Abstract
Genetic and biochemical studies of Schizosaccharomyces pombe and Saccharomyces cerevisiae have identified gene products that play essential functions in both pre-mRNA splicing and cell cycle control. Among these are the conserved, Myb-related CDC5 (also known as Cef1p in S. cerevisiae) proteins. The mechanism by which loss of CDC5/Cef1p function causes both splicing and cell cycle defects has been unclear. Here we provide evidence that cell cycle arrest in a new temperature-sensitive CEF1 mutant, cef1-13, is an indirect consequence of defects in pre-mRNA splicing. Although cef1-13 cells harbor global defects in pre-mRNA splicing discovered through intron microarray analysis, inefficient splicing of the α-tubulin-encoding TUB1 mRNA was considered as a potential cause of the cef1-13 cell cycle arrest because cef1-13 cells arrest uniformly at G2/M with many hallmarks of a defective microtubule cytoskeleton. Consistent with this possibility, cef1-13 cells possess reduced levels of total TUB1 mRNA and α-tubulin protein. Removing the intron from TUB1 in cef1-13 cells boosts TUB1 mRNA and α-tubulin expression to near wild-type levels and restores microtubule stability in the cef1-13 mutant. As a result, cef1-13 tub1Δi cells progress through mitosis and their cell cycle arrest phenotype is alleviated. Removing the TUB1 intron from two other splicing mutants that arrest at G2/M, prp17Δ and prp22-1 strains, permits nuclear division, but suppression of the cell cycle block is less efficient. Our data raise the possibility that although cell cycle arrest phenotypes in prp mutants can be explained by defects in pre-mRNA splicing, the transcript(s) whose inefficient splicing contributes to cell cycle arrest is likely to be prp mutant dependent.
Pre-mRNA splicing and cell cycle regulation have two distinct and apparently nonoverlapping functions for eukaryotic cells. In spite of this, a handful of genes in Saccharomyces cerevisiae and Schizosaccharomyces pombe have been identified in genetic screens for splicing factors (prp screens) and independently in screens for cell cycle regulators (cdc and related screens). These genes include S. cerevisiae PRP3 (also known as DBF5), PRP8 (also known as DBF3), PRP17 (also known as CDC40), and PRP22 and S. pombe prp8+ (also known as cdc28+) and prp2+ (also known as mis11+) (24, 26, 29, 44, 49, 56, 57). Furthermore, several prp mutants in S. pombe display morphologies consistent with defects in cell cycle progression (36, 54). Lastly, two proteins in S. pombe, Cdc5p and Dsk1p, both of which were identified genetically as potential cell cycle regulators, have since been implicated biochemically in pre-mRNA splicing (30, 50). In higher eukaryotes, splicing factors reorganize spatially throughout the cell cycle (19), and CDK2-cyclin E phosphorylates the U2 snRNP protein, SAP155 (43). Collectively, these data raise the possibility that pre-mRNA splicing and cell cycle control are functionally linked in vivo, although the molecular mechanisms underlying this connection remain vague.
The conserved CDC5/Cef1p protein has been implicated in both pre-mRNA splicing and cell cycle control and exemplifies a potential link between these processes. The single temperature-sensitive allele of cdc5+, cdc5-120, arrests during the G2 phase of the cell cycle prior to entry into mitosis (32, 34). cdc5+ encodes an essential protein with homology to the DNA binding domain of the transcription factor c-Myb (34). In its N terminus, Cdc5p contains two classic Myb repeats (R1 and R2) and a third nonclassic Myb-like repeat (MLR3) (33). To date, Cdc5p-related proteins have been isolated from every eukaryotic organism where they have been sought, including the budding yeast S. cerevisiae, where the homologous protein is termed Cef1p (7, 18, 22, 33, 48). The primary structure of CDC5/Cef1p has been conserved throughout evolution, with the Myb-related domains displaying the least divergence (33). We have concluded that this protein is also functionally conserved, since plant, fly, and human CDC5 cDNAs will rescue the temperature-sensitive lethality of S. pombe cdc5-120 (22, 33). Furthermore, inactivation of CDC5/Cef1p in S. cerevisiae (33) and in mammalian cells (6) causes arrest or delay at G2/M.
A major clue to the biochemical function of CDC5/Cef1p proteins came when human CDC5 (hCDC5) (also called CDC5L) was isolated in a biochemical purification of the mammalian spliceosome (31). Several lines of evidence have since established that these proteins play an essential role in pre-mRNA splicing. CDC5 colocalizes with pre-mRNA splicing factors in the nuclei of mammalian cells (11), S. pombe Cdc5p and hCDC5 associate with core components of the splicing machinery (11, 30), S. cerevisiae Cef1p and hCDC5 interact with the spliceosome in vitro (1, 11, 53), and genetic depletion of S. cerevisiae Cef1p or S. pombe Cdc5p causes accumulation of unspliced mRNAs in vivo (11, 30, 53). Lastly, Cef1p and hCDC5 play direct roles in pre-mRNA splicing, because inactivation of Cef1p by antibody interference or immunodepletion of hCDC5 inhibits splicing in vitro (1, 53).
In vivo, all detectable fission yeast Cdc5p is associated with a large (∼40S) multiprotein complex. This particle has been purified by immunoaffinity chromatography, and the identities of 10 Cwf (complexed with cdc5p) proteins have been reported (30). Significantly, most of the Cwf proteins have been directly or indirectly (through homologs in other organisms) implicated in the process of pre-mRNA splicing. S. cerevisiae Cef1p also resides in a protein complex identified through immunoaffinity purification of the splicing factor Prp19p (51, 53). It is likely that the fission yeast Cdc5p- and budding yeast Prp19p-associated protein complexes represent equivalent or related complexes. Lastly, hCDC5 copurifies with many proteins whose identities as known splicing factors were recently reported (1).
Although these data strongly implicate CDC5/Cef1p proteins biochemically and genetically in pre-mRNA splicing, it was unclear how they would also be required for cell cycle progression. Interestingly, phenotypic characterization of S. cerevisiae cef1-null cells revealed that, in addition to harboring splicing and cell cycle defects, these cells also failed to assemble mitotic spindles (33). This observation suggested the possibility that defective microtubules might be mediating cell cycle arrest in these cells.
To investigate the function of CDC5/Cef1p proteins in cell cycle control and pre-mRNA splicing, we analyzed a novel temperature-sensitive allele of CEF1, cef1-13. Like cef1-null cells, cef1-13 cells displayed defects in both processes. Many of the cef1-13 phenotypes, including cell cycle arrest at G2/M, could be suppressed by removing the intron from one of the genes encoding α-tubulin (TUB1). These data indicate that defective pre-mRNA splicing is the primary defect in cef1-13 cells. Removing the TUB1 intron from two other splicing mutants that arrest in G2/M, prp17Δ and prp22-1 strains, only partially suppressed their cell cycle phenotypes. Our data indicate that inefficient splicing of TUB1 is a significant contributor to the G2/M arrest phenotype observed in these splicing mutants. Moreover, our data are consistent with the idea that cell cycle phenotypes of yeast prp mutants can likely be explained as indirect consequences of pre-mRNA splicing defects.
MATERIALS AND METHODS
Strains, growth media, and genetic methods.
All S. cerevisiae strains used in this study are listed in Table 1. Strains produced in our laboratory are derivatives of S288C. prp3-1 (57), prp17 (also known as CDC40Δ; Research Genetics, Huntsville, Ala.), prp18-1 (57), prp22-1 (57), cdc28-1N (38), and sar1Δi (12) strains were obtained from other sources (Table 1). Strains obtained from other laboratories, with the exception of prp3-1, prp18-1, cdc28-1N, and sar1Δi strains, were backcrossed a minimum of three times against YPH98 or YPH252 prior to use. Strains were grown in yeast extract-peptone (YEP) medium supplemented with 2% glucose (YPD) or synthetic minimal medium with the appropriate nutritional supplements. Genetic methods were as described (20). Transformation of S. cerevisiae was performed by the lithium acetate method (25). Permissive temperature for all strains was 25°C, and restrictive temperature was between 35.5 and 37°C.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| YPH98 | MATaade2-101 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 | P. A. Weil |
| YPH252 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 | P. A. Weil |
| KGY1522 | MATacef1-13 ade2-101 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 | This study |
| KGY1760 | MATacdc28-1N | S. Reed |
| KGY1825 | MATaprp3-1 ade1 ade2 ura1 tyr1 his7 lys2 gal1 | B. Rymond |
| KGY1229 | MATaprp18-1 ade2-101 his3Δ200 tyr1 ura3-52 | J. Patton |
| KGY2914 | MATatub1Δi ade2-101 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 | This study |
| KGY2915 | MATacef1-13 tub1Δi ade2-101 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 | This study |
| KGY2818 | MATaprp17/cdc40Δ his3Δ1 leu1Δ0 met15Δ ura3Δ0 | M. Ares |
| KGY2917 | MATaprp17/cdc40Δ tub1Δi his3Δ1 leu1Δ0 met15Δ0 ura3Δ0 | This study |
| KGY2847 | MATaprp22-1 ade2 his3 ura3 | This study |
| KGY2918 | MATaprp22-1 tub1Δi ade2 his3 ura3 | This study |
| CKY569 | MATarse1-1 SAR1-Δi ura3-52 leu2-3,112 | Chris Kaiser |
| KGY3179 | MATarse1-1 SAR1-Δi ura3-52 leu2-3,112 cef1-13 | This study |
Benomyl (Sigma, St. Louis, Mo.) was diluted into YPD agar at the concentrations indicated from a stock solution of 10 mg/ml in dimethyl sulfoxide.
