Abstract
To understand primary cell wall assembly in Arabidopsis, we have focused on identifying and characterizing enzymes involved in xyloglucan biosynthesis. Nine genes (AtFUT2–10) were identified that share between 47% and 62% amino acid similarity with the xyloglucan-specific fucosyltransferase AtFUT1. Reverse transcriptase-PCR analysis indicates that all these genes are expressed. Bioinformatic analysis predicts that these family members are fucosyltransferases, and we first hypothesized that some may also be involved in xyloglucan biosynthesis. AtFUT3, AtFUT4, and AtFUT5 were expressed in tobacco (Nicotiana tabacum L. cv BY2) suspension culture cells, and the resulting proteins did not transfer fucose (Fuc) from GDP-Fuc to tamarind xyloglucan. AtFUT3, AtFUT4, and AtFUT5 were overexpressed in Arabidopsis plants. Leaves of plants overexpressing AtFUT4 or AtFUT5 contained more Fuc than wild-type plants. Stems of plants overexpressing AtFUT4 or AtFUT5 contained more xylose, less arabinose, and less galactose than wild-type plants. We suggest that the AtFUT family is likely to include fucosyltransferases important for the synthesis of wall carbohydrates. A targeted analysis of isolated cell wall matrix components from plants altered in expression of these proteins will help determine their specificity and biological function.
There is increasing interest in the role of the primary cell wall during plant development and in the wall's dynamic nature (Cosgrove, 1997a, 1997b; Kohorn, 2000). Cellulose microfibrils are the major polysaccharide present in primary walls and are synthesized at the plasma membrane in rosette-like structures. These microfibrils are embedded in a matrix of structurally complex polysaccharides (hemicellulose and pectins) that are synthesized in the endomembrane system and then secreted and inserted into the cell wall. The identification of enzymes that synthesize cell wall polysaccharides is necessary to address the regulation and function of wall components and the mechanisms of their assembly. Biochemical approaches to identify the enzymes that synthesize wall components have met with limited success due to a loss of enzymatic activity upon solubilization, the unavailability of soluble acceptor substrates, the absence of important cofactors, and in some cases the possible requirement of multiple enzyme complexes (Kawagoe and Delmer, 1997). However, this approach has been successful in several instances. The genes, which encode a fucosyltransferase that adds the terminal Fuc to xyloglucan from Arabidopsis (AtFUT1, formerly AtFT1; Perrin et al., 1999) and from pea (Pisum sativum; PsFUT1, formerly PsFT1; Faik et al., 2000), were cloned using a biochemical approach. (In previous publications [Perrin et al., 1999; Faik et al., 2000], the abbreviation FTase was used to refer to fucosyltransferase. However, the abbreviation FUT is more widely used in the glycosyltransferase field [for example, see McCurley et al., 1995], and furthermore the abbreviation FTase has been used by others to refer to farnesyltransferase, an unrelated enzyme.) Similarly, a biochemical approach was used to clone the galactosyltransferase involved in galactomannan synthesis in fenugreek (Edwards et al., 1999). Genetic screens have been used to identify mutants altered in cell wall composition (Reiter et al., 1997; Chen et al., 1998) and to identify some cell wall biosynthetic enzymes (for example, see Nickle and Meinke, 1998; Taylor et al., 1999; Favery et al., 2001). However, lethality and redundancy may prevent mutations in many cell wall biosynthetic genes from being recovered. Now that the genome of Arabidopsis is sequenced (Arabidopsis Genome Initiative, 2000), reverse genetic approaches are feasible to characterize the function of candidate cell wall biosynthetic enzymes.
Our main interest is to identify enzymes involved in the synthesis of xyloglucan found in the primary wall of most plants. The identification of the xyloglucan fucosyltransferases AtFUT1 and PsFUT1 (Perrin et al., 1999; Faik et al., 2000) provided a tool for the identification of related genes in Arabidopsis. AtFUT1 and PsFUT1 transfer Fuc from a GDP-Fuc donor to the 2-position of Gal on the xyloglucan acceptor. These proteins are predicted to be Golgi-localized type II membrane proteins containing a short cytoplasmic tail at the amino terminus, followed by a short, single transmembrane domain that is separated from the globular (catalytic) portion of the protein by a variable length stem region. Our preliminary evidence suggests that AtFUT1 is Golgi localized (R. Sarria, V. Kovaleva, K. Keegstra, and N.V. Raikhel, unpublished data).
Nine additional genes that share amino acid identity with AtFUT1 were identified from the Arabidopsis genome database. Because few plant fucosyltransferases have been biochemically and molecularly characterized (Oriol et al., 1999), it is not possible to predict the functions of the enzymes related to AtFUT1 based only on their amino acid sequence. In contrast, many fucosyltransferases have been characterized in bacteria and other eukaryotes because extracellular Fuc is important in cell recognition and signaling. Fucosylation of Gal and GlcNAc residues present on N-linked glycans and on glycolipids generates various types of Lewis epitopes (Oulmouden et al., 1997; Breton et al., 1998; Oriol et al., 1999; Pykari et al., 2000). These epitopes have been shown to be ligands of selectins (adhesion receptors), and are useful markers to diagnose various types of tumors (for review, see Staudacher et al., 1999). Fucosyl residues also have a role in notch receptor signaling (Bruckner et al., 2000; Moloney et al., 2000a, 2000b) and in the symbiotic interaction between legumes and Rhizobium species (Lopez-Lara et al., 1996; Mergaert et al., 1996; Quesada-Vincens et al., 1997; Quinto et al., 1997). Although many of the genes encoding the fucosyltransferases involved in the above processes have been identified, there are numerous putative fucosyltransferase genes present in microorganisms, plants, and animals with unknown biological functions and substrate specificities.
Amino acid sequence comparisons (Breton et al., 1998; Oriol et al., 1999) have been used to place fucosyltransferases into groups based on the Fuc linkage that they catalyze (for example, α-1,2-fucosyltransferases) and sometimes on their acceptor substrate. Members of the same group often share greater than 40% amino acid identity, but there is very little amino acid identity (27% at most) between fucosyltransferase groups.
When the nine additional genes related to AtFUT1 were identified, we considered that some may encode proteins that are redundant to AtFUT1 and transfer Fuc to xyloglucan and that some members of the AtFUT family transfer Fuc to other acceptor substrates in the cell. Thus, we have begun to characterize the new genes related to AtFUT1, and we present the approaches we have taken and the results we have obtained thus far to address the biological functions of individual AtFUT family members.
RESULTS
A Family of Arabidopsis Genes Related to AtFUT1
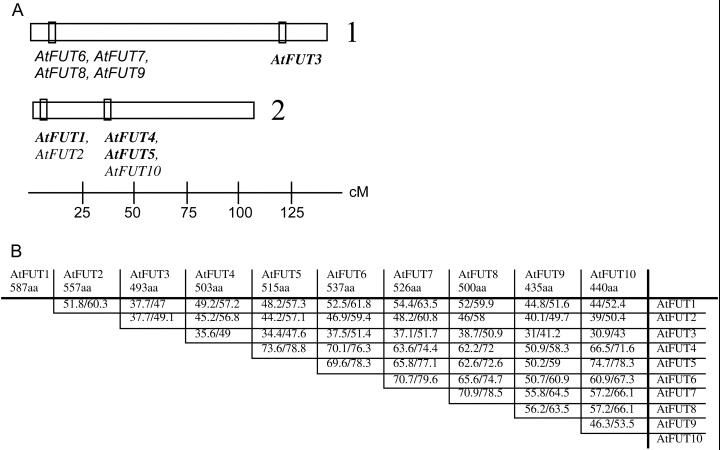
We identified a total of nine sequences from the Arabidopsis database with significant amino acid identity to AtFUT1. All of these genes, named AtFUT2–10 in the order in which they were found, are located on chromosomes 1 or 2 and clustered in four bacterial artificial chromosome (BAC) clones (Fig. 1A).
