Abstract
The retinoblastoma protein, pRb, controls transcription through recruitment of histone deacetylase to particular E2F-responsive genes. We determined the acetylation level of individual nucleosomes present in the cyclin E promoter of RB+/+ and RB−/− mouse embryo fibroblasts. We also determined the effects of pRb on nucleosomal conformation by examining the thiol reactivity of histone H3 of individual nucleosomes. We found that pRb represses the cyclin E promoter through histone deacetylation of a single nucleosome, to which it and histone deacetylase 1 bind. In addition, the conformation of this nucleosome is modulated by pRb-directed histone deacetylase activity. Thus, the repressive role of pRb in cyclin E transcription and therefore cell cycle progression can be mapped to its control of the acetylation status and conformation of a single nucleosome.
The retinoblastoma gene is the prototypical tumor suppressor gene (47). As such, its protein product (pRb) is inactivated in a wide variety of human cancers. Indeed, the pathway containing pRb and proteins that control its activity is deranged in virtually every tumor type. This pathway, which includes cyclins D and E as well as low-molecular-weight inhibitors of cyclin/cyclin-dependent kinase (CDK) activity (e.g., p21 and p16), is critical for progression through the G1 phase of the cell cycle (42, 47). Recently, it has been shown that the repressive (hypophosphorylated) form of pRb associates with members of the histone deacetylase (HDAC) protein family, which may assist pRb in transcriptionally repressing target genes (8, 9, 29, 30).
pRb associates with various transcriptional factors, notably, members of the E2F family, and represses their activity (17, 31). While many genes are regulated by members of the E2F family (31), most likely only a subset of these are regulated by pRb (21, 23, 45). One gene that is critical for cell cycle progression and that is regulated by pRb and E2F is the cyclin E gene (21, 23). Indeed, the cyclin E promoter has been shown to be derepressed in RB−/− mouse embryo fibroblasts (MEFs) during G0 and early G1 (21, 23) and to be under the control of E2F (5, 16, 35). As mentioned above, it has been found that pRb controls transcriptional activity and chromatin structure through recruitment of HDAC activity to particular E2F-responsive genes (9, 29, 30). Notably, it has been shown that the cyclin E promoter is regulated by histone acetylation through its E2F- and pRb-mediated recruitment of HDAC1 (9, 41). The association of pRb with HDAC is disrupted by binding of viral oncoproteins (e.g., simian virus 40 large T antigen) to pRb, by hyperphosphorylation of pRb during cell cycle progression, and obviously by inactivation of the RB gene (9, 29, 30, 51). This disruption leads to derepression of target genes and inappropriate expression, as observed for cyclin E (21, 23).
We sought to investigate the method of pRb repression of the endogenous mouse cyclin E promoter. It was previously shown that embryo fibroblasts derived from mice with a targeted deletion of the RB gene have derepressed cyclin E transcription in G0 and early G1 and display a peak of transcription 4 h earlier in G1 than cells from wild-type littermates (21, 23). Correspondingly, RB−/− cells have a G1 that is shorter by about 4 h (21). It should be noted that RB−/− fibroblasts enter and exit G0 after serum starvation and readdition as readily as RB+/+ cells (12, 13, 18, 19, 21, 23). These MEFs are thus ideally suited for investigations into the role of pRb and associated HDAC activity in the regulation of the cyclin E promoter. We therefore delineated the nucleosomal profile of the cyclin E promoter in these cells and determined the effects of pRb and transcriptional activity on the acetylation status and conformation of individual nucleosomes in the promoter.
Our observations described here suggest that pRb represses the cyclin E promoter through modulation of the level of histone acetylation of a single nucleosome at the transcriptional start site. In addition, pRb and HDAC1 are bound to this nucleosome during periods of transcriptional repression of the gene. Thus, the repressive signal transduction pathway that contains pRb as its most pathologically significant player is now mapped to deacetylation of a single nucleosome. These studies provide basic insight into pRb function and provide a paradigm for the actions of other transcriptional corepressors.
MATERIALS AND METHODS
Cell cultures.
RB−/− and RB+/+ immortalized MEFs were maintained in Dulbecco minimal essential medium supplemented with 10% calf serum. Cells were arrested in G0 by incubation in either medium containing 0.1% serum or serum-free medium containing minimal essential components for 3 days. Cells were induced to late G1 by replacing serum starvation medium with serum-supplemented medium. Due to cell cycle differences between the RB−/− and RB+/+ cells (21), late G1 was found to occur at 16 h after serum addition in RB+/+ cells and at 12 h in RB−/− cells. HDAC inhibition experiments were done with cells arrested in G0 with or without treatment with 100 nM trichostatin A (TSA) for 12 to 16 h. Reverse transcription (RT)-PCR experiments were done with 100 nM TSA for various times.
RT-PCR.
The primers used for RT-PCR amplified the region from +377 to +821 of the murine cyclin E mRNA. rTth RNA PCR (Gene Amp; Perkin-Elmer) was used to amplify 1 μg of total RNA isolated from RB+/+ and RB−/− MEFs. An exhaustive range of PCR cycle numbers was tested in order to ensure that the assay was in the linear range of amplification. The final cycle number (25) was 1 or 2 cycles above that which gave no signal. Amplification from RB+/+ cells was in the linear range, since there was a change after TSA treatment. No differences were observed in the signal from the RB−/− cells RNA after TSA treatment regardless of how many cycles were used.
PCR primer design.
Primer pairs were designed to amplify areas of 80 to 100 bp on the cyclin E promoter. Each PCR product overlaps the neighboring amplified regions by approximately 20 bp. The PCR products were numbered, and the regions amplified were as follows: 13, +101 to +24; 12, +46 to −47; 11, −28 to −108; 10, −119 to −213; 9, −194 to −283; 8, −265 to −365; 7, −347 to −448; 6, −432 to −526; 5, −507 to −592; 4, −583 to −685; 3, −682 to −771; 2, −753 to −835; and 1, −814 to −902.