Construction of cef1-13.
The cef1-13 mutant allele (33) was subcloned by ligating the BamHI/HindIII fragment of pRO148 (pRS415 which harbors the cef1-13 allele [33]) into BamHI/HindIII cleaved pRS406ΔClaI to generate pRO173. pRS406ΔClaI was constructed by linearizing pRS406 with ClaI, treating the linearized vector with Klenow fragment, and ligating the vector on itself. pRO173 was linearized with ClaI and transformed into YPH98 cells with selection applied for Ura prototrophy, and colonies were screened by Southern analysis to identify those transformants that had integrated the plasmid at the CEF1 locus. One such transformant was plated onto 5-fluoroorotic acid (5-FOA) medium to select for excision events, and these colonies were screened for temperature sensitivity. KGY1522 is a single temperature-sensitive clone that arose from these manipulations.
Oligonucleotide arrays.
We use the terms “probe” and “target” where the probe is the known sequence (in this case, the oligonucleotide on the array) and the target is the unknown sequence (in this case, the fluor-labeled cDNA), regardless of which sequence carries the label or which sequence is immobilized. A set of 40-mer oligonucleotide probes carrying 5′ amino linked modifications was synthesized by Weboligos, Inc. Sequences were chosen to be complementary to cDNAs produced by reverse transcription of yeast intron-containing gene transcripts (13; http://www.cse.ucsc.edu/research/compbio/yeast_introns.html). The splice junction oligonucleotide probes span the splice junctions with 20 residues from each exon. The intron and second-exon probes were selected using a set of criteria to match predicted melting temperatures and to contain little internal structure, high information content, and low similarity to secondary sites in the genome. Probes were also synthesized to measure levels of eight intronless yeast mRNAs whose levels do not vary in numerous microarray experiments performed by the Brown laboratory (http://cmgm.stanford.edu/∼ pbrown) for normalization. In experiments with more than 30 different mutations in splicing factor genes, we observe increased levels of intron-containing transcripts and decreased levels of splice junction-containing transcripts, with variable (gene-specific) decreases in second-exon transcripts that indicate general splicing defects such as that exhibited by cef1-13. The details of that work (T. A. Clark, C. W. Sugnet, and M. Ares, Jr., unpublished data) will be presented elsewhere.
Removal of the TUB1 intron from wild-type, cef1-13, prp17Δ, and prp22-1 cells.
The TUB1 gene, including 350 to 550 bp 5′ and 3′ of the open reading frame, was PCR amplified as two overlapping fragments from wild-type genomic DNA and cloned into pBluescript SK (Stratagene, La Jolla, Calif.). The primer pairs TUB1.1 (5′-AACTGCAGCCATTATAACTGACTGTTTCAGATCCTGC-3′)-TUB1.1int (5′-GCAAAGCTTGGTCTTGGGATATCC-3′) and TUB1.2 (5′-AACTGCAGTTACGGGTTCTTGTTTTACCTTCTAATG-3′)-TUB1.2int (5′-GGAATTTGCCGTATACCCTGCTCC-3′) were used to generate the 5′ and 3′ fragments respectively of the TUB1 insert. Both fragments were digested with PstI/HindIII and placed into a three-part ligation with PstI-digested pBluescript SK to create pSKTUB1. The single intron within TUB1 that resides in codon 9 was precisely deleted using the Chameleon double-stranded site-direct mutagenesis kit (Stratagene) with TUB1.3 (5′-CCCAACAGGCATTACCAATCTGACAACCAGCTTGACCGACATTAATAGATATCACTTCTCTCATTGTTTGTTTGC-3′) and TUB1.4 (5′-GCGGCCGCTCTAGAACTAGTGGAACCCCCGGGGTGCAGGAATTCGATATCAAGC-3′) as the mutagenic and selection primers, respectively, to create pSKtubΔi. The mutagenic primer engineered an EcoRV site spanning codons 4, 5, and 6 of TUB1 that conserved the Tub1p amino acid sequence. The insert in pSKtub1Δi was completely sequenced to verify its accuracy.
Transplacement (42) was used to replace the endogenous TUB1 allele with the intronless tub1Δi allele in YPH98, YPH1522, KGY2849, and KGY2818. The tub1Δi allele was subcloned by ligating the SalI/XmaI fragment of pSKtub1Δi into the SalI/XmaI-cleaved integrating vector pRS406 to generate pRS406tub1Δi. The SalI/XmaI fragment from pRS406tub1Δi, which contains all of the DNA sequence downstream of codon 94, was excised from pRS406tub1Δi to create the integrating construct termed “pRS406tub1Δi BclI/BamHI delete.” pRS406tub1Δi BclI/BamHI delete was used as the integrating construct instead of pRS406tub1Δi to decrease the probability that the wild-type locus would be re-created upon excision of the integrated plasmid. pRS406tub1Δi BclI/BamHI delete was cut at the BsaBI site, which is downstream of the exon 1-exon 2 junction, and transformed into the strains listed above. Transformants in which pRS406tub1Δi BclI/BamHI delete had been integrated correctly at the TUB1 locus were identified by Ura auxotrophy and Southern analysis. One such transformant for each parent strain was plated onto 5-fluoroorotic acid medium, and colonies in which TUB1 had been replaced with tub1Δi were identified by Southern blot analysis. For Southern blot analysis, the genomic DNA was digested with EcoRV and probed with the XmaI/SalI fragment from pSKtub1ΔI.
Construction of cef1-13 sar1Δi.
A cef1-13 sar1Δi strain was identified in a tetrad derived from a cross between KGY1522 and KGY2860 in which all four colonies were temperature sensitive. Genomic DNA from each colony in the tetrad was subjected to PCR analysis. The SAR1 and sar1Δi alleles segregated 2:2 in this cross and were identified by the generation of a 463- or 324-bp band, respectively, in PCRs using the SAR15′ (5′-GGCTGGTTGGGATATTTTTGG-3′) and SAR13′ (5′-GCTTCATCAAATCTTTCAGGG-3′) primers. The CEF1 and cef1-13 alleles also segregated 2:2 and were identified by whether the PCR fragment generated with the primers R1R25′ (5′-CGCGGATCCGCATGCCCCCCGTACCAATATACGTGAAAGGCGG-3′) and MLR33′ (5′-CCTCTGCAGTCCTGCTTGCTTTAGCTCACGCCTCTTTTGTAGTTCAGC-3′) were resistant or sensitive, respectively, to digestion with PvuI.
Cell synchronization.
Exponentially growing cells were arrested in G1 by incubation in the presence of α-factor (10 μg/ml; Sigma) for 180 min at 25°C. To release cells from α-factor, cells were washed with 3 to 5 volumes of YPD and resuspended in fresh YPD prewarmed to between 35.5 and 37°C. For synchronization of cells with plasmids, the cells were grown overnight in selective medium and then synchronized and released in YPD medium.
Cytological methods and flow cytometry.
To visualize nuclei, cells were fixed with 70% ethanol overnight at 4°C, washed twice with phosphate-buffered saline, and resuspended in 1 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml. Cells were prepared for indirect immunofluorescence as decribed (37). α-Tubulin was detected using the monoclonal antibody TAT-1 (60) at a dilution of 1:50 followed by an Alexa 594-conjugated goat anti-mouse secondary antibody (Molecular Probes, Eugene, Oreg.).
Cells for flow cytometric analysis were fixed in ice-cold 70% ethanol, sonicated for 15 s in 1 ml of 50 mM sodium citrate (pH 7.0) containing 0.25 mg of RNase A per ml, and incubated overnight at 37°C. Cells were then stained with 1 μM Sytox Green (Molecular Probes) in 1 ml of 50 mM sodium citrate (pH 7.0) for at least 1 h at 4°C in the dark. The DNA content was measured on a Becton Dickinson FACSCalibur as described (15).
Electron microscopy.
Asynchronous, exponentially growing KGY1522 cells were shifted to 35.5°C for 3 h, fixed by high-pressure freezing, and processed for electron microscopy as described (17).
Immunoblot analysis and histone H1 kinase assays.
For immunoblot analyses, protein extracts were prepared from frozen cell pellets by glass bead disruption of cell walls. Lysed cells were heated to 95°C in SDS lysis buffer (10 mM NaPO4 [pH 7.4], 1.0% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol, 1 mM EDTA, 50 mM NaF, 100 μM Na3VO4, leupeptin [4 μg/ml]) for 1 to 3 min and extracted with NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1.0% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 100 μM Na3VO4, leupeptin [4 μg/ml]) supplemented with protease inhibitors (Complete; Boehringer Mannheim, Mannheim, Germany). Lysates were clarified to remove cell debris by microcentrifugation. Bicinchoninic acid assays (Pierce, Rockford, Ill.) were performed, and the total amount of protein in each sample was normalized based on these measurements. SDS sample buffer (5×) was added, and the samples were heated to 95°C prior to electrophoresis.
Protein extracts were fractionated on Novex NuPAGE 4 to 12% bis-Tris gels using NuPAGE MOPS SDS running buffer (Invitrogen, Carlsbad, Calif.) and subsequently transferred to Immobilon P membranes (Millipore Corp., Bedford, Mass.). Proteins were detected using an enhanced chemiluminescence detection system (ECL+Plus; Amersham Pharmacia Biotech, Piscataway, N.J.) and visualized by either film exposure or fluorescence scanning (Storm Phosphorimager; Molecular Dynamics Inc., Sunnyvale, Calif.). α-Tubulin was detected using TAT-1 monoclonal antibody at a dilution of 1:1,000. TATA binding protein (35) and Clb2p (28) were detected using affinity-purified polyclonal antisera at dilutions of 1:10,000 and 1:1,000 respectively. PSTAIRE antibodies (Sigma) were used at a dilution of 1:5,000. Goat anti-rabbit and anti-mouse secondary antibodies (Jackson Immunoresearch Laboratories, Inc.) were used at a dilution of 1:50,000.
