revealed that the two cDNA strands have distinctly different methylation patterns. Dnmt3a prefers CpG sites on a strand in which it is flanked by pyrimidines over CpG sites flanked by purines in vitro. These findings indicate that, unlike Dnmt1, Dnmt3a most likely methylates one strand of DNA without concurrent methylation of the CpG site on the complementary strand. These findings also indicate that Dnmt3a may methylate some CpG sites more frequently than others, depending on the sequence context. Methylation of each DNA strand independently and with possible sequence preference is a novel feature among the known DNA methyltransferases.
CpG methylation is considered to be important for development, X chromosome inactivation, genomic imprinting, and suppression of transposons in mammals. The critical role played by the establishment and maintenance of DNA methylation has been demonstrated by the findings that homozygous disruption of any one of the three known methyltransferases in mice leads to lethality (23, 28). It has also been shown that some methyl-CpG binding proteins associate with histone deacetylase and repress transcription (for a review, see reference 6). The importance of this has been exemplified by the recent identification that a human disease, Rett syndrome, is caused by mutations in one of the methyl-CpG binding proteins, MeCP2 (1). Recently, changes in DNA methylation patterns have also been implicated in carcinogenesis (for a review, see reference 18).
Three vertebrate genes with known methyltransferase function have been identified, namely, Dnmt1, Dnmt3a, and Dnmt3b (4, 27). Dnmt1 has been considered as a maintenance methyltransferase mostly based on its association with the replication foci (22) and its preference for hemimethylated DNA (29, 30, 37). It has been reported that Dnmt1 is capable of methylating fully unmethylated DNA as well as non-CpG sites in vitro (30, 37). Dnmt1 has also been described to have no site preference other than CpGs (37). In Dnmt1 knockout mice, there is still a considerable amount of methylation in the genome (21). This has led to the search for and discovery of two other functional methyltransferases, Dnmt3a and Dnmt3b (27). Since the cloning of murine Dnmt3a and Dnmt3b (27), we and others have demonstrated the methyltransferase activity of these two enzymes in vivo (16, 25, 28). The targeted disruption of Dnmt3a and Dnmt3b in murine ES cells showed that these two proteins are important for mouse development (28). It has been proposed that Dnmt3a and Dnmt3b have some overlapping functions because of the overlapping expression in embryonic ectoderm and because disruption of either one of these genes does not disrupt methylation of proviral sequences as much as disruption of both genes (28). However, these two proteins have some nonoverlapping functions as demonstrated by Dnmt3b methylation of minor satellite sequences (28). Mutations in the DNMT3B gene were identified in a rare human recessive disease, ICF (named for immunodeficiency, centromere instability, and facial anomalies) (11, 28, 35). The classic satellite sequences showed decreased methylation in ICF patients (17). In addition, it has also been reported that some of the CpG islands on the inactive X chromosome and some of the nonsatellite repeats were also hypomethylated in ICF patients (19). These studies provided some information on the targets of these two methyltransferases in DNA methylation.
There is evidence that DNA sequence, promoter strength, and physical location may affect de novo methylation (12). It has been shown in an ES cell study that some sequences are methylated by both Dnmt3a and Dnmt3b, though each enzyme has its own distinct target sites (28). In a previous study using stable episomes as targets of these two methyltransferases in mammalian cells, we have shown that Dnmt3a methylates the EBNA1 region of the Epstein-Barr virus genome better than other sequences on the episome by restriction digestion assay (16). In this study, bisulfite genomic sequencing of plasmid DNA methylated by Dnmt3a in vivo further demonstrated that differential methylation of various regions on the episome was not restricted to the HhaI sites. An important question concerning the de novo methyltransferases is what are the sites that these methyltransferases act upon? We have shown previously that transcriptional activity does not influence de novo methylation by Dnmt3a on the episome in human cells (10). However, de novo methylation observed in vivo may still be complicated by one or a combination of the following: chromatin structure, binding of proteins to the DNA in vivo, activity of other methyltransferases, the concentration of de novo methyltransferases, possible interactions between different methyltransferases, and possible site preferences of the de novo methyltransferases.
The potential site preference of a methyltransferase can only be determined in the absence of all other compounding factors. This can be achieved when naked DNA targets are methylated in vitro and then analyzed. In this study, we used a purified glutathione S-transferase (GST) fusion of wild-type Dnmt3a protein for in vitro assays. Our findings suggest that Dnmt3a does not methylate all CpG sites equally. Sodium bisulfite genomic sequencing of in vitro-methylated plasmid substrates revealed that Dnmt3a generates very distinct methylation patterns on the two cDNA strands in vitro. Analysis of CpG sites frequently methylated by Dnmt3a in vitro revealed that Dnmt3a methylates CpG sites on a strand flanked by pyrimidine-rich sequences more frequently than CpG sites flanked by purine-rich sequences. Experiments using modified oligonucleotide substrates support this finding. This is the first demonstration that Dnmt3a methylates one DNA strand without concurrent methylation on the complementary strand at the same CpG site. This is also the first demonstration of sequence preference for a mammalian methyltransferase in vitro.
MATERIALS AND METHODS
Plasmids for transfection studies.
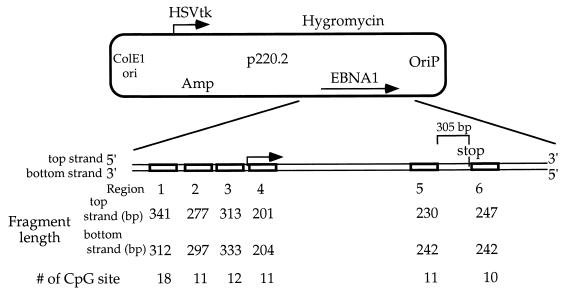
Plasmid pCLH22 was used as methylation target in the transfection studies. pCLH22 (14) bears the following: oriP, the EBNA-1 gene, the luciferase reporter gene driven by the Rous sarcoma virus long terminal repeat (RSV LTR), the hygromycin resistance gene driven by the HSVTk promoter, and the prokaryotic replication origin and the β-lactamase gene selection sequences. pCLH22 was generated by inserting the RSV LTR and the luciferase gene between the oriP and the hygromycin resistance gene on p220.2, such that the only difference between these two plasmids is the lack of the RSV LTR and the luciferase gene on p220.2. p220.2 has been described previously (8).
Cell lines, transfection, and episome recovery.
The 3a-5 cell line has been described previously (16). The calcium phosphate transfection method (14, 34) was used in this study. All transfections were done in duplicate or triplicate for each experiment, and all experiments were performed multiple times. Each time the transfected cells reached confluence, 2.5% of the cells were replated into a 100-mm-diameter plate, and the remaining cells were harvested for plasmid DNA extraction by the Hirt method (13). All the transfection experiments were carried out without any selection for the episomal plasmid.
Plasmids for protein expression.
Plasmid pEBG3a was generated by inserting the Myc-tagged murine wild-type Dnmt3a from pMT3aMyc (16) in-frame with GST into the BamHI and NotI sites of a GST fusion vector, pEBG (26, 33). Plasmid pEBG3amut, which carries the mutant murine Dnmt3a, was constructed similarly using the Myc-tagged mutant Dnmt3a from pMT3aMut (16). The proteins expressed in human cells from these plasmids have a GST domain at the N terminus and a Myc tag at the C terminus. The protein expression was confirmed by immunostaining using the Myc antibody, and both strands of the Dnmt3a insert were sequenced to rule out any mutations.
Protein purification.
The GST-Dnmt3a (GST-3a) and GST-Dnmt3a mutant (GST-3amut) fusion proteins were expressed individually at high levels by transfecting the expression vectors into 293T cells as described previously (33). The transfected cells were harvested 48 h after transfection, washed with phosphate-buffered saline once, and sonicated in a solution containing 25 mM Tris (pH 8.0), 300 mM KCl, 1 mM dithiothreitol (DTT), and 5% glycerol in the presence of protease inhibitors. The extract was centrifuged at 15,000 rpm for 30 min at 4°C in an SS34 rotor. The supernatant was treated with DNase I (20 μg/ml of extract) in the presence of 10 mM MgCl2 for 30 min at room temperature. The extract was then centrifuged at 30,000 rpm in a TLA-100.4 rotor (Beckman) for 1 h at 4°C. The supernatant was incubated with glutathione-agarose beads overnight at 4°C before eluting with a solution containing 100 mM Tris (pH 8.0), 20 mM glutathione, 1 mM DTT, and 20% glycerol. The concentration of the purified protein was estimated on a Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gel by densitometry with various amounts of bovine serum albumin as standards. Approximately 50 μg of the fusion protein can be purified from 10 150-mm-diameter tissue culture dishes of 293T cells transfected with the expression vector. The activity of the methyltransferase was verified by the 3H incorporation assay and by restriction enzyme digestion assay. GST-3a and GST-3aMut all bound to DNA tightly; therefore, each preparation of the protein was analyzed by 3H incorporation assay without addition of DNA substrate to test for and document the absence of DNA contamination in the purified protein.
DNA substrate and in vitro methylation assay.
Plasmid p220.2, used for in vitro methylation assays, has been described previously (8). A 467 bp fragment from HindIII-digested plasmid pOLucOriP (24) was gel purified and used in the restriction digestion assay. In vitro methylation by GST-3a and GST-3aMut was carried out with various amounts of protein and DNA substrates. Unless otherwise specified, a 20 μl reaction with 10 mM Tris (pH 8.0), 1 mM EDTA, 1 mM DTT, and either 80 μM unlabeled S-adenosylmethionine (AdoMet) (for restriction digestion assay) or 1.1 μM [3H]AdoMet (for 3H incorporation assay; New England Nuclear, 14.7 Ci/mmol) was incubated at 37°C overnight (16 h to 20 h). In vitro methylation using either SssI (New England Biolabs) or human Dnmt1 (New England Biolabs) was carried out according to the recommended condition by the manufacturer.
3H incorporation assay and restriction digestion assay.
For the 3H incorporation assay, the reaction was treated with proteinase K at 55°C for 1 h after in vitro methylation before being spotted onto the DE-81 filters and washed as described previously (20). The radioactivity retained on the air-dried DE-81 filters was measured by scintillation counting (Packard Tri Carb 2100TR scintillation counter) with 2 ml of scintillation fluid. For the restriction digestion assay, the DNA was phenol-chloroform extracted and ethanol precipitated after the overnight methylation reaction. Methylation of the DNA was assessed by either digestion with HinPII followed by end labeling with Klenow fragment or by digestion with HhaI followed by Southern blotting. When the Klenow end-labeling method was used, the labeled DNA fragments were fractionated on a 1% agarose gel and exposed to a phosphorimaging screen after the gel was blotted dry. When the Southern blotting method was used, the digested DNA was transferred and probed after fractionation on a 1% agarose gel.
Bisulfite genomic sequencing.
