Abstract
We have constructed an ALDH3a1 null mouse to investigate the role of this enzyme that comprises nearly one-half of the total water-soluble protein in the mouse corneal epithelium. ALDH3a1-deficient mice are viable and fertile, have a corneal epithelium with a water-soluble protein content approximately half that of wild-type mice, and contain no ALDH3a1 as determined by zymograms and immunoblots. Despite the loss of protein content and ALDH3a1 activity, the ALDH3a1−/− mouse corneas appear indistinguishable from wild-type corneas when examined by histological analysis and electron microscopy and are transparent as determined by light and slit lamp microscopy. There is no evidence for a compensating protein or enzyme. Even though the function of ALDH3a1 in the mouse cornea remains unknown, our data indicate that its enzymatic activity is unnecessary for corneal clarity and maintenance, at least under laboratory conditions.
Aldehyde dehydrogenase 3a1 (ALDH3a1) constitutes the major fraction of the water-soluble protein in bovine (up to 40%) and other mammalian corneas (1, 2, 9, 20, 40). ALDHs constitute a large family of enzymes (41) that detoxify the cellular environment by converting highly reactive aldehydes to their corresponding carboxylic acids with concomitant reduction of the cofactor NAD+ (23). Class 3 ALDHs prefer medium-chain lipid and aromatic aldehydes such as hexenal and benzaldehyde (25, 48). Expression of ALDH3a1 in the cornea is particularly relevant because of the presence of aldehydes generated by light-induced lipid peroxidation (12, 13, 16). However, among vertebrate species, only mammals have been found to express ALDH3a1 at high levels in their corneas, although there are exceptions among mammals. For example, rabbit corneas express ALDH1 at high levels, especially within their keratocytes (18). In contrast to ALDH3a1, ALDH1 prefers short-chain aldehydes such as malondialdehyde (23, 24). The limited taxonomic range of expression and its extreme abundance has led to the question of whether ALDH3a1 might be present in amounts greater than its enzymatic need in the cornea and to proposals of additional functions for ALDH3a1 (1, 3, 9, 21, 35, 46, 47).
Examples of enzymes with additional functions can be found in the lenses of some species in which crystallins are enzymes (33, 34). The abundant lens crystallins modify the refractive index and provide the short-range order required for transparency (4, 10, 44). Similar to ALDH3a1 in the cornea, lens enzyme crystallins are taxon specific and represent a diverse set of proteins that include lactate dehydrogenase, argininosuccinate lyase, α-enolase, and, interestingly, ALDH (8, 9, 33–35). Similarly, different proteins have been found to be expressed to crystallin-like levels in the corneas of animals. These include gelsolin in zebrafish, isocitrate dehydrogenase in cow, cyclophilin in chicken, and glutathione S-transferase homologues in squid (33, 43, 49). Among these, the mammalian ALDH3a1 represents a particularly interesting comparison to lens crystallins, since it is a known abundant corneal enzyme that has family members, η-crystallin and Ω-crystallin, represented among the lens crystallins (15, 28, 36, 45, 50).
To begin to investigate ALDH3a1 function in the cornea, we have created an ALDH3a1 knockout mouse. This knockout mouse is viable. Interestingly, although ALDH3a1 comprises nearly one-half of the water-soluble protein fraction in the corneal epithelium, the ALDH3a1 knockout mouse contains no detectable defects in corneal morphology or transparency.
MATERIALS AND METHODS
Generation of the ALDH3a1 targeting vector.
A 16.3-kb region of the ALDH3a1 gene was obtained from a λFIX II 129/Sv mouse genomic library (Stratagene) and subcloned into pBluescript SK(−) [pBS SK(−)] (Stratagene). Cloning of the targeting vector proceeded in the following order. The 859-bp BamHI-ApaI fragment from intron 1 to exon 2 was inserted into pBS SK(−). The PstI-ApaI region in this clone was amplified by PCR with mutations contained on one primer, and the restriction-digested PCR fragment was inserted. Mutations included SgfI and XhoI sites before the ApaI site and the stop codon TAA after the ALDH3a1 initiator methionine codon. A eukaryotic transcriptional terminator (29), the tryptophan attenuator region, was amplified from Escherichia coli DNA with SgfI and XhoI sites included at the primer termini, and the restriction-digested PCR fragment was inserted. A 3.5-kb ApaI-KpnI fragment spanning from exon 2 to intron 6 was inserted. The 1.6-kb pGKneo cassette from the ClaI-XhoI sites was used to replace the 1.2-kb region spanning from within exons 2 to 3. A 4.6-kb ALDH3a1 gene fragment from the upstream SwaI site to the BamHI site within intron I was inserted (pALDH3a1-KO [see Fig. 1A]). Each cloning step was verified by sequencing insert junctions and the entire regions originating from PCR.