Histone H1 kinase assays were performed as described previously (28) using affinity-purified rabbit anti-Clb2p polyclonal antibodies.
Other techniques.
For determination of cell numbers, samples were prepared and analyzed on a Multisizer II device (Beckman Coulter Inc., Fullerton, Calif.) as described (33). For determination of cell viability, cell numbers were assessed at each time point and 300 cells were plated in triplicate onto YPD agar at 25°C. The number of colonies per plate was counted 48 h later. Preparation of RNA and Northern blotting were performed as described (11). The TUB1, TUB2, and TUB3 open reading frames were obtained from Research Genetics.
RESULTS
cef1-13 cells arrest in G2/M with unstable microtubules.
S. cerevisiae cells lacking endogenous CEF1 and harboring a CEN plasmid-borne mutant allele of CEF1, cef1-13, are temperature sensitive for growth (33). Cef1p contains two amino acid substitutions, Trp52 to Gly and Trp84 to Gly, which are located at the third hydrophobic residue in R1 and the second hydrophobic residue in R2, respectively (33). To construct a stable version of the cef1-13 allele, the above mutations were introduced into the chromosomal copy of CEF1 in a wild-type haploid strain (see Materials and Methods). Although cef1-13 cells were unable to grow at the restrictive temperature, a diploid strain carrying one copy of the cef1-13 allele was viable at the same temperature, indicating that the cef1-13 allele is recessive.
To characterize the cell cycle arrest phenotype of cef1-13 cells, we synchronized wild-type and cef1-13 cells in G1 with α-factor and then released the cells at the restrictive temperature. The released cells were monitored for cell number and cell cycle progression as reflected by DNA content. In the time following release, the cell number for the wild-type culture increased exponentially, whereas that for the cef1-13 culture remained unchanged (Fig. 1A). Analysis of DNA content revealed that both wild-type and cef1-13 cells completed a full round of DNA replication (Fig. 1B). Subsequently, wild-type cells progressed normally into the next cell cycle, whereas cef1-13 cells arrested homogeneously in G2/M (Fig. 1B). At 4 h following release, the cell number (Fig. 1A) and DNA profile (data not shown) for the cef1-13 cells were indistinguishable from those at 3 h, indicating that the cells had arrested by 3 h. cef1-13 cells contained high Clb2p levels and Clb2p-associated H1 kinase activity at 3 h following release, demonstrating that cef1-13 cells arrested with activated mitotic CDKs (see below). Evaluation of bud and nuclear morphologies revealed that the majority of cef1-13 cells arrested with large buds and contained a single nucleus in the mother cell (Table 2). Whereas wild-type cells displayed 100% viability at this time, only 45% of cef1-13 cells were viable. These data suggest that cef1-13 cells arrest in the first cell cycle during G2/M prior to anaphase. Like synchronized cultures, asynchronous cef1-13 cultures arrested with large buds in G2/M within 3 h following shift to the restrictive temperature (data not shown).
FIG. 1.
The cef1-13 mutant arrests at G2/M with unstable microtubules. Wild-type (WT) and cef1-13 cells were synchronized in G1 and released to the restrictive temperature. Cell number (A) and fluorescence-activated cell sorter analysis (B) for each strain following release are shown. (C) Staining of microtubules in G2/M-arrested cef1-13 cells. Asynchronous cultures of WT or cef1-13 cells were shifted to the restrictive temperature for 3 h and stained for microtubules using an anti-α-tubulin antibody (TAT-1) (60) followed by an Alexa 594-conjugated secondary antibody. DNA was visualized using DAPI. (D) Electron micrograph analysis of the cef1-13 mutant. An asynchronous culture of cef1-13 cells was shifted to the restrictive temperature for 3 h and processed for electron micrograph analysis by HPS-FS. (Subpanel A) A 50-nm-thin section of an SPB which exhibits a wild-type trilaminar organization. (Subpanel B) Thin section through a short bipolar spindle with both SPBs in the plane of the section. (Subpanels C and D) Two serial semithick (100-nm) sections showing a short bipolar spindle with accompanying involutions of the nuclear membrane. (E) cef1-13 is benomyl sensitive. WT and cef1-13 cells were serially diluted fivefold from a culture at an optical density at 600 nm of 1.0 and spotted onto YPD plates or YPD plates containing the indicated concentrations of benomyl.
TABLE 2.
Quantitation of cell and nuclear morphologies 180 min following release from α-factor
Each morphologic class is shown schematically. Large-budded cells were scored if the bud was equal to or greater than half of the size of the mother. The percentages do not always add up to 100% due to rounding.
n, total number of cells counted.
A notable feature of the cef1-13 arrest phenotype was the presence of large budded cells that had failed to position their nuclei at the mother-bud junction in preparation for mitosis. These defects were seen in approximately 30% of the entire cell population and in 40% of the large-budded cells (Table 2) and were similar to those seen in cef1-null cells (33). Because large buds and nuclear positioning defects are hallmarks of cells with defective microtubules (41), we evaluated the state of microtubules in arrested cef1-13 cells by indirect immunofluorescence following formaldehyde fixation with a monoclonal antibody raised against α-tubulin (60). Approximately 75% of cef1-13 cells lacked visible nuclear and cytoplasmic microtubules, leaving only a small spot of staining near the nucleus (Fig. 1C), most likely at the spindle pole body (SPB). In roughly half of these cells, very short microtubules emanated from the single spot of staining (Fig. 1C). The remaining 25% of cells contained either a short intranuclear spindle with very short cytoplasmic microtubules or an extended mitotic spindle contained entirely within the mother cell (Fig. 1C). The microtubule defects observed in cef1-13 cells were never seen in wild-type cells prepared in parallel (Fig. 1C). cef1-13 cells fixed with methanol also lacked visible microtubules (data not shown). These data suggest that cef1-13 cells are defective in assembling or maintaining stable microtubules.
That a single focus of α-tubulin staining was observed by immunofluorescence suggested that cef1-13 cells contained either one unduplicated SPB or duplicated SPBs that failed to separate. To clarify this issue, we analyzed arrested cef1-13 cells by electron microscopy. Cells were prepared using high-pressure freezing and freeze substitution (HPF-FS) fixation rather than chemical fixation because yeast cells prepared with HPF-FS display less distortion in microtubule structures (17). In seven of seven cells, SPBs had duplicated, were morphologically normal, and had separated to various degrees (Fig. 1D). In all cells observed, intact microtubules emanated from both the nuclear (Fig. 1D) and cytoplasmic (data not shown) faces of SPBs. Surprisingly, five of seven cells contained fully assembled spindles (Fig. 1D, data not shown). However, these spindles were abnormal in that they appeared to exert a force on the nuclear envelope because it was invaginated where the SPBs were inserted. This phenotype is never observed in wild-type cells prepared by HPF-FS (17). We interpret this to mean that spindles assembled in cef1-13 cells are prone to collapse because they are built with unstable microtubules. It is likely that the chemical fixation of cef-13 cells used during processing for indirect immunofluorescence induced a complete collapse of the spindle. Collectively, these data are consistent with the idea that microtubules in cef1-13 cells are inherently unstable.
Because mutants with defective microtubules often display hypersensitivity to the microtubule depolymerizing drug benomyl (41), we determined if cef1-13 cells were benomyl sensitive. Whereas wild-type cells formed colonies on benomyl (15 μg/ml), the cef1-13 mutant was unable to form colonies under the same conditions (Fig. 1E), indicating that the cef1-13 mutant is hypersensitive to benomyl.
cef1-13 cell are defective in pre-mRNA splicing.
Because CDC5/Cef1p proteins have been implicated directly in pre-mRNA splicing (1, 11, 53), we analyzed cef1-13 cells to determine if pre-mRNA splicing defects were present and how widespread the defects were. We utilized splicing-sensitive oligonucleotide microarrays (Clark et al., submitted for publication) for this purpose and compared the levels of spliced and unspliced RNAs from the complete set of intron-containing genes in wild-type and cef1-13 cells. Total RNA was prepared from wild-type and cef1-13 cells soon after they had entered G2 following release from α-factor (60 and 90 min following release for wild-type and cef1-13 cells, respectively; see Fig. 1B) and used these as templates for the synthesis of fluorescent cDNA target sequences by reverse transcription. The target cDNAs were competitively hybridized to oligonucleotide microarrays containing probes for the intron, the splice junction, and the second exon sequence for yeast intron-containing genes (Clark et al., submitted). Out of 204 intron probes, 191 (94%) had signals indicating that their targets were more abundant in cef1-13 cells than in wild-type cells, indicating accumulation of unspliced RNA in cef1-13 cells (Fig. 2). Out of 419 targets detected by the splice junction and second exon probes, 400 (95%) were less abundant in cef1-13 cells, indicating that cef1-13 cells are generally depleted of spliced mRNAs. Transcripts that were most affected by the cef1-13 mutation are listed in Table 3. Cluster analysis of the cef1-13 profile indicated that the defects seen in cef1-13 are similar to those observed upon inactivation of the temperature-sensitive splicing mutant prp4-1 and are more severe than those caused by deletion of most nonessential splicing factors (data not shown).