Bisulfite genomic sequencing was carried out as described previously (7) with minor modifications (15). Corresponding regions of plasmids pCLH22 (14) and p220.2 DNA harvested from transfected Dnmt3a expressing cells, 3a-5 (16), or the same plasmids methylated in vitro were sequenced by this method. Plasmid DNA harvested from transfected cells was digested with RsaI before sodium bisulfite treatment, and in vitro-methylated plasmid DNA was either single digested with RsaI or double digested with RsaI and HhaI before sodium bisulfite treatment. In general, 250 ng of the plasmid DNA was methylated with Dnmt3a in vitro, phenol-chloroform extracted, ethanol precipitated, and then treated with sodium bisulfite. Top and bottom strands of the sodium bisulfite-treated DNA were amplified and ligated into the TOPO TA cloning vector (Invitrogen). Both strands of each clone were sequenced using the EXCEL II sequencing kit (Epicentre) and analyzed on the Li-Cor Sequencer (model 4200L). The primers used for bisulfite sequencing are listed in Table 1. Both strands of the sodium bisulfite-treated DNA were amplified from six regions in and around the EBNA1 gene (Fig. 1).
TABLE 1.
Primers used for bisulfite genomic sequencing
| Primer name | Sequence (5′ → 3′) |
|---|---|
| Luciferase Region 1 topF | TTTAATGTATAGATTTGAAGAAGAGT |
| Luciferase Region 1 topR | TCTCAATAAACCCATATCCTTATC |
| Luciferase Region 2 topF | AATATAAAGGATATTAGGTGGTT |
| Luciferase Region 2 topR | TATAAAAATCTCTCTAATTTTTCTTA |
| EBNA1 Region 1 topF | TGGTGAGTATTTAATTAAGTTATTT |
| EBNA1 Region 1 topR | TCAAAATCACCAAACAAAAATAC |
| EBNA1 Region 1 bottomF | AAAGGAAGAGTATGAGTATTTAAT |
| EBNA1 Region 1 bottomR | CCAAATCATTCTAAAAATAATATATA |
| EBNA1 Region 2 topF | TATTAGGGTTATTGTTTTATGAG |
| EBNA1 Region 2 topR | TCAAAATCACCAAACAAAAATAC |
| EBNA1 Region 2 bottomF | TTAAGGTTATTAGATAGAGATG |
| EBNA1 Region 2 bottomR | TTCAATATTATTAAAACATTTATCA |
| EBNA1 Region 3 topF | TTGAAGAGGAAGAGGAGGGGTTT |
| EBNA1 Region 3 topR | CTAAAAAAAATCAATAAACATCTTTATTAA |
| EBNA1 Region 3 bottomF | ATATTAAAAGAGATTAATAGATATT |
| EBNA1 Region 3 bottomR | ATCTCTATCTAATAACCTTAAAA |
| EBNA1 Region 4 topF | GGAGATATATTTGGATTAGAAGGT |
| EBNA1 Region 4 topR | CCCTTTACAACCAATACAACTTAA |
| EBNA1 Region 4 bottomF | GGGTTTTTTTGTAGTTAATGTAATTTG |
| EBNA1 Region 4 bottomR | AAAACACATCTAAACCAAAAAACTC |
| EBNA1 Region 5 topF | TAAGAGTTTTTTTGGTTAGGAGTT |
| EBNA1 Region 5 topR | ATAACAAACAATAAACTCCCTTAA |
| EBNA1 Region 5 bottomF | AAGATTATGAAATAATAGATAATGGAT |
| EBNA1 Region 5 bottomR | TTAAAAACTCTCCTAACTAAAAATCA |
| EBNA1 Region 6 topF | GAATAAATTTTTAGTAGATATTATG |
| EBNA1 Region 6 topR | TTCCTACTAATTACTCCCATTC |
| EBNA1 Region 6 bottomF | TGTTGGTTGTTTTTATTTTTAG |
| EBNA1 Region 6 bottomR | AATAAATCCCCAATAAACATC |
FIG. 1.
Regions of DNA sequenced using the sodium bisulfite genomic sequencing method. The lengths of the DNA fragments examined and the number of CpG sites in each region are illustrated.
Oligonucleotide substrates and the FdC assay.
The 5-fluoro-2′-deoxycytidine (FdC)-containing oligonucleotides were synthesized by the Microchemical Core facility on site. Duplex DNA substrate containing FdC was generated as described previously (37) with minor modifications using the primer extension method. A primer, 120-rev or 120-for, was reannealed to the 122-mer FdC oligonucleotides at 50°C for 20 min after heating to 96°C for 3 min and allowed to extend for 15 min at 72°C using the Taq polymerase in the presence of [α-32P]dCTP. The sequence of 120-for is 5′-GATACATATTTGAATGTATTTAG-3′, and that of 120-rev is 5′-CCTATTTTTATAGGTTAATGTC-3′. The duplex FdC oligonucleotides generated from 8 pmol of single-stranded oligonucleotide were used in each reaction. The duplex FdC-containing oligonucleotide was incubated with 0.5 pmol of GST-3a with 80 μM AdoMet in a 10-μl reaction mixture containing 10 mM Tris (pH 8.0), 1 mM EDTA, 1 mM DTT, bovine serum albumin (50 μg/ml), 10% glycerol, and proteinase inhibitors for 4 h at 37°C. One unit of SssI methylase (New England Biolabs) was incubated with the oligonucleotides under the conditions recommended by the manufacturer for 4 h at 37°C as a positive control to ensure capture of the methylase by the FdC modification. After the reaction, SDS loading buffer was added (2% final concentration of SDS), and the reaction was heated to 65°C for 10 min. The samples were run on an SDS-6% polyacrylamide gel to separate the bound and unbound oligomers. The gel was dried and autoradiographed using a phosphorimaging screen. A single reaction was loaded on a gel for the selection experiments with random oligonucleotides containing FdC modification to avoid contamination from oligonucleotides in other lanes.
Selection experiment and sequence determination.
A pool of FdC-containing oligonucleotides with random sequences surrounding the site of methylation was used for the selection experiment. The FdC assay was done as described above, and the protein-bound oligomers were excised from the gel and rehydrated in 20 μl of 5 mM Tris (pH 8.0) and 0.5 mM EDTA for 30 min at 37°C. These protein-bound oligonucleotides were amplified using the 120-for and 120-rev primers with the following protocol: 95°C for 3 min; 35 cycles of 94°C for 40 s, 50°C for 40 s, and 72°C for 50 s; and a final cycle of 72°C for 2 min. A fresh aliquot of nucleotides, primers, and Taq polymerase was added, and one single round of amplification at 94°C for 40 s, 50°C for 40 s, and 72°C for 2 min was carried out to minimize heteroduplex formation. The amplified products were ligated into the TOPO-TA cloning vector (Invitrogen). The input oligonucleotides were also amplified and cloned into the TOPO-TA cloning vector with the same conditions as the bound oligonucleotides. After transformation into Escherichia coli, DNA from independent colonies was purified, and sequenced (both strands were sequenced). Any sequence with deletion or ambiguity was eliminated from the analysis.
Determination of methylation frequencies.
The methylation frequency at a given CpG site was calculated by dividing the number of molecules with methylation at the site by the total number of molecules examined for the site. The frequency of overall methylation on each strand of DNA in each region is calculated by dividing the total number of methylated sites on a given strand by the total number of sites examined on that strand. The overall methylation frequency was derived by dividing the total number of methylated sites by the total number of sites examined on molecules from all six regions. A CpG site with a methylation frequency higher than the overall methylation frequency is considered a high-methylation site. A CpG site with a methylation frequency lower than the overall methylation frequency is called a low-methylation site.
Statistical analysis.
A chi-square test was used for testing the null hypothesis that there is no difference between the 33 high-methylation sites and the 113 low-methylation sites sequenced from in vitro-methylated plasmid substrates. For the analysis of the selection experiment, the null hypothesis is that there is no difference between the oligonucleotides covalently linked to the methyltransferase and oligonucleotides that were not treated with any methyltransferase. The chi-square value is calculated as follows. First, the proportions for each nucleotide at each position were calculated from the sum of the two sample groups. The expected value for each nucleotide at each position is calculated by multiplying the proportion of each nucleotide at that position by the number of samples in each sample group. From this, the chi-square value can be calculated by summing the square of the difference between the observed value (O) and the expected value (E) divided by the expected value over the four nucleotides in both sample groups [Σ(E − O)2/E].
The two groups of samples were derived from the same sample pool; therefore, a distribution of chi-square values can be generated by randomly assorting the overall pool of samples into two groups. This is the basis of an effective method for determining the statistical significance of the actual chi-square value calculated from the observed samples and the overall pool of samples (18). We utilize this principle by first generating permutations of the two sample groups; that is, we randomly assort the samples into two groups 10,000 times. For each of these permutations, we calculate the chi-square value as described above. This generates a distribution of chi-square values based on the assumption that the two samples are not different. We then determine the statistical significance (P) of the actual chi-square value by determining where it falls in the distribution of chi-square values generated from the randomly assorted sample groups described above. P is determined by the percentage of chi-square values from the 10,000 simulations that are larger than the actual observed chi-square value. The smaller the P value is, the stronger the evidence is that the two sample groups were different.
RESULTS
Different regions of a stable episome are methylated to different extents by Dnmt3a in human cells.
In our previous study, we followed the acquisition of DNA methylation by a target episome, pCLH22, after transfection into human cells expressing the de novo methyltransferase Dnmt3a. We observed that the EBNA1 gene carried on the episome appears to be more methylated than the luciferase gene based on assays using methylation-sensitive restriction enzymes (16). However, the HhaI enzyme digestion only allowed examination of a small subset of the CpG sites present in these two regions. The sodium bisulfite genomic sequencing method was used to examine more CpG sites in these two regions of the episome after in vivo methylation by Dnmt3a. We chose two regions in the luciferase gene and two near the EBNA1 gene of the episome for detailed analysis. The transfected episomal DNA was only singly digested with RsaI before the sodium bisulfite treatment; therefore, methylation status at the HhaI site on these fragments can be evaluated.
Luciferase region 1 is located approximately 850 bp downstream of the translational start site of the luciferase gene, and region 2 is located approximately 500 bp further downstream from the end of region 1 (Fig. 2A). Luciferase region 1 has 11 CpG sites, and only three of these sites were methylated on three molecules among a total of 28 molecules examined (Fig. 2B). No methylation was detected at the HhaI site (CpG site 6) in luciferase region 1 (Fig. 2B). Luciferase region 2 has 28 CpG sites, and some methylation was detected on each of the 12 molecules analyzed (Fig. 2C). No methylation was detected on any of the 12 molecules at CpG sites 4, 5, 6, 7, 8, 9, 13, 18, and 21, while half of the molecules were methylated at CpG sites 19, 23, and 25 in luciferase region 2 (Fig. 2C). Four of the 12 molecules examined were methylated at the HhaI site (CpG site 22) in luciferase region 2 (Fig. 2C).
FIG. 2.