Figure 1.
A, Diagram showing the chromosomal location of Arabidopsis sequences with amino acid identity to the AtFUT1 protein. Boxes indicate the location of the BAC clones that contain AtFUT genes (F7A19/F16A14, AtFUT6, AtFUT7, AtFUT8, AtFUT9; F1M20, AtFUT3; T18/E12, AtFUT1, AtFUT2; and F26H6, AtFUT4, AtFUT5, AtFUT10). Functions were pursued for the genes in bold font. B, Amino acid identity/similarity between the predicted full-length sequences of the AtFUT family. Pair-wise analysis was done using Clustal W with a gap opening penalty of 10 and a gap extension penalty of 0.1.
With AtFUT1 and PsFUT1, the new genes make up a distinct family of plant glycosyltransferases (no. 37; http://afmb.cnrs-mrs.fr/∼pedro/CAZY; Henrissat and Davies, 2000) and are predicted to be Golgi-localized type II membrane proteins. The gene structure is conserved among family members, each containing a single intron that separates the cytosolic and transmembrane domains from the catalytic domain. The exceptions to this structure are AtFUT4, which lacks an intron, and AtFUT10, which (as currently annotated) is lacking the first exon and intron. Pair-wise comparisons of the amino acid identity/similarity along the entire coding sequence among the family members shows that AtFUT1 shares 37.7% to 54.4% identity with family members (Fig. 1B). The greatest amino acid identity and similarity is in the predicted catalytic domain. When pair-wise comparisons are limited to the second exon, the amino acid identity between AtFUT1 and the other family members increases to 46.9% to 59.2%. In contrast, AtFUT1 shares 56.1% identity along the entire coding length with PsFUT1, and the second exons of PsFUT1 and AtFUT1 are 67% identical, indicating that AtFUT1 shares more amino acid identity with its orthologue in pea than with the Arabidopsis family members.
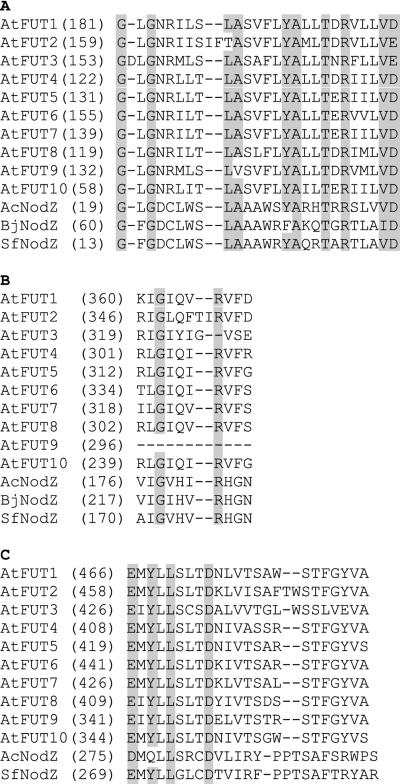
Amino acid sequence comparisons between AtFUT family members and bacterial NodZ α-1,6-fucosyltransferases indicate that the family members contain motifs common to fucosyltransferase proteins (Fig. 2). Figure 2A shows a motif of unknown significance that is present in members of the AtFUT family and in bacterial NodZ, but it is absent in α-1,6-fucosyltransferases from other organisms. Figure 2B shows a motif that is shared between α-1,2- and α-1,6-fucosyltransferases and that is hypothesized to bind GDP-Fuc (Takahashi et al., 2000). The presence of this motif suggests that AtFUT family members are fucosyltransferases. However, AtFUT3 lacks the otherwise invariant Arg residue in this motif that was found to be essential for in vitro activity of human FUT1 (Takahashi et al., 2000), and AtFUT9 is completely lacking this motif. The functional significance of these disruptions in AtFUT3 and AtFUT9 is not known. Faik et al. (2000) noted that AtFUT1 and PsFUT1 combined motifs that were previously thought to distinguish between α-1,2- and α-1,6-fucosyltransferases. Figure 2C shows that all members of the AtFUT family contain this combination of motifs. A phylogenetic tree was generated using the predicted amino acid sequences of the AtFUT family, fucosyltransferases that generate Lewis epitopes (Pykari et al., 2000), enzymes that fucosylate the core of N-linked glycans (Leiter et al., 1999), and enzymes that fucosylate Rhizobium nodulation factors (Lopez-Lara et al., 1996; Mergaert et al., 1996). Figure 3 shows that PsFUT1 and the AtFUT family are distinct from the N-linked glycan fucosyltransferases. The level of amino acid identity between AtFUT family members and these other fucosyltransferases was never higher than 12%. Taken together, this bioinformatic information strongly supports the hypothesis that the AtFUT family is a new group of fucosyltransferases. We predict that, like AtFUT1 and PsFUT1, these new family members are involved in fucosylating cell wall carbohydrates.
Figure 2.
Sequence alignment of Arabidopsis AtFUT proteins with α-1,6-fucosyltransferases from bacteria shows peptide motifs present in the AtFUT family. A, Region strongly conserved between family members. B, Motif shared by α-1,2- and α-1,6-fucosyltransferases hypothesized to bind GDP-Fuc. The shaded Arg (R) was shown to be necessary for human FUT1 activity in vitro. C, Combination of motif III from α-1,6-glycosyltransferases and motif III from α-1,2-glycosyltransferases that is conserved in the AtFUT family. GenBank accession numbers: AF417473, AtFUT3; AF417474, AtFUT4; AF417475, AtFUT5; AC005313, AtFUT2 (At2g03210); AC006920, AtFUT10 (At2g15350); AC007576, AtFUT6 (At1g14080), AtFUT7 (F7A19.15), AtFUT8 (At1g14100), and AtFUT9 (At1g14110). GenBank protein ID numbers: g1293900, Azorhizobium caulinodans NodZ; g404790, Bradyrhizobium japonicum NodZ; g3347912, Sinorhizobium fredii NodZ; and g5231145, AtFUT1.
Figure 3.
Phylogenetic tree showing the relationships of the AtFUT proteins with each other and with fucosyltransferases from other organisms. The tree was generated using CLUSTAL W for alignment, a BLOSOM 45 matrix, gap opening penalty of 13, and gap extension penalty of 0.05. AtFUT1 and PsFUT1 are α-1,2-fucosyltransferases. FUT1 and FUT2 encode α-1,2-fucosyltransferases, NodZ and FUT8 encode α-1,6-fucosyltransferases, and FucT C3 is the enzyme that adds Fuc to the core structure of N-linked glycoproteins in the mung bean (Vigna radiata) plant. FUT3-7 encode α-1,3/1,4-fucosyltransferases that are responsible for many blood group antigens in humans. GenBank protein ID numbers (not in Fig. 2) are: g19526, human FUT1; g12643984, pig FUT1; g1730125, human FUT2; g1730131, rat FUT2; g4758408, human FUT8; g5702039, vigna FucT C3; g4503811, human FUT4; g4503809, human FUT3; g4503815, human FUT6; g1730137, human FUT7; and g7453579, PsFUT1.