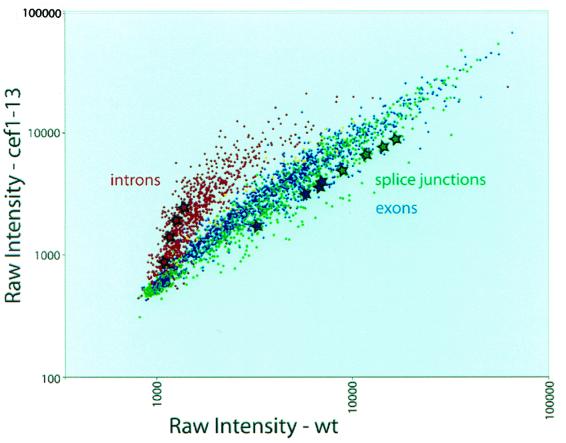
FIG. 2.
Scatter plot of cef1-13 versus wild-type (wt) splicing microarray data. RNA from the cef1-13 mutant was used as a template for reverse transcription in the presence of Cy5-dUTP, and RNA from wt cells was used as a template in the presence of Cy3-dUTP. The labeled target cDNAs were mixed and hybridized to the oligonucleotide probes on a printed microarray (see Materials and Methods). After washing, the array was scanned and the image analyzed to extract the data. Each point in the plot represents raw intensity data from a single probe spot, with the Cy3 (wt) signal on the abscissa and the Cy5 (mutant) signal on the ordinate. Each probe is present four times on the array. Intron probes (specific to unspliced RNA) are labeled in red, splice junction probes (specific to spliced RNA) are labeled in green, exon probes (representing total gene specific RNA) are labeled in blue, and non-intron-containing gene probes used for normalization are labeled in yellow. The four spots representing each of the TUB1-specific oligonucleotides are labeled with stars. Two factors influence the absolute intensity measurements, so it is important to keep in mind that the Cy5/Cy3 ratio is the key indicator of the difference between mutant and wt. First, different oligonucleotides have distinct hybridization properties, making direct comparison of absolute intensities uninformative. Second, since the identical probe is spotted in different positions on the array, their absolute intensities vary due to local hybridization differences. Note that each set of four identical probes falls in a line, indicating a consistent Cy5/Cy3 ratio measurement.
TABLE 3.
Transcripts whose splicing is most defective in cef1-13 cellsa
| Transcript with change in splicing |
|---|
| Increased in cef1-13 |
| Intron |
| QCR10 |
| COF1 |
| ACT1 |
| MATA1-Int1 |
| YNL050c |
| RUB1 |
| SRC1-Int1 |
| SRC1-Int2 |
| RPL39 |
| RPL33A |
| RPL34A |
| RPL27B |
| ARP9 |
| YBL059w |
| ARP2 |
| MMS2 |
| RPL17A |
| YIP3 |
| MATA1-Int2 |
| RPS27B |
| RPS23B |
| RPS19A |
| RPL7B-Int2 |
| RPL25 |
| ERD2 |
| MUD1 |
| RPL33B |
| RPL21A |
| OST5 |
| YDR367w |
| RPL37A |
| YPR063c |
| MAF1 |
| RPL34B |
| RPS30A |
| YDL012c |
| DYN2-Int2 |
| SNC1 |
| RPL27A |
| RIM1 |
| SNR17b |
| YML067c |
| NYV1 |
| RPL23B |
| RPO26 |
| RPS10B |
| QCR9 |
| YGL232w |
| RPS21B |
| IST1 |
| LSM2 |
| RPS30B |
| YIP2 |
| RPS6A |
| RPL29 |
| YLR128w |
| RPL37B |
| RPS7A |
| RPS26A |
| RPS16A |
| TUB1 |
| RPL18A |
| RPL16A |
| RPS10A |
| RPL16B |
| RPL21B |
| RPS8A |
| RPS23A |
| RPL6A |
| RPL40A |
| SAR1 |
| YKL002w |
| UBC13 |
| UBC4 |
| RPL23A |
| NCB2 |
| CNB1 |
| YML056c |
| RPL26B |
| RPL32 |
| SEC14 |
| PMI40 |
| RPL40B |
| Decreased in cef1-13 |
| Intron |
| RPS22B-Int2 |
| Splice junction |
| SEC14 |
| MMS2 |
| GIM5 |
| RPL23A |
| YGL232w |
| YKL158w |
| COF1 |
| QCR10 |
| RPS22B-SJ1 |
| SEC27 |
| YBR230c |
| PFY1 |
| COX4 |
| YLR128w |
| RPL16A |
| MATA1-SJ2 |
| STO1 |
| TUB3 |
| RPL30 |
| RPS22B-SJ2 |
| RPS26B |
| YLR211c |
| TUB1 |
| RPS25B |
| HNT2 |
| NCE101 |
| RPL22B |
| ANC1 |
| NMD2 |
| RPL27B |
| RPL22A |
| YKL002w |
| UBC12 |
| RPL13A |
| RPL35B |
| RPL18B |
| YHR097c |
| RPL27A |
| OST5 |
Total RNA from wild-type and cef1-13 cells that were cell cycle staged at G2/M was competitively hybridized to oligonucleotide microarrays containing probes for the intron and splice junction for yeast intron-containing genes. The transcripts that were increased or decreased twofold or more in cef1-13 cells for each class of probe are listed in decreasing order of fold difference between the mRNA samples. TUB1, TUB3, and GIM5 are shown in boldface type because of their importance for microtubule biogenesis. Raw numbers for the first and last transcript on each list as well as the highlighted transcripts are given here. Fold differences (expressed as cef1-13 relative to wild type) are given in parentheses. The medians for each class are also given. Intron probes: QCR10 (4.87), TUB1 (2.22), RPL40B (2.01); median, 1.84. Splice-junction probes: SEC14 (−4.27), GIM5 (−3.96), TUB3 (−2.36), TUB1 (−2.30), OST5 (−2.00); median, −1.53.
In analyzing these data with respect to the cell cycle, we searched the yeast intron database (13, 46) for intron-containing genes known to have a role in microtubule function and cell cycle progression. Three such genes were considered: TUB1 and TUB3, the two genes encoding α-tubulin (39, 40), and GIM5, a gene required for correct folding of tubulin subunits (16). Although all the spliced forms of the mRNAs from these genes were decreased in the cef1-13 mutant (Table 3), and mutations in each are reported to cause sensitivity to benomyl (16, 40, 41, 47), only TUB1 is an essential gene (40), and therefore, it became the focus of our attention.
cef1-13 cell have less total TUB1 mRNA and α-tubulin protein at the restrictive temperature.
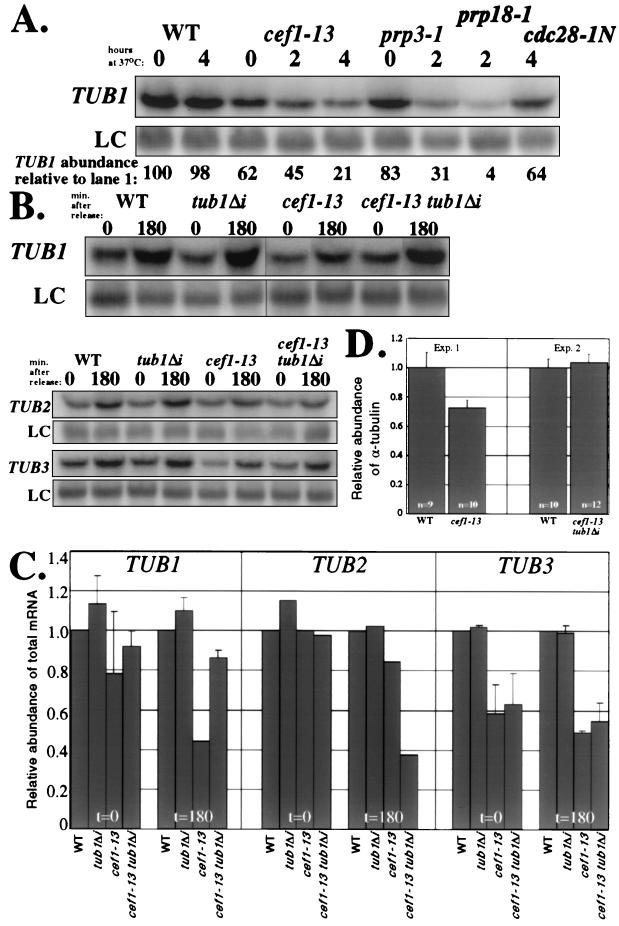
To confirm that TUB1 expression was reduced in cef1-13 cells, we quantified the amount of total TUB1 mRNA by Northern analysis in wild-type and cef1-13 cells shifted to the restrictive temperature. cef1-13 cells contained roughly half the amount of TUB1 message that wild-type cells had (Fig. 3A to C). In shifted asynchronous cells, TUB1 message declined to approximately 20% of the level seen in wild-type cells (Fig. 3A). Two other splicing mutants, prp3-1 and prp18-1, that are defective in the first and second steps of pre-mRNA splicing, respectively, also displayed decreased levels of TUB1 at the restrictive temperature (Fig. 3A). However, a cell cycle mutant that arrests in G2/M, cdc28-1N (38), did not display a significant decrease in total TUB1 (Fig. 3A), demonstrating that loss of TUB1 mRNA is not a consequence of cell cycle arrest in G2/M. Another intron-containing transcript, TUB3, was decreased by approximately 50% in synchronous (Fig. 2C and 3B) and in asynchronous (data not shown) cef1-13 cultures shifted to the restrictive temperature. However, neither of two intronless transcripts, TDH2 (the loading control for all of the Northern blots) and TUB2, was decreased significantly in synchronous cef1-13 cultures shifted to the restrictive temperature (Fig. 3B and C). These data validate the microarray analysis and indicate that TUB1 mRNA levels are reduced in cef1-13 cells. Because the TUB1 intron is very small, it is technically difficult to detect its pre-mRNAs by Northern blot analysis (data not shown). However, using multimerized intron sequences as probes, we were able to conclude that the TUB1 pre-mRNA did not accumulate to the same extent in cef1-13 cells as in the prp3-1 mutant (data not shown). However, it has recently been demonstrated that pre-mRNAs do not accumulate significantly in many pre-mRNA splicing mutants because they are degraded by the exosome complex (8).