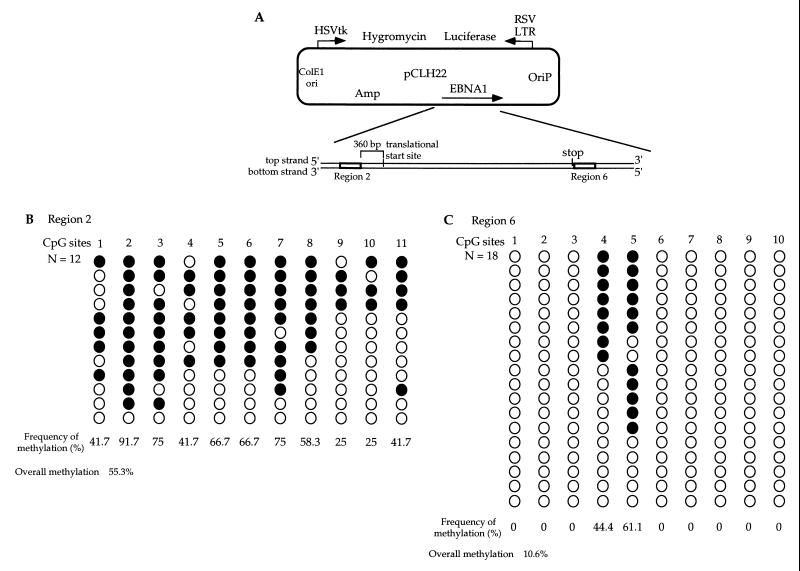
Sodium bisulfite genomic sequencing of luciferase regions 1 and 2 from in vivo-methylated pCLH22 DNA. (A) Illustration of the two regions in the luciferase gene of pCLH22 examined by the sodium bisulfite genomic sequencing method after in vivo methylation by Dnmt3a. (B) Luciferase region 1 DNA (approximately 250 bp in length) with 11 CpG sites was PCR amplified, cloned, and sequenced after sodium bisulfite treatment of pCLH22 DNA which had been harvested from transfected 3a-5 cells. (C) Luciferase region 2 DNA (approximately 290 bp in length) with 28 CpG sites from the same pCLH22 DNA described in panel B was analyzed by the sodium bisulfite genomic sequencing method as described above. The filled circles represent methylated CpG sites, and the open circles represent unmethylated CpG sites. Plasmid DNA methylated and sequenced in independent experiments showed similar results. The frequency of methylation at each site was calculated by dividing the number of molecules with methylation at the site by the total number of molecules examined. The overall methylation frequency of each region was calculated by dividing the total number of sites with methylation by the total number of sites examined. The two strands were methylated similarly, and only the top-strand methylation is shown here.
EBNA1 region 2 corresponds to sequences located upstream of the EBNA1 gene, and region 6 corresponds to sequences located immediately downstream of the EBNA1 stop codon (Fig. 3A). Repetitive sequences are present in the 1.5-kb region in the middle of the EBNA1 gene, and that region was therefore not analyzed. EBNA1 region 2 has 11 CpG sites, and methylation was detected on 11 of the 12 molecules examined (Fig. 3B). Six of the 11 CpG sites analyzed were methylated on more than 60% of the molecules examined, and the least-methylated CpG sites 9 and 10 were methylated on 25% of the molecules (Fig. 3B). Nine of the 12 molecules examined were methylated at the HhaI site (CpG site 3) in EBNA1 region 2 (Fig. 3B). EBNA1 region 6 has 10 CpG sites, and methylation was only observed at CpG sites 4 and 5 (Fig. 3C). Eleven of the 18 molecules sequenced were methylated at the HhaI site (CpG site 5) in EBNA1 region 6 (Fig. 3C).
FIG. 3.
Sodium bisulfite genomic sequencing of EBNA1 regions 2 and 6 from in vivo-methylated pCLH22 DNA. (A) Illustration of the two regions in the EBNA1 gene region of pCLH22 examined by the sodium bisulfite genomic sequencing method after in vivo methylation by Dnmt3a. (B) EBNA1 region 2 DNA (approximately 250 bp long) with 11 CpG sites was PCR amplified, cloned, and sequenced after sodium bisulfite treatment of pCLH22 DNA which had been harvested from transfected 3a-5 cells. (C) EBNA1 region 6 DNA (approximately 250 bp long) with 10 CpG sites from the same pCLH22 DNA described in panel B was analyzed by the sodium bisulfite genomic sequencing method as described above. Plasmid DNA methylated and sequenced in independent experiments showed similar results. The filled circles represent methylated CpG sites, and the open circles represent unmethylated CpG sites. Plasmid DNA methylated and sequenced in independent experiments showed similar results. The frequency of methylation at each site and the overall methylation frequency of each region were calculated as described in the legend to Fig. 2. Only the top-strand methylation is shown here.
Consistent with previous findings using the restriction digestion assay (16), a higher frequency of methylation was detected at the HhaI sites in the EBNA1 region than in the luciferase gene region. Although the HhaI sites in luciferase region 2 were less methylated than the HhaI sites in EBNA1 region 6, most of the CpG sites in EBNA1 region 6 were not methylated, and some CpG sites in luciferase region 2 were methylated on 50% of the molecules examined. This finding indicates that frequent methylation at a given CpG site does not warrant frequent methylation of adjacent CpG sites. The CpG density is the highest in luciferase region 2 (approximately one CpG per 10 bases) among the four DNA regions examined. However, the density of methylation was not the highest in this region. While the CpG density is similar in the other three regions examined, the frequency of methylation observed in each region was widely different, ranging from 1.9% in luciferase region 1 to 55.3% in EBNA1 region 2. The findings here show that the EBNA1 gene region is more methylated than the luciferase gene region as independently demonstrated in the restriction digestion assay in a previous study (16). These results also suggest that Dnmt3a does not necessarily methylate regions with higher CpG density more frequently. This is consistent with the findings of Yates et al. (36). Furthermore, this enzyme does not appear to methylate every CpG site or all DNA regions equally in human cells. Although this is a detailed sequence-level study, findings from in vivo study are complicated by the activity of other methyltransferases and chromatin structure in the cells among other possible cellular factors. The majority of the molecules had unmethylated CpG sites between methylated CpG sites even though many adjacent CpG sites were methylated on the same DNA molecules. This may be the result of other cellular factors disrupting the activity of Dnmt3a or the lack of processivity of Dnmt3a. It is noteworthy that no cytosine methylation was observed at non-CpG sites.
In vitro methylation of DNA by Dnmt3a.
Although the sodium bisulfite sequencing results confirmed that Dnmt3a methylates different regions of the episome with different efficiencies in vivo, it is impossible to discern whether Dnmt3a methylates CpG sites randomly or whether it prefers to methylate some CpG sites over others. Besides the possible site preferences of the de novo methyltransferases, de novo methylation may be complicated by chromatin structure, binding of proteins to the DNA, activity of other methyltransferases, and the concentration of de novo methyltransferases in vivo. When naked DNA targets are methylated in vitro and then analyzed, the potential site preference of the methyltransferase can be determined in the absence of all other compounding factors. To achieve this, we constructed Dnmt3a wild-type and mutant protein expression vectors and purified the overexpressed proteins from transfected human cells for the in vitro assays. SDS-polyacrylamide gel electrophoresis analysis of the purified proteins demonstrates the purity of the proteins (Fig. 4A).
FIG. 4.
GST-3a methylates circular plasmid DNA as well as linear DNA fragments in vitro using a restriction enzyme digestion assay. (A) Purified GST fusion protein of Dnmt3a wild type (GST-3a) and mutant (GST-3aMut). Six units of the DNMT1 (New England Biolabs) and approximately 200 ng each of GST-3a and GST3aMut were loaded on an SDS-7% polyacrylamide gel and stained with Coomassie blue stain. (B) Plasmid p220.0 was incubated (+) with GST-3a or GST3aMut, digested with a 10-fold excess of HinP1I, and labeled with [32P]dCTP using Klenow enzyme as described in Materials and Methods. The complete digestion pattern is shown in the lane that has no protein treatment. The additional bands in the lanes with DNA treated with GST-3a are the HinP1I-resistant bands, indicating DNA methylation. These additional bands are marked by arrowheads. (C) A 467-bp DNA fragment containing three HhaI sites was used as a substrate for GST-3a and GST-3aMut, as described above. The DNA was digested with HhaI before being fractionated on a 1% agarose gel, Southern transferred, and probed with the entire DNA fragment. DNA unmethylated at the HhaI sites gives rise to the 304-bp band after HhaI digestion. The 467-bp band indicates methylation at the three HhaI sites.
The EBNA1 region on the plasmid p220.2 and the HhaI sites within the lacO sequences on plasmid pOLucOriP were known to become methylated in vivo when murine Dnmt3a is overexpressed in 293/EBNA1 cells (16). Therefore, plasmid p220.2 and the 467-bp DNA fragment containing the lacO sequences were used in the in vitro methylation assay. The GST-3a or GST-3aMut protein (40 ng) was incubated with 250 ng of p220.2 or 100 ng of the 467-bp lacO DNA fragment using unlabeled AdoMet at 37°C for 18 h. A mock reaction without any protein was also incubated for 18 h at 37°C as a negative control. The DNA was ethanol precipitated, after phenol and chloroform extraction. The p220.2 DNA was digested with HinP1I (an isoschizomer of HhaI) before labeling with 32P-dCTP using the Klenow end fill-in method. The labeled DNA was fractionated on a 1% agarose gel and exposed to a phosphorimaging screen. Fragments larger than complete HinP1I-digested fragments were observed in the HinP1I-digested p220.2 after GST-3a treatment (Fig. 4B). In contrast, these fragments were not detected in the mock-treated p220.2 or in p220.2 treated with GST-3aMut protein (Fig. 4B). Although the larger HinP1I fragments were observed when the NaCl concentration was at 27.5 mM, the intensity of these fragments appears to be weaker than when the NaCl concentration was at 7.5 mM (Fig. 4B). When the NaCl concentration was increased to 50 mM, no increased-size HinP1I fragment was detected (data not shown). This finding indicates that methylation activity of Dnmt3a is sensitive to salt concentration.
A portion of the 467-bp lacO DNA fragment was digested with HhaI after in vitro methylation with GST-3a, fractionated on a 1% agarose gel with undigested material, and transferred by Southern blotting. A single 467-bp band was observed in the undigested DNA regardless of whether it was incubated with no protein, GST-3a, or GST-3aMut (Fig. 4C). After HhaI-digestion, a single 304-bp band was detected in the mock-treated DNA and the DNA treated with GST-3aMut (Fig. 4C), indicating the lack of methylation at the HhaI sites. In contrast, a 467-bp band and a 304-bp band were observed after HhaI digestion in the DNA treated with GST-3a (Fig. 4C), indicating methylation of HhaI sites on a fraction of the molecules. These findings clearly demonstrate that GST-3a can methylate linear as well as circular DNA substrates in vitro and that higher NaCl concentrations inhibit in vitro methylation activity of Dnmt3a.
Basic biochemical features of Dnmt3a.