PCR nucleosome mapping.
Nuclei were isolated from RB−/− and RB+/+ cells in G0 and late G1. Chromatin was digested to mononucleosomal form with micrococcal nuclease. The digestion was stopped by adding EDTA to a final concentration of 50 mM. Nuclei were lysed in 1% sodium dodecyl sulfate (SDS) lysis buffer. Lysates were treated with 0.1 mg of proteinase K/ml overnight at 37°C. The DNA was purified by phenol extraction followed by ammonium acetate-isopropanol precipitation. DNA pellets were resuspended in Tris-EDTA buffer and subsequently analyzed by agarose gel electrophoresis. The mononucleosomal DNA fraction was gel extracted using Spin-X centrifuge tube filters (Corning), and 100 ng was used as a template in each PCR. Undigested genomic DNA was used as the positive control. No template was added to the PCR negative control. In order to deal with the different amplification efficiencies of the primer sets, each was extensively and independently characterized to determine appropriate cycle numbers. Primer sets from nucleosome-free regions were designated as such because they never gave signals above the background regardless of the cycle number.
ChIP and thiol pulldown assays.
Chromatin immunoprecipitation (ChIP) and thiol pulldown experiments were performed by using a modified version of previously described methods (15, 36). Cellular protein was cross-linked to DNA by adding formaldehyde directly to the cell culture media to a final concentration of 0.75%. Nuclei were isolated, and chromatin was digested to mononucleosomal form with micrococcal nuclease. The digestion was stopped by adding EDTA to a final concentration of 50 mM. Nuclei were pelleted by brief centrifugation and resuspended in 1% SDS lysis buffer. In order to ensure complete lysis, samples were sonicated for 10 s. In some experiments (see Fig. 4), cells were harvested and sonicated for 30 s in order to produce larger fragments of undigested cross-linked DNA. A small portion of the lysate was removed from each sample, and the extracted DNA was analyzed by agarose gel electrophoresis to ensure complete digestion. The lysate was also analyzed by spectrophotometry to obtain the DNA concentration.
FIG. 4.
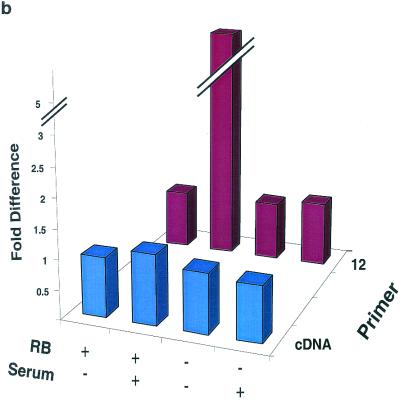
ChIP analysis of histone H3 and histone H4 acetylation on the mouse cyclin E promoter and gene in RB+/+ and RB−/− MEFs. Cells were arrested in G0 (−serum) or induced to late G1 (+serum) prior to harvest. (a) PCR results labeled +1 were obtained by using cell lysates digested with micrococcal nuclease to produce DNA fragments approximately 150 bp long, and immunoprecipitated DNA was amplified by using primer set 12 (+46 to −47) of the cyclin E promoter. PCR results labeled Transcribed region were obtained by using cell lysates sonicated to produce DNA fragments of approximately 500 to 1,500 bp, and the immunoprecipitated DNA was amplified by using a primer set in the coding region of the cyclin E gene (+223 to +316). Cntl, control; IP, immunoprecipitation. (b) Graphic representation of the results shown in panel a. See the legend to Fig. 3 for details.
Cellular lysate containing the same amount of DNA per sample was diluted 1:10 in 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl and added to antibodies to acetylated histone H3 or H4 (Upstate Biotechnology), pRb (PharMingen; clone G3-245), or HDAC1 (Affinity Bioreagents; catalog no. PA1-860) and protein A/G-agarose beads or activated thiol-Sepharose 4B beads (Amersham Pharmacia). The acetylated histone antibodies are specific for tetra acetylated histone H4 (28) and diacetylated histone H3 (4, 7). The immunoprecipitation negative control samples contained lysate and protein A or G beads in the absence of antibody. It is possible that any material that has not been completely cross-linked by the formaldehyde treatment and that is disrupted by the 1% SDS lysis will randomly reform chromatin complexes during overnight incubation. However, these complexes will contain all forms of acetylated histones distributed randomly throughout all nucleosomes reformed. This being the case, any signal obtained from these reassembled nucleosomes will not confound interpretation of our results but will at most temper them. However, we are confident that the vast majority of chromatin is cross-linked, since cycle numbers similar to those used in the non-cross-linking mapping experiments were used in these ChIP and thiol pulldown experiments.
Following overnight incubation, the beads were washed extensively, and the cross-linked histone-DNA complexes were eluted. Proteinase K was added, and the samples were heated at 65°C to reverse the cross-link. The DNA was purified by phenol extraction followed by sodium acetate-ethanol precipitation. The purified DNA was used as a template in PCRs with the previously described primers. Each PCR was repeated exhaustively using various cycle numbers to ensure that the results were within the linear range of amplification. PCR positive and negative controls consisted of plasmid containing the cyclin E promoter (39) and no template, respectively. Samples were run on polyacrylamide gels and stained with ethidium bromide.
RESULTS AND DISCUSSION
Cyclin E transcription is regulated by HDAC activity and pRb.
