Abstract
Lesquerella fendleri seed oil contains up to 60% hydroxy fatty acids, nearly all of which is the 20-carbon hydroxy fatty acid lesquerolic acid (d-14-hydroxyeicos-cis-11-enoic acid). Previous work suggested that lesquerolic acid in L. fendleri was formed by the elongation of the 18-carbon hydroxy fatty acid, ricinoleic acid. To identify a gene encoding the enzyme involved in hydroxy fatty acid elongation, an L. fendleri genomic DNA library was screened using the coding region of the Arabidopsis Fatty Acid Elongation1 gene as a probe. A gene, LfKCS3, with a high sequence similarity to known very long-chain fatty acid condensing enzymes, was isolated. LfKCS3 has a 2,062-bp open reading frame interrupted by two introns, which encodes a polypeptide of 496 amino acids. LfKCS3 transcripts accumulated only in the embryos of L. fendleri and first appeared in the early stages of development. Fusion of the LfKCS3 promoter to the uidA reporter gene and expression in transgenic Arabidopsis resulted in a high level of β-glucuronidase activity exclusively in developing embryos. Seeds of Arabidopsis plants transformed with LfKCS3 showed no change in their very long-chain fatty acid content. However, when these Arabidopsis plants were crossed with the transgenic plants expressing the castor oleate 12-hydroxylase, significant amounts of 20-carbon hydroxy fatty acids accumulated in the seed, indicating that the LfKCS3 condensing enzyme specifically catalyzes elongation of 18-carbon hydroxy fatty acids.
Hydroxy fatty acids are a component of the seed oils of a small number of diverse plant species (Badami and Patil, 1981; van de Loo et al., 1993). The most familiar of these is castor bean (Ricinus communis), which produces a seed rich in oil (35%–50% of seed weight) containing 85% to 90% ricinoleic acid (d-12-hydroxyoctadec-cis-9-enoic acid; 18:1-OH). This fatty acid is a valuable industrial raw material and castor is widely grown as an oil crop in countries such as India, Brazil, and China. Ricinoleic acid is formed in the developing endosperm of the castor bean by the direct hydroxylation of oleic acid (18:1Δ9) esterified to the sn-2 position of the membrane lipid phosphatidylcholine (Bafor et al., 1991). The hydroxylase responsible for this activity has been cloned (van de Loo et al., 1995) and has high homology to plant endoplasmic reticulum (ER) membrane-bound fatty acid desaturase enzymes.
In the Brassicaceae, members of the genus Lesquerella also produce hydroxy fatty acids in their seed oil. In most species of Lesquerella, the predominant hydroxy fatty acids have a chain length of 20 carbons (C20) with the hydroxy group on carbon 14 (Δ14) and one or two double bonds at the Δ11 and Δ17 positions (Hayes et al., 1995). The seeds of L. fendleri, for example, can contain nearly 60% lesquerolic acid (d-14-hydroxyeicos-cis-11-enoic acid; 20:1-OH) and virtually no hydroxy fatty acids with an 18-carbon chain length. However, some members of the genus do accumulate significant amounts of the C18 hydroxy fatty acids such as ricinoleic acid and densipolic acid (d-12-hydroxyoctadec-cis-9,cis-15-dienoic acid and 18:2-OH). Both of these fatty acids have the hydroxy group on carbon 12 with the first double bond between carbons 9 and 10 (Δ9). The positions of these functional groups suggest that in L. fendleri, the C20 hydroxy fatty acids are formed by the elongation of C18 hydroxy precursors.
Biochemical studies conducted on developing embryos from a number of Lesquerella species have shown that they hydroxylate oleic acid at the Δ12 position to form ricinoleic acid and can then desaturate and elongate this fatty acid to form densipolic, lesquerolic, and auricolic (d-14-hydroxyeicos-cis-11,cis-17-dienoic acid; 14-OH, 20:2-OH) acids (Engeseth and Stymne, 1996; Reed et al., 1997). Isolation of the gene encoding the seed-specific fatty acid hydroxylase from L. fendleri (Broun et al., 1998) and characterization of the gene product has demonstrated that the enzyme is a bifunctional Δ12-oleate hydroxylase:desaturase. It is highly homologous to the hydroxylase enzyme from castor bean and plant ER Δ12 fatty acid desaturases. Because the predominant hydroxy fatty acid accumulating in L. fendleri is lesquerolic acid, not ricinoleic acid, it appears that the plant very efficiently elongates C18 hydroxy fatty acid to C20.