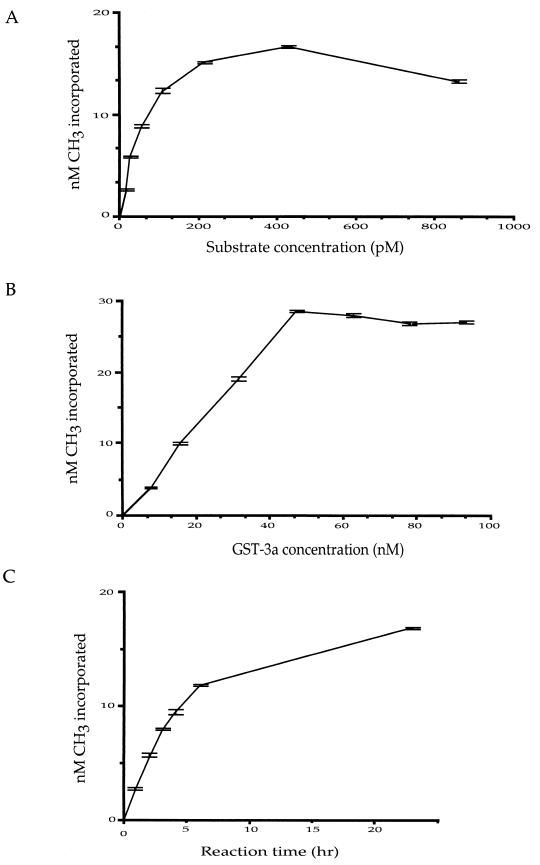
The plasmid p220.2 was used as a DNA methylation target to determine some basic enzymatic features of Dnmt3a in a 3H incorporation assay. To study the effect of DNA substrate concentration on the incorporation of 3H, various concentrations of DNA were incubated with purified GST fusion protein of Dnmt3a in the presence of [3H]AdoMet overnight (14 to 18 h) at 37°C. A reaction with no protein and a reaction with no DNA substrate were included as controls for each experiment. The substrate concentration and the 3H incorporation showed a nearly linear relationship, within a substrate concentration range of 27 pM to 214 pM, when p220.2 was used in the presence of a 15.7 nM concentration of GST-3a (Fig. 5A). With increased substrate concentration beyond 214 pM with a 15.7 nM concentration of GST-3a, very little increase of 3H incorporation was observed (Fig. 5A). This indicates that 0.31 pmol of GST-3a reaches its maximum activity with 4.3 fmol of DNA substrate, p220.2, in the presence of 1.1 μM AdoMet. A twofold increase of AdoMet was tested to verify whether the AdoMet is limiting the activity of Dnmt3a. No increase in 3H incorporation was observed (data not shown). We also found no difference using circular or linearized p220.2 as a substrate (data not shown).
FIG. 5.
Basic enzymatic features of GST-3a. (A) Relationship between substrate concentration and 3H incorporation at an enzyme concentration of 15.7 nM (in a 20-μl reaction mixture). (B) Relationship between enzyme concentration and 3H incorporation at a substrate concentration of 108 pM. (C) Time course of methylation catalyzed by GST-3a at a substrate concentration of 108 pM and an enzyme concentration of 15.2 nM.
Based on the above findings, various concentrations of GST-3a and 108 pM p220.2 DNA (in 20-μl reaction mixtures) were incubated overnight at 37°C. The 3H incorporation results showed linearity of the reaction progression for GST-3a concentrations between 7.8 and 47.2 nM (Fig. 5B). Based on the number of CpG sites on the plasmid target and the amount of 3H incorporated, the percentage of CpG sites that become methylated can be estimated. It appears that approximately 60% of the CpG sites on the plasmid were not methylated, even at the highest level of 3H incorporation on p220.2 DNA when the GST-3a concentration was at 47.2 nM or higher. This finding suggests that GST-3a does not methylate every CpG site on the plasmid.
A linear progression of the reaction was also observed between 1 and 4 h of reaction time with a GST-3a concentration of 15.7 nM and a DNA concentration of 108 pM (Fig. 5C). The 3H incorporation at the 4-h time point was approximately 75% of the overnight reaction. To determine whether the stability of AdoMet can affect the reaction progression after the 4-h time point, 1.1 μM [3H]AdoMet was incubated with 108 pM DNA substrate in the reaction buffer for 2, 3, 4, and 6 h at 37°C before GST-3a was added to a concentration of 15.7 nM. After the addition of GST-3a, incubation of the reaction mixtures was continued at 37°C for 16 h. All reactions showed the same end point 3H incorporation (data not shown), suggesting there was enough AdoMet in the reaction for methylation to take place after 6 h of incubation. These results indicate that the methylation by Dnmt3a is very slow in vitro, and the enzyme appears to remain active for longer than 4 h at 37°C.
Dnmt3a generates a strand-specific methylation pattern on plasmid substrates in vitro.
In order to assess potential site preferences by Dnmt3a unambiguously, we used purified Dnmt3a protein in an in vitro assay and analyzed the resulting methylated sites by the bisulfite genomic sequencing method. Under these conditions, there is no possible influence of the chromatin structure or interference by any other proteins. Similarly, the impact of substrate length, number of CpG sites, and density of CpG sites on the reaction should be minimal using p220.2, an 8.9-kb plasmid containing 459 CpG sites distributed in various densities in different regions.
As described above and in the previous study (16), the EBNA1 region was methylated more frequently by Dnmt3a in vivo than the luciferase gene region. We examined regions 2 and 6 in the EBNA1 region on p220.2 methylated by Dnmt3a in vitro. Each of these regions includes approximately 250 bp of DNA containing 10 or 11 CpG sites on each strand. Bisulfite genomic sequencing allows the amplification of each strand of DNA independently; therefore, the methylation status of each CpG on each DNA strand can be assessed using this assay. The in vitro-methylated p220.2 was double digested with RsaI and HhaI before the sodium bisulfite treatment to enrich for methylated molecules even though methylation at the HhaI site does not warrant methylation at adjacent CpG sites based on data from in vivo methylation.
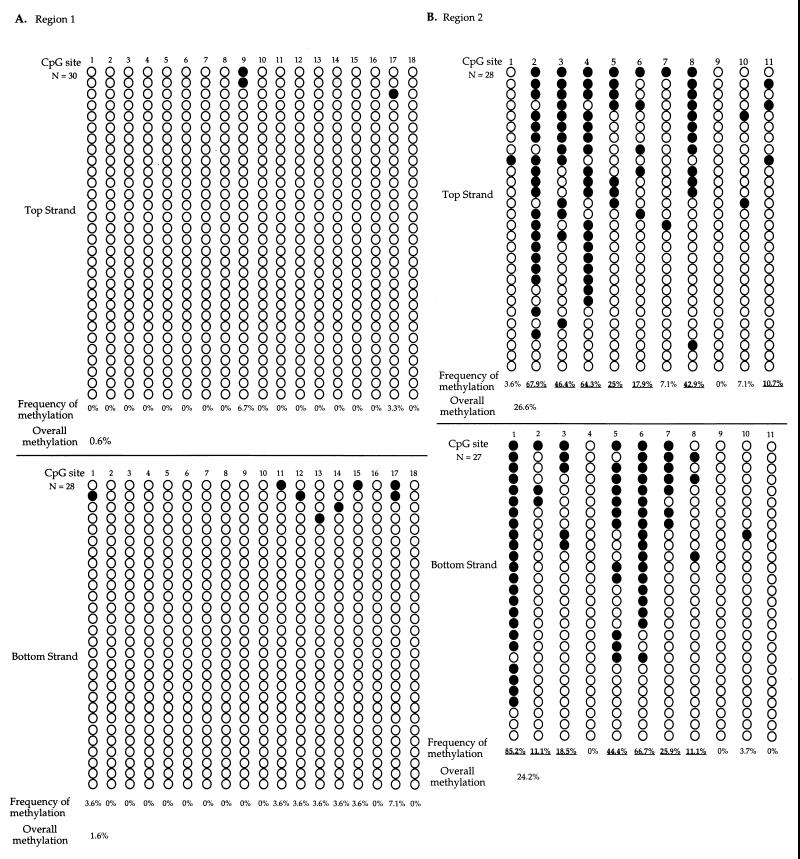
Results obtained for region 2 are summarized in Fig. 6A. CpG sites 2, 3, 4, and 8 were methylated much more frequently than other sites on the top strand. In contrast, CpG sites 1, 5, 6, and 7 were more frequently methylated than other CpG sites by Dnmt3a on the bottom strand in region 2. The methylation patterns of the two strands are distinctly different. The contrast is best exemplified with the CpG sites mentioned above, for which sites more frequently methylated on one strand were much less frequently methylated on the complementary strand. This strand difference was not observed when the same DNA fragment from the in vivo-methylated episome was sequenced by the sodium bisulfite sequencing method with the same primers and conditions. Therefore, bias due to sodium bisulfite treatment or PCR amplification is not the cause of this observation. Furthermore, the HhaI digestion prior to the sodium bisulfite treatment should eliminate nearly all molecules unmethylated at CpG site 3 within the HhaI recognition site. If the CpG site on the cDNA strand was methylated either at the same time or immediately after the methylation of the CpG site on the first strand, CpG site 3 should have equal or similar methylation frequencies on the two DNA strands. However, CpG site 3 was the most methylated CpG site on the top strand, while it was mostly unmethylated on the bottom strand. Similar to in vivo-methylated DNA, many CpG sites remained unmethylated in between methylated CpG sites on the same molecule, indicating the lack of processivity of Dnmt3a in vitro. Based on the action of Dnmt1 at hemimethylated sites, CpG sites methylated by Dnmt3a on only one strand will become methylated or unmethylated on both strands after DNA replication in vivo. Therefore, the more highly methylated CpG sites on either strand observed in vitro should be the more highly methylated CpG sites in vivo unless other cellular factors protect these sites from methylation in vivo. If one sums the methylation observed on both strands for each CpG site, sites 9 and 10 were the least methylated sites in EBNA1 region 2 in vitro. Importantly, these two CpG sites also were the least-methylated sites in EBNA1 region 2 in vivo, among 11 sites examined.
FIG. 6.
Sodium bisulfite genomic sequencing of EBNA1 regions 2 and 6 from in vitro-methylated p220.2 DNA. (A) EBNA1 region 1 DNA (approximately 250 bp in length) with 11 CpG sites was PCR amplified, cloned, and sequenced after sodium bisulfite treatment of p220.2 DNA methylated by GST-3a in vitro. CpG site 3 is an HhaI site. (B) EBNA1 region 2 DNA (approximately 250 bp long) with 10 CpG sites from in vitro Dnmt3a methylated p220.2 was analyzed by the sodium bisulfite genomic sequencing method, as described above. CpG site 5 is an HhaI site. Plasmid DNA methylated and sequenced in independent experiments showed similar results. The filled circles represent methylated CpG sites, and the open circles represent unmethylated CpG sites. Plasmid DNA methylated and sequenced in independent experiments showed similar results. The frequency of methylation at each site and the overall methylation frequency of each region were calculated as described in the legend to Fig. 3.