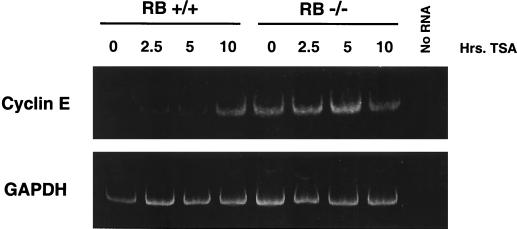
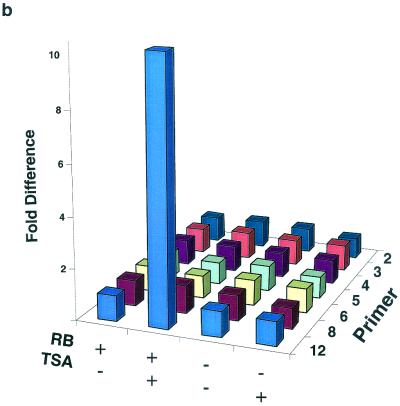
As mentioned above, the cyclin E promoter is a candidate for investigating the role of pRb in altering histone acetylation in a specific chromosomal location. We first determined if histone deacetylation could control endogenous cyclin E promoter activity. Figure 1 shows an analysis of RNA isolated from G0-arrested RB+/+ and RB−/− MEFs after TSA treatment. Cyclin E mRNA levels are increased in RB+/+ cells upon the addition of TSA, suggesting that promoter repression in G0 is achieved through histone deacetylation. In contrast, TSA treatment does not affect cyclin E mRNA levels in RB−/− cells. This result suggests that repression of the cyclin E promoter in G0 and early G1 is directed by pRb-dependent histone deacetylation.
FIG. 1.
Effects of the deacetylase inhibitor TSA on cyclin E mRNA levels in RB+/+ and RB−/− MEFs. TSA was added to G0-arrested cells for the indicated times before lysis and RNA purification. RT-PCR was performed in order to visualize the small amounts of cyclin E mRNA present in arrested RB+/+ cells. The primers used were from either cyclin E or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA sequences.
Mapping of nucleosomal regions of the cyclin E promoter.
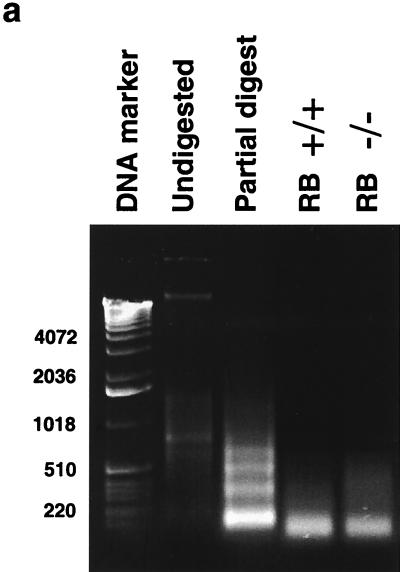
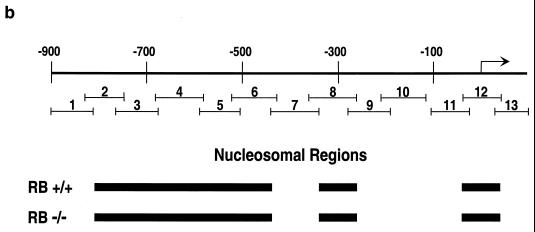
We next determined the nucleosomal regions of ∼1 kb of the cyclin E promoter. This was accomplished by using a novel PCR-based method. Briefly, nuclei isolated from RB+/+ and RB−/− MEFs were digested with micrococcal nuclease. Shorter digestion times led to nucleosomal laddering, while longer treatment times resulted in the production of only mononucleosome-size DNA fragments (Fig. 2a). A mononucleosome-size fragment was excised from the gel and used as a template for PCR amplification of cyclin E promoter regions. DNA that is not assembled into nucleosomes is digested to completion with the nuclease and is not amplified (A. J. Morrison and R. E. Herrera, unpublished observation). This method was validated through investigation of the c-fos promoter, for which the position of nucleosomes was previously determined by using standard indirect end-labeling techniques (20). In the c-fos promoter, primers from the region of the serum response element are not amplified (Morrison and Herrera, unpublished), consistent with the observation that this region is nucleosome free (20, 22). However, primers from the region centered at −170, which has been determined to be assembled into a nucleosome (20, 22), are amplified (Morrison and Herrera, unpublished). In addition, this technique was recently used by others to map nucleosomal regions (6). We next proceeded to map the nucleosomal regions of the cyclin E promoter using this technique. Thirteen PCR primer pairs from the cyclin E promoter were chosen to amplify overlapping regions of ∼100 bp each to ensure representation in the mononucleosomal band if the region was assembled into nucleosomes (Fig. 2b).
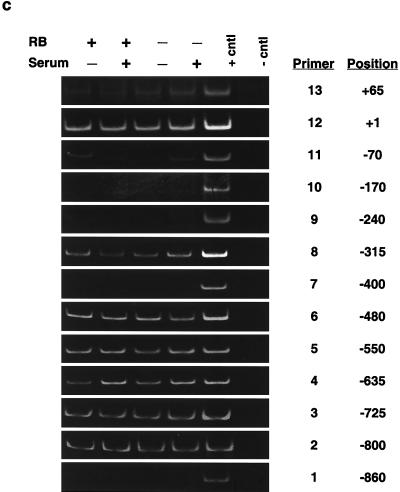
FIG. 2.
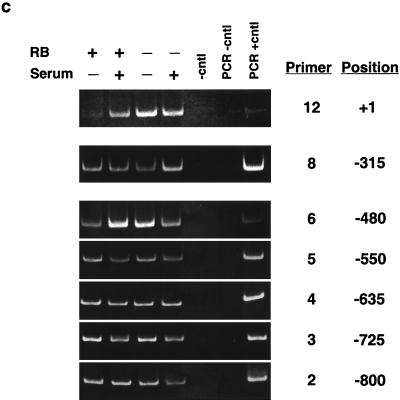
Determination of nucleosome positioning on the mouse cyclin E promoter in RB+/+ and RB−/− MEFs. (a) Micrococcal nuclease digestion of RB+/+ and RB−/− MEF nuclei to produce mononucleosomal DNA. Samples were from formaldehyde-cross-linked chromatin that either was not digested with micrococcal nuclease (undigested) or was subjected to micrococcal nuclease digestion to produce partially digested chromatin (partial digest) or predominantly mononucleosome-size chromatin (RB+/+ and RB−/−). Marker sizes are in base pairs. (b) Illustration of the cyclin E promoter. The amplified region of each PCR product is shown with its corresponding number (see the text). The resulting nucleosomal regions are diagrammed as solid black bars. (c) Results of PCR analyses, with the region of the cyclin E promoter and the number corresponding to the amplified PCR product listed. RB+/+ and RB−/− MEFs were arrested in G0 (−serum) or induced to late G1 (+serum). cntl, control.