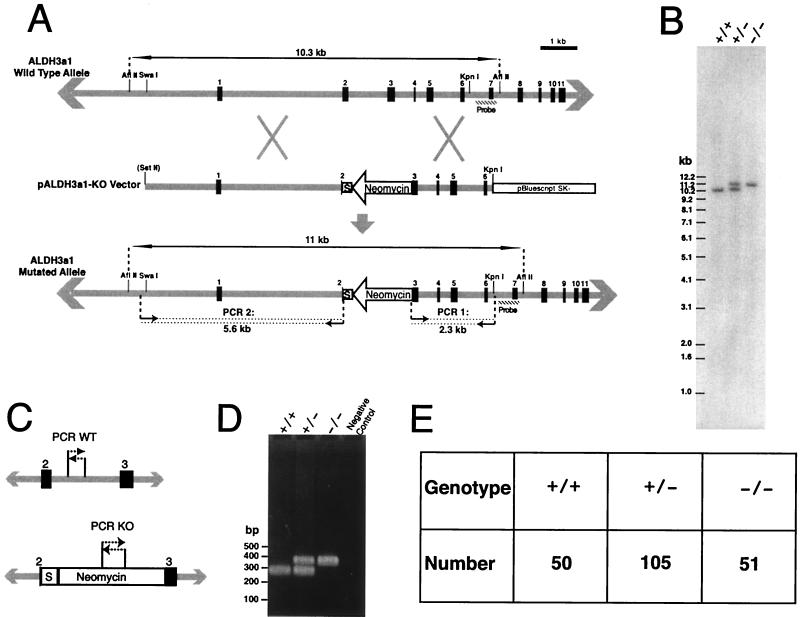
FIG. 1.
Generation of ALDH3a1-deficient mice. (A) The mouse ALDH3a1 gene locus is represented by gray lines, with exons identified by the number above the black boxes. Restriction maps are shown that were used for cloning, Southern blot analysis, and linearization of the vector at the polylinker. Double-ended arrows delineate the genomic fragments from AflII digests for Southern blot analysis; the hatched boxes indicate regions used as probes. The SstII site is in parentheses to indicate that it was present only in the polylinker of pBS SK(−). The small arrows followed by dotted lines delineate amplified regions in PCR analyses with predicted sizes shown below arrows. The neomycin cassette is labeled, and the arrow indicates the direction of its transcription. The box marked S indicates sequences that contained translation termination codons and a putative transcriptional terminator (see the text). (B) Southern blot analysis of wild-type (+/+), heterozygote (+/−), and homozygote (−/−) animals for the ALDH3a1 allele. For wild-type and homozygote lanes, 1 μg of DNA was loaded, and for the heterzygote lane, 2 μg of DNA was loaded. (C) Regions for PCR analysis of wild-type and knockout ALDH3a1 alleles in progeny from matings between heterozygotes. (D) Results of PCR analyses for wild-type (+/+), heterozygote (+/−), and knockout (−/−) ALDH3a1 alleles in mice. (E) Distribution of ALDH3a1 genotypes for 206 progeny from matings of ALDH3a1 heterozygotes.
Generation of ALDH3a1-deficient mice.
The embryonic cell line J1 was maintained in the presence of leukemia inhibitor factor at all times (22). A total of 107 cells were electroporated with 17 μg of SstII-linearized targeting vector (5 nM) in 400 μl of 1× Hanks’ balanced salt solution (Gibco BRL 14185-052) supplemented with 10 mM HEPES (pH 7.0) by using a Bio-Rad Gene Pulser set to 240 V and 500 μF with a 0.2-cm electrode gap. The cell clones were selected on medium supplemented with 200 μg of G418 per ml and then expanded for cryopreservation and DNA isolation. DNA from clonal cell lines was isolated with DNAzol (Gibco BRL) and used in two PCR screens and a Southern blot analysis to assay for proper gene targeting (see Fig. 1A).
Chimeric embryos were made by injecting ALDH3a1+/− cells into C57BL/6 blastocysts for implantation into pseudopregnant CD1 female mice. Chimeric male mice were mated with 129/SvEvTac females, and tail DNA from offspring was tested for the presence of the knockout allele by PCR (see Fig. 1C and D) (6). ALDH3a1 gene targeting was confirmed in the ALDH3a1−/− mice by Southern blot analysis (see Fig. 1A and B).
SDS-PAGE, immunoblotting, and zymography.
Corneal epithelial sheets were obtained by treatment with EDTA (42). The two epithelial sheets from individual mice were extracted by five freeze-thaw cycles and disruption with a mini-homogenizer in 50 μl of 10% glycerol-63 mM Tris-HCl (pH 7.5)-1.4 mM β-mercaptoethanol solution with proteinase inhibitors (Mini complete; Boehinger Mannheim). Cell debris was removed by centrifugation at 14,000 × g for 5 min at 4°C, and supernatant was collected as the water-soluble fraction.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10% Bis-Tris gels and morpholinepropanesulfonic acid (MOPS) running buffer and transfers to membranes were performed as described by the manufacturer (Novex, Invitrogen). Except where noted, 10 μg of protein was loaded per lane. Gels were stained with SyproRed and membranes for immunoblots were stained with SyproRuby (Molecular Probes), and their digital images were obtained under UV light with a Chemilmager 4000 (Alpha Innotech Corp.). Band signals were quantitated with the ChemiImager software (Alpha Innotech Corp.).
For immunoblotting, membranes blocked with a 5% solution of dry milk were incubated with polyclonal rabbit anti-rat ALDH3a1 or anti-human ALDH1 antibodies (kind gifts of R. Lindahl, Department of Biochemistry, University of South Dakota, Vermillion, SD) at dilutions of 1:7,000 and 1:10,000, respectively (12, 18). Secondary-antibody incubations with horseradish peroxidase-linked donkey anti-rabbit immunoglobulin (Amersham Pharmacia Biotech) were performed at a 1:3,000 dilution of antibody. Detection was performed by enhanced chemiluminescence as described by the manufacturer (Amersham Pharamcia Biotech).
