Abstract
A new model for grass functional genomics is described based on Brachypodium distachyon, which in the evolution of the Pooideae diverged just prior to the clade of “core pooid” genera that contain the majority of important temperate cereals and forage grasses. Diploid ecotypes of B. distachyon (2n = 10) have five easily distinguishable chromosomes that display high levels of chiasma formation at meiosis. The B. distachyon nuclear genome was indistinguishable in size from that of Arabidopsis, making it the simplest genome described in grasses to date. B. distachyon is a self-fertile, inbreeding annual with a life cycle of less than 4 months. These features, coupled with its small size (approximately 20 cm at maturity), lack of seed-head shatter, and undemanding growth requirements should make it amenable to high-throughput genetics and mutant screens. Immature embryos exhibited a high capacity for plant regeneration via somatic embryogenesis. Regenerated plants display very low levels of albinism and have normal fertility. A simple transformation system has been developed based on microprojectile bombardment of embryogenic callus and hygromycin selection. Selected B. distachyon ecotypes were resistant to all tested cereal-adapted Blumeria graminis species and cereal brown rusts (Puccinia reconditia). In contrast, different ecotypes displayed resistance or disease symptoms following challenge with the rice blast pathogen (Magnaporthe grisea) and wheat/barley yellow stripe rusts (Puccinia striformis). Despite its small stature, B. distachyon has large seeds that should prove useful for studies on grain filling. Such biological characteristics represent important traits for study in temperate cereals.
The past two decades have witnessed an explosion in the use of model eukaryotic organisms to aid studies on species of significant commercial or biological interest. Historically, specific eukaryotes (e.g. Saccharomyces cerevisiae and Caenorhabditis elegans) have attained the status of “models” because they reflect the individual characteristics of species of medical, industrial, or agricultural interest and are often small, easy to work with in large numbers, and cheap to maintain. More recently, the power of model species has been augmented by the development of whole genome sequencing programs. The new field of “functional genomics” provides further challenges for model organisms in the drive to understand the function of each gene in any genome. This requires a plethora of tools to allow an integrative examination of complex biological problems and relate phenotype to genotype. There are two distinct categories of technology that need to be in place to exploit fully any proposed model organism in a functional genomics program (Table I). The technology associated with “physical genomics” is often standardized, generally organism independent, and can be developed theoretically for any species, given sufficient investment of time and resources. In contrast, the “biological genomics” capability associated with any particular organism is species dependent reflecting genome size, organization and degree of redundancy, breeding behavior, and in many cases, the development of gene transfer procedures and mutagenesis strategies (e.g. transposon tagging and gene trapping technology). Pragmatically, the utility of a model organism is dependent also on its possession of a range of biological features, for instance, small physical size, rapid life cycle, and undemanding growth requirements that make it amenable to high-throughput screening routines. Such characteristics allow easy adoption by many laboratories internationally, which accelerates the process of discovery and the development of bioinformatic tools (e.g. sequence databases) that are essential to the success of a functional genomic program.
Table I.
Model species requirements for functional genomics analysis
| Physical Genomicsa | Biological Genomicsb |
|---|---|
| Large insert libraries | Small, diploid genome |
| Genome sequencing | Tractable genetics and mapping strategies |
| Expressed sequence tag databases | Mutagenesis techniques |
| Transcriptome, proteome, and metabolome data sets | Easily transformable and low-cost logistics for growth and maintenance |
| Bioinformatics tools | Exhibition of important biological traits |
Technological infrastructure necessary to employ multidimensional analysis of a biological facet of the model species.
Biological attributes of a species that are required to be a valuable model species.
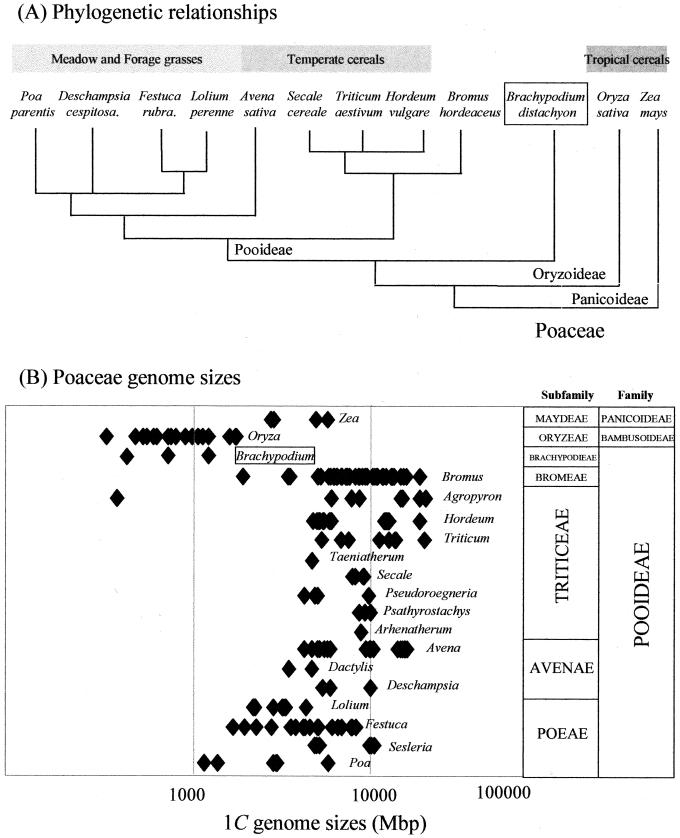
All these features are exemplified in what is without doubt the most highly developed plant model system, the dicotyledon Arabidopsis (Meyerowitz and Sommerville, 1994; Meinke et al., 1998). This species is a small crucifer that is an inbreeding annual with a rapid life cycle (Redei, 1970). The Arabidopsis nuclear genome size has been estimated by flow cytometry to be 164 Mbp (Bennett et al., 2000), whereas a recent calculation following completion of the whole genome sequence (The Arabidopsis Genome Initiative, 2000) has indicated a figure as low as a 125 Mbp. However, phylogenetically, Arabidopsis is only distantly related to the Poaceae, which includes all of the world's major cereals crops and forage grasses (Keller and Feuillet, 2000). Hopes that Arabidopsis could serve as an “anchor” genome to help locate important chromosomal locations in cereal species have not been substantiated by recent studies (Bennetzen et al., 1998; Devos et al., 1999). Thus, it is clear that grass (Poaceae) model systems are a key requirement for the future identification of genes of agronomic interest from cereals and forage grasses.
With its international status as a staple food source and many years of intensive plant breeding, rice (Oryza sativa), with its compact genome (approximately 441 Mbp; Bennett et al., 2000) has been promoted as a model for cereal genomics (for review, see Havukkala, 1996; Goff, 1999). Considerable international effort has developed rice genetic maps, expressed sequence tag programs, and assembled a considerable germplasm collection (highlighted in McCouch, 1998), and the completed rice genomic sequence has just been announced (Dickson and Cyranoski, 2001). Furthermore, the representation of all grass genome sectors by less than 30 linkage blocks (with centromeric sites often defining the breakpoints) within a consensus grass map “anchored” by reference to the rice genome offers the prospect of easing mapping in cereals with larger genomes (Gale and Devos, 1998). However, the value of rice as a model for the temperate cereals and forage grasses in the relatively distantly related Pooideae subfamily (see Fig. 1A) may be, on occasion, questionable. For instance, to effectively use the rice genomic map to isolate a corresponding genic region in the larger genomes of temperate cereals, “microsynteny” will have to be conserved at the size of DNA inserts found in bacterial artificial chromosome (BAC) or yeast artificial chromosome (YAC) clones (approximately 100 kb and 0.5 Mb, respectively). This does not always appear to be the case. For example, although the Sh2 locus appears to be colinear in many cereals (Chen et al., 1997), regions flanking Adh1 locus from maize (Zea mays), sorghum (Sorghum bicolor), and rice were dissimilar (Bennetzen et al., 1998), and molecular analysis has detected multiple small rearrangements (Tikhonov et al., 1999). Also the “physical and biological genomics” infrastructure, though impressive, is incomplete. For instance, large scale, organized, and publicly available insertion mutagenesis resources (Izawa et al., 1997; Enoki et al., 1999) are not yet available, and a routine transgenic capability, even after decades of concerted effort, is restricted to a relatively few laboratories. Furthermore, the simple logistics of handling rice (as a large, outbreeding plant with a long life cycle and demanding growth requirements) will complicate the future development of rice as a model species for high-throughput functional genomics. Finally, and crucially, rice does not necessarily exhibit all the traits that are relevant to study in temperate crops, especially forage grasses. For example, resistance to specific types of pathogens, overwintering and freezing tolerance, vernalization, perenniality (including meristem dormancy mechanisms), wear and injury tolerance (amenity grasses), sward behavior, and post-harvest biochemistry of silage are all important areas of research relevant to temperate grasses, which are rarely studied in rice.
Figure 1.
A, Schematic phylogenetic relationship of B. distachyon to other Poaceae adapted from the data presented by Catalan et al. (1995, 1997; Catalan and Olmstead, 2000). B, Nuclear genome sizes (Mbp/1C) in different grass genera. Data derived from The Kew Gardens Angiosperm C- value database (http://www.rbgkew.org/cval/homepage.html).