C20 and other very long-chain fatty acids (VLCFAs) are common components of the seed oils of members of the Brassicaceae. They are synthesized by a microsomal fatty acid elongation (FAE) system that involves four enzymatic reactions: condensation of malonyl-coenzyme A (CoA) with a long-chain fatty acyl-CoA; reduction to β-hydroxyacyl-CoA; dehydration to an enoyl-CoA; and reduction of the enoyl-CoA, resulting in acyl-CoA elongation by two carbons (von Wettstein-Knowles, 1982; Fehling and Mukherjee, 1991). Of the four enzymes of the FAE, only genes encoding condensing enzymes have been cloned and characterized. Expression of FAE1, the condensing enzyme involved in VLCFA synthesis in Arabidopsis seeds, in transgenic plants and yeast (Saccharomyces cerevisiae) has demonstrated that the substrate specificity and the extent of elongation are a function of this enzyme (Millar and Kunst, 1997; Millar et al., 1999). In Arabidopsis, FAE1 catalyzes the elongation of oleic acid (18:1) to eicosenoic acid (20:1) and, less efficiently, of eicosenoic acid by a further two carbons. Expression of the castor bean and L. fendleri oleate hydroxylases in transgenic Arabidopsis has shown that FAE1 also has a low level of activity with ricinoleic acid and can elongate this hydroxy fatty acid to form lesquerolic acid (Broun et al., 1998; Smith et al., 2000).
In the seed oil of L. fendleri, oleic acid (18:1) accounts for as much as 10% to 15% of the total fatty acids, but eicosenoic acid (20:1) content is less than 1%. Conversely, most of the hydroxy fatty acids exist as lesquerolic (20:1-OH) and auricolic (20:2-OH), whereas the ricinoleic acid (18:1-OH) content is less than 1% of the total fatty acids. This implies that only the hydroxy fatty acids are elongated in L. fendleri, and therefore, the condensing enzyme is highly specific for hydroxy fatty acids. Here, we report the isolation and characterization of LfKCS3, the gene encoding the condensing enzyme responsible for lesquerolic acid production in L. fendleri. In addition, we present results showing that the LfKCS3 promoter is seed specific and active from an early stage of embryo development in transgenic Arabidopsis.
RESULTS
Cloning and Characterization of a Gene Encoding a VLCFA Condensing Enzyme
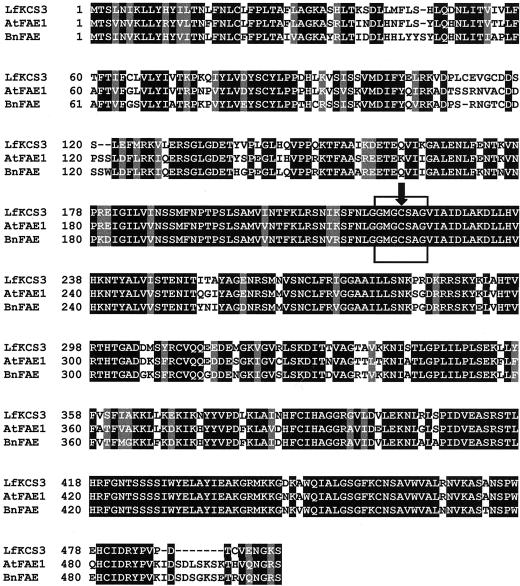
A genomic clone of the putative condensing enzyme was isolated by probing a genomic library of L. fendleri with the coding region of the Arabidopsis FAE1 gene (James et al., 1995). An EcoRI fragment subcloned into the plasmid pMHS15 was fully sequenced and a 4,313-bp consensus sequence was assembled using the GCG program (Genetics Computer Group, Madison, WI; Edelman et al., 1994). The 4,313-bp genomic DNA included a 573-bp 5′-flanking region, a 2,062-bp open reading frame, and a 1,678-bp 3′-flanking region. The nucleotide sequence of the clone was deposited in GenBank as accession number AF367052. A sequence comparison between the L. fendleri genomic DNA and Arabidopsis FAE1 using BCM Search Launcher: Multiple Sequence Alignments (Smith et al., 1996) revealed two introns in the L. fendleri genomic DNA separated by 421 bp. The deduced amino acid sequence was obtained after removing the introns with the aid of a translation tool, “Translate” (http://www.expasy.ch/tools/dna.html).This revealed that the gene, LfKCS3, encoded a polypeptide (LfKCS3) of 496 amino acids with an estimated molecular mass of 55.3 kD. Sequence comparisons (Fig. 1) showed that LfKCS3 shared 80% identity with the Arabidopsis FAE1 (James et al., 1995), 76% identity with the oilseed rape (Brassica napus) FAE1 (Clemens and Kunst, 1997), and approximately 50% identity with the Arabidopsis CUT1/CER6 (Millar et al., 1999; Fiebig et al., 2000) and Simmondsia chinensis jojoba KCS (Lassner et al., 1996).