Expression Profiles of the AtFUT1-Like Genes
Early in this study, cDNAs for three of the AtFUT1-like family members were identified. AtFUT3 and AtFUT4 were found in the Arabidopsis expressed sequence tag collection, and 5′-RACE was used to determine the 5′ end of these genes (cDNA sizes: 1,482 and 1,512 bp, respectively). For AtFUT3, 5′-RACE consistently generated a sequence 195 bp shorter than the sequence predicted in the Arabidopsis database and resulted in a 65-amino acid shortening of the predicted polypeptide. An AtFUT5 cDNA was obtained by direct PCR amplification from the seedling cDNA library CD4-14. Sequencing of the PCR amplified fragment revealed that it corresponded to the intron-less version of the gene and was 1,548 bp long.
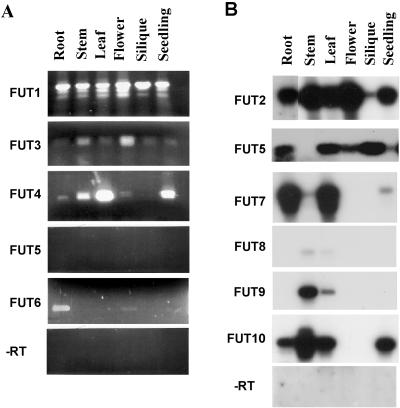
To determine whether the AtFUT genes were differentially expressed, we used reverse transcriptase (RT)-PCR with gene specific primers to characterize their expression (Fig. 4, A and B). AtFUT1 was used as a positive control because it is expressed in all of the organs examined. For these RT-PCR experiments, only amplification of AtFUT3, AtFUT4, and AtFUT6 generated products visible in ethidium bromide-stained gels (Fig. 4A). AtFUT3 is expressed in all tissues analyzed; however, transcript levels were higher in flowers and stem tissues. AtFUT4 is expressed in all tissues with higher transcript accumulation in stems, leaves, and 7-d-old seedlings (Fig. 4A). AtFUT6 was expressed in roots and in flowers (Fig. 4A). Lack of AtFUT5 amplification was unexpected (Fig. 4A), considering that this gene was cloned by direct PCR amplification from an Arabidopsis cDNA library. Southern-blot analysis of AtFUT5 RT-PCR reactions revealed PCR products in roots, flower, siliques, and leaves (Fig. 4B). The same analysis was extended to AtFUT2, AtFUT7, AtFUT8, AtFUT9, and AtFUT10, revealing that these genes were expressed at very low levels (Fig. 4B). It is interesting that some of the genes that were clustered on the same BAC have similar expression patterns. AtFUT2 was expressed in the same tissues as AtFUT1. AtFUT8 and AtFUT9 were both expressed in stems and leaves, although AtFUT8 was barely detectable. The pattern of AtFUT10 expression was similar to AtFUT4 expression except that AtFUT10 was not expressed in flower tissue. AtFUT7 transcripts were present primarily in root and leaf tissues with lower accumulation of transcripts in stem and 7-d-old seedlings. The response of the AtFUT genes to biotic or abiotic stresses was not addressed. In conclusion, all members of the AtFUT family are expressed and there are both distinct and overlapping expression patterns among family members.
Figure 4.
RT-PCR showing the expression pattern of the AtFUT gene family. A, RT-PCR products stained by ethidium bromide. AtFUT1 was used as a positive control because its RT-PCT product was present in all tissues analyzed. Negative controls (without reverse transcriptase) are blank, indicating that there was no DNA contamination. B, Southern analysis of RT-PCR products was done for genes that did not produce visible bands on ethidium bromide-stained gels.
Biochemical Assays for Fucosyltransferases
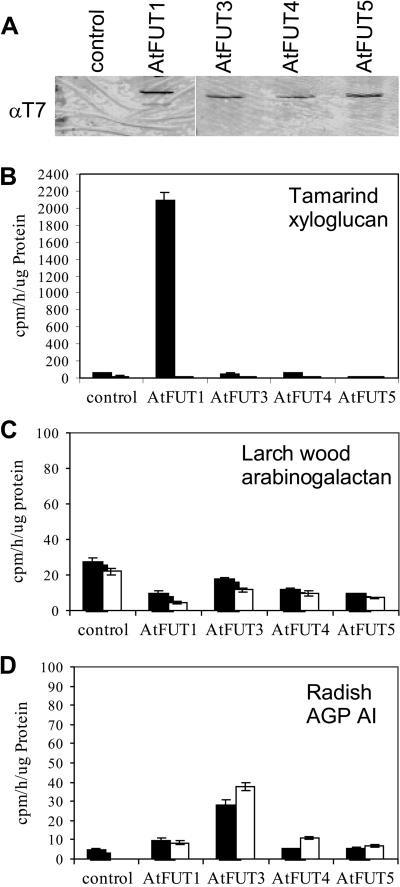
Next, we pursued a biochemical approach to find a function for AtFUT3, AtFUT4, and AtFUT5. Because AtFUT1 and PsFUT1 are very specific for the acceptor substrate xyloglucan (Faik et al., 2000), it was worthwhile to test AtFUT3-5 for similar activity. Tobacco (Nicotiana tabacum L. cv BY2) xyloglucan lacks Fuc (York et al., 1996); therefore, we used BY2 cell cultures as an expression system to produce T7/His-tagged versions of AtFUT3, AtFUT4, and AtFUT5 (Fig. 5A). AtFUT3, AtFUT4, and AtFUT5 did not exhibit any activity compared with AtFUT1 (Fig. 5B) using naturally occurring, non-fucosylated tamarind xyloglucan as an acceptor substrate and GDP 14C-Fuc as a donor substrate (Perrin et al., 1999). Thus, AtFUT3-5 may not be involved in xyloglucan biosynthesis.
Figure 5.
Activity assays for AtFUT3, AtFUT4, and AtFUT5 produced in tobacco cells. A, Western blot (α T7) showing the tobacco lines expressing tagged versions of AtFUT1, AtFUT3, AtFUT4, and AtFUT5. B, Assay showing GDP-14C Fuc incorporation into tamarind xyloglucan. AtFUT1 was used as a positive control. C and D, Activity assays using larch wood arabinogalactan and radish (Raphanus sativus) arabinogalactan protein (AGP)-AI as acceptor substrates for GDP-14C Fuc incorporation, respectively. Results are the average of two separate experiments. For all graphs, black bars indicate assay done in the presence of an acceptor and white bars indicate absence of acceptor.