Figure 2b and c show the nucleosomal region analysis and a summary of ∼1 kb of the cyclin E promoter. Nucleosome-free regions are centered at about positions +65, −170, −400, and −860, while the remainder of the regions are nucleosomal. The nucleosomal regions remain constant whether the gene is repressed in G0 or is active in late G1 in both RB+/+ and RB−/− MEFs (Fig. 2c). This technique maps only nucleosomal regions, not the positions of single nucleosomes. That is, while single nucleosomes can be identified if they are separated from neighboring nucleosomes by more than ∼100 bp (such as the nucleosome identified at the transcriptional start site of the cyclin E promoter), adjacent nucleosomes cannot be resolved (such as those between positions −400 and −860). However, the amplified products from primer pairs 11, 12, and 13 are small (80, 93, and 77 bp, respectively) and overlap by about 20 bp each. In total, they cover an area of 209 bp. Therefore, the area is large enough to accommodate only one nucleosome centered near the start site.
It remains a formal possibility that sequences are not detected in the mononucleosome fraction because they are hypersensitive or resistant to micrococcal nuclease for reasons other than the presence or absence of nucleosomes. This possibility could be due to sequence bias in the cleavage preferences of micrococcal nuclease. Because of this possibility, sequences that we detect as nucleosome free might actually be nucleosomal. We believe this possibility to be unlikely, since the ChIP experiments with antibodies against acetylated histones (see below) were done with micrococcal nuclease-digested chromatin from which mononucleosomal DNA had not been gel purified, as detailed in Materials and Methods. Therefore, any micrococcal nuclease-resistant sequences would be present in the immunoprecipitations. However, primers from regions determined to be nucleosome free in these mapping experiments did not produce a signal above background, suggesting the absence of histones and therefore nucleosomes in these regions.
After micrococcal nuclease digestion and DNA purification, equal amounts of DNA were subjected to PCR analysis to determine nucleosomal regions. Equal numbers of cycles were performed on samples from both RB+/+ and RB−/− cells. There is generally no difference in band intensities from the different samples. Therefore, there is no preferential release of nucleosomes from the RB−/− cells, despite the chromatin of these cells being more sensitive to micrococcal nuclease in a global manner and characterized by increased levels of histone H1 phosphorylation (18). The analysis in Fig. 2 enabled us to determine the effects of pRb and transcriptional activity on the acetylation status and conformation of individual nucleosomes positioned on the cyclin E promoter.
pRb controls the acetylation status of a single nucleosome in the cyclin E promoter.
In order to determine the effects of pRb and transcription on the acetylation status of the nucleosomes mapped above, we used a modified version of the ChIP method described previously (14, 15, 36, 38). We digested formaldehyde-cross-linked chromatin with micrococcal nuclease to obtain only mononucleosome-size DNA fragments for use as substrates in the PCR amplification step of the ChIP assay (Fig. 2a). This procedure enables us to use the assay to achieve single-nucleosome resolution and to determine the relative acetylation status of individual nucleosomes positioned on the cyclin E promoter. This modification was recently successfully used in various studies to determine the acetylation status of nucleosomal regions (6, 33). In addition, such analysis has also been reported for chicken glyceraldehyde-3-phosphate dehydrogenase, carbonic anhydrase, β-globin, and ovalbumin genes with non-cross-linked chromatin (32). Figure 3 displays ChIP experiments done with antibodies against acetylated H3 and H4 and with cyclin E promoter regions in G0-arrested, TSA-treated, and serum-stimulated RB+/+ and RB−/− MEFs.
FIG. 3.
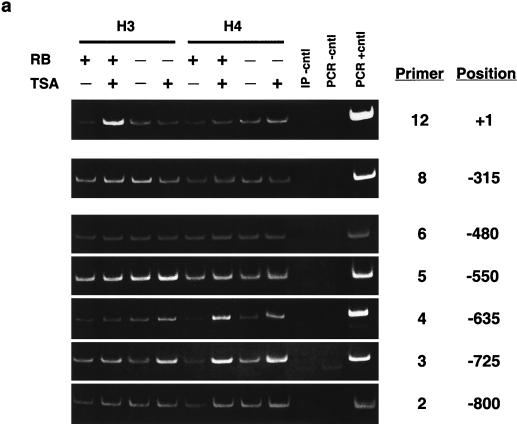