Sites 2, 3, 4, and 5 were the most-methylated CpG sites on the top strand, while sites 1, 8, and 10 were more frequently methylated than other sites on the bottom strand in EBNA1 region 6 (Fig. 6B). Also, the CpG site 5 within the HhaI recognition site was methylated on the top strand in nearly 80% of the molecules, while the same CpG site was only methylated on the bottom strand in 10% of the molecules. In contrast to the in vivo-methylated plasmid, many other CpG sites were methylated at a high frequency in addition to sites 4 and 5. This may be due to cellular factors protecting these sites from methylation in vivo. The distinctly different methylation pattern on the top and the bottom strand of DNA in EBNA1 region 6 was similar to what was observed in EBNA1 region 2. The difference in methylation pattern on the two strands was not likely due to sodium bisulfite treatment or PCR bias, because this difference was not observed in the in vivo-methylated episome using the same analysis. Again, many unmethylated CpG sites were observed in between methylated sites on the same molecule.
The complementary strands of the same CpG site were methylated at similar frequencies (data not shown) on the in vivo-methylated pCLH22, resulting in very similar methylation patterns on the top and the bottom strands. The lack of strand difference in the sodium bisulfite sequencing of in vivo-methylated episomes indicates the absence of bias in sodium bisulfite treatment, PCR, or sequencing. However, the following controls were done to independently rule out this possibility. Plasmid pCLH22 DNA was in vitro methylated using the prokaryotic methylase, SssI, which methylates Cs at all CpGs without any known preferences. The DNA was digested, sodium bisulfite treated, PCR amplified, cloned, and sequenced. Nearly all of the CpG sites examined were methylated, and no difference in methylation was observed on the two DNA strands (data not shown). This finding indicates that SssI does not methylate the two cDNA strands differently in vitro. To rule out possible sodium bisulfite treatment or PCR bias, we sodium bisulfite treated a mixture of unmethylated and SssI-methylated plasmid DNA. The sodium bisulfite-treated DNA was PCR amplified, cloned, and sequenced. The same fraction of the molecules was methylated among all the molecules examined from each strand (data not shown). These results further confirm that the observed distinct methylation pattern generated by Dnmt3a in vitro on the two DNA strands was not likely due to a bias occurring in the sodium bisulfite treatment, PCR amplification, cloning, or sequencing steps. These results also indicate that the prokaryotic methylase, SssI, does not generate a strand specific methylation pattern as Dnmt3a does. As commonly believed, SssI does not appear to have a strong preference for some CpG sites over others, since nearly all CpG sites were methylated by the enzyme in vitro.
It has been reported that supercoiling may affect the sequence specificity of a DNA methyltransferase (3). To assess whether supercoiling is responsible for the asymmetrical methylation pattern on the two strands observed, we repeated the previous experiment using linearized p220.2 as a substrate in an in vitro methylation assay. The results from bisulfite genomic sequencing were similar to those described above for the supercoiled p220.2 substrate (data not shown). This finding indicates that supercoiling does not influence the methylation patterns generated by Dnmt3a.
Sequences surrounding the CpG dinucleotide play a significant role in site choice by Dnmt3a in vitro.
Although the methylation pattern at HhaI recognition sites strongly indicates that the DNA is hemimethylated at the two HhaI sites examined, it is important to rule out possible biases generated by the existence of HhaI sites in these two regions. Furthermore, the frequency of methylation at the HhaI sites is likely to be biased when the DNA was digested with HhaI prior to sodium bisulfite treatment. To discern the unbiased frequency of methylation at each of the CpG sites examined, methylated DNA singly digested with RsaI was sequenced by the sodium bisulfite genomic sequencing method to avoid the possible bias by HhaI digestion. Plasmid DNA from the same methylation reaction used in the above experiment was used here.
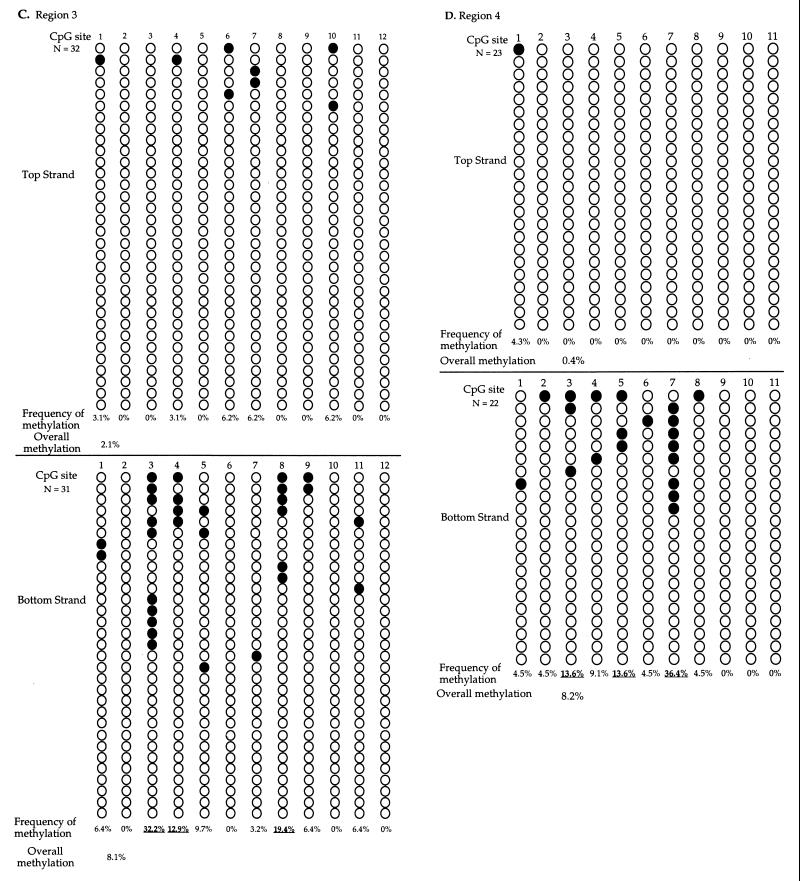
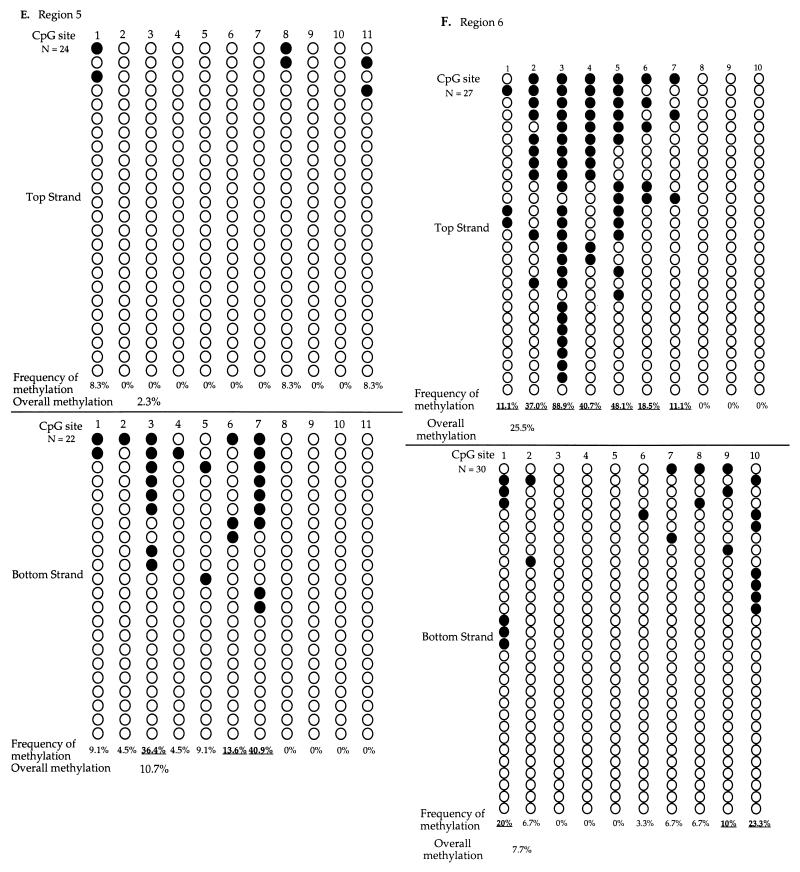
The sodium bisulfite genomic sequencing results and the methylation frequency of each CpG site in six regions sequenced are summarized in Fig. 7. The methylation pattern of the top strand was distinctly different from that of the bottom strand for all six regions sequenced. This finding is consistent with the findings above. The distribution of methylation across CpG sites in regions 2 and 6 (Fig. 7B and F) is very similar to what was observed, although the methylation frequency is approximately 10% lower when DNA was not digested with HhaI before sodium bisulfite treatment (Fig. 6). These findings indicate that although the HhaI digestion may enrich the molecules methylated at the HhaI site in regions containing the HhaI site, it does not affect the distinct difference of methylation pattern on the two cDNA strands. The methylation frequency was the lowest in region 1, though this region has the highest CpG density among the six regions sequenced. This finding is consistent with observations in vivo that CpG density does not influence sites of methylation by Dnmt3a (Fig. 2 and 3). No cytosine methylation at non-CpG sites was detected in any of the clones sequenced.
FIG. 7.
Sodium bisulfite genomic sequencing of EBNA1 region from in vitro-methylated p220.2 DNA. Molecules from regions 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), and 6 (F). After in vitro methylation, DNA was digested with RsaI, sodium bisulfite treated, PCR amplified, cloned, and sequenced. The methylation patterns of the top and the bottom strands are clearly different in all regions examined. Plasmid DNA methylated and sequenced in independent experiments showed similar results. Each row presents an independent molecule sequenced. The filled circles represent methylated CpG sites, and the open circles represent unmethylated CpG sites. The frequency of methylation at each site was calculated by dividing the number of molecules with methylation at the site by the total number of molecules examined. The overall methylation frequency was calculated by dividing the total number of sites with methylation by the total number of sites examined. Underlining indicates the sites with higher methylation frequency, compared to the average methylation frequency of all six regions.
Methylation frequency and sequence content of each of the 146 CpG sites from six regions were analyzed to discern whether sequence surrounding the CpG site plays a role in Dnmt3a site choice. The frequency of methylation by Dnmt3a in vitro at a given site on each strand is calculated by dividing the number of molecules with methylation at the site by the total number of molecules examined for that site. For example, site 2 on the top strand of region 2 was methylated at 67.9% frequency (19 methylated molecules in 28 molecules sequenced [Fig. 7B]). The frequency of overall methylation on each strand of DNA in each region can be calculated by dividing the total number of methylated sites on a given strand by the total number of sites examined on the strand within the region. For example, there was 26.6% methylation on the top strand of region 2 (82 methylated sites among 308 sites analyzed [Fig. 7B]). An overall methylation frequency of 9.8% was derived by dividing the total number of methylated sites by the total number of sites examined on molecules from all six regions. The CpG sites that became methylated at a higher frequency than the overall methylation frequency of 9.8% were called high-methylation sites. The CpG sites that were methylated at a lower frequency than the overall frequency of methylation were designated low-methylation sites.