Aside from xyloglucan, fucosyl residues are also present in the pectins rhamnogalacturonan I (RG-I) and rhamnogalacturonan II (RG-II; for review, see Mohnen, 1999; Fransen et al., 2000; Vidal et al., 2000) and in some AGPs (Nakamura et al., 1984; Tsumuraya et al., 1984; Ogura et al., 1985; Misawa et al., 1996). In addition, Fuc is also present in N-linked glycoproteins (Fitchette-Laine et al., 1997; Fitchette et al., 1999). However, the proteins related to AtFUT1 are distinct from fucosyltransferases involved in N-linked glycosylation. An α-1,3-fucosyltransferase (GlcNAc) gene has been cloned from mung bean (Leiter et al., 1999), and this fucosyltransferase and three related proteins from Arabidopsis (Wilson et al., 2001) are in glycosyltransferase family 10 (http://afmb.cnrs-mrs.fr/∼pedro/CAZY), a separate family from AtFUT1. We assayed the ability of AtFUT3-5 to fucosylate larch wood arabinogalactan (Sigma, St. Louis) and radish root AGP-AI (gift from Y. Hashimoto, Department of Biochemistry, Saitama University, Urawa, Japan; Fig. 5, C and D), but no detectable amounts of 14C-Fuc were incorporated into these AGPs.
Overexpression Phenotypes and Sugar Composition Analysis of the Cell Walls
We expressed the T7-tagged versions of AtFUT3, AtFUT4, and AtFUT5 driven by the 35S promoter in Arabidopsis to look for morphological phenotypes and changes in cell wall carbohydrate composition. Lines segregating for a single insertion were selected and expression of the transgenes was confirmed by northern and western analysis (data not shown). Lines overexpressing AtFUT4 or AtFUT5 were phenotypically indistinguishable from wild type. In contrast, homozygous AtFUT3 plants (Fig. 6) were stunted and had small, curly leaves and shorter petioles compared with wild-type plants. These plants did not survive to set seeds in culture medium or on soil. This phenotype was found in multiple lines and the severity of this phenotype varied among plants from the same line. Thus, overexpression of AtFUT3 was detrimental to plant development.
Figure 6.

Phenotype of Arabidopsis plants overexpressing the AtFUT3 gene. At 10 d, lines overexpressing AtFUT3 segregated for dwarf plants compared with wild type. The dwarf phenotype was not fertile; thus, it could not be confirmed genetically that dwarf plants were homozygous for the transgene. However, homozygous lines could not be generated from phenotypically normal plants.
To determine whether overexpression of these genes altered cell wall carbohydrates, leaf and stem tissues from two independent lines for each gene were analyzed for cell wall neutral glycosyl residue composition (Tables I and II). Leaf tissue from plants that overexpressed AtFUT3, AtFUT4, and AtFUT5 showed significant differences in cell wall composition compared with wild-type leaves (Table I). However, these differences were not always observed in both independent lines. It is interesting that leaves from plants that overexpressed AtFUT4 and AtFUT5 consistently showed 33% more Fuc than wild type. In addition to Fuc, the levels of other sugars were altered in plants that overexpressed AtFUT4 and AtFUT5. For example, compared with wild-type leaves, AtFUT4-2 and AtFUT5-1 also showed a 19% increase in Rha, a 13% decrease in Ara, and a 6% to 10% decrease in Gal (Table I). Because both lines that overexpress AtFUT3 contain plants that show abnormal leaf development, and because AtFUT3 is predicted to be Golgi localized, we expected to see changes in leaf cell wall carbohydrate for plants that overexpressed AtFUT3. However, only one AtFUT3 line showed a change in cell wall carbohydrate composition (Table I). In conclusion, overexpression of AtFUT4 and AtFUT5 increased the amount of Fuc in the leaf cell wall.
Table I.
Neutral glycosyl residue compositions (mol %)a of alcohol insoluble residue from Arabidopsis leaves overexpressing AtFUT genes (average of three analyses ± sd)
| Plant | Rha | Fuc | Ara | Xyl | Man | Gal |
|---|---|---|---|---|---|---|
| Wild type | 16 ± 0.6 | 3 ± 0 | 23 ± 0.6 | 21 ± 1.0 | 5 ± 0 | 32 ± 1.2 |
| AtFUT3-1 | 15 ± 0.6 | 4 ± 0.6 | 24 ± 0.6 | 22 ± 0.6 | 5 ± 0.6 | 30 ± 1.2 |
| AtFUT3-2 | 18 ± 0.6 | 3 ± 1.0 | 22 ± 1.2 | 19 ± 1.7 | 6 ± 0.6 | 32 ± 1.5 |
| AtFUT4-1 | 18 ± 1.0 | 4 ± 0 | 22 ± 1.2 | 22 ± 1.7 | 5 ± 0 | 29 ± 1.5 |
| AtFUT4-2 | 19 ± 0.6 | 4 ± 0 | 20 ± 0.6 | 22 ± 1.5 | 5 ± 0.6 | 30 ± 0.6 |
| AtFUT5-1 | 19 ± 1.7 | 5 ± 1.0 | 20 ± 0.6 | 21 ± 1.5 | 6 ± 0.6 | 29 ± 1.2 |
| AtFUT5-2 | 16 ± 1.5 | 4 ± 0 | 21 ± 1.0 | 24 ± 3.0 | 5 ± 0 | 30 ± 2.0 |
Values in bold indicate a significant difference (P ≤ 0.05) from the wild-type samples by the Student's t test.
All samples were shown by gas chromatography (GC)-mass spectrometry (MS) to contain small amounts (<0.1%) of 2-O-methyl Fuc.
Table II.
Neutral glycosyl residue compositions (mol %)a of transformed Arabidopsis stem alcohol-insoluble residue (average of three analyses ± sd)
| Plant | Rha | Fuc | Ara | Xyl | Man | Gal |
|---|---|---|---|---|---|---|
| Wild type | 10 ± 0.6 | 2 ± 0.6 | 21 ± 0 | 35 ± 1.2 | 6 ± 0.6 | 26 ± 0.6 |
| AtFUT3-1 | 10 ± 0.6 | 3 ± 0.6 | 25 ± 0 | 32 ± 2.0 | 4 ± 0 | 26 ± 2.3 |
| AtFUT3-2 | 12 ± 0.6 | 3 ± 0 | 21 ± 0.6 | 34 ± 2.0 | 5 ± 0 | 25 ± 1.5 |
| AtFUT4-1 | 10 ± 1.0 | 3 ± 0.6 | 17 ± 1.0 | 44 ± 3.0 | 5 ± 0.6 | 21 ± 1.0 |
| AtFUT4-2 | 10 ± 1.0 | 3 ± 0.6 | 15 ± 0.6 | 48 ± 1.0 | 6 ± 0.6 | 18 ± 1.0 |
| AtFUT5-1 | 11 ± 0.6 | 3 ± 0.6 | 18 ± 0.6 | 43 ± 2.5 | 6 ± 1.0 | 19 ± 1.5 |
| AtFUT5-2 | 11 ± 1.5 | 3 ± 0.6 | 17 ± 0 | 45 ± 3.2 | 6 ± 0.6 | 18 ± 0.6 |
Values in bold indicate a significant difference (P ≤ 0.05) from the wild-type samples by the Student's t test.
All samples were shown by GC-MS to contain small amounts (<0.1%) of 2-O-methyl Fuc.