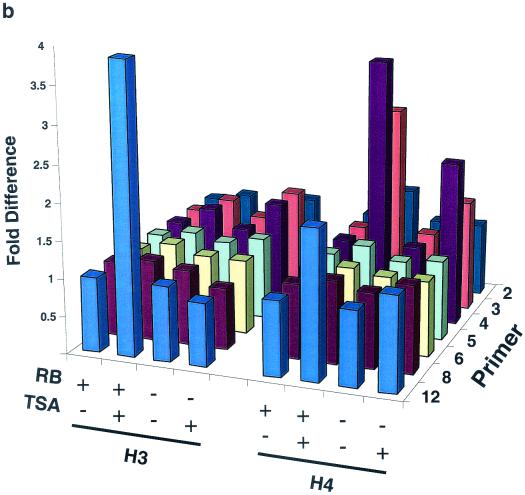
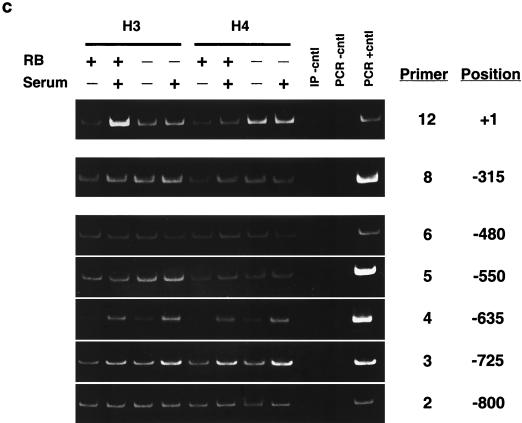
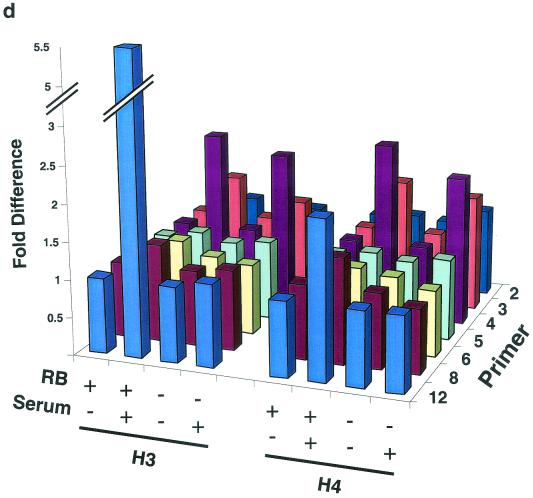
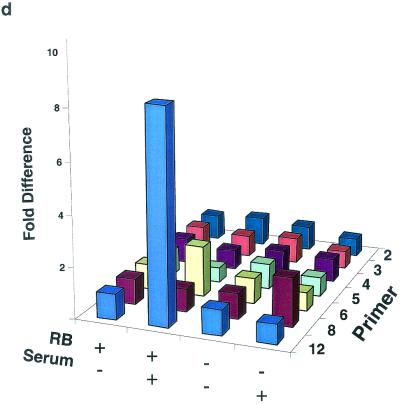
ChIP analysis of histone H3 and histone H4 acetylation on the mouse cyclin E promoter in micrococcal nuclease-digested RB+/+ and RB−/− MEF lysates. (a) Cells were arrested in G0 and treated with either TSA or vehicle alone. The position of the nucleosomal region of the cyclin E promoter is shown, as is the designated primer set number for that region. Cntl, control; IP, immunoprecipitation. (b) Graphic representation of the results shown in panel a. This graph was obtained by performing density scans of PCR results using NIH Image 1.62 software. Data are presented as the fold difference, obtained by setting the value for the untreated (−TSA) samples within each cell line to 1. (c) Cells were arrested in G0 (−serum) or induced to late G1 (+serum). The position of the nucleosomal region of the cyclin E promoter is shown, as is the designated primer set number for that region. (d) Graphic representation of the results shown in panel c.
Figure 3a and b show anti-acetylated histone H3 and H4 ChIP experiments completed after TSA treatment of G0-arrested MEFs as well as their graphic representations. The acetylation status of nucleosomes positioned at −800, −550, −480, and −315 of the cyclin E promoter does not change after TSA treatment, suggesting that they are not targets of HDAC activity. However, histone H4 of nucleosomes positioned at −725 and −635 displays an increase in acetylation in both RB+/+ and RB−/− MEFs after TSA treatment. This result suggests that these two nucleosomes are substrates of pRb-independent HDAC activity. In contrast, the relative acetylation of histones H3 and H4 in the nucleosome positioned at +1 increases after TSA treatment only in RB+/+ cells. Therefore, histones H3 and H4 in this nucleosome alone are targets of pRb-directed HDAC activity. Binding sites for the main repressor complex of the cyclin E promoter as well as E2F are contained within the DNA of this nucleosome (5, 27).
We next examined the relative levels of histone H3 and H4 acetylation in nucleosomes of the cyclin E promoter during maximal transcriptional activity in middle or late G1 (Fig. 3c and d). This phase of the cell cycle occurs 12 and 16 h after the addition of serum to G0-arrested RB−/− and RB+/+ cells, respectively (21). Like those seen after TSA treatment, the acetylation levels of nucleosomes positioned at −800, −550, −480, and −315 of the cyclin E promoter do not change after activation of the gene, while histone H4 of nucleosomes positioned at −725 and −635 displays an increase in acetylation levels in both cell types. In addition, histone H3 acetylation levels in the nucleosome positioned at −635 also increase after transcriptional activation of the gene in RB+/+ as well as RB−/− MEFs. This result suggests that histone H3 of this nucleosome is a target of pRb-independent transcriptionally initiated histone acetylase activity, since it displays no change in acetylation after TSA treatment. As was observed after TSA treatment, the relative histone H3 and H4 acetylation levels in the nucleosome positioned at +1 increase after activation of the gene only in RB+/+ cells. This result further supports the conclusion that histones H3 and H4 in this nucleosome alone are targets of pRb-directed HDAC activity.
The overall levels and cell cycle regulation of histone H3 and H4 acetylation are identical in RB+/+ and RB−/− MEFs (R. E. Herrera, unpublished observation). We point out that our conclusions are derived only from the difference in signal within a given cell line. The only valid interpretations from our ChIP experiments are qualitative and can be drawn only between samples derived from the same cell type and obtained after different treatments, that is, before and after TSA and serum addition. While it is possible that the differences between RB+/+ and RB−/− MEFs are informative, ChIP analyses alone cannot tell us if this is the case.