Against this background, it would seem appropriate to reexamine the Pooideae to identify a grass species that has the potential to be developed into a useful model representative of important temperate cereals and forage grasses. The Kew Gardens Angiosperm C- value Database (http://www.kew.org/cval/homepage.html) shows that the genus Brachypodium is distinct from other genera in the Pooideae in that all species examined to date have a narrow range of genome sizes with the smallest 1-C values being very similar to that of rice (Fig. 1B). Various molecular phylogenetic analyses have demonstrated that the genus Brachypodium diverged from the ancestral stock of Pooideae immediately prior to the radiation of the modern “core pooids” (Triticeae, Bromeae, Poeae, and Aveneae; see Fig. 1A), which includes the majority of important temperate cereals and forage grasses (Shi, 1991; Shi et al., 1993; Catalan et al., 1995, 1997; Catalan and Olmstead, 2000). Its phylogenetic position and small genome size prompted Moore and colleagues (Aragon-Alcaide et al., 1996) to use Brachypodium sylvaticum (2n = 18) in a search for archetypal grass centromere sequences by screening for repetitive DNA conserved between wheat (Triticum aestivum), maize, and rice and Brachypodium. Brachypodium species have small chromosomes with a variable base number (x = 5, 7, 8, or 9), making them unusual in the Pooideae that tend to have large chromosomes and a base number of 7 (Shi et al., 1993). An analysis of ribosomal DNA structure revealed that Brachypodium had the smallest (150 bp) 5S rDNA spacer region found in the grasses (Shi, 1991; Catalan et al., 1995) and a very simple rDNA repeat unit with a low degree of methylation (Shi et al., 1993). Further characterization revealed that genomes of Brachypodium species contained typically less than 15% highly repeated DNA (Shi, 1991; Catalan et al., 1995). Therefore, we consider that the genus Brachypodium has several attributes in relation to its ancestry and genome characteristics that make it a useful focus for studies relating to the evolution of form and function in the Pooideae. The present paper describes the development of B. distachyon, the only true annual species within the genus Brachypodium, as a potential model species for functional genomics and a representative of the temperate cereals and forage grasses.
RESULTS
Diploid Ecotypes of B. distachyon Have the Smallest Reported Genome Size in the Poaceae
The genome size of several species of Brachypodium was estimated in preliminary studies by scanning densitometry of Feulgen-stained nuclei prepared from root tip squashes (Table II). Representative accessions of each species had a 1C nuclear genome size of less than 0.5 pg, equivalent to ≤410 Mbp. Of particular note was the large discrepancy between the genome sizes of two ecotypes of B. distachyon ABR99 and ABR1 (0.3 pg/1C and 0.15 pg/1C, respectively).
Table II.
Estimation of genome size in Brachypodium species
| Species | Ecotype | 2n | Microdensitometry
|
Flow Cytometrya
|
||||
|---|---|---|---|---|---|---|---|---|
| N | IC/pg | Mbpb | N | IC/pg | Mbp | |||
| B. sylvaticum | B28 | 18 | 31 | 0.22 (0.026)c | 180 (21) | 6 | 0.27 (0.01) | 221 (9) |
| B. pinnatum | B144 | 18 | 28 | 0.27 (0.019) | 221 (16) | –d | – | – |
| B11 | 28 | 30 | 0.49 (0.067) | 402 (55) | 6 | 0.57 (0.01) | 468 (9) | |
| B. phoenicoides | B39 | 28 | 40 | 0.45 (0.077) | 328 (26) | – | – | – |
| B. arbuscula | B96 | 18 | 51 | 0.35 (0.039) | 287 (32) | – | – | – |
| B. distachyone | ABR99 | ?f | 20 | 0.30 (0.024) | 246 (19) | – | – | – |
| ABR100 | 30 | – | – | – | 6 | 0.65 (0.01) | 530 (12) | |
| ABR1 | 10 | 43 | 0.15 (0.023) | 123 (18) | 6 | 0.21 (0.001) | 172 (1) | |
| ABR2 | 10 | 20 | 0.15 (0.033) | 123 (27) | 6 | 0.21 (0.004) | 172 (3) | |
| ABR3 | – | – | – | – | 6 | 0.21 (0.002) | 165 (2) | |
| ABR5 | – | – | – | – | 6 | 0.19 (0.004) | 158 (3) | |
For the flow cytometry, genome sizes were calibrated from the standard genomes listed in “Materials and Methods” from which the following formula was derived: Genome size (Mbp) = Flow cytometer peak −7.5846/0.0265 (R2 = 0.9976). Given this formula our results gave the genome size of Arabidopsis = 165 (±9 se) n = 6. Results for genome sizes in Mbp are rounded up to the nearest whole number.
Scanning microdensitometry data (pg/IC) were converted to Mbp on the basis of 0.1 pg = 82 Mbp (Bennett et al., 2000).
Nos. in parentheses are the se.
–, Not done.
B. distachyon is the only annual species.
ABR99 could not be revived from stocks and was replaced with ABR100, which had a chromosome complement of 30.
Propidium iodide was used to stain nuclei isolated from our collection of 52 ecotypes of B. distachyon, and their genome size was determined by flow cytometry (Dolezel et al., 1989, 1998). To estimate genome size, the flow cytometer was calibrated against stained nuclei prepared from representative species with known genome sizes (Bennett et al., 2000). This analysis (Table II) revealed that the genome size in several B. distachyon accessions was indistinguishable from that of Arabidopsis, which is taken to be 164 Mbp/1C.
Diploid B. distachyon Has a Simple, Distinctive Karyotype and Displays High Levels of Recombination
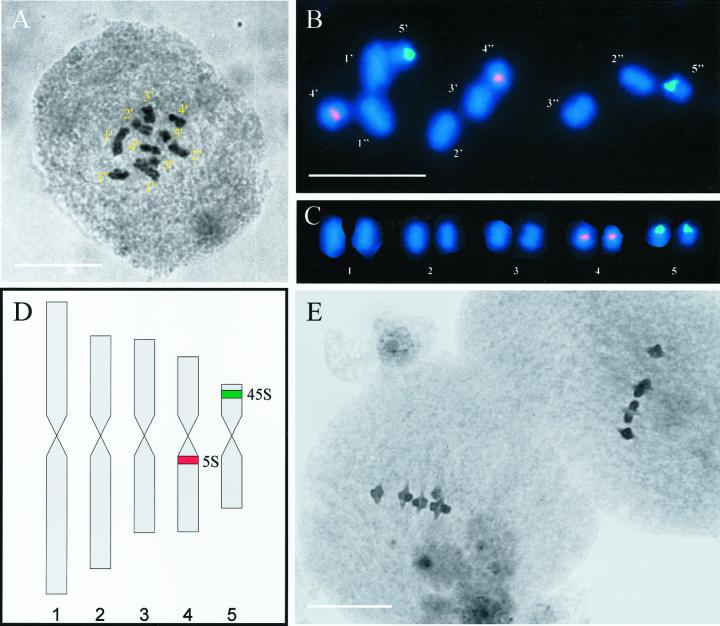
Somatic metaphase chromosomes were examined in seven ecotypes of B. distachyon (ABR1–ABR7), which had all been found to have a genome size of less than 175 Mbp. The analysis revealed that their karyotypes were morphologically the same (Fig. 2A), which justified the construction of a consensus karyotype for this species. The chromosomes have been arranged in descending order of short-arm length and were designated 1 to 5, in accordance with normal conventions. Chromosome 1 is submetacentric, distinctly the largest of the complement, and unlikely to be confused with chromosome 2, which is more acrocentric and considerably smaller. The short-arms of chromosomes 2 and 3 are of similar length, but 3 is shorter and characteristically the only metacentric of the complement. Chromosome 4 shares similar features with 3, which could ordinarily be the source of potential mislabeling. However, FISH reveals that chromosome 4 only has a major 5S rDNA locus, which is proximally located in its long-arm (Fig. 2, B and C). Chromosome 5 is acrocentric and by far the smallest of the complement. In addition, it is the only chromosome to bear a major 45S rDNA locus, which occupies a distal site in the short arm (Fig. 2, B and C). In summary, each chromosome has diagnostic features enabling unequivocal identification of every chromosome of the complement (Fig. 2D).
Figure 2.
Somatic and meiotic chromosomes of B. distachyon ABR1 showing light micrograph of the somatic chromosome complement (A) in which the five pairs of chromosomes are readily identified and fluorescence in situ hybridization (FISH; B) of 5S rDNA (red) and 45S rDNA (green) to the five pairs of somatic chromosomes. C, Karyogram derived from the image shown in B. D, Idiogram showing the distinctive and diagnostic shapes and lengths of the haploid set of chromosomes. The sites of the two rDNA loci are indicated. E, Light micrograph of two pollen mother cells at metaphase I. Note the five ring bivalents in each. Bars = 10 μm.
Examination of 20 pollen mother cells at metaphase I of one ecotype (ABR1) revealed that 16 form five ring bivalents and four form four ring bivalents and one rod involving chromosome 5 in all cases. Two example cells are presented in Figure 2E, which also clearly shows that chiasmata are not strictly localized to any particular region of the chromosomes.