Zymograms used the ALDH3a1-specific substrates NADP+ and benzaldehyde (14). Protein samples were not heated in SDS-PAGE loading buffer to avoid a small loss of activity noted in our experiments. Gels were washed four times for 20 min each in 10 mM phosphate buffer (pH 7.0)-1 mM dithiothreitol-2.5% Triton X-100 and then twice for 20 min in 10 mM phosphate buffer. ALDH3a1 bands were developed for 15 min at 37°C with 10 ml of a 10 mM phosphate buffer (pH 7.0) solution containing 20 mg NADP+, 8 mg of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, 0.4 mg of phenazine methosulfate, and 20 μl of benzaldehyde. The reaction was stopped by washing the gel with double-dionized water.
Determinations of water-soluble protein content in the corneal epithelium.
ALDH3a1−/− and wild-type 129/SvEvTac mice, matched in age to within 1 week, were used for each set of analyses. Their ages varied from 5 to 8 weeks. The corneal epithelial sheets were removed from three mice, and each was washed gently in 1× phosphate-buffered saline before extraction. Extractions were maintained separately for each mouse and were conducted by a freeze-thaw process as described above, except that the protease inhibitors were excluded from the extraction buffer. The nuclear pellets generated after centrifugation of the extracts were used to determine DNA contents with the fluorescent dye picogreen (5). Protein determinations were made using the fluorescent dye NanoOrange (Molecular Probes).
Microscopic analyses.
Mice were euthanized with CO2, and their eyes were surgically excised, fixed overnight in neutral fresh 4% paraformaldehyde, placed into phosphate-buffered saline, and embedded into glycol methacrylate. Cross sections (2 μm thick) were cut and stained with hematoxylin and eosin by standard methods.
For electron microscopy, the eyes were surgically excised and fixed at room temperature in 2.5% glutaraldehyde-6% sucrose in sodium cacodylate buffer (pH 7.2) for 24 h or more. Portions of corneas (0.5 by 1.0 mm) were processed for electron microscopy and embedded in expoxy resin. Micrographs of uranyl acetate- and lead citrate-stained ultrathin sections were obtained with a JEM-100CX electron microscope (JEOL USA, Inc.).
Excised eyes were analyzed for transparency under a Zeiss model Stemi 2000-C dissecting microscope. Images were obtained with an RT color SPOT digital camera (Diagnostic Instruments, Inc). Slit lamp biomicroscopy was performed with a HAI SL-5000 slit lamp microscope (Hightech American Industrial Laboratories, Inc.). For this, live mice were anesthetized by intraperitoneal injection with 2.5% Avertin at 0.06 to 0.01 ml per g of body weight (30).
RESULTS
ALDH3a1−/− mice.
The ALDH3a1 gene was targeted in embryonic J1 stem cells with a construct engineered with multiple disruptive sequences (Fig. 1A). The region spanning from within exon 2 through part of exon 3 was replaced with the neomycin resistance cassette in the reverse transcriptional orientation. Also, an in-frame TAA stop codon was inserted immediately after the initiator methionine that is located in exon 2. Additionally, a putative eukaryotic transcriptional terminator, the E. coli tryptophan attenuator (29), which also contains stop codons in all three reading frames, was inserted in the region between the inserted stop codon and the neomycin gene (S in Fig. 1A). Homologous recombination of the knockout vector with the mouse genome was shown by a PCR assay for J1 cell lines and by Southern blot analysis for cell lines and mice (Fig. 1A and B). To assess the viability of ALDH3a1−/− mice, the genotypes were tracked for the first 206 progeny from matings of ALDH3a1+/− mice. A tabulation of the numbers of knockout, heterozygous, and wild-type progeny revealed a nearly perfect 1:2:1 Mendelian distribution of mice as scored by a multiplex PCR assay (Fig. 1C to E). Homozygotes appear normal and have lived as long as 2 years.
Analysis of protein expression in corneal epithelia.
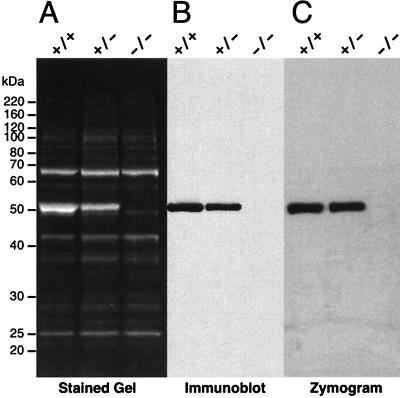
Water-soluble proteins were extracted from corneal epithelia of ALDH3a1−/−, ALDH3a1+/−, and wild-type siblings produced from a mating between ALDH3a1+/− mice, and the protein samples were subjected to SDS-PAGE. ALDH3a1 (51 kDa) was missing from the knockout mice and was approximately 50% less abundant in the heterozygous mice than in the wild-type mice (Fig. 2A). In contrast, the other proteins, including the abundant transketolase (38), remained unchanged in their expression among wild-type, heterozygous, and knockout mice (Fig. 2A). The same results were apparent when a 10 to 20% polyacrylamide SDS-Tricine gel was used to resolve smaller protein species (data not shown) or when mice ranging in age from 9 to 57 weeks were used. Analyses of band signal intensities indicated that the ALDH3a1 represented 44% of the total SyproRed-stained protein in samples from wild-type mice after SDS-PAGE.
FIG. 2.