FIG. 3.
cef1-13 cells have less TUB1 mRNA and α-tubulin protein unless the TUB1 intron is deleted. (A) Northern analysis of total TUB1 mRNA abundance in asynchronous cultures shifted to the restrictive temperature. Total RNA was prepared from wild-type (WT), cef1-13, prp3-1, prp18-1, and cdc28-1N cells incubated at the permissive temperature or following shift to the restrictive temperature for the indicated number of hours. The levels of TUB1 mRNA in each sample were quantified by Northern blot analysis using a probe to the entire open reading frame of TUB1. The TUB1 signal in each lane was normalized to that of the non-intron-containing loading control TDH2 (LC) and expressed as a percentage of TUB1 in the first lane. (B) Northern analysis of total TUB1, TUB2, and TUB3 mRNA abundance following synchronization in G1 and release to the restrictive temperature. Total RNA was prepared from WT, tub1Δi, cef1-13, and cef1-13 tub1Δi cells 0 and 180 min following release and blotted with probes for the entire open reading frames for TUB1, TUB2, and TUB3. The non-intron-containing TDH2 transcript was used as an LC. (C) Quantification of Northern analysis shown in panel B. The levels of TUB mRNAs in panel B were normalized to that of the LC TDH2 and expressed as a proportion of lane 1. The relative amounts of each transcript are shown for each strain according to the time point following release from G1. (D) Examination of α-tubulin protein in cef1-13 strains. WT, cef1-13, and cef1-13 tub1Δi cells were synchronized with mating pheromone and released to the restrictive temperature. At 180 min following release, multiple pellets (optical density = 10) were collected. Strains for each experiment were prepared twice. Protein lysates were made from each pellet, and bicinchoninic assays were conducted to allow normalization of protein concentrations between all samples. Equivalent amounts of protein were blotted with an α-tubulin monoclonal antibody (α) and polyclonal antisera raised against TATA-binding protein (encoded by a non-intron-containing gene) as a loading control. The immunoblots were quantitated using a Molecular Dynamics Storm instrument, and the results of α-tubulin protein relative to TATA-binding protein were graphed. The bars indicate the average and standard deviation (error bars). The number of independent analyses for each strain is indicated.
To determine whether the decrease in TUB1 mRNA translated to a decrease in protein levels, α-tubulin protein levels in wild-type and cef1-13 cells were determined by immunoblotting 3 h following shift to the restrictive temperature. Given that the TUB1 mRNA was decreased by only 50% on average, we did not expect to detect a large decrease in α-tubulin protein levels. For this reason, we examined α-tubulin protein levels multiple times in two independent experiments and present the results of these immunoblotting experiments graphically. Following quantitation, we determined that α-tubulin protein levels in cef1-13 cells were 72% of the level found in wild-type cells (Fig. 3D). This relatively modest reduction in protein levels is consistent with the known exquisite sensitivity of cells to reduced levels of α-tubulin protein. Diploid S. cerevisiae cells are not viable with a single TUB1 allele (27, 39). Due to the contribution of TUB3, such cells have been predicted to contain 60% of the wild-type level of α-tubulin protein (27, 39).
cef1-13 cells progress through mitosis when the single intron is removed from TUB1.
If the cef1-13 arrest phenotype arises, in whole or in part, because the TUB1 pre-mRNA is not properly spliced, then removing the TUB1 intron in the cef1-13 strain should rescue all or some of the its phenotypes. To test this prediction, an intronless TUB1 allele, designated tub1Δi, was created by precisely deleting the single intron from TUB1 (see Materials and Methods). The genomic copies of TUB1 were replaced with tub1Δi in wild-type and cef1-13 cells (see Material and Methods) to create the tub1Δi and cef1-13 tub1Δi strains. Removing the tub1Δi intron did not alter colony-forming efficiency (data not shown), cell cycle progression (Fig. 4A), or growth (Fig. 4B).
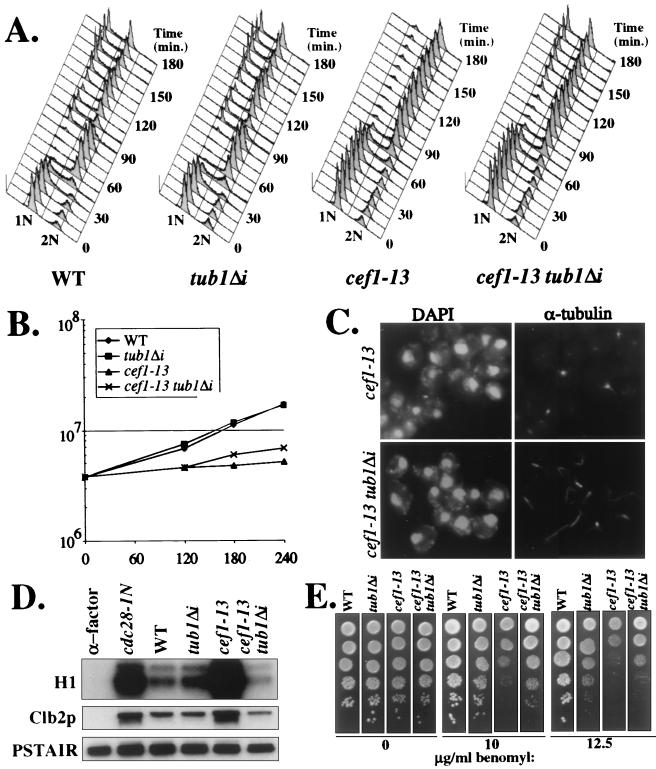
FIG. 4.
Removing the intron from TUB1 allows cef1-13 cells to progress through mitosis. Wild-type (WT), tub1Δi, cef1-13, and cef1-13 tub1Δi cells were synchronized in G1 with α-factor and released to the restrictive temperature. Following their release, cultures were monitored for cell number (B) and cell cycle progression as reflected by DNA content (A). (C) At 2 h following release, the status of microtubule structures in cef1-13 and cef1-13 tub1Δi cells was evaluated with indirect immunofluorescence. (D) At 3 h following release, the levels of the mitotic cyclin Clb2p and its associated H1 kinase activity were evaluated. G1-arrested WT cells and G2/M-arrested cdc28-1N cells were used as controls for low and high Clb2p and kinase activity, respectively. G1-arrested cells were obtained by treating WT cells with α-factor. G2/M-arrested cells were obtained by synchronizing cdc28-1N cells with α-factor and releasing them to the restrictive temperature for 3 h. (E) Tenfold serial dilutions of WT, tub1Δi, cef1-13, and cef1-13 tub1Δi cells were spotted on YPD agar with the indicated concentrations of benomyl and incubated at 25°C for 3 days prior to being photographed.
To determine if removing the intron from TUB1 alters its expression, quantitative Northern blotting was conducted on RNA from wild-type, tub1Δi, cef1-13, and cef1-13 tub1Δi cells following synchronization and release to the restrictive temperature (Fig. 3B and C). TUB1 expression was similar between wild-type and tub1Δi strains at all time points analyzed. As stated previously and consistent with the cef1-13 intron microarray profile, cef1-13 cells contained roughly half the amount of TUB1 message that wild-type cells had, and removing the TUB1 intron restored expression of TUB1 in cef1-13 cells to near wild-type levels (Fig. 3B and C). Specifically, at each time point there was a >79% increase in TUB1 expression following removal of its intron. Further, the level of α-tubulin protein was restored to the level seen in wild-type cells when the TUB1 intron was removed from cef1-13 cells (Fig. 3D). These data indicate that the decrease in TUB1 expression observed in cef1-13 cells requires the presence of the TUB1 intron.
To demonstrate that removing the TUB1 intron does not restore expression of another intron-containing gene, we analyzed expression of the intron-containing TUB3 transcript. TUB3 levels were equivalent in wild-type and tub1Δi cells but decreased by approximately 50% in both of the cef1-13 and cef1-13 tub1Δi strains (Fig. 3B and C). Lastly, we analyzed expression of the TUB2 transcript, which does not contain an intron. TUB2 expression was roughly equivalent between all strains at both time points analyzed (Fig. 3B and C), with the notable exception being that arrested cef1-13 tub1Δi cells displayed decreased expression. Since TUB2 expression peaks in G2 (45) and cef1-13 tub1Δi cells arrest in late mitosis and G1/S (see below), it would be predicted that these cells would contain TUB2 mRNA in lower abundance.