Diploid B. distachyon Ecotypes Display Growth and Life Cycle Characteristics Suitable for High-Throughput Genetics
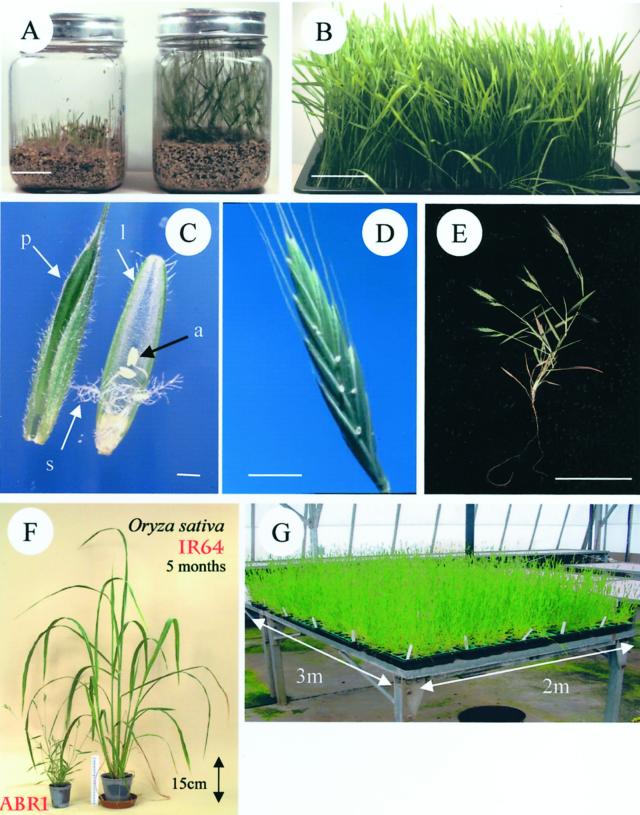
Preliminary experiments established that all ecotypes performed well under standardized conditions (see “Materials and Methods”). The B. distachyon ecotypes were then characterized in terms of size at maturity, general growth habit, vernalization requirement, duration of life cycle, and amount of seed set. ABR1 was typical and exhibited undemanding maintenance requirements, growing successfully under sterile conditions in glass jars (Fig. 3A) on vermiculite supplemented with 0.5× Hoagland solution (Draper et al., 1988) or at high-density (2,000 seedlings 18- × 30-cm tray−1) in compost (Fig. 3B). With only two exceptions (ABR13 and ABR15), all of the diploid ecotypes (including ABR1) benefited from a standard vernalization treatment (6 weeks at 5°C) to ensure synchronous induction of flowering and synchronized embryo development. Floral spikelet emergence occurs around 3 to 4 weeks after removal from the cold room (Fig. 3C). By 4 to 5 weeks following vernalization, visible anthesis occurs (Fig. 3D) with each spikelet eventually containing typically around 10 to 12 seeds. At maturity (4–5 mo), the diploid ecotypes ranged from 15 to 30 cm in height and very rarely shed any seed prematurely, thus aiding easy harvesting (Fig. 3E; Table IV). For all polyploids and two diploid ecotypes (ABR13 and ABR15), flowering is accelerated by approximately 6 weeks, as no vernalization stage is required (see Table IV). Furthermore, the smaller stature of B. distachyon compared with, for instance, rice (Fig. 3F), allows typical planting densities of at least 300 plants square meter−1 in ordered arrays (Fig. 3G) for M2 mutant seed production or M3 mutant screening.
Figure 3.
Growth habit and anatomy of the diploid B. distachyon ecotype ABR1 grown in glass jars (A) on vermiculite supplemented with 0.5× Hoagland solution (Draper et al., 1988) at 1 week (left jar) and 2 weeks (right jar), and at high density (B) in 30- × 18-cm trays grown on 1:1 mixes of Levington's:gravel. Bar = 5 cm. Flower morphology (C) prior to dehiscence and indicating hairy palea (p) and lemea (l), stigma (s), and anthers (a). Bar = 1 mm. Each plant has 6 to 10 flowering spikes that actually set seed and seed heads (D) have a “brome-like” appearance and typically carry 10 viable seeds. Bar = 4 mm. At maturity (E), ABR1 plants were 15 cm high with short nonrhizomatous roots. Bar = 5 cm. F, B. distachyon ABR1 (8 weeks post-sowing) is compared in size with rice cv IR64 (20 weeks post-sowing). G, 2,000 M1 progeny of γ-irradiated (at the Vienna Atomic Energy Institute) B. distachyon seeds grown under typical temperate greenhouse conditions.
Table IV.
Comparison of “biological genomic” traits in established model plants and B. distachyon
| Arabidopsis | B. distachyon (ABR1) | Rice | |
|---|---|---|---|
| Plant family | Crucifereae | Pooideae | Oryzoideae |
| Chromosome no. | 10 (2n) | 10 (2n) | 24 (2n) |
| Genome size (1C) | 164 Mbpa | 160 Mbp | 441 Mbpa |
| Amount of repetitive DNA | 16%b | 12%–15%c | ∼20%d |
| Breeding strategy | Self fertile | Self fertile | Outbreeder |
| Life cycle (weeks)e | 8–10f | 11–18g | 20–30h |
| Height at maturity (m) | ∼0.2 | ∼0.2 | 1–1.2 |
| Planting density for M2 seed collection (plants m−2) | ∼300 | ∼300 | 8–10 |
| Seed yield plant−1 | >1000 | 80–200 | >1,000 |
| Seed yield m−2 | >300,000 | 24–60,000 | >8,000 |
| Growth requirements | Simple | Simple | Relatively specializedi |
| Transformation efficiency | 0.5%–4%j | ∼5 | 3–8k |
Life cycle defined as time from seed germination to harvesting of first seed.
Depending on vernalization requirement.
Observed at the UW Aberystwyth.
To achieve fast growth and controllable flowering in temperate latitudes, many rice varieties require to be grown at high light intensity in a heated greenhouse with purpose-built irrigation/flooding systems. Specialized environmentally controlled growth cabinets are often required for the production of somatic embryos suitable for tissue culture.
Transformation frequency in Arabidopsis is presented as the proportion of transgenic to wild-type seeds recovered from florally “dipped” plants (Cough and Bent, 1998).
The comparative data concerning rice callus transformation is derived from Li et al. (1993) and Biswas et al. (1998) and is expressed as the number of transgenic plants produced 1 g embryogenic tissue−1 bombarded with microprojectiles.
B. distachyon Is Readily Responsive to Tissue Culture
The diploid ecotype ABR1 was chosen for initial tissue culture studies as it was relatively compact and offered a high throughput of immature embryos for minimal growth space. A detailed analysis of embryo development in individual B. distachyon spikelets (Fig. 4A) revealed, as expected, a variation in maturity, with more mature embryos present in the center at 10 to 15 d post-visible anthesis (data not shown). Isolated immature embryos were divided into one of five size classes (types 1–5; Fig. 4, B and C) and were placed on callus induction media (LS 2.5 and N6 2.5; Bablak et al., 1995), and typically after 10 to 15 d had formed a mass of callus on the surface of the scutellum (see Fig. 4, D and E). This included large areas of creamy-white, type II embryogenic tissue (dry, friable, and pale) easily identified by scanning electron microscopy (Fig. 4F). Immature embryos with the greatest potential for somatic embryogenesis were in the size range of 0.3 to 0.7 mm (see Table IIIA) and were generally off-white with a translucent scutellum margin and were suspended in a soft endosperm (see Fig. 4, B, class 2 and C, class 3). LS 2.5 medium proved superior for the induction of embryogenic callus, with around 45% of isolated embryos responding within this size range (Table IIIA). Type II callus could be separated from callus growing on the original explants and maintained on LS 2.5 for up to 9 to 12 mo after which it was normally discarded.
Figure 4.
Tissue culture and transformation of B. distachyon. A, B. distachyon ABR1 seed head at 15 d postanthesis (bar = 4 mm) from which (B) isolated seeds have had their paleas and lemmas removed (embryos arrowed, bar = 0.5 mm) or (C) the embryos are isolated from endosperm to indicate embryo development (classes 1–5, bar = 0.5 mm). D, Scanning electron micrograph showing immature embryo structure. Indicated are the coleoptile (col), scutellum (scut), radical (rad), and coleorhiza (chz). Bar = 0.1 mm. E, Embryogenic callus (c) formation around the edge of the scutellum (scut) after 15 d of culture (bar = 0.1 mm), which (F) scanning electron micrography revealed to contain many organized structures with a distinctive embryoid shape (arrowed; coleoptilar pore; bar = 20 μm). G, Propagated embryogenic callus of ABR100 stained for GUS activity 24 h following biolistic bombardment with pACt1GUSHm. Bar = 1 mm. H, Selection of ABR100 transgenic tissue on Hm- (40 μg mL−1) selective media. Bar = 3 mm. I, The second subculture of regenerating plants from bombarded embryogenic callus of ABR100 on selective media. Bar = 5 mm. J, Isolated Hm-resistant plantlet of ABR100 exhibiting tiller formation. Bar = 5 mm. K, Hm-resistant ABR100 shoot stained for GUS activity (bar = 1 cm). L, Rooted T0 transgenic lines in soil producing viable seeds. M, The PCR amplification of hgh gene internal sequences (516-bp product) from five independent T1 transgenic lines (1–5). N, Southern blot of BamHI-digested genomic DNA from T1 transgenic lines (1–5) and wild-type ABR100 probed with hgh sequences. Indicated is the 1.07-kb BamHI internal hgh gene fragment.
Table III.