ALDH3a1 expression. (A) SDS-PAGE (10% polyacrylamide) of 10 μg of water-soluble protein from the corneal epithelium of ALDH3a1 wild-type (+/+), heterozygote (+/−), and knockout (−/−) mice. The size and position of markers are shown on the left. The major 68-kDa band is transketolase (38), and the major 51-kDa band is ALDH3a1. (B) Immunoblot probed with anti-ALDH3 antibody. SDS-PAGE was performed as in panel A, except that 1 μg of protein was loaded in each lane. (C) Zymogram for ALDH3a1 activity using benzaldehyde and NADP+ as substrates. SDS-PAGE was performed as in Panel A.
Further experiments verified the loss of ALDH3a1 and its activity. Immunoblot analysis using a rabbit anti-rat ALDH3a1 antibody confirmed that the corneal epithelium of knockout mice lacked ALDH3a1 (Fig. 2B). Zymography was used to test for ALDH3a1 enzymatic activity. Following SDS-PAGE, the gels were incubated in several changes of phosphate buffer to remove SDS and allow recovery of the protein’s native state. The gels were developed with the ALDH3a1 preferred substrates benzaldehyde and NADP+, and the reduction of NADP+ was colorimetrically visualized. The zymograms revealed ALDH3a1 activity at 51 kDa in lanes containing corneal epithelial extracts from wild-type and heterozygous mice but not in lanes containing extracts from ALDH3a1−/− mice (Fig. 2C). As a control, when the ALDH3a1-specific substrate NADP+ was not included, no colormetric development was observed in any lane (data not shown).
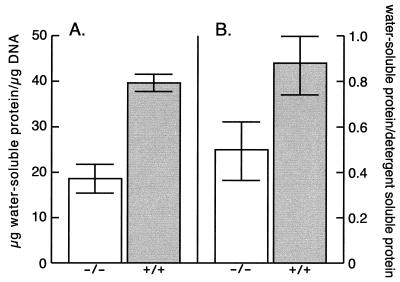
We decided to investigate further whether there is a compensating increase in the amounts of water-soluble proteins in the corneas of knockout mice. To quantify the water-soluble protein content against an internal cellular component, the water-soluble protein content per microgram of DNA was determined for isolated corneal epithelia from three wild-type and ALDH3a1−/− mice. These analyses revealed that the ALDH3a1−/− mice contained 53% less soluble protein in the corneal epithelium than did the wild-type 129/Sv mice (Fig. 3). In a similar type of experiment, the water-soluble protein content was normalized against the insoluble protein content. This analysis revealed a loss of 43% of the soluble protein in the ALDH3a1−/− mice (Fig. 3). These decreases in total protein content are in agreement with the selective loss of ALDH3a1 as judged from its band signal intensity in water-soluble protein extracts of the wild-type cornea after SDS-PAGE given above.
FIG. 3.
Water-soluble protein content in corneal epithelial cells. (A) The water-soluble protein was normalized to the DNA content of corneal epithelial cells from ALDH3a1 knockout (−/−) and wild-type (+/+) mice. (B) The corneal epithelial water-soluble protein was normalized against detergent-soluble protein from ALDH3a1 knockout (−/−) and wild-type (+/+) mice.
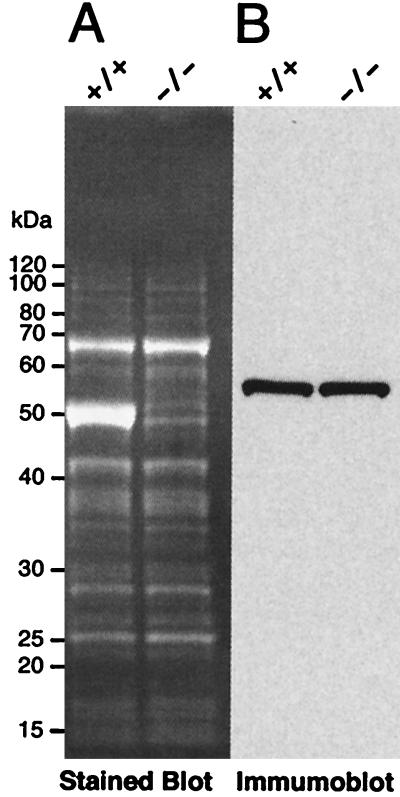
Although ALDH3a1 is the most highly expressed soluble protein in the mouse cornea, rabbit corneas instead express ALDH1 at high levels (18). Since there is some overlap in substrate specificity of ALDH1 and ALDH3a1 (23), soluble extracts from the corneal epithelia of ALDH3a1 knockout and 129/Sv wild-type mice were compared by immunoblot analysis performed with a rabbit anti-rat ALDH1 antibody. An approximately 55-kDa protein did show antigenic response to this antibody; however, no difference in its expression was detected between ALDH3a1 knockout and 129/Sv wild-type mice (Fig. 4). Taken together, our data indicate that there has been no compensation for the loss of water-soluble protein or ALDH3a1 activity by additional gene expression in the ALDH3a1 null mice.
FIG. 4.
Immunoblot analysis to test for compensation by ALDH1. (A) For SDS-PAGE, 10 μg of water-soluble protein from wild-type (+/+) and knockout (−/−) corneal epithelia were loaded onto each lane; after SDS-PAGE, the blots were stained with SyproRuby to reveal proteins. (B) Immunoblot probed with anti-ALDH1 antibody.
Analysis of ALDH3a1−/− cornea by light and electron microscopy.