Significant differences were also seen for stem cell wall carbohydrate composition from plants that overexpressed AtFUT3, AtFUT4, and AtFUT5 (Table II). Overexpressing lines for both AtFUT4 and AtFUT5 showed similar alterations of normal cell wall carbohydrate composition (Table II). Xyl increased 31% and 25%, Ara content was reduced to 24% and 17%, and Gal content was reduced 25% and 29% in AtFUT4 and AtFUT5 plants, respectively, compared with wild type. The stem cell wall carbohydrate composition of lines overexpressing AtFUT3 was different from the wild type and lines overexpressing AtFUT4 and AtFUT5 (Table II). However, the two AtFUT3 lines also showed different cell wall carbohydrate composition. This analysis suggests that AtFUT4 and AtFUT5 are functionally similar to each other and distinct from AtFUT3.
DISCUSSION
The plant cell wall is a complex dynamic structure and identifying the many genes involved in primary cell wall synthesis is necessary for our understanding of plant growth and development. To date, genetic and biochemical approaches have identified only a limited number of genes with confirmed functions. With the Arabidopsis genome sequenced, and sequencing projects of other plant genomes in progress, reverse genetic approaches may also be employed to identify genes involved in cell wall biosynthesis. Our studies demonstrated both the potential and the limitations of functional genomics and reverse genetics in characterizing complex biochemical pathways, in this case the addition of Fuc during the biosynthesis of cell wall polymers. Although the specific lessons learned from these studies are unique to this particular family of putative FUT genes, the results also have broader implications. Many plant genes are part of gene families with several related, but different, members. As with the FUT genes, many of the members of these gene families may perform related, but distinctly different, biochemical reactions. As with the FUT gene products, it will often be difficult to establish biochemical assays to examine the functions of each gene product, in part because one can only guess at the appropriate substrates, and in part because the substrates are not commercially available. As with the FUT genes, reverse genetics may often produce plants with subtle or no phenotype. In some cases, this may be due to redundancy among members of the gene family, but in other cases detailed molecular studies will be needed to determine very subtle phenotypes.
Bioinformatic approaches predict that the nine additional Arabidopsis genes related to AtFUT1 are fucosyltransferases. AtFUT family members contain motifs that are present in α-1,6- and α-1,2-fucosyltransferases, and seven of the new AtFUT proteins contain a motif that is proposed to bind GDP-Fuc (Takahashi et al., 2000). AtFUT1-10 and PsFUT1 have been assigned to glycosyltransferase family 37, which is distinct from the fucosyltransferases that have been identified from fungi, animals, and bacteria (http://afmb.cnrs-mrs.fr/∼pedro/CAZY). Amino acid comparisons (Fig. 1B) and phylogenetic trees (Fig. 3) indicate that the AtFUT family could be subdivided further: AtFUT1, AtFUT2, AtFUT3, each in their own group, AtFUT9, in its own group or included in a larger group containing AtFUT4, -5, -6, -7, -8, and -10. AtFUT1 shares more amino acid identity with PsFUT 1 (56.1%) than with other Arabidopsis family members. AtFUT4 shares a relatively high amount of identity (62.2%–73.6%) with AtFUT5, -6, -7, -8, and -10. However, bioinformatics cannot predict with absolute certainty the donor or acceptor substrates for these enzymes. Many glycosyltransferases show greater conservation of protein secondary structure than of amino acid sequence. As more glycosyltransferases are crystallized and their three-dimensional structures solved, it should be possible to make stronger predictions based on bioinformatics alone (Charnock et al., 2001).
All members of the AtFUT gene family were found to be expressed by RT-PCR. AtFUT2, AtFUT5, AtFUT7, AtFUT8, AtFUT9, and AtFUT10 showed extremely low expression levels, but higher expression levels may not be necessary to produce the amount of protein necessary for activity, given that Fuc is a quantitatively minor constituent of the extracellular matrix. In addition, the encoded proteins may be very stable or have high specific activities (as is the case for AtFUT1). Also, the possibility exists that some of the AtFUT genes are redundant or encode similar enzymes that are expressed in tissue-specific patterns. The expression pattern of AtFUT2 is interesting because AtFUT2 is only 360 bp downstream from the coding region of AtFUT1 and is expressed at low levels in the same tissues as AtFUT1. AtFUT2 expression may be driven by a small promoter present in the spacer region or by the AtFUT1 promoter. In addition, potential intron/exon splice sites are present that could result in an alternative splice variant between AtFUT1 and AtFUT2, although there is no evidence for this. Of the subfamilies predicted by bioinformatics, AtFUT1, AtFUT2, and AtFUT3 were expressed in all the tissues analyzed, perhaps indicating that their respective acceptor substrate is present throughout the plant. In contrast, AtFUT9 showed a more restrictive expression pattern. When combined, the family members of the large subgroup of AtFUT were expressed in all analyzed samples with overlapping expression patterns in roots, stems, and leaves.
AtFUT3, AtFUT4, and AtFUT5 were expressed in tobacco cell lines to produce protein for biochemical assays. These proteins did not fucosylate xyloglucan in vitro. A preliminary analysis of an AtFUT1 knockout mutant showed that the mutant is devoid of Fuc on its xyloglucan (T.A. Wagner, R. Sarria, K. Keegstra, and N.V. Raikhel, unpublished data). If so, this would confirm our prediction that AtFUT1 is the only family member involved in xyloglucan synthesis.
The inability of the AtFUT1-like proteins to transfer Fuc to xyloglucan (Fig. 5B) makes it likely that these proteins fucosylate other acceptors. Several fucosyltransferases would be necessary to fucosylate pectins: an α-1,2-fucosyltransferase to form α-l-Fuc-(1→2)-β-d-Galp-(1→) that is present on RG-I, and an α-1,2-fucosyltransferase to form α-l-Fuc-(1→2)-β-d-Galp-(1→) and an α-1,4-fucosyltransferase to form α-l-Fuc-(1→4)-β-l-Rhap-(1→) (Rhap) that are present in RG-II (Mohnen, 1999; Vidal et al., 2000). It was shown recently that a terminal α-l fucosyl residue is linked to GalA on soybean (Glycine max) pectin (Fransen et al., 2000), which would require an additional fucosyltransferase. Some AGPs are fucosylated (Nakamura et al., 1984; Tsumuraya et al., 1984; Ogura et al., 1985), and an α-1,2-fucosyltransferase has been reported to generate α-l-Fuc-(1→2)-α-l-Araf(1→) that is present on radish root AGPs (Misawa et al., 1996). AGPs show very specific expression patterns (Schultz et al., 2000), and it remains to be determined if these patterns correlate with the patterns we have seen with some AtFUTs.
The current lack of suitable substrates limited our ability to determine biochemically the acceptor substrates for these enzymes. AtFUT3, AtFUT4, and AtFUT5 did not fucosylate radish AGP or larch wood arabinogalactan. However, it is not known if these acceptors had fucosylation sites available. Nevertheless, members of the AtFUT family, except AtFUT1, are candidates for fucosylating RG-I and RG-II. It is possible that AtFUT6 is an orthologue of the radish root AGP fucosyltransferase (Misawa et al., 1996) because of its relatively strong expression in roots.