Next, we evaluated whether removing the TUB1 intron would alter the arrest phenotype of cef1-13 cells. Wild type, tub1Δi, cef1-13, and cef1-13 tub1Δi strains were synchronized in G1 with α-factor and released to the restrictive temperature. Following their release, all four strains completed a full round of DNA replication (Fig. 4A). Subsequently, wild-type and tub1Δi cells progressed into the next cell cycle with identical kinetics (Fig. 4A). Following release from G1 and completion of DNA replication, cef1-13 cells remained arrested at G2/M (Fig. 4A). In contrast, cef1-13 tub1Δi cells did not arrest homogeneously in G2/M but instead progressed into the next cell cycle as evidenced by the reappearance of a G1 peak later in the time course (Fig. 4A). Cell number determinations made throughout the time course indicated that removing the TUB1 intron allowed more cef1-13 cells to undergo cell division (at 3 h, cef1-13 and cef1-13 tub1Δi had undergone 0.35 and 0.80 doublings, respectively). The cell number (Fig. 4B) and DNA content profiles (data not shown) for the cef1-13 tub1Δi strain were identical at 3 and 4 h, indicating that a terminal arrest phenotype had been reached within 3 h. Analysis of the arrest phenotype for cef1-13 tub1Δi cells confirmed that removing the TUB1 intron allowed cef1-13 cells to progress through mitosis (Table 2). Whereas the majority of cef1-13 cells arrested as large-budded cells with a single nucleus, the majority of cef1-13 tub1Δi arrested as unbudded, small-budded, and large-budded cells with two nuclei. The predominant phenotype present in cef1-13 cells was not highly represented in the cef1-13 tub1Δi population (Table 2), demonstrating that removal of the TUB1 intron allowed cef1-13 cells to undergo anaphase and proceed into the next cell cycle.
That cef1-13 tub1Δi cells were progressing further into the cell cycle was confirmed biochemically by analyzing the levels of Clb2p and Clb2p-associated H1 kinase activity. In contrast to cef1-13 cells, which arrested with high levels of Clb2p and its associated kinase activity, cef1-13 tub1Δi cells contained low levels of both, indicative of cells that have progressed through mitosis (Fig. 4D) (61). Removing the TUB1 intron from cef1-13 cells did not decrease the restrictive temperature for cef1-13 (data not shown), but it did restore the viability of cef1-13 cells to near wild-type levels (90% for cef1-13 tub1Δi versus 45% for cef1-13 at 3 h following release from α-factor). Taken together, these data demonstrate that cef1-13 cells arrest prior to anaphase in large measure because the TUB1 mRNA levels become limiting, probably due to inefficient pre-mRNA splicing.
Based on the bud and nuclear morphologies, we predicted that, contrary to cef1-13 cells, cef1-13 tub1Δi cells should contain stable microtubules. To confirm this, we visualized α-tubulin in wild-type, tub1Δi, cef1-13, and cef1-13 tub1Δi cells by indirect immunofluorescence 2 h following release from α-factor. Wild-type and tub1Δi cells displayed indistinguishable microtubule arrays (data not shown). Consistent with previous experiments, cef1-13 cells did not display visible microtubule structures, whereas cef1-13 tub1Δi cells contained normal microtubule structures including spindles (Fig. 4C).
Lastly, we determined what effect removing the TUB1 intron would have on cef1-13 cell benomyl sensitivity. Wild-type, tub1Δi, cef1-13, and cef1-13 tub1Δi cells were plated on agar plates containing increasing concentrations of benomyl. The tub1Δi strain formed an equal number of colonies to the wild-type strain at each concentration (Fig. 4E). Removing the TUB1 intron in cef1-13 cells modestly increased this strain’s resistance to benomyl (Fig. 4E). These data indicate that inefficient splicing of TUB1 contributes to the cef1-13 strain’s sensitivity to benomyl. The residual benomyl sensitivity in cef1-13 tub1Δi cells is likely a function of reduced TUB3 and GIM5 mRNAs. Although neither of these genes is essential, mutations in either cause hypersensitivity to benomyl (16, 40).
An alternative explanation for the suppression of the cef1-13 cell cycle arrest is that removing the TUB1 intron decreases demand on the splicing machinery. To test this, we created a cef1-13 strain that lacked the intron for another gene, SAR1. The SAR1 mRNA is present at a greater copy number per cell than the TUB1 mRNA and is transcribed at a greater rate (5.8 or 2.6 copies per cell and 15.1 or 6.1 mRNAs/h for SAR1 and TUB1, respectively [23]). Following synchronous release to the restrictive temperature, cef1-13 sar1Δi cells displayed growth characteristics and a G2/M arrest phenotype matching those of cef1-13 cells (data not shown). Furthermore, removing the TUB1 intron allowed cef1-13 cells to segregate their nuclei, whereas removing the SAR1 intron did not (Table 4). These data indicate that removing the SAR1 intron did not allow cef1-13 cells to progress through mitosis, demonstrating that the effect of removing the TUB1 intron is specific.
TABLE 4.
Quantitation of cell and nuclear morphologies 180 min following release from α-factor
Each morphologic class is shown schematically. Large-budded cells were scored if the bud was equal to or greater than half of the size of the mother. The percentages do not always add up to 100% due to rounding.
n, total number of cells counted.
An extra copy of wild-type TUB1 allows cef1-13 cells to progress through mitosis.
If inefficient splicing of TUB1 causes the cef1-13 phenotype, then it would be predicted that, like removing the TUB1 intron, introducing an extra copy of the wild-type TUB1 gene into cef1-13 cells might allow progression through mitosis. To test this prediction, wild-type and cef1-13 cells were transformed with three plasmids: an empty CEN vector (pRS415), the same vector containing the wild-type TUB1 gene (pRS415TUB1), or the same vector containing the intronless tub1Δi allele (pRS415tub1Δi). All six transformants were synchronized and released to the restrictive temperature. The three wild-type transformants were indistinguishable with respect to cell number (data not shown), cell cycle progression (data not shown), and bud and nuclear morphologies (Table 5). As expected, the cef1-13 strain carrying the empty vector arrested in the first cell cycle with a minimal increase in cell number (Fig. 5), homogenous arrest in G2/M (Fig. 5), and a predominance of large-budded cells with a single nucleus (Table 5). In contrast, cef1-13 cells carrying the wild-type TUB1 allele were able to reenter the next cell cycle as evidenced by the reappearance of a G1 peak late in the time course (Fig. 5). Furthermore, these cells displayed a greater increase in cell number (Fig. 5) and a disappearance of the large-budded phenotype compared to cef1-13 cells carrying the empty vector (Table 5). These data indicate that an extra copy of TUB1 alone will allow cef1-13 cells to progress through mitosis. cef1-13 cells carrying the plasmid-borne tub1Δi allele were similarly able to progress through mitosis (Fig. 5; Table 5). These cells showed the greatest increase in cell number of all of the cef1-13 strains, which indicates that the intronless tub1Δi allele is more efficient than the wild-type allele at allowing mitotic progression in these conditions.
TABLE 5.
Quantitation of cell and nuclear morphologies 180 min following release from α-factor
Each morphologic class is shown schematically. Large-budded cells were scored if the bud was equal to or greater than half of the size of the mother. The percentages do not always add up to 100% due to rounding.
n, total number of cells counted.
WT, wild type.
FIG. 5.
An extra copy of TUB1 allows cef1-13 cells to progress through mitosis. cef1-13 cells carrying an empty CEN vector or the same vector containing the wild-type TUB1 allele or the intronless tub1Δi allele were synchronized in G1 and released to the restrictive temperature. Released cultures were monitored for cell number (upper left) and cell cycle progression as reflected by DNA content.
Removing the intron from TUB1 allows prp17 and prp22 mutants to segregate their nuclei.
To determine if other prp mutants that arrest at G2/M do so because of a failure to splice TUB1 pre-mRNAs, we evaluated the effects of removing the TUB1 intron from temperature-sensitive mutants of prp17 (56) and prp22 (24, 57), the only other splicing mutants reported to arrest at G2/M. PRP17 encodes a protein involved in the second catalytic step of the splicing reaction, and the temperature-sensitive prp17-null (prp17Δ) cells arrest at G2/M without microtubules (56). PRP22 encodes a DEAH box protein required for mRNA release from the spliceosome (58), and temperature-sensitive alleles of PRP22 were identified in a screen for mutants that arrest in mitosis (24).
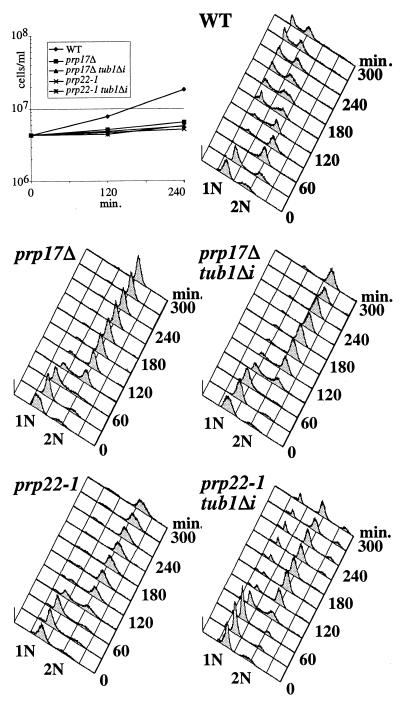
We removed the TUB1 intron from the genomic TUB1 locus in prp17Δ and prp22-1 mutants to determine if, as for cef1-13, the intronless TUB1 would allow for progression through mitosis. These cells (along with their isogenic TUB1 intron-containing counterparts) were synchronized in G1 with α-factor and released to the restrictive temperature. Cell numbers were essentially constant throughout the course of the experiment for all four mutant strains (Fig. 6). Following DNA replication, all of the mutant strains arrested predominantly with a 2N content of DNA, although a small G1 peak reappeared in the prp22-1 tub1Δi strain (Fig. 6). Analysis of bud and nuclear morphologies 3 h following release revealed that like cef1-13 cells, prp17Δ and prp22-1 cells arrested with a significant percentage of large-budded cells with a single nucleus (Table 6). Removing the TUB1 intron from either mutant decreased the percentage of cells with this arrest phenotype and increased the percentage of large-budded cells with segregated nuclei (Table 6). These data indicate that removing the TUB1 intron from two other splicing mutants allowed for nuclear division. Therefore, inefficient splicing of TUB1 can explain in part or in full why certain splicing mutants arrest with unsegregated chromosomes.