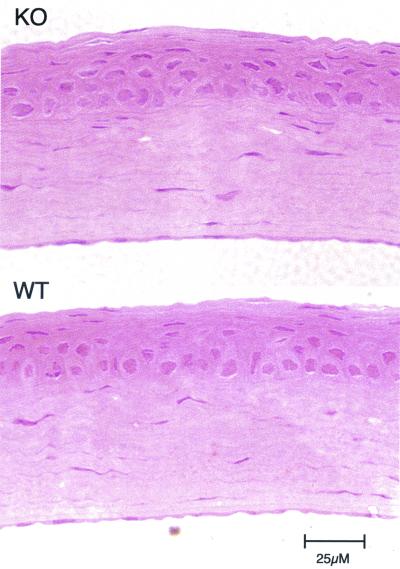
Histology and electron microscopy were performed to compare cell and tissue morphology of the corneas of ALDH3a1 knockout and wild-type mice. Corneas from wild-type, ALDH3a1+/− and ALDH3a1−/− mice appeared identical when fixed, sectioned, and stained with hematoxylin and eosin (Fig. 5). Similarly, the ultrastructures of ALDH3a1−/− corneas were indistinguishable from those of wild-type corneas (Fig. 6). Cell nuclei were absent from most of the superficial squamous epithelial cells but were present in the basal epithelial cells in the corneas of both wild-type and ALDH3a1 knockout mice. Also, the keratocytes that intercalate the stroma appeared normal, as did the corneal basal lamina, hemidesmosomes, and Descemet’s membrane of the ALDH3a1−/− mice.
FIG. 5.
Hematoxylin-and-eosin-stained cross sections of cornea from a 42-day-old ALDH3a1 knockout mouse (KO) and a 129/Sv wild-type mouse (WT) showing normal tissue stratification. Micrographs reveal intact epithelial, stromal, and endothelial layers in both corneas.
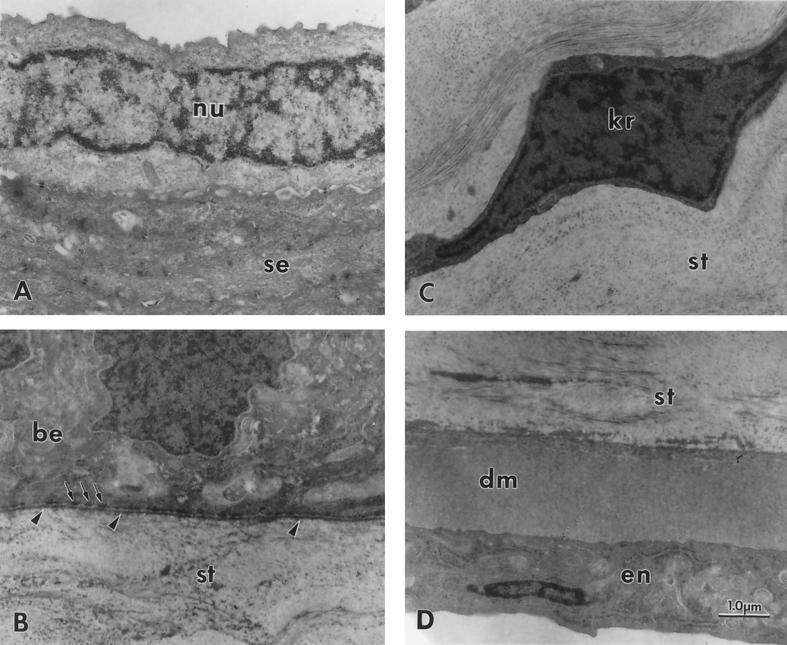
FIG. 6.
Transmission electron microscopy of a transected cornea from a 51-day-old ALDH3a1 knockout mouse showing normal structure. (A) Superficial epithelium consisting mainly of nucleus-free squamous epithelium (se) but occasionally showing a nucleated cell (nu); (B) basal portion of the basal epithelum (be) and adjacent anterior stroma (st) with normal structures for a mouse, including basal lamina (arrowheads) and adjacent hemidesmosomes (arrows), but no Bowman’s membrane; (C) normal keratocyte (kr) in the central stroma (st); (D) normal-appearing posterior stroma (st). Descemet’s membrane (dm), and endothelium (en). The corneas of heterozygote and homozygote wild-type mice were similarly normal in structure. Magnification, ×15,000. Calibration bar, 1.0 μm for all micrographs.
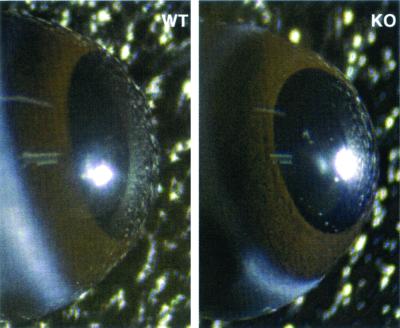
To observe the transparency of the corneas, mice were euthanized, the eyes were surgically removed, and images were obtained through a dissecting microscope. Due to the normal corneal transparency, the main structures observable were the brown iris beneath the cornea, the central black pupil, and the lens (Fig. 7). Corneas of knockout mice appeared clear and unaffected by the loss of ALDH3a1. In addition, slit lamp biomicroscopy revealed transparent corneas in vivo in both knockout and wild-type mice (data not shown).
FIG. 7.
Wild-type (WT) and ALDH3a1 knockout (KO) eyes are shown. The eyes were imaged immediately after dissection. The brown iris is observable underneath the cornea of both eyes. Each lens is observable through the pupil.