In order to further confirm the notion that pRb controls the acetylation status of this single nucleosome only, we examined the acetylation levels for nucleosomes in the transcribed region of the gene. It has been shown that nucleosomal positioning in the transcribed regions of genes is disrupted after their activation (20, 50). Because of this finding, we were unable to map individual nucleosomes in the transcribed region of the cyclin E gene after its activation using the nuclease digestion step of the ChIP protocol used for the promoter region. Therefore, we examined the levels of acetylation of nucleosomes in the transcribed region using sonicated chromatin, as is generally used in the ChIP protocol. This procedure gives a more general view of the acetylation levels for nucleosomes in a region of ∼500 to 1,500 bp. Figure 4 shows a ChIP experiment done with primers that amplify the region from +223 to +316 of the cyclin E gene compared to primers that amplify the region of the single nucleosome at +1. The acetylation status of histone H3 in the transcribed region is not altered by transcriptional activity or pRb status. Similar results were obtained regarding the acetylation status of histone H4 (data not shown). Unaltered levels of histone acetylation in actively transcribed regions have been observed in vivo for the beta interferon promoter and in vitro as directed by p300 (26, 38). These data suggest that the acetylation status of histones H3 and H4 in the nucleosome positioned at +1 is modulated directly by pRb-regulated HDAC activity and not simply by the transcriptional activity of the gene.
pRb and HDAC1 localize to the nucleosome positioned at the transcriptional start site of the cyclin E promoter.
It was previously shown that the primary repressor complex of the cyclin E promoter maps to the region containing the nucleosome shown above to be the target of pRb-directed HDAC activity (27). In addition, E2F has also been shown to bind this region (5). Together, these data suggest that pRb and HDAC bind to the same nucleosome that they modify by modulating levels of histone acetylation. It was therefore of interest to determine if pRb and HDAC were indeed bound to the nucleosome positioned at the transcriptional start site of the cyclin E promoter.
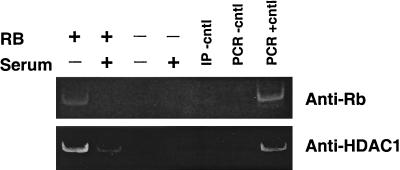
Figure 5 shows the results of a ChIP experiment done to determine if pRb and HDAC1 were bound to this region in RB+/+ but not RB−/− MEFs. Lysates were prepared from cells arrested in G0 or during maximal transcriptional activity of the gene. pRb and HDAC1 associate with the region at the start site in G0-arrested RB+/+ cells but not in cells lacking pRb, as expected. After serum stimulation and during maximal activity of the gene, the levels of both pRb and HDAC1 associated with this region are dramatically reduced. These data suggest that pRb and HDAC1 are associated with the nucleosome that they regulate through histone deacetylation.
FIG. 5.
ChIP analysis of pRb and HDAC1 binding to the mouse cyclin E promoter in RB+/+ and RB−/− MEFs. Cells were arrested in G0 (−serum) or induced to late G1 (+serum) prior to harvest. Lysates were digested with micrococcal nuclease and immunoprecipitated using anti-pRb or anti-HDAC1 antibody. Primers amplified the region of the cyclin E promoter from +46 to −47 (position +1 and primer set 12). Cntl, control; IP, immunoprecipitation.
pRb controls the conformation of a single nucleosome in the cyclin E promoter.
It was previously shown that increased histone acetylation leads to a change in chromatin structure and facilitates transcription (25, 34). In order to correlate single-nucleosome acetylation status with a change in nucleosome structure, we further modified the ChIP assay. Cross-linked mononucleosomal chromatin was incubated with Sepharose beads linked to activated thiol groups as an alternative to the immunoprecipitation step of the assay. Cysteine 110 of histone H3 is accessible to external thiol reactivity only when nucleosomes are in a more open conformation, as observed previously for transcriptionally active regions and for isolated nucleosomes exposed to high ionic strengths (2, 10, 11, 43, 49). The results of the thiol reactivity assay enabled us to determine which nucleosomes positioned in the cyclin E promoter are in an open conformation and to correlate this information with acetylation, pRb, and transcriptional status.
Figure 6 shows thiol pulldown experiments completed after treatment of G0-arrested MEFs with TSA and serum. In RB+/+ cells, the only nucleosome that displays a change in thiol reactivity after TSA treatment or serum induction is the one centered at +1. In contrast, nucleosomes positioned at the cyclin E promoter of RB−/− cells display no changes in thiol reactivity after either treatment. These results suggest that only the nucleosome at position +1 undergoes a structural transition after inhibition of deacetylase activity or transcriptional activation and that this transition is dependent on the presence of pRb. These results correlate with the pRb control of histone acetylation status observed in Fig. 3. However, a relative increase in histone acetylation does not necessarily lead to increased thiol reactivity, since the latter does not change in nucleosomes centered at positions −550, −635, and −725 after TSA or serum treatment.
FIG. 6.
Thiol pulldown analysis of the chromatin structure on the mouse cyclin E promoter in micrococcal nuclease-digested RB+/+ and RB−/− MEF lysates. (a) Cells were arrested in G0 (−serum) and treated with either TSA or vehicle alone. The position of the nucleosomal region of the cyclin E promoter is shown, as is the designated primer set number for that region. Cntl, control. (b) Graphic representation of the results shown in panel a. See the legend to Fig. 3 for details. (c) Cells were arrested in G0 (−serum) or induced to late G1 (+serum). The position of the nucleosomal region of the cyclin E promoter is shown, as is the designated primer set number for that region. (d) Graphic representation of the results shown in panel c.
The experiments shown in Fig. 6 were performed with a formaldehyde cross-linking step. Therefore, any cysteine-containing protein associated with the nucleosome either directly or indirectly may be pulled down, confounding our interpretation of a change in the conformation of the nucleosome. To address this point, we performed the entire thiol pulldown experiment without formaldehyde cross-linking. Extraction and pulldown were performed under stringent conditions (1% Triton X-100, 0.1% SDS, 1% deoxycholate, 0.5 M NaCl). These conditions have been shown to disrupt non-histone protein association with nucleosomes and to retain only those nucleosomes in which a conformational change has occurred, rendering the thiol groups of H3 accessible to SH reagents (46). The increase in the thiol reactivity of the nucleosome at the transcriptional start site after serum treatment is identical to that seen after cross-linking as in the original design (data not shown). Since the pulldown experiment was performed with micrococcal nuclease-treated samples, we are convinced that what we are pulling down is histone H3 that is part of a nucleosome with an open conformation.