FIG. 6.
Cell cycle arrest of prp17 and prp22 mutants. Wild-type (WT), prp17Δ, prp17Δtub1Δi, prp22-1, and prp22-1 tub1Δi cells were synchronized in G1 and released to the restrictive temperature. Released cultures were montitored for cell number (upper left) and cell cycle progression as reflected by DNA content.
TABLE 6.
Quantitation of cell and nuclear morphologies 240 min following release from α-factor
Each morphologic class is shown schematically. Large-budded cells were scored if the bud was equal to or greater than half of the size of the mother. The percentages do not always add up to 100% due to rounding.
n, total number of cells counted.
DISCUSSION
Pre-mRNA splicing and cell cycle control have independent functions for eukaryotic cells. However, these processes might be linked, because yeast proteins have been identified through genetic criteria as splicing factors and independently as essential for cell cycle progression (10). The conserved Myb-related CDC5/Cef1p proteins carry essential functions in both processes, because their genetic depletion in the fission (30) and budding (11) yeasts causes accumulation of pre-mRNAs and cell cycle arrest at G2/M. Through characterization of a novel temperature-sensitive allele of S. cerevisiae CEF1, cef1-13, we have demonstrated that G2/M cell cycle arrest results from inefficient splicing of the TUB1 pre-mRNA that encodes the major yeast α-tubulin. TUB1 intron deletion also partially suppresses the G2/M arrest observed in mutants of the splicing proteins Prp17p and Prp22p.
cef1-13 cells arrest homogeneously in G2/M prior to the first encountered anaphase when released to the restrictive temperature as a synchronous culture from G1. The arrest phenotype includes many features indicative of defects in microtubules: nuclear positioning defects, depolymerization of microtubules upon chemical fixation, and benomyl sensitivity (41). Because CDC5/Cef1p proteins have been implicated in pre-mRNA splicing, we hypothesized that the cef1-13 phenotype might arise because a specific nascent transcript was not processed efficiently at the restrictive temperature.
We used splicing-sensitive oligonucleotide microarrays to search for those transcripts not spliced efficiently in cef1-13 cells and whose inefficient splicing might account for the G2/M arrest phenotype. We determined that cef1-13 was generally defective in pre-mRNA splicing, confirming the conclusion that CEF1 is a bona fide splicing factor (1, 11, 53). Furthermore, of essential transcripts, TUB1 was the most affected. Despite the fact that cef1-13 cells contained higher levels of the TUB1 precursor as detected with the microarrays, we did not detect a temperature-dependent accumulation of this species in cef1-13 cells by Northern analysis (data not shown). This feature of the cef1-13 phenotype is common among splicing mutants. Many pre-mRNAs do not accumulate in temperature-sensitive splicing mutants despite a strong reduction in mRNA levels, because the precursors are degraded by a pathway for nuclear pre-mRNA turnover (8).
Despite the general defect in splicing, our attention was drawn to the TUB1 gene. Of essential intron-containing transcripts, the microarray analysis indicated that TUB1 was the most affected in cef1-13 cells. TUB1 encodes the major α-tubulin protein in S. cerevisiae, and tub1 mutants can display phenotypes similar to those of cef1-13 cells, including unstable microtubules, nuclear positioning defects, and G2/M arrest (39–41). Therefore, we tested the hypothesis that inefficient splicing of TUB1 was responsible for the cell cycle phenotypes displayed by cef1-13 cells.
Uncoupling the production of mature TUB1 mRNA from splicing of its pre-mRNA was accomplished by removing the single intron from the genomic TUB1 locus. Consistent with our hypothesis, removal of the TUB1 intron allowed cef1-13 cells to progress through mitosis, as evidenced by nuclear and cell division. Removing the intron from TUB1 did not rescue the growth defect of the cef1-13 strain, indicating that another transcript(s) becomes rate limiting for growth after the cells have passed through mitosis. Removing the intron from another gene, SAR1, did not alleviate the cell cycle phenotype in cef1-13 cells, indicating that the effect of removing the TUB1 intron is specific.
The twofold reduction in total TUB1 mRNA in cef1-13 cells at the restrictive temperature translated to a 30% reduction in the steady-state levels of α-tubulin protein. Given that α-tubulin is a stable (4) and moderately abundant (21) protein, it is understandable why decreased synthesis would take generation times to fully manifest itself with an equivalent reduction in the total pool of α-tubulin protein. However, it is not surprising that a modest decrease in α-tubulin protein would have a profound effect on microtubule dynamic instability in vivo and therefore on cell growth. In S. cerevisiae, it has been well established that increasing the ratio of β- to α-tubulin through mutations in the α-tubulin-encoding TUB1 (40, 41) or overproduction of the β-tubulin-encoding TUB2 (9, 27, 59) will cause cells to arrest in G2/M with unstable microtubules, large buds, and single nuclei. Furthermore, diploid cells hemizygous with respect to TUB1 (these cells would be predicted to contain 60% of α-tubulin levels, with the extra 10% coming from TUB3 [27]) cannot be isolated without an extra copy of chromosome 13, which carries both TUB1 and TUB3 (27, 39). Since the β-tubulin mRNA (encoded by the non-intron-containing TUB2 gene, see above) and protein levels (data not shown) are unaffected in cef1-13 cells, the decrease in α-tubulin protein would be predicted to decrease the ratio of α- to β-tubulin and result in several features of the cef1-13 phenotype. In light of this, it is not surprising that extra copies of intron-containing TUB1 allowed cef1-13 cells to progress through mitosis, similar to removal of its intron.
In addition to altering the ratio of β- to α-tubulin, we would predict a significant decrease in the pool of newly synthesized α-tubulin protein in cef1-13 cells that is presumably unpolymerized. Since the rate of microtubule polymerization in vitro is highly dependent on the concentration of tubulin (14), a reduction in the levels of unpolymerized tubulin could have dramatic consequences on the stability of polymerized microtubules in vivo. Indeed, cef1-13 cells have unstable microtubules, and removing the intron from TUB1 restores microtubule stability. Furthermore, because other genes involved in tubulin biogenesis contain introns and are not spliced properly in cef1-13 cells (such as GIM5), it is likely that cef1-13 cells have a lower tolerance than wild-type cells for a reduction in α-tubulin production. Lastly, our data do not rule out the possibility that an alteration in the production of TUB1 message directly influences microtubule stability. To our knowledge, however, there are no data in the literature indicating that such a pathway might exist.
In addition to TUB1, there are two other intron-containing genes that are known to be important for microtubule function: TUB3, which encodes the minor α-tubulin (39, 40), and GIM5, which encodes a protein that is required for correct folding of tubulin subunits (16). Although neither gene is essential for growth, deletion of either one causes benomyl sensitivity (16, 40, 47). Furthermore, synthetic lethal interactions between GIM5 and mutations in TUB1 have been reported (55). Therefore, it is possible that a cell cycle block could be caused by inefficient production of a combination of these different mRNAs. The spliced forms of the mRNAs from TUB3 and GIM5 were decreased in the cef1-13 mutant (Table 3). However, because removal of the TUB1 intron effectively eliminates a cell cycle phenotype, it is likely that a significant impact of splicing inefficiency on cell cycle progression in cef1-13 cells is mediated through TUB1. In the case of mutations in two other splicing factors, Prp17p and Prp22p, that cause G2/M arrest (5, 24), we found that removal of the TUB1 intron allowed increased efficiency of nuclear division and, to a lesser extent, cell division. Suppression may not be as complete in these cases as it is with cef1-13 if the prp17 and prp22 mutations have stronger effects on the processing of TUB3 and GIM5. Whether removal of the introns from these genes in addition to that of TUB1 will eliminate the cell cycle arrest phenotype of the prp17 and prp22 mutants is under investigation.
It is likely that the CEF1 ortholog in S. pombe, cdc5+, is also required for splicing the intron-containing α-tubulin gene, nda2+ (52), in this evolutionarily distinct yeast. The temperature-sensitive cdc5-120 mutant exhibits sensitivity to the microtubule-destabilizing drug thiabendazole and displays defects in cell polarity when combined with mutations in β-tubulin (R. Ohi and K. L. Gould, unpublished results), both characteristics of defective microtubules in fission yeast. However, because the majority of cdc5-120 cells arrest prior to entry into mitosis (34), it seems unlikely that reduced expression of the mature nda2+ transcript is as significant in their arrest phenotype. Compared to S. cerevisiae, S. pombe contains a greater percentage of genes with introns (approximately 45% [62]), and there are a large number of candidate genes whose inefficient splicing might contribute to the G2 arrest of cdc5-120 cells.
To determine whether cell cycle arrest is a common feature of S. cerevisiae splicing mutants, we have evaluated the cell cycle phenotypes of 10 splicing mutants (prp2-1, prp3-1, prp4-1, prp5-1, prp17-1, prp17/cdc40Δ, prp18-1, prp19-1, prp20-1, and prp22-1 strains) following release to the restrictive temperature from an α-factor block. We found that some mutants display cell cycle phenotypes although they differ, and some mutants arrest heterogeneously throughout the cell cycle (C. G. Burns and K. L. Gould, unpublished results). Only mutations in PRP17 and PRP22 caused a first cell cycle arrest in G2/M.