Tissue culture and transformation in B. distachyon
| A. The responsiveness of immature ABR1 embryos to culture
| |||||
|---|---|---|---|---|---|
| Embryo classa | 1 | 2 | 3 | 4 | 5 |
| Size (mm) | <0.3 | 0.3–0.4 | 0.5–0.69 | 0.7–0.89 | >0.9 |
| Medium | Percentage of embryos producing type II embryogenic callus | ||||
| LS 2.5 | 4 | 41 | 54 | 16 | 5 |
| N6 2.5 | 16 | 43 | 34 | 12 | 2 |
| B. Plant regeneration from ABR1 embryogenic callus on different mediab | |||||
| Medium | Murashige and Skoog Salts (MSO) | 190-2 | Rice Regeneration Medium (N6 0) | MWW | RM1 |
| No. of plants g callus−1 | 66 | 25 | 71 | 43 | 40 |
| C. Results of typical transformation experiments with ABR100 callus | |||||
| Experiment | No. of Independent Hm-Resistant calli g tissue−1 | Percentage of Calli- Producing Plants | Transformation Frequency (no. of plants g tissue−1) |
|---|---|---|---|
| 1 | 8 | 25 | 3 |
| 2 | 6 | 50 | 7 |
| 3 | 7 | 30 | 5 |
| Average | 7 | 35 | 5 |
Embryo classes are illustrated in Fig. 4, B and C. 190-2, Lolium regeneration medium; MWW, winter wheat regeneration medium; RM1, barley regeneration medium (all are described in Bablak et al., 1995).
The regeneration potential of type II callus after subculture on LS 2.5 (approximately 6 weeks) was examined by inoculating onto a range of established regeneration media. Plantlets were produced on all media tested, with the highest regeneration potential realized on MS0 and N60 (Table IIIB). Although embryogenic callus could be obtained at a high frequency when immature embryos were placed on LS 2.5 or N6 2.5 media, albino shoots were recovered at a much higher frequency from tissues maintained on N6 2.5 (45% compared with 7% on LS2.5), and thus LS 2.5 was used for all future work on transformation.
Transgenic Plants Can Be Recovered at a High Frequency from Embryogenic Callus Using Microprojectile Technology and Hygromycin (Hm) Selection
Tissue from the hexaploid accession ABR100 remained highly embryogenic when maintained on LS 2.5 medium and so was used to bulk up material for development of transformation technology. Embryogenic callus from ABR100 was bombarded with microprojectiles loaded with the plasmid pACt1-GUSHm (pAGH). A small sample of bombarded callus tissue with well-dispersed cauliflower mosaic virus 35S-β-glucuronidase (GUS) activity is shown in Figure 4G. One week after bombardment, callus was subcultured to LS 2.5 medium containing 40 mg L−1 Hm where it rapidly became brown and necrotic, but after 2 weeks, small nodes of actively growing cells were visible. Once these had developed to a diameter of 1 to 2 mm, these tissues were transferred twice at weekly intervals for further growth on selection medium containing 40 mg L−1 Hm. In typical experiments, an average of seven Hm-resistant clones were recovered per gram of target tissue (Table IIIC).
Hm-resistant calli were removed from selection plates and were transferred onto regeneration medium (see “Materials and Methods”) containing 30 mg L−1 Hm and they produced plantlets within 7 to 10 d (Fig. 4H). Callus containing young shoots and viable somatic embryos were transferred subsequently to germination medium (MS0) and were incubated in the light. An average of five Hm-resistant plants were recovered per gram of bombarded tissue (Table IIIC). The formation of roots was initially rather poor on the regeneration medium (Fig. 4I), but a well-developed root system could be produced following further subculture on the same hormone-free media once the plantlets had been separated from necrotic tissue (Fig. 4J). Histochemical staining with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid of regenerated plantlets surviving on Hm revealed clear evidence of GUS activity (Fig. 4K). All rooted plants that were established successfully in soil produced fertile flowers and set seed (Fig. 4L). T1 progeny of five independent transgenic lines were examined for the presence of the Hm gene by PCR and Southern blotting to confirm that stably transformed lines had been generated. PCR amplified a 516-bp fragment from all transgenic lines (Fig. 4M), which hybridized to a hgh probe following Southern blotting (data not shown). In addition, probing BamHI-digested genomic DNA extracted from these transgenic lines with a hgh probe indicated hybridization to a 1.07-kb fragment that corresponded to the size predicted from the plasmid used for the initial bombardment (Fig. 4N).
B. distachyon Displays Traits That Are Important for Temperate Cereal Research
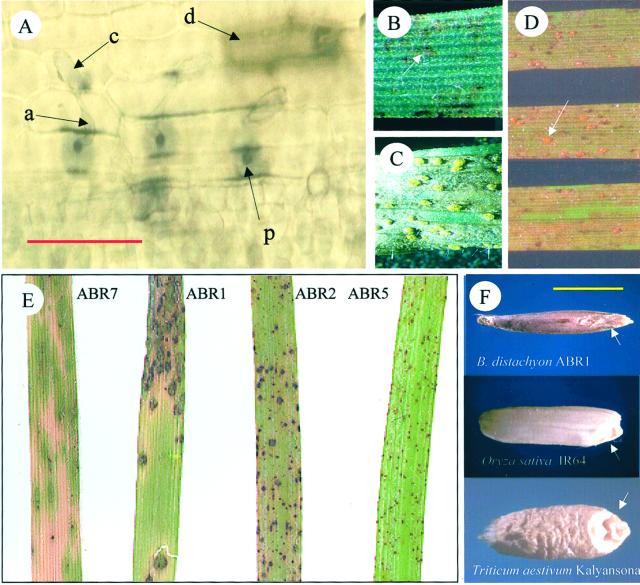
We have characterized the responses of B. distachyon ecotypes to challenge with a range of pathogens that are the cause of significant crop loss in cereals. Blumeria graminis is the causal agent of powdery mildew on cereals and occurs in several forma specialis (f. sp.). Isolates of B. graminis f.s.p hordei, avenae, and tritici were used to challenge ecotypes (ABR1–ABR7 and ABR100) of B. distachyon and failed to elicit disease symptoms that were observable to the naked eye. However, light microscopic examination revealed single epidermal cell death and also papillae-based resistance that are typical of a hypersensitive response (Fig. 5A). Attempts to isolate a Blumeria sp. capable of establishing an infection on B. distachyon by exposing plants to the outdoor environment have, to date, been unsuccessful.
Figure 5.
Targets for B. distachyon research. Responses to pathogen challenge and grain development. A, The responses of ABR1 to challenge with condiophores of Blumeria graminis f. sp. Triticae. Bar = 0.1 mm. Arrowed are the condiophore (c), appressorial germtube (a) and papillae formation (p) and single cell-death (d) in the host cell. Challenging with Puccinia striformis f. sp. triticae (wheat yellow stripe rust) elicits (B) localized necrotic flecking on ABR105 and (C) yellow uridea (pustule) formation on ABR100 that develop from (D) extensive areas of necrotic tissue. (Uridea forming within the necrotic tissue are arrowed.) E, Variable responses by B. distachyon ecotypes to challenge with M. grisea Guy-11. F, Comparison of mature seed size and morphology of B. distachyon ABR1 with rice and wheat cv Kalyansona). Embryos are arrowed on each seed. Bar = 2 mm.
The responses of B. distachyon ecotypes (ABR1–ABR7, and ABR100, 101, 105, and 113) were examined following challenge with the rust pathogens Puccinia reconditia f. sp. hordei and f. sp. triticae (barley and wheat brown rust, respectively) as well as P. striformis f. sp. hordei and f. sp. triticae (barley and wheat yellow stripe rust, respectively). ABR100 displayed a “brown flecking” (BF) following challenge with wheat brown rust strains with all other tested ecotypes eliciting no macro- or microscopic responses. Ecotypes ABR1, ABR3, ABR100, and ABR105 also exhibited BF symptoms with barley brown rust strains. In contrast, all ecotypes exhibited some form of visible response to wheat and barley yellow stripe rusts ranging from BF (ABR1–ABR7, and 101 and 111; Fig. 5B) or the more extensive “brown tissue” response. It is interesting that ABR100 and ABR105 displayed uridinea (raised pustule) formation (Fig. 5C), indicating that the fungus has successfully colonized the host, though these formed within areas of extensive necrosis (Fig. 5D).
A range of symptoms were also elicited (Fig. 5E) following challenge of B. distachyon ecotypes (ABR1–ABR7) with the causal agent of rice blast, Magnaporthe grisea (strain Guy-11). On ABR7 and ABR1, an extensive and spreading necrosis was observed that was reminiscent of blast symptoms on rice. In contrast, highly localized necrotic lesions formed on ABR5 that did not change significantly in phenotype over time and were consistent with the exhibition of full resistance to M. grisea Guy-11.
Seed development in B. distachyon has yet to be extensively analyzed. However, in terms of overall size and external anatomy, mature seeds of the diploid ABR1 are very similar to those of the major grain crops rice and wheat, the only major structural difference being a smaller endosperm volume in B. distachyon (Fig. 5F).
DISCUSSION
New “post-genomic” techniques are offering increasingly powerful ways to link plant phenotype to the transcriptome, proteome (Gura, 2000), or the metabolome (Fiehn et al., 2000), allowing the development of a “blueprint” of plant form and function (Chory et al., 2000). Optimal exploitation of these technologies is dependent on possession of suitable model species. It is now becoming clear that Arabidopsis cannot be considered as an ideal model for the grasses (Devos et al., 1999; Tikhonov et al., 1999; Keller and Feuillet, 2000), and there is significant international effort to develop rice as a model species more suitable for the Poaceae (Havukkala, 1996; Goff, 1999). However, against this background, the limitations of rice to provide such a model and the complementary opportunities offered by other species needs to be recognized.