Conclusions.
It was recently shown that, of the numerous cell cycle and E2F-regulated genes identified, the cyclin E gene is distinctive in that it is regulated by HDAC activity (41) and pRb (21, 23, 45, 48) and not by other members of the RB family of proteins (23, 45). The cyclin E promoter has been implicated as the primary target of pRb during cell cycle progression (21, 23, 45). The observations described here suggest that pRb represses the cyclin E promoter through modulation of the level of histone acetylation of a single nucleosome to which it and HDAC1 are bound during periods of transcriptional repression. In addition, we have found that the conformation of this nucleosome is also modulated by pRb-directed HDAC activity. Therefore, the RB pathway, which controls cell cycle progression, is now mapped to the modulation of histone acetylation levels and the conformation of a single nucleosome. These studies provide basic insight into pRb function as well as transcriptional regulation in general.
The observation that pRb represses the cyclin E promoter through modulation of the acetylation status of a single nucleosome was not predicted. It has not yet been determined if transcriptional repressors or activators control chromatin structure only very locally or over entire transcribed regions (37, 44). Our results show that, for pRb, the effect is propagated only at the most local level, that of a single nucleosome. This finding is in agreement with studies of the beta interferon promoter that show an increase in histone acetylation only near the transcriptional start site after promoter activation (38) as well as with in vitro studies showing that acetylation is directed by p300 associated with GAL4-VP16 (26). In addition, studies with yeast cells have shown that the Esa1 histone acetylase is recruited to specific genes, where it directs H4 acetylation of nucleosomal regions in those genes (40). Finally, a very recent study showed that Myc binds to the cyclin D promoter and controls the acetylation status of a single nucleosome (6). Together, these studies suggest that the acetylation of particular nucleosomes may be of critical importance in regulating transcriptional activity. Future investigations are needed to determine why these single nucleosomes are targeted for regulation via histone acetylation and what the consequences of that regulation are.
As shown here, the acetylation of nucleosomes in the cyclin E promoter is altered in pRb-dependent and -independent ways in response to the transcriptional status of the gene. However, this acetylation correlates with an open conformation only on the nucleosome that pRb regulates. As previously shown, the presence of acetylated histones in transcribed regions of DNA does not cause thiol reactivity within the core histone (24). Additionally, HDAC inhibitor treatment has little effect on the thiol reactivity of total nucleosomes isolated from nuclear extracts (3). These data, combined with the observations reported here, support the idea that transcriptional activity, in particular, transcriptional initiation, may occur by modification of a nucleosome at the start site. It was also previously shown that transcription factor binding is enhanced on templates that contain acetylated nucleosomes (1, 34). It is possible that the very localized nucleosomal acetylation and conformational change that occur on the cyclin E promoter occur to facilitate transcriptional initiation, perhaps by allowing RNA polymerase II entry and/or initiation on the chromatin template.
It was recently shown that pRb also controls the methylation status of the same nucleosome in the cyclin E promoter as that shown here to be the target of pRb-directed deacetylase activity (33). This action is accomplished by SUV39H1 binding to pRb and leads to the subsequent binding of HP1 to H3 of this nucleosome. The interplay between pRb-associated HDAC and H3 methylase activities is not understood at this point, but investigations into their regulation of the cyclin E promoter should yield a more complete understanding of how histone modification controls transcriptional activity.
Acknowledgments
We thank G. Chamness, J. Rosen, D. R. Garza, and A. Contreras for helpful comments on the manuscript.
This work was supported by the American Cancer Society and the V Foundation.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667–678. [DOI] [PubMed] [Google Scholar]
- 2.Allegra, P., R. Sterner, D. F. Clayton, and V. G. Allfrey. 1987. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J. Mol. Biol. 196:379–388. [DOI] [PubMed] [Google Scholar]
- 3.Boffa, L. C., J. Walker, T. A. Chen, R. Sterner, M. R. Mariani, and V. G. Allfrey. 1990. Factors affecting nucleosome structure in transcriptionally active chromatin. Histone acetylation, nascent RNA and inhibitors of RNA synthesis. Eur. J. Biochem. 194:811–823. [DOI] [PubMed] [Google Scholar]
- 4.Boggs, B. A., B. Connors, R. E. Sobel, A. C. Chinault, and C. D. Allis. 1996. Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma 105:303–309. [DOI] [PubMed] [Google Scholar]
- 5.Botz, J., K. Zerfass-Thome, D. Spitkovsky, H. Delius, B. Vogt, M. Eilers, A. Hatzigeorgiou, and P. Jansen-Durr. 1996. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol. Cell. Biol. 16:3401–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, C., O. Dittrich, A. Kiermaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein, M., R. E. Sobel, C. D. Allis, B. M. Turner, and J. R. Broach. 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16:4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24:142–145. [DOI] [PubMed] [Google Scholar]
- 9.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597–601. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T. A., and V. G. Allfrey. 1987. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc. Natl. Acad. Sci. USA 84:5252–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen-Cleland, T. A., L. C. Boffa, E. M. Carpaneto, M. R. Mariani, E. Valentin, E. Mendez, and V. G. Allfrey. 1993. Recovery of transcriptionally active chromatin restriction fragments by binding to organomercurial-agarose magnetic beads. A rapid and sensitive method for monitoring changes in higher order chromatin structure during gene activation and repression. J. Biol. Chem. 268:23409–23416. [PubMed] [Google Scholar]