There were a total of 33 high-methylation sites and 113 low-methylation sites in the 146 sites studied. Nucleotide contents of the six nucleotides upstream and six nucleotides downstream of the CpG site from high-methylation sites and from low-methylation sites were compared and analyzed (Table 2). A chi-square test was used to test the null hypothesis that there is no difference between the high-methylation sites and the low-methylation sites. The chi-square values indicated that the nucleotide contents of the high-methylation sites are significantly different than those of the low-methylation sites at positions −2 and +1 (Table 2). Examination of the nucleotide content at these positions revealed that C and T residues were present much more often at the −2 and +1 positions in the high-methylation sites than expected (Table 2). All of the other positions showed much less bias (Table 2). It is also noteworthy that only 1 of the 33 high-methylation sites (3%) and 40 of the 113 low-methylation sites (35.4%) had purines at both the −2 and the +1 positions while the theoretical frequency of having purines at both of these positions should be 25%. These findings suggest that Dnmt3a prefers 5′-PyNCGPy-3′ sites for methylation in vitro, although purine-rich sequences were not completely excluded from the sites with high methylation.
TABLE 2.
Nucleotide contents of six positions downstream of each CpG site examineda
| Position | No. of sites containing nucleotide
|
χ2 | P (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| High-methylation sites | All sites | |||||||||
| A | C | G | T | A | C | G | T | |||
| −6 | 8 | 9 | 7 | 9 | 29 | 40 | 42 | 35 | 1.45 | 71.5 |
| −5 | 7 | 5 | 9 | 12 | 33 | 29 | 40 | 44 | 1.06 | 80.0 |
| −4 | 4 | 8 | 5 | 16 | 36 | 27 | 32 | 51 | 6.61 | 8.6 |
| −3 | 8 | 12 | 3 | 10 | 27 | 45 | 38 | 36 | 6.43 | 9.7 |
| −2 | 0↓ | 14↑ | 6↓ | 13↑ | 24 | 39 | 53 | 30 | 22.18 | 0.01 |
| −1 | 7 | 7 | 7 | 12 | 37 | 39 | 35 | 35 | 3.64 | 31.7 |
| 1 | 3↓ | 12↑ | 2↓ | 16↑ | 34 | 36 | 40 | 36 | 22.96 | 0.01 |
| 2 | 8 | 14 | 6 | 5 | 31 | 51 | 39 | 25 | 2.13 | 56.1 |
| 3 | 6 | 13 | 5 | 9 | 35 | 39 | 45 | 27 | 8.34 | 3.6 |
| 4 | 9 | 8 | 7 | 9 | 52 | 34 | 27 | 33 | 1.43 | 71.7 |
| 5 | 9 | 11 | 4 | 9 | 45 | 37 | 27 | 37 | 2.25 | 54.5 |
| 6 | 7 | 10 | 8 | 8 | 33 | 44 | 43 | 26 | 1.42 | 72.4 |
High-methylation sites are the 33 CpG sites with higher-than-average frequency of methylation. “All sites” refers to the 146 CpG sites examined from the six regions presented in Fig. 1. The chi-square value is calculated as described in the Materials and Methods and is used to compare the 33 high-methylation sites to the 113 low-methylation sites. The statistical significance of the chi-square value (P) is determined as described in Materials and Methods. Arrows indicate increase (↑) or decrease (↓) from the expected value.
Preference for flanking pyrimidines can be detected using random oligonucleotides containing FdC at the CpG site.
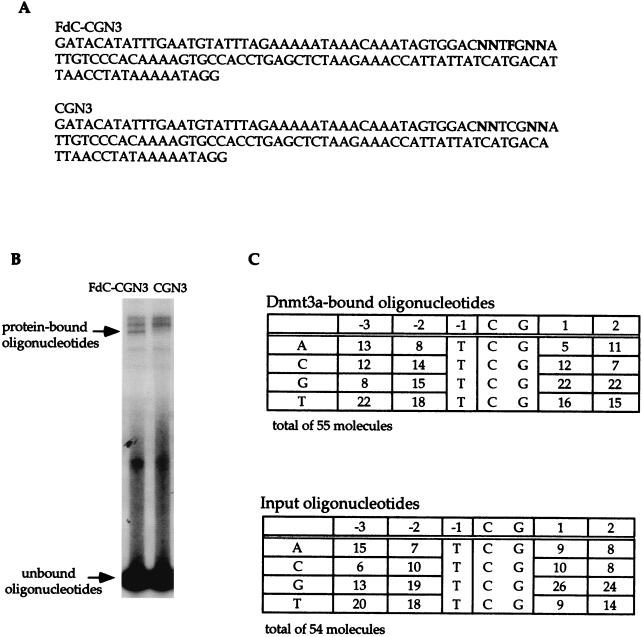
To test the finding that Dnmt3a has a preference for pyrimidine-rich sequences with an independent method, a selection experiment using random oligonucleotides containing FdC at the CpG site was done. The FdC modification at the CpG site captures the methyltransferase covalently linked to the DNA substrate after the transfer of the methyl group is completed (31, 37). If Dnmt3a has a site preference, addition of Dnmt3a to FdC-containing oligonucleotides with random nucleotides placed at key positions should allow the retrieval of its preferred targets. Generally, the selection experiment is carried out using multiple rounds of a binding assay using random oligonucleotides to determine specific sequences that bind to the protein of interest. After incubation of the protein and random oligonucleotides under the appropriate conditions, the protein-bound oligonucleotides are separated from the unbound oligonucleotides on a gel. The mobility-retarded band containing the protein-bound oligonucleotides is excised, and the oligonucleotides are amplified, and this material is used as the substrate for the next round of binding, gel shift, and amplification. After several rounds of selections, the amplified products are cloned and sequenced. The vast excess of unbound oligonucleotide in the reaction often contaminates the shifted gel band in electrophoresis. Therefore, several rounds of this selection are usually required to identify the consensus sequence for protein binding. However, only one round of this selection can be done in this study due to the inability to make the FdC modification at the CpG site after the amplification of the protein bound oligonucleotides. Without the FdC modification at the CpG site, the oligonucleotides cannot capture the methyltransferase after methylation in the presence of AdoMet; therefore, multiple rounds of selection are not possible. With this severe limitation, it is understood that this approach may only provide an initial suggestive trend of the site preference and not a definitive answer.
An oligonucleotide pool, FdC-CGN3 (Fig. 8A), with random nucleotides placed at positions −3, −2, +1, and +2 and a thymidine placed at position −1 relative to the FdCpG site, was used. The +2 position was included as a position of randomness to serve as a negative control site. A control oligonucleotide pool, CGN3 (Fig. 8A), without the FdC modification was used as a negative control for covalent linkage. Three slow-migrating bands were routinely detected on the gel (Fig. 8B). The two bands on the top were observed regardless of the presence of FdC on the oligonucleotides (Fig. 8B) or the presence of CpG on the oligonucleotide (see next section for details). Although the cause was not determined, these two bands are most likely due to nonspecific binding. In contrast, the fastest-migrating band among these three bands was only present when oligonucleotides containing FdC were used (Fig. 8B). Quantitation of the radioactivity in the unbound and the bound oligonucleotides revealed that less than 0.5% of the FdC-CGN3 oligonucleotide was bound to Dnmt3a.
FIG. 8.
Selection experiment using FdC-containing random oligonucleotides. (A) Sequences of oligonucleotides with random nucleotides at four positions surrounding the CpG site. FdC-CGN3 has FdC at the CpG site, while CGN3 does not have the FdC modification. (B) Separation of unbound and protein-bound oligonucleotides. The band specific for protein-bound FdC-containing oligonucleotides is as indicated. The two other slower-migrating bands were most likely nonspecific (see also Fig. 9). The protein-bound oligonucleotide band was excised, amplified, cloned, and sequenced. (C) Sequencing results on Dnmt3a-bound oligonucleotides and the input oligonucleotides.
The input FdC-CGN3 oligonucleotides and the FdC-CGN3 oligonucleotides that bound to Dnmt3a were amplified, cloned, and sequenced. Sequences of a total of 54 oligonucleotides from the input oligonucleotides and a total of 55 oligonucleotides from the GST-3a-bound oligonucleotides were obtained. The nucleotide contents of these clones at each of the four random positions are summarized in Fig. 8C. The chi-square values were not significant for rejecting the null hypothesis that protein-bound oligonucleotides are the same as unbound oligonucleotides. However, a small increase of pyrimidine residues and a decrease of purine residues were observed at positions −3, −2, and +1 in the Dnmt3a-bound oligonucleotides, while almost no difference in the nucleotide content between unbound and Dnmt3a-bound oligonucleotides at position +2 was observed (Fig. 8C). In several experiments, when other oligonucleotides with different nucleotide contents at two of the nonrandom positions were loaded in another lane on the same gel, 25 to 33% of the clones were contaminants from oligonucleotides in the other lane (data not shown). It is likely that contamination of the band containing protein-bound oligonucleotides by the unbound oligonucleotide from the same lane of the gel might have been greater than 30%. In similar experiments using HhaI-methylase, this contamination did not appear to be a problem and all the clones recovered bear the HhaI site at the FdC site (data not shown). However, approximately 20-fold more of the oligonucleotides shifted with HhaI-methylase than with Dnmt3a in these experiments. Therefore, this problem is much more significant when the fraction of oligonucleotide shifted is small, as for Dnmt3a, rather than large, as for Dnmt1 or HhaI-methylase. Taking this into account, the small increases of pyrimidine and decreases of purines at positions −3, −2, and +1 are suggestive, and they support the result from bisulfite genomic sequencing of plasmid methylated with Dnmt3a in vitro.
Pyrimidines surrounding the CpG Site on the FdC-containing oligonucleotide can influence Dnmt3a methylation.