The source of variability in the timing of arrest and appearance of cell cycle phenotypes among splicing mutants is unclear. The cell cycle defect (or lack thereof) might be a function of the severity of the splicing defect. For instance, mutants that arrest early after release from a G1 block might have a more severe splicing defect than those that arrest later or continue to undergo one or two divisions and arrest without a homogeneous phenotype. An alternative explanation would be that protein factors are differentially required for efficient processing of single transcripts or a subset of transcripts. If this is true, then multiple rate-limiting transcripts (and, therefore, cell cycle phenotypes) would arise from mutating different splicing factors. For example, it is possible that CEF1, PRP17, and PRP22 have more important roles in splicing transcripts critical for G2/M progression and that is why mutations in these proteins cause G2/M arrest. That protein factors perform specialized roles in splicing specific transcripts would be analogous to the role of the TAF proteins in regulating transcription. Of the TAF proteins in S. cerevisiae, yTAF145, TSM1, and yTAF90 are required for transcriptional activation of a subset of genes necessary for cell cycle progression (2, 3, 23, 60). A complete list of transcriptional targets for many of the TAFs has been identified through genome-wide expression analysis of TAF mutants (23). A similar approach would be valuable to determine if protein factors play specialized roles in pre-mRNA splicing and which transcripts require specialized regulation.
Acknowledgments
We thank K. Gull, D. C. Kaiser, D. Kellogg, J. Patton, S. Reed, B. Rymond, and P. A. Weil for their generous gifts of strains, plasmids, and antibodies. We are grateful to D. McFarland and Michelle Thornsberry for flow cytometric analysis. Mary Morphew (Boulder Laboratory for 3D Fine Structure) kindly prepared specimens for electron micrograph analysis. J. Flick and all members of the Gould laboratory are acknowledged for providing useful discussions.
This work was supported by NIH grant GM47728 to K.L.G. Microarray work was supported by a grant from the W. M. Keck Foundation to the RNA Center at the University of California at Santa Cruz. C.G.B. was supported by NIH Medical Scientist Training Program grant GM07347. K.L.G. is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ajuh, P., B. Kuster, K. Panov, J. C. Zomerdijk, M. Mann, and A. I. Lamond. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19:6569–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1–13. [DOI] [PubMed] [Google Scholar]
- 3.Apone, L. M., C. M. Virbasius, J. C. Reese, and M. R. Green. 1996. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 10:2368–2380. [DOI] [PubMed] [Google Scholar]
- 4.Archer, J. E., L. R. Vega, and F. Solomon. 1995. Rbl2p, a yeast protein that binds to beta-tubulin and participates in microtubule function in vivo. Cell 82:425–434. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Yehuda, S., I. Dix, C. S. Russell, M. McGarvey, J. D. Beggs, and M. Kupiec. 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156:1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, H. S., and S. R. Coughlin. 1998. A mammalian homolog of fission yeast Cdc5 regulates G2 progression and mitotic entry. J. Biol. Chem. 273:4666–4671. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, H. S., and S. R. Coughlin. 1997. Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J. Biol. Chem. 272:5833–5837. [DOI] [PubMed] [Google Scholar]
- 8.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765–775. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D., P. Gasdaska, and L. Hartwell. 1989. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns, C. G., and K. L. Gould. 1999. Connections between pre-mRNA processing and regulation of the eukaryotic cell cycle, p.59–82. In S. L. Chew (ed.), Post-transcriptional regulation of gene expression and its importance to the endocrine system, vol. 25. Karger, Basel, Switzerland. [DOI] [PubMed]
- 11.Burns, C. G., R. Ohi, A. R. Krainer, and K. L. Gould. 1999. Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96:13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, E. J., A. R. Frand, E. Chitouras, and C. A. Kaiser. 1998. A link between secretion and pre-mRNA processing defects in Saccharomyces cerevisiae and the identification of a novel splicing gene, RSE1. Mol. Cell. Biol. 18:7139–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, C. A., L. Grate, M. Spingola, and M. Ares, Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28:1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, A., and T. J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- 15.Epstein, C. B., and F. R. Cross. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6:1695–1706. [DOI] [PubMed] [Google Scholar]
- 16.Geissler, S., K. Siegers, and E. Schiebel. 1998. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 17:952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giddings, T. H. J., E. T. O’Toole, M. Morphew, D. N. Mastronarde, J. R. McIntosh, and M. Winey. 2001. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groenen, P. M., G. Vanderlinden, K. Devriendt, J. P. Fryns, and W. J. Van de Ven. 1998. Rearrangement of the human CDC5L gene by a t(6;19)(p21;q13.1) in a patient with multicystic renal dysplasia. Genomics 49:218–229. [DOI] [PubMed] [Google Scholar]
- 19.Gui, J. F., W. S. Lane, and X. D. Fu. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369:678–682. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif. [PubMed]
- 21.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirayama, T., and K. Shinozaki. 1996. A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93:13371–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717–728. [DOI] [PubMed] [Google Scholar]
- 24.Hwang, L. H., and A. W. Murray. 1997. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol. Biol. Cell 8:1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston, L. H., and A. P. Thomas. 1982. The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 186:439–444. [DOI] [PubMed] [Google Scholar]
- 27.Katz, W., B. Weinstein, and F. Solomon. 1990. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol. Cell. Biol. 10:5286–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellogg, D. R., and A. W. Murray. 1995. NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol. 130:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundgren, K., S. Allan, S. Urushiyama, T. Tani, Y. Ohshima, D. Frendewey, and D. Beach. 1996. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell 7:1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald, W. H., R. Ohi, N. Smelkova, D. Frendewey, and K. L. Gould. 1999. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19:5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46–50. [DOI] [PubMed] [Google Scholar]
- 32.Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178. [DOI] [PubMed] [Google Scholar]
- 33.Ohi, R., A. Feoktistova, S. McCann, V. Valentine, A. T. Look, J. S. Lipsick, and K. L. Gould. 1998. Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol. 18:4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohi, R., D. McCollum, B. Hirani, G. J. Den Haese, X. Zhang, J. D. Burke, K. Turner, and K. L. Gould. 1994. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 13:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon, D., and P. A. Weil. 1993. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J. Biol. Chem. 268:15325–15328. [PubMed] [Google Scholar]
- 36.Potashkin, J., D. Kim, M. Fons, T. Humphrey, and D. Frendewey. 1998. Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet. 34:153–163. [DOI] [PubMed] [Google Scholar]
- 37.Pringle, J., A. E. M. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast, p. 565–601. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 38.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schatz, P. J., L. Pillus, P. Grisafi, F. Solomon, and D. Botstein. 1986. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol. Cell. Biol. 6:3711–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schatz, P. J., F. Solomon, and D. Botstein. 1986. Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol. 6:3722–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schatz, P. J., F. Solomon, and D. Botstein. 1988. Isolation and characterization of conditional-lethal mutations in the TUB1 alpha-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics 120:681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherer, S., and R. W. Davis. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76:4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seghezzi, W., K. Chua, F. Shanahan, O. Gozani, R. Reed, and E. Lees. 1998. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol. Cell. Biol. 18:4526–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shea, J. E., J. H. Toyn, and L. H. Johnston. 1994. The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res. 22:5555–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spingola, M., L. Grate, D. Haussler, and M. Ares, Jr. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stearns, T., M. A. Hoyt, and D. Botstein. 1990. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics 124:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stukenberg, P. T., K. D. Lustig, T. J. McGarry, R. W. King, J. Kuang, and M. W. Kirschner. 1997. Systematic identification of mitotic phosphoproteins. Curr. Biol. 7:338–348. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, K., H. Yamada, and M. Yanagida. 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 5:1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang, Z., M. Yanagida, and R. J. Lin. 1998. Fission yeast mitotic regulator Dsk1 is an SR protein-specific kinase. J. Biol. Chem. 273:5963–5969. [DOI] [PubMed] [Google Scholar]
- 51.Tarn, W. Y., C. H. Hsu, K. T. Huang, H. R. Chen, H. Y. Kao, K. R. Lee, and S. C. Cheng. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13:2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toda, T., Y. Adachi, Y. Hiraoka, and M. Yanagida. 1984. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different α-tubulin genes in Schizosaccharomyces pombe. Cell 37:233–242. [DOI] [PubMed] [Google Scholar]
- 53.Tsai, W. Y., Y. T. Chow, H. R. Chen, K. T. Huang, R. I. Hong, S. P. Jan, N. Y. Kuo, T. Y. Tsao, C. H. Chen, and S. C. Cheng. 1999. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem. 274:9455–9462. [DOI] [PubMed] [Google Scholar]
- 54.Urushiyama, S., T. Tani, and Y. Ohshima. 1996. Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol. Gen. Genet. 253:118–127. [DOI] [PubMed] [Google Scholar]
- 55.Vainberg, I. E., S. A. Lewis, H. Rommelaere, C. Ampe, J. Vandekerckhove, H. L. Klein, and N. J. Cowan. 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93:863–873. [DOI] [PubMed] [Google Scholar]
- 56.Vaisman, N., A. Tsouladze, K. Robzyk, S. Ben-Yehuda, M. Kupiec, and Y. Kassir. 1995. The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol. Gen. Genet. 247:123–136. [DOI] [PubMed] [Google Scholar]
- 57.Vijayraghavan, U., M. Company, and J. Abelson. 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 3:1206–1216. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, J. D., E. Jankowsky, M. Company, A. M. Pyle, and J. N. Abelson. 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17:2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstein, B., and F. Solomon. 1990. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol. Cell. Biol. 10:5295–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woods, A., T. Sherwin, R. Sasse, T. H. MacRae, A. J. Baines, and K. Gull. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500. [DOI] [PubMed] [Google Scholar]
- 61.Yeong, F. M., H. H. Lim, C. G. Padmashree, and U. Surana. 2000. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell 5:501–511. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, M. Q., and T. G. Marr. 1994. Fission yeast gene structure and recognition. Nucleic Acids Res. 22:1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]