Fuc is also present in low-Mr glycosides found in plants. For example, it is an integral sugar in the saponins, glycosylated triterpenoids that have been characterized from Quillaja saponaria bark, so an α-1-fucosyltransferase is necessary for saponin synthesis (Guo et al., 2000). It is likely that these molecules are fucosylated by soluble enzymes, not members of the AtFUT family. A family of soluble glycosyltransferases that act on low-Mr glycosides has been described (Li et al., 2001). There is also a possibility that epidermal growth factor repeats found in plants are fucosylated similar to what has been described for the Notch receptor protein (Moloney et al., 2000b). If so, this would necessitate another fucosyltransferase.
The synthesis of complex N-linked glycans in plants requires an α-1,3-fucosyltransferase (GlcNAc) to produce the typical structure, and an α-1,4-fucosyltransferase (GlcNAc) to make a Lewis a epitope (Fitchette-Laine et al., 1997; Fitchette et al., 1999). Available bioinformatic information (http://afmb.cnrs-mrs.fr/∼pedro/CAZY) and the phylogenetic analysis (Fig. 3) makes it improbable that AtFUT family members are involved in N-linked glycosylation.
The tagged versions of AtFUT3, AtFUT4, and AtFUT5 were also expressed in Arabidopsis, and we examined these plants for morphological phenotypes and for alterations in cell wall carbohydrate (neutral glycosyl residue) composition. Plants overexpressing AtFUT3 segregated for stunted and infertile plants, and this may suggest that AtFUT3 inhibits development. However, the alterations in carbohydrate composition of leaf and stem cell walls were not seen in both of the independent lines that overexpressed AtFUT3. In contrast, plants overexpressing AtFUT4 and AtFUT5 looked normal. Overexpression of these proteins in leaves led to an increase in Fuc, and increase in Rha, decrease in Gal, and (less consistently) a decrease in Ara. Overexpression of AtFUT4 and AtFUT5 in stems led to an increase in Xyl, a decrease in Ara, and a decrease in Gal in the cell walls for both genes compared with wild type. In the case of AtFUT4 and AtFUT5 overexpression, the alterations that were detected for multiple cell wall neutral glycosyl residues may indicate that extra fucosylation of a wall polysaccharide (not dectable in stems) had an indirect effect on total wall polysaccharide composition, perhaps by creating additional glycosylation sites or by triggering changes in the polysaccharide composition of stems. Although results from overexpression are difficult to interpret because of the potential for mis-localization of overexpressed protein, these results are consistent with our bioinformatic predictions that suggest that AtFUT4 and AtFUT5 share a similar activity that is distinct from AtFUT3.
For the overexpressing plants, sugar analysis of total cell walls was not sufficient to determine the biological function of these genes. Further analysis of individual cell wall matrix components (RG-I, RG-II, and AGPs) is necessary to determine which, if any, of the Arabidopsis complex carbohydrates are affected by AtFUT4 and AtFUT5 overexpression and to enrich for Fuc-containing components in those cases where no changes in sugar composition were observed. Our preliminary analysis shows that a knockout of AtFUT5 did not show a phenotype and no alterations in cell wall carbohydrate composition was observed (T.A. Wagner, M.A. O'Neill, R. Sarria, K. Keegstra, and N.V. Raikhel, unpublished data). Extensive knockout analysis of multiple AtFUT genes, simultaneously and in multiple combinations (using RNAi or insertional mutations), is required to address the biological function of these genes.
In conclusion, the AtFUT family members appear to be fucosyltransferases by bioinformatic predictions. Using multiple approaches, the results that we have obtained for AtFUT3, -4, and -5 suggest that these AtFUT proteins are not functionally redundant with AtFUT1. Although we are not able to assign specific functions to each of the new family members, the data also suggest that there are at least three distinct biochemical activities encoded by members of this family. A great deal of additional work is still required to determine the precise biochemical and biological function encoded by each of the AtFUT genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis ecotypes Columbia and Wassilewskija were grown in soil at 22°C and 80% relative humidity with 16 h of light. T-DNA-mutagenized seeds of Wassilewskija ecotype were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). BY2 tobacco (Nicotiana tabacum L. cv BY2) cell suspensions were maintained as described (Nagata et al., 1992).
Overexpression Constructs and Plant Transformation
The 35S-cauliflower mosaic virus promoter and the 3′nos termination signal were used to express the AtFUT1, 3, 4, and 5 cDNAs with amino-terminal T7 and His tags. AtFUT1 was cloned into pET28a (Novagen, Madison, WI) as a BamHI/SalI fragment. The SacI/XbaI fragment containing the tagged AtFUT1 gene was cloned into pCAMBIA1300 (Cambia, Canberra, Australia; modified to contain those sites by A. Sanderfoot, personal communication). Similar constructs for AtFUT3, 4, and 5 were generated by replacing AtFUT1 from the above construct with BamHI/SalI fragments of AtFUT3, 4, and 5. The constructs were sequenced and introduced into Agrobacterium tumefaciens by triparental mating and then transformed into Arabidopsis (Sanderfoot et al., 1999). Seeds harvested from transformed plants were grown on Murashige and Skoog selection plates (Murashige and Skoog, 1962). Vancomycin (500 mg L−1) was used to control bacterial growth and hygromycin (50 mg L−1) was used for selection of transgenic plants. T2 homozygous plants were selected based on segregation analysis of the transgenes, and the resulting T3 seed was used for experiments. The same constructs were used to introduce AtFUT1, -3, -4, and -5 into tobacco BY2 cells (Matsuoka et al., 1995).
Nucleic Acids and Southern and Northern Analysis
Genomic DNA was isolated using hexadecyltrimethylammonium bromide extraction (Sanderfoot et al., 2001) or the DNeasy plant mini-kit (Qiagen, Valencia, CA). RNA isolations were done as described (Puissant and Houdebine, 1990) or using the RNeasy plant mini kit (Qiagen). Southern and northern analyses were performed as described in Sambrook et al. (1989). 32P-dATP labeled probes, from gene-specific PCR products (see below), were generated using the Prime it II system (Stratagene, La Jolla, CA) following the manufacturer's recommendations.