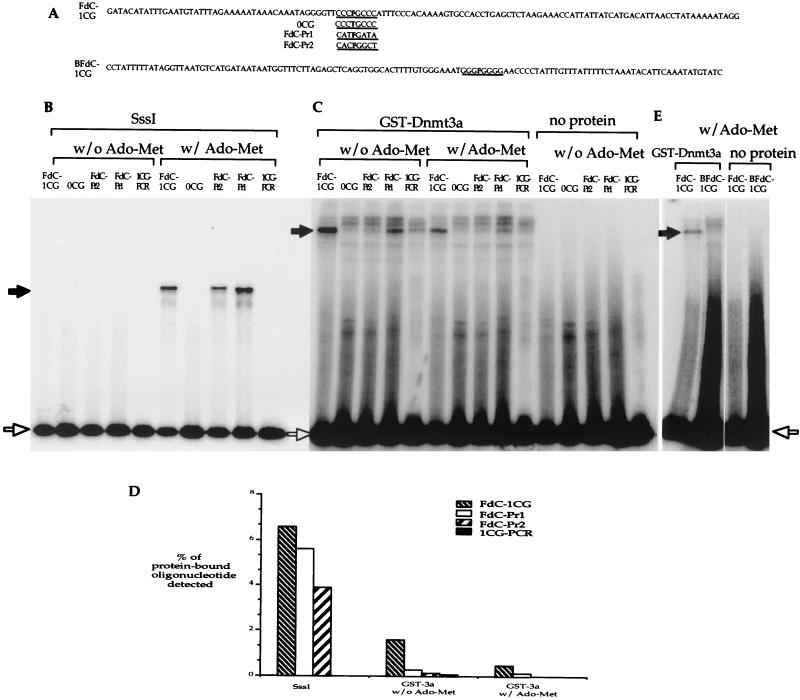
Selection experiments using random oligonucleotides only provide a weak suggestion of the site preference. Specific oligonucleotides with FdC modification at the CpG site were used to test whether an oligonucleotide with pyrimidine-rich sequence surrounding the CpG site is preferred by Dnmt3a over an oligonucleotide with purine-rich sequences adjacent to the CpG site. If Dnmt3a does not methylate all CpG sites equally, FdC-containing oligonucleotides with different nucleotide sequences surrounding the FdC site will show different efficiencies in capturing the methyltransferase. Three FdC-containing oligomers with different nucleotides at the three bases upstream of the CpG site and the three bases downstream of the CpG site were used in the experiment (Fig. 9A). An oligonucleotide, 0CG, identical to the FdC-1CG oligomer but with a T at the FdC site, was used as a no-CpG site control. All the single-stranded oligonucleotides were made into double-stranded substrates as described in Materials and Methods. A double-stranded DNA fragment, 1CG-PCR, generated by PCR amplification using FdC-1CG as the template was also included in the experiment as a no-FdC modification control. The parallel experiments were carried out using the prokaryotic methylase SssI. When the reactions were carried out in the absence of AdoMet, none of the oligonucleotides captured any SssI (Fig. 9B). In the presence of AdoMet, all three FdC-containing oligonucleotides, FdC-1CG, FdC-Pr1, and FdC-Pr2, captured SssI enzyme while oligonucleotides with no CpG or no FdC did not capture any SssI enzyme (Fig. 9B). These findings were consistent with previous reports on prokaryotic methylases (31, 37) and validated the oligonucleotides and the assay. Unlike Dnmt1 (37) and SssI, FdC-1CG and FdC-Pr1 oligonucleotides as well as the 1CG-PCR DNA fragment clearly captured Dnmt3a in the absence of AdoMet (Fig. 9C). In the presence of AdoMet, only FdC-1CG and FdC-Pr1 oligonucleotides clearly showed capturing of Dnmt3a, and FdC-1CG showed a stronger shifted band than FdC-Pr1 (Fig. 9C). Many repeated experiments showed the same results (data not shown). These results indicated that Dnmt3a could be captured by the FdC modification at the CpG site as can other cytosine-5-methyltransferases. However, unlike other cytosine-5-methyltransferases known to date, the presence of AdoMet is not required for covalent association between the oligonucleotides and Dnmt3a.
FIG. 9.
FdC assay using specific oligonucleotides to test the Dnmt3a site preference. (A) Sequences of oligonucleotides with different sequence contents at the three bases upstream and three bases downstream of the CpG site. Oligonucleotide 0CG has no CpG but was otherwise identical to the FdC-1CG oligonucleotide. Both FdC-Pr1 and FdC-Pr2 have purines at the key positions. The duplex DNA molecules generated from oligonucleotides FdC-1CG and BFdC-1CG were identical except that the FdC modification was located on the opposite strands. A substrate without FdC modification, 1CG-PCR, was generated by PCR amplification of the FdC-1CG oligonucleotide using primers 120-for and 120-rev as described. (B) FdC assays using the prokaryotic methylase, SssI. The oligonucleotide used in the reaction is indicated above the lane. Reactions with or without the addition of AdoMet are as indicated by the brackets. (C) FdC assays using GST-Dnmt3a. The oligonucleotides used in the reaction are indicated above the lanes. Reactions with or without the addition of AdoMet are indicated with brackets. Two more slowly migrating bands were observed in all reactions when protein was added. These two bands were most likely nonspecific, because these bands were present even when no CpG site was on the oligonucleotide. (D) Illustration of the percentage of protein-bound oligonucleotides detected in the reaction in panel C. The percentage of protein-bound oligonucleotides in each lane was derived by dividing the radioactivity in the band containing protein-bound oligonucleotides by the total radioactivity in the band containing unbound oligonucleotides and the band containing protein-bound oligonucleotides. (E) Dnmt3a does not bind the same CpG site on the complementary strands of DNA equally. While a shifted band is detected in the reaction using FdC-1CG oligonucleotide, no protein-bound BFdC-1CG oligonucleotide was detected. In panels B, C, and E, the protein-bound oligonucleotides are indicated with a solid arrow, and the unbound oligonucleotides are indicated with an open arrow.
Quantitation of the radioactivity in the shifted bands and the unbound oligonucleotides revealed that 6.6% of the FdC-1CG oligonucleotide, 5.6% of the FdC-Pr1 oligonucleotide, and 3.9% of the FdC-Pr2 oligonucleotide were bound to the SssI enzyme (Fig. 9D). The abilities of these three oligonucleotides to capture SssI enzyme appear to be similar. The same analysis indicated that 1.6% of the FdC-1CG oligonucleotide, 0.3% of the FdC-Pr1 oligonucleotide, 0.17% of the FdC-Pr2 oligonucleotide, and 0.14% of the 1CG-PCR DNA fragment were bound to Dnmt3a in the absence of AdoMet (Fig. 9D). In the presence of AdoMet, 0.5% of the FdC-1CG oligonucleotide, 0.15% of the FdC-Pr1 oligonucleotide, and no detectable FdC-Pr2 oligonucleotide were bound to the GST-3a (Fig. 9D). The FdC-1CG oligonucleotide captured Dnmt3a 5 times more often than the FdC-Pr1 oligo and almost 10 times more often than the FdC-Pr2 oligonucleotide in the absence of AdoMet. In the presence of AdoMet, FdC-1CG oligonucleotide captures Dnmt3a three times more often then the FdC-Pr1 oligonucleotide. Based on the results from sodium bisulfite genomic sequencing of the in vitro-methylated plasmid, a G residue may be more biased against than an A residue at the +1 position (Table 2). This may explain the difference observed between the FdC-Pr1 and the FdC-Pr2 oligonucleotides. These findings indicate that sequence surrounding the CpG site plays a role in Dnmt3a methylation. The same site preference was observed regardless of the presence or absence of AdoMet, suggesting that the site preference by Dnmt3a most likely is exerted at the binding step and not at the catalytic step.
If the sequence surrounding the CpG site can affect methylation by Dnmt3a, the same CpG site on the two complementary strands may be methylated differently by Dnmt3a. To test this possibility, an oligonucleotide, BFdC-1CG, having the complementary sequence of FdC-1CG and FdC modification at the only CpG site was used. The double-stranded oligonucleotides generated from the FdC-1CG and the BFdC-1CG oligonucleotides are identical except that the FdC modification is on the other strand of the molecule. If Dnmt3a does not discriminate sequence surrounding the CpG sites, these two substrates should have similar capability in capturing the enzyme in the reaction. It is clear that the BFdC-1CG oligonucleotide does not capture Dnmt3a nearly as effectively as the FdC-1CG oligonucleotide, even though the BFdC-1CG oligonucleotide was labeled threefold better than the FdC-1CG oligonucleotide (Fig. 9E). This finding further strengthens the results described above that Dnmt3a does not methylate all CpG sites equally and that the sequence surrounding the CpG site plays a determinant role in Dnmt3a methylation. This result also indicates that the stronger shift observed with the FdC-1CG oligonucleotide was not due to any covalent linkage of the protein to the complementary strand without FdC modification. Results from these specific FdC-containing oligonucleotides strongly support the findings above that Dnmt3a prefers CpG sites surrounded by pyrimidine-rich sequences.
DISCUSSION
In this study, we showed that Dnmt3a methylates some regions of the episome, pCLH22, more frequently than other regions and methylates some CpG sites more frequently than others in the same region in vivo. Dnmt3a does not preferentially methylate regions with higher CpG density. In order to assess whether Dnmt3a methylates CpG sites randomly, we purified the GST fusion protein of Dnmt3a, GST-3a, expressed in human cells for in vitro analysis. We showed that GST-3a has methyltransferase activity in vitro by two different assays. Analyses of in vitro-methylated DNA using the bisulfite genomic sequencing method revealed that Dnmt3a methylates the two cDNA strands differently. This asymmetrical pattern strongly indicates that Dnmt3a does not methylate hemimethylated sites immediately after the first strand methylation, as Dnmt1 does. Analysis of the sites methylated at high frequency by Dnmt3a in vitro revealed that CpG sites with pyrimidines at the −2 position upstream and the +1 position downstream (PyNCGPy) are preferred by this enzyme. Selection experiments using FdC-containing random oligonucleotides showed the same trend for Dnmt3a site preference even though there are many limitations of this experiment. Using specific FdC-containing oligonucleotides, a clear difference in pyrimidine-rich and purine-rich sequences was evident. The site choice by Dnmt3a in vitro is most likely to occur at the binding step rather than the catalytic step of the reaction. Unlike Dnmt1 or SssI enzymes, AdoMet is not required for FdC oligonucleotides to capture Dnmt3a in vitro. No cytosine methylation at non-CpG sites was detected in any of the clones sequenced. The sodium bisulfite sequencing data also indicate that Dnmt3a is not processive, at least under the conditions used in vitro. This is the first report of a mammalian methyltransferase that methylates two complementary strands differently due to site preference.
It is believed that after Dnmt1 de novo methylates an unmethylated CpG site, it immediately methylates the complementary strand due to its high affinity for hemimethylated CpGs. Unlike Dnmt1 and the prokaryotic methylase SssI, Dnmt3a generates very distinct methylation patterns on two cDNA strands of the plasmid in vitro. This strongly suggests that Dnmt3a does not methylate hemimethylated sites efficiently. This is consistent with previous observations (9, 27) and our own observation (data not shown) that Dnmt3a does not have a higher affinity for hemimethylated DNA. Two possible explanations could account for this strand bias. Dnmt3a could have a very low affinity for hemimethylated DNA; therefore, it will not methylate a hemimethylated CpG site. However, this should result in a completely random pattern of methylation due to random site choice. Therefore, we favor the second possible cause: site preference of Dnmt3a. We further ruled out that supercoiling or PCR bias can be the cause of this strand specificity.