Analysis of the B. distachyon Genome Will Provide a Valuable Resource for Physical Genomics in Temperate Cereals and Forage Grasses
Differing analytical techniques pose different problems when attempting to estimate genome size. The commonly used flow cytometry technique is dependent on accurate calibration, and even apparently definitive DNA sequencing approaches do not easily include regions of highly repeated sequence and may therefore underestimate genome sizes. Nevertheless, we have taken The Arabidopsis Genome Initiative (2000) estimate (approximately 125 Mbp) as the more accurate and, given that we failed to distinguish any size difference between diploid B. distachyon ecotypes and Arabidopsis genomes by flow cytometry (Table IV), we propose that they should be considered to be approximately the same. This genome size is somewhat at odds with a previous report (4C = 3 pg; Bennett and Leitch, 1995). However, our present data suggest the presence of a polyploid series within B. distachyon (2x = 10, 4x = 20, and 6x = 30) and, therefore, the discrepancy can be explained by the fact that these authors used a hexaploid accession for their determination (e.g. ABR100 2n = 6x = 30, 4C approximately 2.6 pg, derived from Table II). The flow cytometry screen identified 12 diploids, eight tetraploids, and 32 hexaploids in our germplasm collection.
We recognize that the utility of B. distachyon will be augmented by the development of genetic and physical chromosome markers. Given the small chromosome size in this species, there is a surprisingly high chiasma frequency of at least 9.8 cell−1. Furthermore, this chiasma frequency is likely to be an underestimate because of the technical problem of resolving more than one chiasma per chromosome arm at this meiotic stage. Thus, we expect that genetic maps of B. distachyon could be quickly generated given its high crossover frequency and given the development of high-throughput markers such as single nucleotide repeats (Ellis, 2000). In diploid B. distachyon ecotypes, the five pairs of chromosomes are sufficiently dissimilar to enable positive identification in routine squash preparations of somatic tissue. Meiotic prophase chromosomes are amenable to study in B. distachyon, and provide longer substrates for the hybridization in situ of fluorescent probes. Thus, it should be possible also to physically map DNA probes from large insert libraries (such as BACs) to meiotic prophase chromosomes and DNA fibers, as has been achieved in tomato (Lycopersicon esculentum; Zhong et al., 1999). If the chromosomes are probed simultaneously with landmarks such as rDNA and pericentromeric repeats originally isolated from B. sylvaticum (Abbo et al., 1995), it should be possible to construct rapidly physical linkage maps and assign contiguous megabase tracts of DNA to particular chromosome arms. Such an approach may aid future programs requiring the targeted sequencing of specific regions of the B. distachyon genome. As publicly available rice genomic sequences from the International Rice Genome Sequencing Project are only available for around 25% of the rice genome, with the more complete sequence only available through “research contracts” with Syngenta (Dickson and Cyranoski, 2001), then the rapid generation of sequence information from the B. distachyon genome could have great utility in the academic community.
The genus Brachypodium is considered a sister group to the “core pooid” clade, which contains all of the important temperate cereals, fodder grasses, and amenity grasses. We further propose that B. distachyon, by occupying this key phylogenic position and by virtue of its exceedingly “compact genome” (allowing representation by a very small genomic library), could provide a significant resource for the analysis of the much bigger genomes possessed by important temperate grasses. For example, to aid long-distance chromosome walking and quantitative trait loci analysis in cereals with large genomes, mapping information from one grass genome can theoretically be used to locate a likely syntenic (or colinear) region represented in BAC/YAC clone contigs from a cereal with a smaller genome (Moore et al., 1993). In reality, there are as yet very few examples of this approach being successful (e.g. Chen et al., 1997), which is due largely to the breakdown of genome colinearity at the microsyntenic level (e.g. 50 Kb–1 Mb) where the species are relatively distantly related (Foote et al., 1997). However, considering the phylogenetic position of Brachypodium in the Pooideae, microsynteny is likely to be relatively more conserved within this subfamily and thus B. distachyon could provide an archetypal map specifically for pooids to which other colinear genomes could be aligned. In practical terms, physical markers linked only loosely to important traits in the large genomes of species such as wheat and barley (Hordeum vulgare) might be located to relevant B. distachyon genomic library clones to provide rapid access to syntenic genomic regions.
Biological Characteristics Indicate That B. distachyon May Be Amenable for Large-Scale Mutagenesis Programs
Mutagenesis is commonly used to relate genotype to phenotype. Large-scale mutagenic techniques may employ chemical, physical, or genetic mutagens, and the latter especially has proved a powerful tool for gene tagging in Arabidopsis (for example, Tissier et al., 1999). Successful mutagenesis of a plant species is dependent on a small diploid genome, available mapping strategies, ease of crossing, transformability, and not inconsequential features such as a small physical stature, a rapid life cycle, and good seed yield, which eases mutant screening and collecting selected lines (Vizir et al., 1996). Table IV illustrates how Arabidopsis fulfills all these requirements, but B. distachyon also scores highly. As stated above, genetic maps now can be generated relatively quickly, and so the only remaining unfavorable feature is the smaller seed yield compared with Arabidopsis. However, compared with rice, B. distachyon has a more rapid life cycle, smaller stature, and an inbreeding reproductive strategy with anthers and stigmas enclosed tightly within the palea and lemma. The latter makes crossing different plants slightly more demanding technically. Floral spikelet emergence proved to be the best stage to remove the three anthers and apply pollen to the stigmas when conducting crossing experiments. However, this inbreeding reproductive behavior and the absence of seed head “shatter” in most B. distachyon ecotypes means that F1 seed can be collected without the need for hand pollination or the time-consuming bagging of flowering plants. Thus, individual plants do not have to be isolated when grown in large populations to avoid outcrossing, which is a great advantage for mutagenesis studies. We have already generated γ-irradiated populations, and transposon-based mutagenic approaches are under development (J. Draper, L.A.J. Mur, G. Jenkins, and P. Bablak, unpublished data).
B. distachyon, A Potential High-Throughput Transformation System
A key technology for a model plant species is the availability of a facile transformation system. For many dicot species, high efficiency Agrobacterium tumefaciens-mediated transformation is well established, but though the approach has been used to generate transgenic rice (Chan et al., 1992), wheat (Mooney et al., 1991), maize (Gould et al., 1991), and barley (Tingay et al., 1997), this technique is far from routine. For Poaceae, particle bombardment of embryogenic callus followed by selection of transgenic tissue (for review, see Potrykus, 1990; Hansen and Wright, 1999) has proven robust and has been used to transform maize (Klein et al., 1989; Fromm et al., 1990), wheat (Vasil et al., 1991), and rice (Cao et al., 1992). Nevertheless, the number of publications describing the use of transgenic cereals or forage grasses for basic studies in biology is still very limited in comparison with dicots. The success of the transformation techniques is dependent upon the optimal culture of highly embryogenic callus (Hansen and Wright, 1999). This is derived commonly from immature zygotic embryos, as this tissue is very responsive to in vitro culture and has a high plant regenerative capacity. Immature embryos from B. distachyon were not an exception. Large numbers of immature embryos can be isolated from B. distachyon plants grown under simple greenhouse conditions, without the need for specialist environmentally controlled growth chambers. For example, a single 18- × 30-cm tray containing 24 flowering plants can provide sufficient embryos to supply embryogenic cultures for a single researcher for 1 mo. The low frequency of gross morphological abnormalities (e.g. albinism and male sterility) suggested that significant somaclonal variation did not occur under the culture conditions used. Based on this behavior in tissue culture, we report a transformation system for a hexaploid ecotype of B. distachyon that is comparable with the best rice systems currently available in terms of ease of use and final transformation efficiency (Tables IIIC and IV; Li et al., 1993; Biswas et al., 1998). As testing a particular transgene in wheat can still take up to 5 years (Dunwell, 2000), given the rapid life cycle of B. distachyon, our development of a facile, low-cost transformation system will offer a rapid “test bed” for transgenic studies in grasses. We also demonstrate that embryogenic callus can be generated routinely from several diploid ecotypes, and current experiments aim to develop transposon tagging and gene trapping resources in diploid ecotypes of B. distachyon (J. Draper, L.A.J. Mur, G. Jenkins, and P. Bablak, unpublished data).
B. distachyon Displays Many Traits That Are Relevant for Cereal and Forage Grass Improvement
In considering a potential model for temperate cereals, the value of any plant species obviously depends on whether the particular organism displays key traits representing those targeted currently in plant breeding and biotechnology programs. Crop loss through pathogen attack is significant and represents a key aspect for research. We also note that in simple practical terms, a small plant, which is easy to maintain and which will support several classes of important cereal diseases may find utility in fungicide screening programs. All ecotypes of B. distachyon tested appeared to exhibit single cell death and papillae formation following challenge with powdery mildew pathogens adapted to cereals. These responses are symptomatic of resistance in other grass/cereal species (Carver et al., 1995) and may be indicative of lack of race structure. The molecular basis of such “non-host” HR is being investigated in model dicots (e.g. Lauge et al., 2000), and B. distachyon could serve in a similar role for the Poaceae, where race-specific resistance exhibits little longevity in the field. Different ecotypes of B. distachyon exhibited great variability in response to infection with individual rust species ranging from no visible symptoms or BF associated with a HR, to pustule formation (albeit associated with necrotic tissue). These data suggest that B. distachyon may prove to be a useful host for future studies on the molecular biology and genetics of these important plant-pathogen interactions. In contrast, we observed clear evidence of susceptibility and resistance exhibited by different ecotypes to challenge with Magnaporthe grisea Guy-11 and additionally, the phenotypes closely resembled those observed on rice cultivars (data not shown). M. grisea is undoubtedly one of the most virulent and economically devastating plant pathogens, causing losses of between 11% and 30% of the annual world crop and representing up to 157 million tons of rice (Baker et al., 1997). Thus, we are extensively characterizing the B. distachyon/M. grisea pathosystem to identify key resistance determinants and to understand defense-associated signaling (A.P.M. Routledge, L.A.J. Mur, and G. Shelley, unpublished data).