PCR and RT-PCR
PCR reactions for amplifications of genomic or cDNA templates were performed in 1.25 mm dNTPs, 5 μm each of the primers, 1× Taq buffer (Roche, Indianapolis), and 2 units of Taq polymerase (Roche) in a volume of 50 μL. The amplification program used consisted of an initial 94°C cycle for 1 min followed by 30 cycles of 92°C, 30 s; 58°C, 1 min; and 72°C, 90 s, and a final extension at 72°C for 5 min. Reverse transcription reactions for RT-PCR were done using the Superscript II reverse transcriptase system following recommendations from the manufacturer (Gibco-BRL, Rockville, MD). Gene-specific primers were used to generate first-strand cDNAs and that primer was used as the reverse primer in the PCR reaction (Gibco-BRL). The primer pairs used for RT-PCR were: AtFUT1, 5′-GGAGGGCTACTTGCTTCTGGTTTT-3′ and 5′-TCCCGATGAATGTTTGGTCTCCTT-3′; AtFUT2, 5′-TCTTTGCACCGTCTCTTTTCTTGATTTC-3′ and 5′-AGGTGGATTTGGGTTGGTTTGATTCTCT-3′; AtFUT3, 5′-TACTGTGTGAAAGCAAGATCAA-3′ and 5′-GAAAAGAATTTAGAACTCGATC-3′; AtFUT4, 5′-GTTTGGGATATCGTCACTA-3′ and 5′-GGCGGATCAGGAGCTTTGTTA-3′; AtFUT5, 5′-CCCGGATCCAGATGTATCAAAAATTTCAGATC-3′ and 5′-CCCTCGAGCTAAAATTCATCAT-ATAGCTT-3′; AtFUT6, 5′-CCCGTCGACCTATAACTCATCAAATAG-3′ and 5′-CCCGGATCCAGAT-GTATCACATCTTTCAG-3′; AtFUT7, 5′-CCCGGATCCAGATGAAGACAAAGCTCATG-3′ and 5′-CCCCTCGAGCTATAACTCATTTTTGGT-3′; AtFUT8, 5′-CCCGGATCCAGATGCAACTCA-TTCTTC-3′ and 5′-CCCGTCGACCTAATTTGAATCAACTAG-3′; AtFUT9, 5′-CCCGGATCCA-GATGATAAAGCTCACGATA-3′ and 5′-CCCGTCGACTCAAAGTTCATCTGAAAC-3′; and AtFUT10, 5′-CCGTCGACCTAAAAGTCATCATATAGCTT-3′ and 5′-CCCGGATCCAGATGCCTTCGGAATATCTCGTC-3′.
Protein Purification and Fucosyltransferase Assays
Microsomal protein was isolated from tobacco cell suspensions as described by Faik et al. (1997). Protein extracts were treated with 1% (w/v) Triton prior to assays of crude extracts or before affinity purification. His tagged AtFUT1, -3, -4, and -5 were affinity purified using an Ni+ affinity chromatography column (Novagen). For western-blot analysis, 20 μg of microsomal protein or 2 μg of affinity purified protein was separated by SDS-PAGE, subsequently transferred to nitrocellulose, and probed with T7-monoclonal antibodies (Novagen; Sanderfoot et al., 1999). Xyloglucan fucosyltransferase assays were conducted according to the procedure developed by Gordon and Maclachlan (1989) and Faik et al. (1997). Assay conditions for radish root AGP-AI fucosylation were as described (Tsumuraya et al., 1988), but using 0.4% (w/v) radish root AGP. Similar conditions were used for larch wood arabinogalactan fucosylation. A control with no added protein was also included to determine the background level.
Preparation of Alcohol-Insoluble Residue (AIR)
Frozen plant tissue was ground to a powder in liquid nitrogen. The powder was then homogenized in aqueous 80% (v/v) ethanol (10 mL) using a Polytron blender, and the suspension was centrifuged (1,000g). The AIR was washed with 80% (v/v) ethanol (10 mL), and then with absolute ethanol (10 mL). The residue was suspended in chloroform:methanol (1:1 [v/v]; 5 mL), and stirred for 1 h at room temperature. The suspension was centrifuged again, and the AIR was washed with acetone (10 mL) and then vaccum dried at room temperature.
Glycosyl Residue Composition Analysis
AIR (1–2 mg) was suspended in 2 m trifluoroacetic acid (0.5 mL) and heated at 120°C for 1.5 h. The released glycoses were converted to their corresponding alditols by treatment for 2 h at room temperature with 1 m NH4OH containing 300 μL of NaB2H4 (10 mg mL−1) as described (York et al., 1985) and then analyzed by GC and GC-MS. GC was performed with an HP5880A gas chromatograph with a flame ionization detector (Hewlett Packard, Avondale, PA) and a 30-m SP2330 column (Supelco Inc., Bellefonte, PA) operated at 235°C. GC-MS was performed with an HP5890 GC interfaced to an HP5970 mass selective detector operated in the electron impact mode and a 30-m SP2330 column operated at 235°C.
Footnotes
This work was supported by the National Science Foundation Plant Genome Program (grant no. DBI–9975815), by the Department of Energy Biosciences program (grant no. DE–FG0201ER to K.K. and N.V.R.), and by the Department of Energy (grant nos. DE–FG05–93ER20115 and DE–FG09–93ER20097 to M.A.O.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010596.
LITERATURE CITED
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Breton C, Oriol R, Imberty A. Conserved structural features in eukaryotic and prokaryotic fucosyltransferases. Glycobiology. 1998;8:87–94. doi: 10.1093/glycob/8.1.87. [DOI] [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- Charnock SJ, Henrissat B, Davies GJ. Three-dimensional structures of UDP-sugar glycosyltransferases illuminate the biosynthesis of plant polysaccharides. Plant Physiol. 2001;125:527–531. doi: 10.1104/pp.125.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Carpita NC, Reiter WD, Wilson RH, Jeffries C, McCann MC. A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J. 1998;16:385–392. doi: 10.1046/j.1365-313x.1998.00301.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol. 1997a;13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell. 1997b;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JS. Molecular characterization of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999;19:691–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- Faik A, Bar-Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K. Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem. 2000;275:15082–15089. doi: 10.1074/jbc.M000677200. [DOI] [PubMed] [Google Scholar]
- Faik A, Chileshe C, Sterling J, Maclachlan G. Xyloglucan galactosyl- and fucosyltransferase activities from pea epicotyl microsomes. Plant Physiol. 1997;114:245–254. doi: 10.1104/pp.114.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001;15:79–89. doi: 10.1101/gad.188801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchette AC, Cabanes-Macheteau M, Marvin L, Martin B, Satiat-Jeunemaitre B, Gomord V, Crooks K, Lerouge P, Faye L, Hawes C. Biosynthesis and immunolocalization of Lewis a-containing N-glycans in the plant cell. Plant Physiol. 1999;121:333–344. doi: 10.1104/pp.121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchette-Laine AC, Gomord V, Cabanes M, Michalski JC, Saint Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L. N-Glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- Fransen CT, Haseley SR, Huisman MM, Schols HA, Voragen AG, Kamerling JP, Vliegenthart JF. Studies on the structure of a lithium-treated soybean pectin: characteristics of the fragments and determination of the carbohydrate substituents of galacturonic acid. Carbohydr Res. 2000;328:539–547. doi: 10.1016/s0008-6215(00)00130-0. [DOI] [PubMed] [Google Scholar]
- Gordon R, Maclachlan G. Incorporation of UDP-[14C] glucose into xyloglucan by pea membranes. Plant Physiol. 1989;91:373–378. doi: 10.1104/pp.91.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Falk E, Kenne L, Ronnberg B, Sundquist BG. Triterpenoid saponins containing an acetylated branched D-fucosyl residue from Quillaja saponaria Molina. Phytochemistry. 2000;53:861–868. doi: 10.1016/s0031-9422(99)00422-7. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Davies GJ. Glycoside hydrolases and glycosyltransferases: families, modules, and implications for genomics. Plant Physiol. 2000;124:1515–1519. doi: 10.1104/pp.124.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe Y, Delmer DP. Pathways and genes involved in cellulose biosynthesis. Genet Eng. 1997;19:63–87. doi: 10.1007/978-1-4615-5925-2_4. [DOI] [PubMed] [Google Scholar]