DISCUSSION
The present investigation shows that complete elimination of ALDH3a1 by targeted disruption in mice has no obvious effect on animal viability or corneal phenotype, even though enzymatically active ALDH3a1 comprises approximately 40 to 50% of the water-soluble protein of the normal wild-type mouse corneal epithelium. The absence of an obvious phenotype in the ALDH3a1 null mice raises the possibility that a compensatory mechanism exists to offset the loss of ALDH3a1. However, our SDS-PAGE analyses, immunoblots, and total water-soluble protein concentration measurements in the cornea revealed that the ALDH3a1 null mouse contains no elevated protein mass or ALDH enzymatic activity to compensate for the loss of ALDH3a1. Examination of histological sections also failed to show any difference in the number or size of corneal epithelial cells between the ALDH3a1−/− and wild-type mice that might indicate a decrease in cell volume that would compensate for the loss of protein mass in the ALDH3a1−/− mouse. Indeed, it is remarkable that the corneal epithelial cells of these mice have approximately 50% less water-soluble protein than do normal wild-type cells without any obvious differences in either clarity or structure.
Our results are consistent with earlier investigations showing a transparent cornea in the SWR/J mouse strain (11). This strain expresses normal ALDH3a1 transcript levels but carries four amino acid mutations (39) that lead to a decreased amount of ALDH3a1 in the SWR/J mouse cornea (11, 31). The literature contains conflicting reports on the corneal content of ALDH3a1 in the SWR/J mouse (11, 31). Our immunoblots reveal a 51-kDa species in corneal extracts from SWR/J corneas that cross-reacts with the rabbit anti-rat ALDH3a1 antibody. Furthermore, the signal intensity of the 51-kDa band in SDS-PAGE analysis indicates that the ALDH3a1 content is approximately 7% of that in the wild-type 129/Sv mouse cornea (unpublished data) The present experiments show that ALDH3a1 is not necessary for corneal transparency, at least in the laboratory setting, thus extending the previous observations on the SWR/J mouse that has lower than wild-type levels of ALDH3a1. Our results also show that ALDH3a1 is unnecessary for the development and maintenance of corneal structure and cell morphology. The SWR/J mouse cornea is more susceptible to hazing than is the wild-type mouse cornea when subjected to significant doses of UV radiation (11). The nature of the corneal damage, and the cellular mechanisms underlying this event, have not been explored.
Corneal ALDH3a1 might have multiple functions in a phenomenon called gene sharing, which is common in lens crystallins (33). For example, the water-soluble proteins of the cornea contribute to its UV light-filtering capacity (27). Thus, corneal ALDH3a1 may have a significant function in directly absorbing UV radiation in addition to its enzymatic detoxifying role on UV lipid peroxidation products (1). In addition, several functions for corneal ALDH3a1 analogous to those of lens crystallins have not been ruled out. Crystallins modify the lens refractive index (44). Also, it has been suggested that ALDH3a1 may play a role in thiol regulation (46), a function in which crystallins have been implicated (19, 26). α-Crystallins have an additional chaperone role to prevent the aggregation of partially denatured proteins (17, 37), and, indeed, a chaperone function has been postulated for ALDH3a1 in the cornea (47). It is interesting that, similar to our present findings, knockout of the αB-crystalin gene in mouse does not alter the phenotype of the lens, its site of highest expression (7).
In addition to the situation with ALDH3a1 in the mouse, the corneas of other species contain water-soluble proteins whose high proportion remains unexplained (32). This has been compared to the taxon-specific accumulation of lens crystallins and has been the basis for speculating that these abundant corneal proteins are similar to lens crystallins (8, 9, 32, 33). As in the lens, the cornea accumulates certain enzymes, often but not always related to detoxification, as well as other proteins. An example of the latter is gelsolin in zebrafish cornea (49). Accumulation of ALDH in mammalian corneas is of interest, since η-crystallin of the elephant shrew lens (15) and Ω-crystallin of the cephalopod (28, 45, 50) and scallop (36) lenses are also members of the ALDH family. Further experiments are required to determine the roles for the different, abundant, water-soluble proteins in the cornea, including ALDH3a1 in the mouse and other mammals.
Acknowledgments
We thank Dionne Davis and R. Steven Lee for their helpful technical assistance.