An even more important trait than pathogenic interactions for a Pooid model is the analysis of genetic and biochemical events underlying grain-filling and quality. The large size of the seeds in the genus Brachypodium has been highlighted previously (Catalan et al., 1997) and there have already been some basic studies on seed storage proteins (Khan and Stace, 1999). Thus, it is envisaged that resource allocation, grain filling, and endosperm development in particular will be a valuable focus for mutagenesis program in B. distachyon. Further features, including freezing tolerance, perenniality, repetitive injury (mowing and trampling) tolerance, meristem dormancy mechanisms, post-harvest biochemistry of silage and hay, mycorrhizae, and sward ecology—all of which are important to temperate forage grasses and many temperate cereals—are traits that are not exhibited by or are difficult to study in rice, but are possible targets for functional genomics in Brachypodium.
In summary, the present data demonstrate that B. distachyon has great potential to be adapted for high-throughput genetics. In many aspects of its biology such as genome size, chromosome number, height, planting density, breeding system, and duration of life cycle, it is very similar to Arabidopsis. We propose that B. distachyon is best seen as a complementary model to rice offering, as it does, the opportunity to study grass traits that are not well exhibited by rice. In addition, the simpler logistics of handling large B. distachyon populations, as compared with rice, may well make B. distachyon an attractive prospect where investigative resources, such as access to controlled growth environments, are limited.
MATERIALS AND METHODS
Plant Growth Conditions
Full details of all Brachypodium distachyon ecotypes can be found at http://www.aber.ac.uk/plantpathol/Brachyomics. Seed samples of different Brachypodium ecotypes were obtained from Brachyomics (Botany Gardens, Aberystwyth, Ceredigion, UK) under a “Research Only” Materials Transfer Agreement. All B. distachyon ecotypes were grown under a 16-h light period at 23°C ± 2°C. The plants were illuminated with 55W (Osram, Sylvania, Munich) high-frequency lighting tubes (4,580 lumen output) and were supplemented with 2 × 30W clear tube cooled lighting (Osram) and placed between 60 and 100 cm of the light bank. Light intensities always exceeded 10 μmol m−2 s−1. Plants were grown routinely on Levington's Universal Compost (Levington Horticulture, Suffolk, UK) supplemented with gravel (the longest axis was approximately 0.5 cm) to approximately 50% (v/v) to improve drainage. Plants were usually watered at 2-d intervals and were never allowed to stand in water.
Nuclear Genome Size Estimations: Scanning Microdensitometry and Flow Cytometry
For microdensitometry, root tips were fixed in 4% (w/v) formaldehyde for 2 h, and were then washed with distilled water prior to acid-hydrolysis by treatment with 5 m HCl at 20°C for 45 min. Following washing with distilled water, Feulgen stain was added and the samples were kept in the dark for 45 min. The stain was washed off with SO2 water, and then root tip squashes were prepared. The density of well-stained prophase nuclei was scanned using an M86 scanning microdensitometer (Vickers Instruments, York, UK). The relative amount of DNA in each nucleus was calculated using the following formula: absorbance units of test species/absorption units of a standard species × 4C value of standard species = 4C value of test species. Mung bean (Vigna radiata var. Berken Mung Bean; 2C = 1.05 pg) was used as the standard species (seeds supplied by W. Atley Burpee Seed Company, Leicester, UK).
Nuclei were isolated for flow cytometry using the chopping technique developed by Galbraith et al. (1983) using LB01 buffer at pH 7.5 (Dolezel et al., 1989, 1998) supplemented with 50 μg mL−1 propidium iodide and 50 μg mL−1 RNase (Sigma, Poole, UK). The fluorescence of stained nuclei was analyzed using PA or III flow cytometers (Partec, Münster, Germany). Calibration standards (Bennett et al., 2000) used to estimate the B. distachyon 1C genome size were Lotus japonicus (approximately 445 Mbp; Bennett and Smith, 1976), Aesculus hippocastanum (approximately 110 Mbp; Bennett et al., 1982), pea (Pisum sativum; approximately 4,150 Mbp; Bennett and Leitch, 1997), Arabidopsis (approximately 164 Mbp; Bennett et al., 2000), rice subsp. indica cv IR34 (approximately 441 Mbp; Bennett et al., 2000), and barley cv Sultan (approximately 4,116 Mbp; Bennett et al., 2000).
Karyotyping and FISH
After flow cytometry experiments, seeds of seven putative diploid accessions of B. distachyon (ABR1–ABR7) were germinated on filter paper moistened with tap water at 22.5°C in the dark. Whole seedlings with roots of about 1 cm long were incubated in a saturated solution of 1-bromonaphthalene at 0°C overnight, or in 2 mm aqueous 8-hydroxyquinoline for 1 to 2 h at room temperature. After washing and fixing for at least 2 h in a 3:1 mixture of methanol:acetic acid, root-tips destined for bright-field microscopy were hydrolyzed in 1 m HCl at 60°C for 10 min, stained in Feulgen solution, and mounted in 2% (w/v) orcein in 45% (w/v) propionic acid. Material for FISH was washed after fixation in 10 mm citrate buffer and was digested for 1.5 to 2 h at 37°C in a mixture comprising 1% (w/v) cellulase (Calbiochem, La Jolla, CA), 1% (w/v) cellulase (Onozuka RS, Yakult Honsha C. Ltd., Tokyo), and 20% (v/v) pectinase (Sigma, St. Louis). After washing, root-tips were detached and their meristems were dissected out into 45% (w/v) acetic acid and squashed onto microscope slides. Coverslips were removed by freezing and the preparations were post-fixed in 3:1 ethanol:acetic acid, dehydrated in absolute ethanol, and air dried. Immature inflorescences were fixed in Carnoy's solution. Anthers at first metaphase of meiosis were macerated in propionic orcein and squashed under coverslips. Somatic and meiotic chromosomes were photographed onto Imagelink HQ microfilm (Eastman-Kodak, Rochester, NY) with an MC100 camera attached to a Axioplan microscope (Zeiss, Welwyn Garden City, UK), and they were electronically scanned and processed in CorelDraw (Corel Corporation, Ottawa, Ontario, Canada).
The 5S rDNA probe was amplified and labeled with rhodamine-4-dUTP (Amersham Pharmacia, Uppsala) from the wheat clone pTa 794 (Gerlach and Dyer, 1980), using PCR with universal M13 sequencing primers under the following conditions: 94°C for 1 min, 35 cycles of 94°C for 40s, 55°C for 40s, 72°C for 1 min, and 1 cycle of 72°C for 5 min. The 45S rDNA probe was obtained by nick translation with digoxigenin-11-dUTP (Roche, Basel) of a 2.3-kb subclone of the 25S rDNA unit of Arabidopsis (Unfried and Gruendler, 1990).
FISH was adapted with some modifications from Schwarzacher and Heslop-Harrison (2000). In short, slides were pre-treated with RNAse for 1 h at 37°C, post-fixed in 1% (w/v) aqueous formaldehyde in phosphate-buffered saline buffer for 10 min, and dehydrated in an ethanol series. Probe DNA was mixed to a concentration of 100 ng slide−1 with 50% (w/v) deionized formamide, 10% (w/v) Dextran sulfate, 2× SSC, and 1% (w/v) SDS. The probe DNA was allowed to hybridize with the chromosome preparations overnight at 37°C in an Omnislide in situ hybridization system (Thermo Hybaid, Ashford, Kent, UK). Slides were washed stringently in 20% (w/v) deionized formamide in 0.1% (w/v) SSC at 42°C, followed by detection of digoxigenin by fluorescein isothiocyanate-conjugated anti-digoxigenin antibodies. The chromosomes were counterstained with 4′,6-diamino-phenylindole (2 μg mL−1), mounted in Vectashield (Vector Laboratories, Burlingame, CA), and photographed onto Provia 400 color reversal film (Fuji Photo Film, Tokyo) with an MC100 camera attached to a Axioplan epifluorescence microscope (Zeiss, Jena, Germany). Images were scanned electronically and processed using Micrografx Picture Publisher software.
Tissue Culture and Transformation
All media were as described previously by Bablak et al. (1995) except, when required, maltose was substituted for Suc at a level of 30g L−1. Immature embryos or callus material was viewed by scanning electron microscopy as described previously (Bablak et al., 1995). Seedlings were germinated and grown as described above, and immature embryos were isolated from sterilized seeds (Bablak et al., 1995) under a dissecting microscope approximately 14 d after anthesis and their size was estimated (when required) using an eye-piece graticule. Approximately 50 embryos were cultured in the dark in a 9- × 1.5-cm petri dish containing 25 mL of LS 2.5 medium (Bablak et al., 1995). After 4 weeks, the developed calli were detached from the explants and were subcultured onto the same medium. In several experiments, LS 2.5 was substituted with N6 2.5 or SH 2.5 (Bablak et al., 1995). Type II callus (creamy-white, dry, and friable) was constantly selected at each subculture to bulk up tissue for regeneration and transformation studies. Media used for regeneration were MS0; 190–2 (Lolium regeneration medium), MWW (Winter wheat regeneration medium), and RM1 (Barley regeneration media; Bablak et al., 1995).