- 12.Classon, M., B. K. Kennedy, R. Mulloy, and E. Harlow. 2000. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 97:10826–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Classon, M., S. Salama, C. Gorka, R. Mulloy, P. Braun, and E. Harlow. 2000. Combinatorial roles for pRB, p107, and p130 in E2F-mediated cell cycle control. Proc. Natl. Acad. Sci. USA 97:10820–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davie, J. R., and V. A. Spencer. 1999. Control of histone modifications. J. Cell. Biochem. 75(Suppl. 32–33):141–148. [DOI] [PubMed] [Google Scholar]
- 15.Dedon, P. C., J. A. Soults, C. D. Allis, and M. A. Gorovsky. 1991. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal. Biochem. 197:83–90. [DOI] [PubMed] [Google Scholar]
- 16.Geng, Y., E. N. Eaton, M. Picon, J. M. Roberts, A. S. Lundberg, A. Gifford, C. Sardet, and R. A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12:1173–1180. [PubMed] [Google Scholar]
- 17.Harbour, J. W., and D. C. Dean. 2000. Rb function in cell-cycle regulation and apoptosis. Nat. Cell Biol. 2:E65–E7. [DOI] [PubMed] [Google Scholar]
- 18.Herrera, R. E., F. Chen, and R. A. Weinberg. 1996. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc. Natl. Acad. Sci. USA 93:11510–11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrera, R. E., T. P. Makela, and R. A. Weinberg. 1996. TGF beta-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol. Biol. Cell 7:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera, R. E., A. Nordheim, and A. F. Stewart. 1997. Chromatin structure analysis of the human c-fos promoter reveals a centrally positioned nucleosome. Chromosoma 106:284–292. [DOI] [PubMed] [Google Scholar]
- 21.Herrera, R. E., V. P. Sah, B. O. Williams, T. P. Makela, R. A. Weinberg, and T. Jacks. 1996. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 16:2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera, R. E., P. E. Shaw, and A. Nordheim. 1989. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature 340:68–70. [DOI] [PubMed] [Google Scholar]
- 23.Hurford, R. K., Jr., D. Cobrinik, M. H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447–1463. [DOI] [PubMed] [Google Scholar]
- 24.Imai, B. S., P. Yau, J. P. Baldwin, K. Ibel, R. P. May, and E. M. Bradbury. 1986. Hyperacetylation of core histones does not cause unfolding of nucleosomes. Neutron scatter data accords with disc shape of the nucleosome. J. Biol. Chem. 261:8784–8792. [PubMed] [Google Scholar]
- 25.Krajewski, W. A., and P. B. Becker. 1998. Reconstitution of hyperacetylated, DNase I-sensitive chromatin characterized by high conformational flexibility of nucleosomal DNA. Proc. Natl. Acad. Sci. USA 95:1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551–561. [DOI] [PubMed] [Google Scholar]
- 27.Le Cam, L., J. Polanowska, E. Fabbrizio, M. Olivier, A. Philips, E. Ng Eaton, M. Classon, Y. Geng, and C. Sardet. 1999. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 18:1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, R., J. W. Leone, R. G. Cook, and C. D. Allis. 1989. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J. Cell Biol. 108:1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473. [DOI] [PubMed] [Google Scholar]
- 30.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601–605. [DOI] [PubMed] [Google Scholar]
- 31.Muller, H., and K. Helin. 2000. The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta 1470:M1–M12. [DOI] [PubMed] [Google Scholar]
- 32.Myers, F. A., D. R. Evans, A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 2001. Targeted and extended acetylation of histones H4 and H3 at active and inactive genes in chicken embryo erythrocytes. J. Biol. Chem. 276:20197–20205. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O’Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561–565. [DOI] [PubMed] [Google Scholar]
- 34.Nightingale, K. P., R. E. Wellinger, J. M. Sogo, and P. B. Becker. 1998. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 17:2865–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 11:205–214. [DOI] [PubMed] [Google Scholar]
- 37.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471–475. [DOI] [PubMed] [Google Scholar]
- 38.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125–129. [DOI] [PubMed] [Google Scholar]
- 39.Polanowska, J., E. Fabbrizio, L. Le Cam, D. Trouche, S. Emiliani, R. Herrera, and C. Sardet. 2001. The periodic down regulation of Cyclin E gene expression from exit of mitosis to end of G(1) is controlled by a deacetylase- and E2F-associated bipartite repressor element. Oncogene 20:4115–4127. [DOI] [PubMed] [Google Scholar]
- 40.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297–1307. [DOI] [PubMed] [Google Scholar]
- 41.Sambucetti, L. C., D. D. Fischer, S. Zabludoff, P. O. Kwon, H. Chamberlin, N. Trogani, H. Xu, and D. Cohen. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J. Biol. Chem. 274:34940–34947. [DOI] [PubMed] [Google Scholar]
- 42.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672–1677. [DOI] [PubMed] [Google Scholar]
- 43.Sterner, R., L. C. Boffa, T. A. Chen, and V. G. Allfrey. 1987. Cell cycle-dependent changes in conformation and composition of nucleosomes containing human histone gene sequences. Nucleic Acids Res. 15:4375–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599–606. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 46.Walker, J., T. A. Chen, R. Sterner, M. Berger, F. Winston, and V. G. Allfrey. 1990. Affinity chromatography of mammalian and yeast nucleosomes. Two modes of binding of transcriptionally active mammalian nucleosomes to organomercurial-agarose columns, and contrasting behavior of the active nucleosomes of yeast. J. Biol. Chem. 265:5736–5746. [PubMed] [Google Scholar]
- 47.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323–330. [DOI] [PubMed] [Google Scholar]
- 48.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong, N. T., and E. P. Candido. 1978. Histone H3 thiol reactivity as a probe of nucleosome structure. J. Biol. Chem. 253:8263–8268. [PubMed] [Google Scholar]
- 50.Wu, C., Y. C. Wong, and S. C. Elgin. 1979. The chromatin structure of specific genes II. Disruption of chromatin structure during gene activity. Cell 16:807–814. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79–89. [DOI] [PubMed] [Google Scholar]