REFERENCES
- 1.Abedinia, M., T. Pain, E. M. Algar, and R. S. Holmes. 1990. Bovine corneal aldehyde dehydrogenase: the major soluble corneal protein with a possible dual protective role for the eye. Exp. Eye Res. 51:419–426. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, R. J., B. Silverman, and W. L. Henley. 1981. Isolation and characterization of BCP 54, the major soluble protein of bovine cornea. Exp. Eye Res. 32:205–216. [DOI] [PubMed] [Google Scholar]
- 3.Algar, E. M., M. Abedinia, J. L. VandeBerg, and R. S. Holmes. 1991. Purification and properties of baboon corneal aldehyde dehydrogenase: proposed UVR protective role. Adv. Exp. Med. Biol. 284:53–60. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim, F. A., and E. L. Siew. 1983. Effect of change in concentration upon lens turbidity as predicted by the random fluctuation theory. Biophys. J. 41:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaheta, R. A., B. Kronenberger, D. Woitaschek, S. Weber, M. Scholz, H. Schuldes, A. Encke, and B. H. Markus. 1998. Development of an ultrasensitive in vitro assay to monitor growth of primary cell cultures with reduced mitotic activity. J. Immunol. Methods 211:159–169. [DOI] [PubMed] [Google Scholar]
- 6.Blin, N., and D. W. Stafford. 1976. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 3:2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady, J. P., D. Garland, D. E. Green, E. Tamm, F. J. Giblin, and E. Wawrousek. 2001. αB-crystallin in lens development and muscle integrity: a gene knockout approach. Investig. Ophthalmol. Visual Sci. 42:2924–2934. [PubMed] [Google Scholar]
- 8.Cooper, D. L., N. R. Isola, K. Stevenson, and E. W. Baptist. 1993. Members of the ALDH gene family are lens and corneal crystallins. Adv. Exp. Med. Biol. 328:169–179. [DOI] [PubMed] [Google Scholar]
- 9.Cuthbertson, R. A., S. I. Tomarev, and J. Piatigorsky. 1992. Taxon-specific recruitment of enzymes as major soluble proteins in the corneal epithelium of three mammals, chicken, and squid. Proc. Natl. Acad. Sci. USA. 89:4004–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaye, M., and A. Tardieu. 1983. Short-range order of crystallin proteins accounts for eye lens transparency. Nature 302:415–417. [DOI] [PubMed] [Google Scholar]
- 11.Downes, J. E., P. G. Swann, and R. S. Holmes. 1994. Differential corneal sensitivity to ultraviolet light among inbred strains of mice. Correlation of ultraviolet B sensitivity with aldehyde dehydrogenase deficiency. Cornea 13:67–72. [DOI] [PubMed] [Google Scholar]
- 12.Evces, S., and R. Lindahl. 1989. Characterization of rat cornea aldehyde dehydrogenase. Arch. Biochem. Biophys. 274:518–524. [DOI] [PubMed] [Google Scholar]
- 13.Feeney, L., and E. R. Berman. 1976. Oxygen toxicity: membrane damage by free radicals. Investig. Ophthalmol. 15:789–792. [PubMed] [Google Scholar]
- 14.Gondhowiardjo, T. D., N. J. van Haeringen, R. Hoekzema, L. Pels, and A. Kijlstra. 1991. Detection of aldehyde dehydrogenase activity in human corneal extracts. Curr. Eye Res. 10:1001–1007. [DOI] [PubMed] [Google Scholar]
- 15.Graham, C., J. Hodin, and G. Wistow. 1996. A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of η-crystallin. J. Biol. Chem. 271:15623–15628. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, R. S., and J. L. VandeBerg. 1986. Ocular NAD-dependent alcohol dehydrogenase and aldehyde dehydrogenase in the baboon. Exp. Eye Res. 43:383–396. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz, J. 2000. The function of α-crystallin in vision. Semin. Cell Dev. Biol. 11:53–60. [DOI] [PubMed] [Google Scholar]
- 18.Jester, J. V., T. Moller-Pedersen, J. Huang, C. M. Sax, W. T. Kays, H. D. Cavangh, W. M. Petroll, and J. Piatigorsky. 1999. The cellular basis of corneal transparency: evidence for ‘corneal crystallins’. J. Cell Sci. 112:613–622. [DOI] [PubMed] [Google Scholar]
- 19.Kannan, R., B. Ouyang, E. Wawrousek, N. Kaplowitz, and U. P. Andley. 2001. Regulation of GSH in αA-expressing human lens epithelial cell lines and in αA knockout mouse lenses. Investig. Ophthalmol Visual Sci. 42:409–416. [PubMed] [Google Scholar]
- 20.Kays, W. T., and J. Piatigorsky. 1997. Aldehyde dehydrogenase class 3 expression: identification of a cornea-preferred gene promoter in transgenic mice. Proc. Natl. Acad. Sci. USA. 94:13594–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konishi, Y., and Y. Mimura. 1992. Kinetic properties of the bovine corneal aldehyde dehydrogenase (BCP 54). Exp. Eye Res. 55:569–578. [DOI] [PubMed] [Google Scholar]
- 22.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915–926. [DOI] [PubMed] [Google Scholar]
- 23.Lindahl, R. 1992. Aldehyde dehydrogenases and their role in carcinogenesis. Crit. Rev. Biochem. Mol. Biol. 27:283–335. [DOI] [PubMed] [Google Scholar]
- 24.Lindahl, R., and S. Evces. 1984. Rat liver aldehyde dehydrogenase. I. Isolation and characterization of four high Km normal liver isozymes. J. Biol. Chem. 259:11986–11990. [PubMed] [Google Scholar]
- 25.Lindahl, R., and D. R. Petersen. 1991. Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem. Pharmacol. 41:1583–1587. [DOI] [PubMed] [Google Scholar]