For microprojectile bombardment, 1.0-μm gold particles were coated with the plasmid and “fired” using the procedure described in the PDS1000/He biolistic gun instruction manual (Bio-Rad, Hercules, CA). The plasmid used was pACt1GUSHm (McElroy et al., 1991), which contained an Hm-resistance gene (hgh) driven by a cauliflower mosaic virus 35S promoter (35S) for transformant selection and a 35S-GUS gene for transient transformation assays. In preliminary transformation experiments, it was discovered that the substitution of Suc with maltose increased the regeneration potential of tissue bombarded with microprojectiles. The bombardment procedure was optimized by selecting conditions that achieved routinely >1,000 GUS-positive regions per target plate when examined by histochemical staining. Approximately 1.0 g fresh weight of embryogenic callus was centered in petri dishes containing LS 2.5 supplemented with 140 g L−1 maltose and 2.5 g L−1 of Phytagel and incubated for 2 h. Calli were bombarded with a single microprojectile firing using a PDS1000 particle acceleration device (Bio-Rad) with a helium pressure of 1,300 psi, under a chamber pressure of 27 mm Hg at a distance of 13 cm below the microprojectile stopping plate. For transient assays, calli were stained for GUS activity (Warner et al., 1993) after 24 to 36 h, and only callus batches that exhibited 1,000 blue GUS spots or more were used for stable transformation experiments. For stable transformation, all target materials were bombarded once with pACt1GUSHm and were then transferred to LS 2.5 (with maltose as a carbon source) for 1 week in the dark at 25°C. Bombarded calli were then transferred to the same medium supplemented with 40 mg L−1 Hm (Duchefa, Haarlem, The Netherlands). Two weeks later, growing tissue was picked out and transferred to fresh medium containing 40 mg L−1 Hm, a process that was repeated twice at 7 d intervals. Resistant calli were transferred to regeneration medium (LS containing 0.2 mg L−1 kinetin, 2.5 g L−1 Phytagel, and 30 g L−1 maltose) containing 30 mg L−1 Hm. After 5 to 7 d, the callus sections containing germinating shoots and viable embryos were transferred onto MS0 medium (Bablak et al., 1995) supplemented with 30 mg L−1 Hm and were placed in the light. When plantlets had reached a size of 3 to 4 cm in height, they were separated gently from any remaining necrotic callus and removed to one-half strength MS0 to enhance root development. The transformation frequency was expressed in terms of average number of transgenic plants derived per gram of tissue. After rooting, plants were transferred to soil as described previously (Bablak et al., 1995).
Genomic Analysis of Transgenic B. distachyon Lines
Total genomic DNA was isolated from leaves using a modified cetyl-trimethyl-ammonium bromide protocol (Murray and Thompson, 1980) and was quantified after RNase treatment. Plants were screened using PCR amplification of the introduced hgh gene. PCR primers (5′-CCTGAACTCACCGCGAC-3′ and 3′-GCTCATCGAGA-GCCTGC-5′) were used to amplify a 516-bp fragment encoding the hgh gene, which was analyzed by electrophoresis in 0.8% (w/v) agarose/ethidium bromide gels. For Southern analysis, BamHI-digested genomic DNA (10 μg lane−1) was separated on an agarose gel, blotted onto a membrane, and probed with a radioactive probe following a standard protocol (Sambrook et al., 1989). The radioactive probe was prepared by the random primer method (Feinberg and Volgstein, 1983), and consisted of a 1.07-kb BamHI fragment containing the coding sequence of the hgh gene.
Challenge with Fungal Pathogens
All fungal infections were performed on batches of 24 B. distachyon plants with each challenge repeated at least three times on seedlings aged from 3 to 4 weeks post-germination. Diseased cereal plants with confirmed infections of Blumeria graminis f. sp. avenae, hordei, or triticae were used to infect B. distachyon ecotypes by simply shaking the condiophores on the recipient plants. Challenging with the rust fungal pathogens Puccinia striformis f. sp. hordei (strains BWR 80-1 and PB 60-7), f. sp. triticae (strains WYR 95-6 and IPO 86053; yellow stripe rust), and Puccinia recondita f. sp. hordei (strains BBR 79-1 and PB60-2-2), f. sp. triticae (strains WYRP96-8 and WBRP90-25; brown leaf rust) involved powder spraying a talc/urideospore suspension on to 3- to 4-week-old B. distachyon seedlings. Plants were then bagged for 24 h in high humidity and were incubated at 10°C, after which the bag was removed and the temperature was increased to 24°C. Symptoms were first observed by 16 to 20 d following challenge. M. grisea strain Guy-11 was cultured on Potato Dextrose Agar (Sigma) and the spores were harvested into 0.2% (w/v) gelatin and diluted to 105 spore mL−1. The spore suspension was sprayed onto B. distachyon 3- to 4-week-old seedlings to run off and the challenged material was bagged for 24 h to maintain a high humidity. Symptoms were first observed 3 to 4 d after spraying.
ACKNOWLEDGMENTS
We acknowledge the unpublished data in relation to Brachypodium genome sizes estimated by scanning densitometry provided by Shi Ying, John Bailey, and Clive Stace (University of Leicester, Leicester, UK). We thank Pilar Catalan (Universidad de Zaragoza, Zaragoza, Spain) for discussions in relation to Brachypodium phylogeny, and we thank Nick Talbot (University of Exeter, Exeter UK), Tim Carver (Institute of Grassland and Enviromental Research, Aberystwyth, UK), and Lesley Boyd (John Innes Centre, Norwich Research Park, Norwich, UK) for supplying fungal pathogens and aid in challenging B. distachyon ecotypes. We thank Derek Fallding for excellent technical assistance with the molecular cytogenetics. Thanks are due also to David Summers, Pat Causton, and Mark Levy who provided the plants used in this research. We acknowledge the preliminary analysis of pathogen interactions in Brachypodium provided by Greg Shelley and Joel Smith. Kathryn Bailey and Joel Smith provided assistance with optimization of the transformation conditions and preliminary analysis of transgenic plants. We would also like to thank Danny Thorogood, Ian King, Iain Donnison, Judith Webb, Howard Thomas, and Chris Pollock (Institute of Grassland and Enviromental Research) for their advice and support of the B. distachyon project.
Footnotes
This work was supported in part by the Gatsby Foundation (grant to J.D.) and by the Biotechnology and Biological Sciences Research Council (PhD studentship to A.P.M.R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010196.