A study using FdC substrates also showed that Dnmt1 has a bias against, but not absolute elimination of, G at the −1 position as a de novo methyltransferase (37). However, it has been suggested that structural features may be more important than sequence context for site choice by the de novo methylation activity of Dnmt1 (4, 5). Dnmt3a appears to be different than Dnmt1 in several aspects. FdC-containing oligonucleotides cannot capture Dnmt1 in the absence of AdoMet (31). It has been suggested that while Dnmt1 can bind to DNA first, it typically binds AdoMet before it binds DNA (2). In contrast, FdC-containing oligonucleotides can capture Dnmt3a in the absence of AdoMet, indicating that the DNA substrate can bind to Dnmt3a first and that AdoMet is not required for the capturing of the enzyme. This is consistent with the finding that Dnmt3a was captured less efficiently by oligonucleotides with purine-rich sequence surrounding the CpG site, regardless of whether AdoMet was present. If the site choice by Dnmt3a were to take place at the catalytic step, then all the FdC-containing oligonucleotides should capture the enzyme with similar efficiency in the absence of AdoMet. Therefore, this finding would also indicate that the site choice by Dnmt3a most likely occurs at the binding step rather than the catalytic step.
Dnmt3a appears to prefer pyrimidine (bias against purine) at multiple positions surrounding the CpG site. Although the most-distinctive preference for pyrimidine is at positions −2 and +1 and the top and bottom strand sequences can both be preferred by Dnmt3a, there is an overall preference for pyrimidine-rich sequence in sites surrounding the CpG. The complementary strand of pyrimidine-rich sequences is typically purine rich, and the CpG site surrounded by purine-rich sequences is clearly not a preferred substrate for Dnmt3a. It is important to note that the site preference of Dnmt3a is not all-or-none; rather, the less favorable CpG sites are simply methylated at a lower frequency. This preference would result in more frequent methylation of only one DNA strand at most of the CpG sites, with some CpG sites being methylated well on both strands and some CpG sites not being methylated well on either strand. This is consistent with the finding that Dnmt3a can methylate hemimethylated DNA as well as unmethylated DNA (27; L. Han and C.-L. Hsieh, unpublished results) in vitro, and can generate strand-specific methylation patterns in vitro.
Many limitations exist in the selection experiment using random oligonucleotides containing FdC. First, the effect of the FdC moiety on site recognition by the enzyme is uncertain. Second, this selection experiment can only be carried out for a single round, whereas selection experiments determining noncovalent protein binding preferences are generally carried out for at least five rounds. The small fraction of the oligonucleotides that captured Dnmt3a can be easily contaminated with unbound oligonucleotides within the same lane on the gel. Despite these severe limitations, the single-round selection results were still suggestive, albeit weakly. The site preference derived from the bisulfite genomic sequencing data is probably a more accurate reflection of the preference of the enzyme than that observed in the selection experiments. Despite the limitations of the selection experiments, the general conclusion from these experiments supports the bisulfite genomic sequencing result. Experiments using specific FdC-containing oligonucleotides showed that oligonucleotides with pyrimidine-rich sequence surrounding the CpG site captured Dnmt3a more efficiently than oligonucleotides with purine-rich sequence surrounding the CpG site. Strengthened by the results from experiments using specific oligonucleotides containing FdC, it is reasonable to conclude that Dnmt3a prefers pyrimidine at the −2 position upstream and at the +1 position downstream of the CpG site.
Although it is clear that Dnmt3a methylates the two cDNA strands differently in vitro, correlation between the in vitro and in vivo findings remains difficult. Methylation patterns on the two complementary strands are mostly identical in vivo due to methylation by other methyltransferases and/or demethylation due to the action of the hemidemethylase. For some CpG sites, the in vivo finding reflects the in vitro results. In vivo, the least-methylated CpG site was site 9 in EBNA1 region 2. In vitro, site 9 was also the least-methylated CpG site in EBNA1 region 2 when the methylation sites on the top and the bottom strands were combined. For some other sites, the in vivo and in vitro methylation patterns were very different. Methylation was only found at two CpG sites in EBNA1 region 6 in vivo, while methylation was detected at every CpG site when both the top and bottom strands were taken into account. This difference suggests that most of the CpG sites in EBNA1 region 6 are not accessible to Dnmt3a in vivo. The in vivo targets for Dnmt3a can only be absolutely determined in cells lacking all other methyltransferases if such cells can be generated. Although one can argue that the in vitro analysis does not reflect the biological role of Dnmt3a, it provides a clear view of the targets of the enzyme without the influence of other factors. When in vivo analysis becomes possible in the future, the in vitro and in vivo methylation differences can help us dissect the role of other factors in determining DNA methylation patterns.
It has been proposed that a cis-acting element is needed to signal de novo methylation in vivo and that methylation can spread from a methylation center to the surrounding region (for a review, see reference 32). It is possible that de novo methylation targeting and spreading may require interaction between different DNA methyltransferases. The most preferred and accessible CpG sites become methylated more frequently while the less favorable sites become methylated less often or not at all. Over time, more and more CpG sites can become methylated, giving the overall impression of methylation spreading from a central site; that site may be the most-favored and -accessible site. The expression level of the methyltransferases may also play a role in the number of sites that become methylated, as well as how fast the spreading appears. It is clear that Dnmt3a methylates one DNA strand and leaves these methylated sites hemimethylated without methylating the same CpG site on the complementary strand immediately in vitro. This is most likely due to the site preference of the enzyme. However, the consensus of preferred CpG sites and the interplay between Dnmt3a and other methyltransferases in establishing and changing the DNA methylation pattern in the cells remain to be explored.
Acknowledgments
We thank S. Tavare for guidance on statistical analysis. We thank T. Bestor for suggesting the selection experiment using FdC-modified oligonucleotides and for providing information on technical aspects of the experiment. We also thank F. Chedin, R. Irvine, K. Yu, Y. Ma, and M. R. Lieber, for helpful discussion and critical reading of the manuscript.
This work was supported by NIH grants GM60237 and GM54781.
REFERENCES
- 1.Amir, R. E., I. B. Van den Veyver, M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185–188. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla, A., S. Pradhan, J. E. Larson, R. J. Roberts, and R. D. Wells. 2001. Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J. Biol. Chem. 276:18605–18613. [DOI] [PubMed] [Google Scholar]
- 3.Bestor, T. H. 1987. Supercoiling-dependent sequence specificity of mammalian DNA methyltransferase. Nucleic Acids Res. 15:3835–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor, T. H., A. P. Laudano, R. Mattaliano, and V. M. Ingram. 1988. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203:971–983. [DOI] [PubMed] [Google Scholar]
- 5.Bestor, T. H., and B. Tycko. 1996. Creation of genomic methylation patterns. Nat. Genet. 12:363–367. [DOI] [PubMed] [Google Scholar]
- 6.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression—belts, braces, and chromatin. Cell 99:451–454. [DOI] [PubMed] [Google Scholar]
- 7.Clark, S., J. Harrison, C. Paulm, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DuBridge, R. B., P. Tang, H. C. Hsia, P.-M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle vector. Mol. Cell. Biol. 7:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowher, H., and A. Jeltsch. 2001. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites. J. Mol. Biol. 309:1201–1208. [DOI] [PubMed] [Google Scholar]
- 10.Han, L., I. G. Lin, and C.-L. Hsieh. 2001. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol. Cell. Biol. 21:3416–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen, R. S., C. Wijmenge, P. Luo, A. M. Stanek, T. K. Canfield, C. M. R. Weemaes, and S. M. Gartler. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96:14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertz, J. M., G. Schell, and W. Doerfler. 1999. Factors affecting de novo methylation of foreign DNA in mouse embryonic stem cells. J. Biol. Chem. 274:24232–24240. [DOI] [PubMed] [Google Scholar]
- 13.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cultures. J. Mol. Biol. 26:365–369. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh, C.-L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh, C.-L. 1999. Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol. 19:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh, C.-L. 1999. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19:8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeanpierre, M., C. Turleau, A. Aurias, M. Prieur, F. Ledeist, A. Fischer, and E. Viegas Pequignot. 1993. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 2:731–735. [DOI] [PubMed] [Google Scholar]
- 18.Jones, P. A., and P. W. Larid. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163–167. [DOI] [PubMed] [Google Scholar]
- 19.Kondo, T., M. P. Bobek, R. Kuick, B. Lamb, X. Zhu, A. Narayan, D. Bourc’his, E. Viegas-Pequignot, M. Ehrlich, and S. M. Hanash. 2000. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 9:597–604. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., X. Cheng, J. W. Pflugrath, and R. J. Roberts. 1992. Purification, crystallization, and preliminary X-ray diffraction analysis of an M.HhaI-AdoMet complex. Biochemistry 31:8648–8653. [DOI] [PubMed] [Google Scholar]
- 21.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195–3205. [DOI] [PubMed] [Google Scholar]
- 22.Leonhardt, H., A. W. Page, H.-U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–873. [DOI] [PubMed] [Google Scholar]
- 23.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915–926. [DOI] [PubMed] [Google Scholar]
- 24.Lin, I. G., T. J. Tomzynski, Q. Ou, and C.-L. Hsieh. 2000. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol. 20:2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyko, F., B. H. Ramsahoye, H. Kashevsky, M. Tudor, M.-A. Mastrangelo, T. L. Orr-Weaver, and R. Jaenish. 1999. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 23:363–366. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219–220. [DOI] [PubMed] [Google Scholar]
- 28.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methylationtransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257. [DOI] [PubMed] [Google Scholar]
- 29.Pradhan, S., D. Talbot, M. Sha, J. Benner, L. Hornstra, E. Li, R. Jaenisch, and R. J. Roberts. 1997. Baculovirus-mediated expression and characterization of the full-length murine DNA methyltransferase. Nucleic Acids Res. 25:4666–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pradhan, S., A. Bacolla, R. D. Wells, and R. J. Roberts. 1999. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274:33002–33010. [DOI] [PubMed] [Google Scholar]
- 31.Pradhan, S., and R. J. Roberts. 2000. Hybrid mouse-prokaryotic DNA (cytosine-5) methyltransferases retain the specificity of the parental C-terminal domain. EMBO J. 19:2103–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turker, M. S. 1999. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin. Cancer Biol. 9:329–337. [DOI] [PubMed] [Google Scholar]
- 33.Weis-Garcia, F., E. Besmer, D. J. Sawchuk, W. Yu, Y. Hu, S. Cassard, M. C. Nussenzweig, and P. Cortes. 1997. V(D)J recombination: in vitro coding joint formation. Mol. Cell. Biol. 17:6379–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wigler, M., R. Sweet, G. K. Sim, B. Wold, A. Pellicer, E. Lacy, T. Maniatis, S. Silverstein, and R. Axel. 1979. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell 16:777–785. [DOI] [PubMed] [Google Scholar]
- 35.Xu, G.-L., T. H. Bestor, D. Bourc’his, C.-L. Hsieh, N. Tommerup, M. Bugge, M. Hulten, X. Ou, J. J. Russo, and E. Viegas-Pequignot. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187–191. [DOI] [PubMed] [Google Scholar]
- 36.Yates, P. A., R. W. Burman, P. Mummaneni, S. Krussel, and M. S. Turker. 1999. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 274:36357–36361. [DOI] [PubMed] [Google Scholar]
- 37.Yoder, J. A., N. S. Soman, G. L. Verdine, and T. H. Bestor. 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270:385–395. [DOI] [PubMed] [Google Scholar]