- 26.Lou, M. F. 2000. Thiol regulation in the lens. J. Ocul. Pharmacol. Ther. 16:137–148. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, J., and R. J. Cenedella. 1995. Quantitation of ultraviolet light-absorbing fractions of the cornea. Cornea 14:266–272. [DOI] [PubMed] [Google Scholar]
- 28.Montgomery, M. K., and M. J. McFall-Ngai. 1992. The muscle-derived lens of a squid bioluminescent organ is biochemically convergent with the ocular lens. Evidence for recruitment of aldehyde dehydrogenase as a predominant structural protein. J. Biol Chem. 267:20999–21003. [PubMed] [Google Scholar]
- 29.Nakagoshi, H., Y. Ueno, and F. Imamoto. 1991. Effects of prokaryotic termination signals on RNA polymerase II transcription in HeLa cells. J. Biochem. (Tokyo). 110:159–162. [DOI] [PubMed] [Google Scholar]
- 30.Papaioannou, V. E., and J. G. Fox. 1993. Efficacy of tribromoethanol anesthesia in mice. Lab. Anim. Sci. 43:189–192. [PubMed] [Google Scholar]
- 31.Pappa, A., N. A. Sophos, and V. Vasiliou. 2001. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem. Biol. Interact. 130–132:181–191. [DOI] [PubMed] [Google Scholar]
- 32.Piatigorsky, J. 2001. Enigma of the abundant water-soluble cytoplasmic proteins of the cornea: the “refraction” hypothesis. Cornea 32:853–858. [DOI] [PubMed] [Google Scholar]
- 33.Piatigorsky, J. 1998. Gene sharing in lens and cornea: facts and implications. Prog. Retin. Eye Res. 17:145–174. [DOI] [PubMed] [Google Scholar]
- 34.Piatigorsky, J. 1998. Multifunctional lens crystallins and corneal enzymes. More than meets the eye. Ann. N. Y. Acad. Sci. 842:7–15. [DOI] [PubMed] [Google Scholar]
- 35.Piatigorsky, J. 2000. Review: a case for corneal crystallins. J. Ocul. Pharmacol. Ther. 16:173–180. [DOI] [PubMed] [Google Scholar]
- 36.Piatigorsky, J., Z. Kozmik, J. Horwitz, L. Ding, E. Carosa, W. G. Robison, Jr., P. J. Steinbach, and E. R. Tamm. 2000. Ω-Crystallin of the scallop lens. A dimeric aldehyde dehydrogenase class 1/2 enzyme-crystallin. J. Biol. Chem. 275:41064–41073. [DOI] [PubMed] [Google Scholar]
- 37.Rao, P. V., Q. L. Huang, J. Horwitz, and J. S. Zigler, Jr. 1995. Evidence that α-crystallin prevents non-specific protein aggregation in the intact eye lens. Biochim. Biophys. Acta 1245:439–447. [DOI] [PubMed] [Google Scholar]
- 38.Sax, C. M., C. Salamon, W. T. Kays, J. Guo, F. X. Yu, R. A. Cuthbertson, and J. Piatigorsky. 1996. Transketolase is a major protein in the mouse cornea. J. Biol. Chem. 271:33568–33574. [DOI] [PubMed] [Google Scholar]
- 39.Shiao, T., P. Tran, D. Siegel, J. Lee, and V. Vasiliou. 1999. Four amino acid changes are associated with the Aldh3a1 locus polymorphism in mice which may be responsible for corneal sensitivity to ultraviolet light. Pharmacogenetics 9:145–153. [PubMed] [Google Scholar]
- 40.Silverman, B., R. J. Alexander, and W. L. Henley. 1981. Tissue and species specificity of BCP 54, the major soluble protein of bovine cornea. Exp. Eye Res. 33:19–29. [DOI] [PubMed] [Google Scholar]
- 41.Sophos, N. A., A. Pappa, T. L. Ziegler, and V. Vasiliou. 2001. Aldehyde dehydrogenase gene superfamily: the 2000 update. Chem. Biol. Interact. 130–132:323–337. [DOI] [PubMed] [Google Scholar]
- 42.Spurr, S. J., and I. K. Gipson. 1985. Isolation of corneal epithelium with Dispase II or EDTA. Effects on the basement membrane zone. Investig. Ophthalmol. Visual Sci. 26:818–827. [PubMed] [Google Scholar]
- 43.Sun, L., T.-T. Sun, and R. M. Lavker. 1999. Identification of a cytosolic NADP+-dependent isocitrate dehydrogenase that is preferentially expressed in bovine corneal epithelium. A corneal epithelial crystallin. J. Biol. Chem. 274:17334–17341. [DOI] [PubMed] [Google Scholar]
- 44.Tardieu, A. 1998. α-Crystallin quaternary structure and interactive properties control eye lens transparency. Int. J. Biol. Macromol. 22:211–217. [DOI] [PubMed] [Google Scholar]
- 45.Tomarev, S. I., R. D. Zinovieva, and J. Piatigorsky. 1991. Crystallins of the octopus lens. Recruitment from detoxification enzymes. J. Biol. Chem. 266:24226–24231. [PubMed] [Google Scholar]
- 46.Uma, L., J. Hariharan, Y. Sharma, and D. Balasubramanian. 1996. Corneal aldehyde dehydrogenase displays antioxidant properties. Exp. Eye Res. 63:117–120. [DOI] [PubMed] [Google Scholar]
- 47.Uma, L., J. Hariharan, Y. Sharma, and D. Balasubramanian. 1996. Effect of UVB radiation on corneal aldehyde dehydrogenase. Curr. Eye Res. 15:685–690. [DOI] [PubMed] [Google Scholar]
- 48.Vasiliou, V., A. Bairoch, K. F. Tipton, and D. W. Nebert. 1999. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 9:421–434. [PubMed] [Google Scholar]
- 49.Xu, Y. S., M. Kantorow, J. Davis, and J. Piatigorsky. 2000. Evidence for gelsolin as a corneal crystallin in zebrafish. J. Biol. Chem. 275:24645–24652. [DOI] [PubMed] [Google Scholar]
- 50.Zinovieva, R. D., S. I. Tomarev, and J. Piatigorsky. 1993. Aldehyde dehydrogenase-derived Ω-crystallins of squid and octopus. Specialization for lens expression. J. Biol. Chem. 268:11449–11455. [PubMed] [Google Scholar]