LITERATURE CITED
- Abbo S, Dunford RP, Foote TN, Reader SM, Flavell RB, Moore G. Organization of retro-element and stem-loop repeat families in the genomes and nuclei of cereals. Chromosome Res. 1995;3:5–15. doi: 10.1007/BF00711156. [DOI] [PubMed] [Google Scholar]
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromeric sequence. Chromosoma. 1996;10:261–268. doi: 10.1007/BF02524643. [DOI] [PubMed] [Google Scholar]
- Bablak P, Draper J, Davey MR, Lynch PT. Plant regeneration and micropropagation of Brachypodium distachyon. Plant Cell Tissue Organ Culture. 1995;42:97–107. [Google Scholar]
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses: 807 new estimates. Ann Bot. 2000;86:859–909. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Ann Bot. 1995;76:113–176. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts and genome size in angiosperms: 583 new estimates. Ann Bot. 1997;80:169–196. [Google Scholar]
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Phil Trans Soc London. 1976;B274:227–345. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB, Heslop-Harrison JS. Nuclear DNA amounts in angiosperms. Proc Royal Soc London. 1982;B216:179–199. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, San Miguel P, Chen M, Tikhonov A, Francki M, Avramova Z. Grass genomes. Proc Natl Acad Sci USA. 1998;95:1975–1978. doi: 10.1073/pnas.95.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas GCG, Chen DF, Elliott MC. A routine system for generation of transgenic rice (Oryza sativa L.) plants by microprojectile bombardment of embryogenic cell clusters. Plant Sci. 1998;133:203–210. [Google Scholar]
- Cao J, Duan XL, McElroy D, Wu R. Regeneration of herbicide resistant transgenic rice plants following microprojectile-mediated transformation of suspension-culture cells. Plant Cell Rep. 1992;11:586–591. doi: 10.1007/BF00233098. [DOI] [PubMed] [Google Scholar]
- Carver TLW, Ingerson-Morris SM, Thomas BJ, Zeyen RJ. Early interactions during powdery mildew infection. Can J Bot. 1995;73:632–639. [Google Scholar]
- Catalan P, Kellogg EA, Olmstead RG. Phylogeny of Poaceae subfamily Pooideae based on chloroplast ndhF gene sequences. Mol Phylogen Evol. 1997;8:150–166. doi: 10.1006/mpev.1997.0416. [DOI] [PubMed] [Google Scholar]
- Catalan P, Olmstead RG. Phylogenetic reconstruction of the genus Brachypodium P-Beauv. (Poaceae) from combined sequences of chloroplast ndhF gene and nuclear ITS. Plant System Evol. 2000;220:1–19. [Google Scholar]
- Catalan P, Shi Y, Armstrong L, Draper J, Stace CA. Molecular phylogeny of the grass genus Brachypodium P-Beauv based on RFLP and RAPD analysis. Bot J Linnean Soc. 1995;117:263–280. [Google Scholar]
- Chan MT, Lee TM, Chang HH. Transformation of Indica rice (Oryza sativa L.) mediated by Agrobacterium tumefaciens. Plant Cell Physiol. 1992;33:577–583. [Google Scholar]
- Chen M, San Miguel P, de Oliveira AC, Woo SS, Zhang H, Wing RA, Bennetzen JL. Microcolinearity in sh2-homologous regions of the maize rice and sorghum genomes. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Ecker JR, Briggs S, Caboche M, Coruzzi GM, Cook D, Dangl J, Grant S, Guerinot ML, Henikoff S. National Science Foundation-Sponsored Workshop Report: “The 2010 Project” functional genomics and the virtual plant: a blueprint for understanding how plants are built and how to improve them. Plant Physiol. 2000;123:423–426. doi: 10.1104/pp.123.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Deshpande VG, Ranjekar PK. Repetitive DNA in three Gramineae species with low DNA content. Hoppe Seylers Z Physiol Chem. 1980;361:1223–1233. doi: 10.1515/bchm2.1980.361.2.1223. [DOI] [PubMed] [Google Scholar]
- Devos KM, Beales J, Nagamura Y, Sasaki T. Arabidopsis-rice: will colinearity allow gene prediction across the eudicot-monocot divide? Genome Res. 1999;9:825–829. doi: 10.1101/gr.9.9.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson D, Cyranoski D. Commercial sector scores success with whole rice genome. Nature. 2001;409:551. doi: 10.1038/35054705. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Binarova P, Lucretti S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant. 1989;31:113–120. [Google Scholar]
- Dolezel J, Greilhubers J, Lucretti S, Meister A, Lysak MA, Nardi L, Obermayers R. Plant Genome size estimates by flow cytometry: inter-laboratory comparison. Ann Bot. 1998;82:17–26. [Google Scholar]
- Draper J, Scott R, Armitage P, Walden R. Plant Genetic Transformation and Gene Expression: A Laboratory Manual. Oxford: Blackwell Scientific Publications; 1988. [Google Scholar]
- Dunwell JM. Transgenic approaches to crop improvement. J Exp Bot. 2000;51:487–496. doi: 10.1093/jexbot/51.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- Ellis MC. “Spot-on” SNP genotyping. Genome Res. 2000;10:895–897. doi: 10.1101/gr.10.7.895. [DOI] [PubMed] [Google Scholar]
- Enoki H, Izawa T, Kawahara M, Komatsu M, Koh S, Kyozuka J, Shimamoto K. Ac as a tool for the functional genomics of rice. Plant J. 1999;19:605–613. doi: 10.1046/j.1365-313x.1999.00549.x. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Volgstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Foote T, Roberts M, Kurata N, Sasaki T, Moore G. Detailed comparative mapping of cereal chromosome regions corresponding to the Ph1 locus in wheat. Genetics. 1997;147:801–807. doi: 10.1093/genetics/147.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm ME, Morrish F, Armstrong C, Williams R, Thomas J, Klein TM. Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Bio/Technology. 1990;8:833–839. doi: 10.1038/nbt0990-833. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell-cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Dyer TA. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980;8:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA. Rice as a model for cereal genomics. Curr Opin Plant Biol. 1999;2:86–89. doi: 10.1016/S1369-5266(99)80018-1. [DOI] [PubMed] [Google Scholar]
- Gould J, Devey M, Hasegawa O, Ulian EC, Peterson G, Smith RH. Transformation of Zea mays L. using Agrobacterium tumefaciens and the shoot apex. Plant Physiol. 1991;95:426–434. doi: 10.1104/pp.95.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gura T. Reaping the plant gene harvest. Science. 2000;287:412–414. doi: 10.1126/science.287.5452.412. [DOI] [PubMed] [Google Scholar]
- Hansen G, Wright MS. Recent advances in the transformation of plants. Trends Plant Sci. 1999;4:226–231. doi: 10.1016/s1360-1385(99)01412-0. [DOI] [PubMed] [Google Scholar]
- Havukkala IJ. Cereal genome analysis using rice as a model. Curr Opin Genet Dev. 1996;6:711–714. doi: 10.1016/s0959-437x(96)80025-6. [DOI] [PubMed] [Google Scholar]
- Izawa T, Ohnishi T, Nakano T, Ishida N, Enoki H, Hashimoto H, Itoh K, Terada R, Wu C, Miyazaki C. Transposon tagging in rice. Plant Mol Biol. 1997;35:219–229. [PubMed] [Google Scholar]
- Keller B, Feuillet C. Colinearity and gene density in grass genomes. Trends Plant Sci. 2000;5:246–251. doi: 10.1016/s1360-1385(00)01629-0. [DOI] [PubMed] [Google Scholar]
- Khan MA, Stace CA. Breeding relationships in the genus Brachypodium (Poaceae: Pooideae) Nordic J Bot. 1999;19:257–269. [Google Scholar]
- Klein TM, Kornstein L, Sanford JC, Fromm ME. Genetic transformation of maize by particle bombardment. Plant Physiol. 1989;91:440–444. doi: 10.1104/pp.91.1.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauge R, Goodwin PH, De Wit PJ, Joosten MH. Specific HR-associated recognition of secreted proteins from Cladosporium fulvum occurs in both host and non-host plants. Plant J. 2000;23:735–745. doi: 10.1046/j.1365-313x.2000.00843.x. [DOI] [PubMed] [Google Scholar]
- Li LC, Qu RD, Dekochko A, Fauquet C, Beachy RN. An improved rice transformation system using the biolistic method. Plant Cell Rep. 1993;12:250–255. doi: 10.1007/BF00237129. [DOI] [PubMed] [Google Scholar]
- McCouch S. Toward a plant genomics initiative: thoughts on the value of cross-species and cross-genera comparisons in the grasses. Proc Natl Acad Sci USA. 1998;95:1983–1985. doi: 10.1073/pnas.95.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Blowers AD, Jenes B, Wu R. Construction of expression vectors based on the rice actin1 (Act1) 5′ region for use in monocot transformation. Mol Gen Genet. 1991;231:150–160. doi: 10.1007/BF00293832. [DOI] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:679–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E, Sommerville CR. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Mooney PA, Goodwin PB, Dennis ES, Llewellyn DJ. Agrobacterium tumefaciens gene transfer into wheat tissues. Plant Cell Tissue Organ Cult. 1991;25:209–218. [Google Scholar]
- Moore G, Gale MD, Kurata N, Flavell RB. Molecular analysis of small grain cereal genomes: current status and prospects. Bio/Technology. 1993;11:584–589. [Google Scholar]
- Murray HG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus I. Gene transfer to cereals: an assessment. Bio/Technology. 1990;8:535–542. [Google Scholar]
- Pruitt RE, Meyerowitz EM. Characterization of the genome of Arabidopsis thaliana. J Mol Biol. 1986;187:169–183. doi: 10.1016/0022-2836(86)90226-3. [DOI] [PubMed] [Google Scholar]
- Redei GP. Arabidopsis thaliana (L) Heynh: a review of the genetics and biology. Biblio Genet. 1970;20:1–153. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Practical in situ hybridization. Oxford: BIOS; 2000. pp. 90–124. [Google Scholar]
- Shi Y. Molecular studies of the evolutionary relationships of Brachypodium (Poaceae). PhD thesis. Leicester, UK: University of Leicester; 1991. [Google Scholar]
- Shi Y, Draper J, Stace C. Ribosomal DNA variation and its phylogenetic implications in the genus Brachypodium (Poaceae) Plant Systematics Evol. 1993;188:125–138. [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Tikhonov AP, San Miguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramova Z. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingay S, McElroy D, Kalla R, Fieg S, Wang MB, Thornton S, Brettell R. Agrobacterium tumefaciens-mediated barley transformation. Plant J. 1997;11:1369–1376. [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JD. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell. 1999;11:1841–1852. doi: 10.1105/tpc.11.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi AK, Mohanty A. Rice transformation for crop improvement and functional genomics. Plant Sci. 2000;158:1–18. doi: 10.1016/s0168-9452(00)00325-3. [DOI] [PubMed] [Google Scholar]
- Unfried I, Gruendler P. Nucleotide sequence of the 5.8S and 25S rDNA genes and the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990;18:4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Brown SM, Re D, Fromm ME, Vasil IK. Stably transformed callus lines from microprojectile bombardment of cell suspension cultures of wheat. Bio/Technology. 1991;9:734–747. [Google Scholar]
- Vizir I, Thorlby G, Mullingan B. Foster GD, Twell D, eds, Plant Gene Isolation: Principles and Practice. New York: John Wiley & Sons; 1996. Classical mutagenesis and genetic analysis; pp. 214–245. [Google Scholar]
- Warner SAJ, Scott R, Draper J. Isolation of an Asparagus intracellular PR gene (AoPR1) wound-responsive promoter by the inverse polymerase chain-reaction and its characterization in transgenic tobacco. Plant J. 1993;3:191–201. doi: 10.1046/j.1365-313x.1993.t01-11-00999.x. [DOI] [PubMed] [Google Scholar]
- Zhong X-B, Bodeau J, Fransz PF, Williamson VM, van Kammen A de, Jong JH, Zabel P. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L respectively. Theor Appl Genet. 1999;98:365–370. [Google Scholar]