- Kohorn BD. Plasma membrane-cell wall contacts. Plant Physiol. 2000;124:31–38. doi: 10.1104/pp.124.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter H, Mucha J, Staudacher E, Grimm R, Glossl J, Altmann F. Purification, cDNA cloning, and expression of GDP-L-Fuc:Asn-linked GlcNAc α-1,3-fucosyltransferase from mung beans. J Biol Chem. 1999;274:21830–21839. doi: 10.1074/jbc.274.31.21830. [DOI] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- Lopez-Lara IM, Blok-Tip L, Quinto C, Garcia ML, Stacey G, Bloemberg GV, Lamers GE, Lugtenberg BJ, Thomas-Oates JE, Spaink HP. NodZ of Bradyrhizobium extends the nodulation host range of Rhizobium by adding a fucosyl residue to nodulation signals. Mol Microbiol. 1996;21:397–408. doi: 10.1046/j.1365-2958.1996.00644.x. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Bassham D, Raikhel NV, Nakamura K. Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting achineries in tobacco cells. J Cell Biol. 1995;130:1307–1318. doi: 10.1083/jcb.130.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley RS, Recinos A, 3rd, Olsen AS, Gingrich JC, Szczepaniak D, Cameron HS, Krauss R, Weston BW. Physical maps of human alpha (1,3) fucosyltransferase genes FUT3-FUT6 on chromosomes 19p13.3 and 11q21. Genomics. 1995;1:142–146. doi: 10.1016/0888-7543(95)80094-3. [DOI] [PubMed] [Google Scholar]
- Mergaert P, D'Haeze W, Fernandez-Lopez M, Geelen D, Goethals K, Prome JC, Van Montagu M, Holsters M. Fucosylation and arabinosylation of Nod factors in Azorhizobium caulinodans: involvement of NolK, NodZ as well as NoeC and/or downstream genes. Mol Microbiol. 1996;21:409–419. doi: 10.1046/j.1365-2958.1996.6451366.x. [DOI] [PubMed] [Google Scholar]
- Misawa H, Tsumuraya Y, Kaneko Y, Hashimoto Y. α-L-fucosyltransferases from radish primary roots. Plant Physiol. 1996;110:665–673. doi: 10.1104/pp.110.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. Biosynthesis of pectins and galactomannans. In: Pinto BM, editor. Carbohydrates and Their Derivatives Including Tannins, Cellulose, and Related Lignins. Vol. 3. Oxford: Elsevier Science LTD; 1999. pp. 497–527. [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000a;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem. 2000b;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962;15:473–497. [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- Nakamura K, Tsumuraya Y, Hashimoto Y, Yamamoto S. Arabinogalactan-proteins reacting with eel Anti-H agglutinin from leaves of cruciferous plants. Agric Biol Chem. 1984;48:753–760. [Google Scholar]
- Nickle TC, Meinke DW. A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 1998;15:321–332. doi: 10.1046/j.1365-313x.1998.00212.x. [DOI] [PubMed] [Google Scholar]
- Ogura K, Tsumuraya Y, Hashimoto Y, Yamamoto S. An arabinogalactan-protein from rape leaves. J Biol Chem. 1985;49:2851–2857. [Google Scholar]
- Oriol R, Mollicone R, Cailleau A, Balanzino L, Breton C. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology. 1999;9:323–334. doi: 10.1093/glycob/9.4.323. [DOI] [PubMed] [Google Scholar]
- Oulmouden A, Wierinckx A, Petit JM, Costache M, Palcic MM, Mollicone R, Oriol R, Julien R. Molecular cloning and expression of a bovine α(1,3)-fucosyltransferase gene homologous to a putative ancestor gene of the human FUT3-FUT5-FUT6 cluster. J Biol Chem. 1997;272:8764–8773. doi: 10.1074/jbc.272.13.8764. [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- Puissant C, Houdebine L-M. An improvement of the single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Pykari M, Toivonen S, Natunen J, Niemela R, Salminen H, Aitio O, Ekstrom M, Parmanne P, Valimaki M, Alais J. The acceptor and site specificity of α 3-fucosyltransferase: V. High reactivity of the proximal and low of the distal, Gal beta 1-4GlcNAc unit in i-type polylactosamines. J Biol Chem. 2000;275:40057–40063. doi: 10.1074/jbc.M007922200. [DOI] [PubMed] [Google Scholar]
- Quesada-Vincens D, Fellay R, Nasim T, Viprey V, Burger U, Prome JC, Broughton WJ, Jabbouri S. Rhizobium sp. strain NGR234 NodZ protein is a fucosyltransferase. J Bacteriol. 1997;179:5087–5093. doi: 10.1128/jb.179.16.5087-5093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinto C, Wijfjes AH, Bloemberg GV, Blok-Tip L, Lopez-Lara IM, Lugtenberg BJ, Thomas-Oates JE, Spaink HP. Bacterial nodulation protein NodZ is a chitin oligosaccharide fucosyltransferase which can also recognize related substrates of animal origin. Proc Natl Acad Sci USA. 1997;94:4336–4341. doi: 10.1073/pnas.94.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR. Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 1997;12:335–345. doi: 10.1046/j.1365-313x.1997.12020335.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. The t-SNARE atVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 1999;121:929–938. doi: 10.1104/pp.121.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Pilgrim M, Adam L, Raikhel NV. Disruption of individual members of Arabidopsis syntaxin gene families indicates each has essential functions. Plant Cell. 2001;13:659–666. doi: 10.1105/tpc.13.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A. The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell. 2000;12:1751–1768. doi: 10.1105/tpc.12.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher E, Altmann F, Wilson IB, Marz L. Fucose in N-glycans: from plant to man. Biochim Biophys Acta. 1999;1473:216–236. doi: 10.1016/s0304-4165(99)00181-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Ikeda Y, Tateishi A, Yamaguchi Y, Ishikawa M, Taniguchi N. A sequence motif involved in the donor substrate binding by 1,6-fucosyltransferase: the role of the conserved arginine residues. Glycobiology. 2000;10:503–510. doi: 10.1093/glycob/10.5.503. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumuraya Y, Hashimoto Y, Yamamoto S, Shibuya N. Structure of L-arabino-D-galactan-containing glycoproteins from radish leaves. Carbohydr Res. 1984;134:215–228. [Google Scholar]
- Tsumuraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S. Arabinogalactan proteins from primary and mature roots of radish (Raphanus sativus L.) Plant Physiol. 1988;86:155–160. doi: 10.1104/pp.86.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S, Doco T, Williams P, Pellerin P, York WS, O'Neill MA, Glushka J, Darvill AG, Albersheim P. Structural characterization of the pectic polysaccharide rhamnogalacturonan II: evidence for the backbone location of the aceric acid-containing oligoglycosyl side chain. Carbohydr Res. 2000;326:277–294. doi: 10.1016/s0008-6215(00)00036-7. [DOI] [PubMed] [Google Scholar]
- Wilson IB, Rendic D, Freilinger A, Dumic J, Altmann F, Mucha J, Muller S, Hauser Cloning and expression of cDNAs encoding alpha 1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim Biophys Acta. 2001;1527:88–96. doi: 10.1016/s0304-4165(01)00151-9. [DOI] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]
- York WS, Kumar Kolli VS, Orlando R, Albersheim P, Darvill AG. The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res. 1996;285:99–128. doi: 10.1016/s0008-6215(96)90176-7. [DOI] [PubMed] [Google Scholar]