Figure 1.
Alignment of deduced amino acid sequences determined for LfKCS3 and two other VLCFA condensing enzymes from members of the Brassicaceae. Identical amino acids are highlighted on a black background; similar amino acids are shown on a gray background. The boxed region with an arrow indicates a highly conserved region among all the condensing enzymes involved in VLCFA biosynthesis. The active site Cys (Ghanevati and Jaworski, 2001) is marked with an arrow. GenBank accession numbers for LfKCS3, AtFAE1, and BnFAE are AF367052, U29142, and AF009563, respectively.
Analysis of LfKCS3 Expression and Hydroxy Fatty Acid Accumulation in L. fendleri
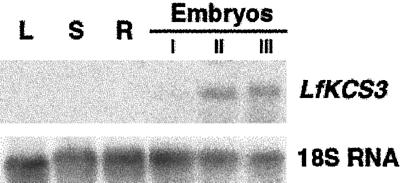
Lesquerolic acid accumulates only in the seed of L. fendleri. However, several condensing enzymes involved in the production of VLCFAs in vegetative tissues (Millar et al., 1999; Todd et al., 1999; Yephremov et al., 1999) also show a high degree of similarity with LfKCS3. For this reason, the expression pattern of LfKCS3 in L. fendleri was examined by northern-blot analysis. A 461-bp probe was used that consisted of 367 bp of the 5′ end of the coding region together with 94 bp of the adjacent 5′ genomic sequence. This region was chosen because it showed the least similarity when compared with the sequences of other condensing enzymes. Northern-blot analysis revealed a single band of approximately 1.8 kb in RNA from developing embryos (Fig. 2). No detectable hybridization was observed in leaves, stems, or roots.
Figure 2.
Northern-blot analysis of LfKCS3 expression in L. fendleri. A 32P-labeled probe corresponding to 461 bp of the 5′ region of the LfKCS3 genomic clone was hybridized to 10 μg of total RNA from leaves (L), stems (S), roots (R), and developing embryos (Embryos). Developing embryos were divided into three stages based on their size and color: early (I), mid- (II), and late (III) developmental stages (see Fig. 3). For loading control, the same blot was stripped and rehybridized with a 32P-labeled probe made from the Arabidopsis 18S rRNA (18S RNA; Unfried et al., 1989).
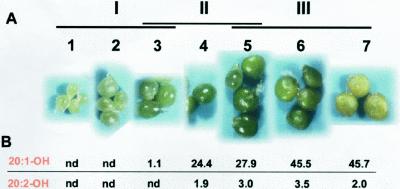
To examine a developmental pattern of accumulation of the very long-chain hydroxy fatty acids in the embryos of L. fendleri, embryos were harvested and separated into seven stages according to their size and color (Fig. 3A). Analyses of fatty acid composition of the individual embryos of different stages indicated that the C20 hydroxy fatty acids, mainly as lesquerolic acid (20:1-OH), appeared in stage three embryos, followed by a rapid increase in lesquerolic acid content in subsequent stages (Fig. 3B). This result was consistent with the data previously reported by Reed et al. (1997). In addition, there was a good correlation between the accumulation of the very long-chain hydroxy fatty acids (Fig. 3B) and LfKCS3 transcript accumulation (Fig. 2) in L. fendleri embryos throughout development. LfKCS3 transcripts were detected at low abundance in the early stages of development (group I = stages 1, 2, and 3) and abruptly increased at mid stage (group II = stages 3, 4, and 5).
Figure 3.
Development of L. fendleri embryos. A, Developing embryos of L. fendleri were harvested and divided into seven stages based on their color and size. Stages grouped for the northern-blot experiment (see Fig. 2) are indicated: I, embryos at the early stage of development; II, mid-stage; and III, late developmental stage. B, The C20 hydroxy fatty acid content (weight percentage) of the embryos at each stage was determined by gas chromatography. 20:1-OH, Lesquerolic acid; 20:2-OH, auricolic acid; nd, not detected.
The LfKCS3 Promoter Is Seed Specific and Active from an Early Stage of Embryo Development in Transgenic Arabidopsis
To determine whether the 5′-promoter region of LfKCS3 was sufficient to confer the tissue specificity and the developmental pattern of LfKCS3 expression seen in L. fendleri, a 573-bp 5′ fragment was placed upstream of the uidA gene encoding β-glucuronidase (GUS) and was introduced into Arabidopsis plants (ecotype Columbia). Histochemical staining was performed on leaves, stems, inflorescences, roots, and siliques at different stages of development for more than 30 independent primary transgenic plants. No staining was observed in any tissue other than developing embryos (data not shown) where an appearance of an intense blue color indicated that GUS was exclusively expressed in this tissue. The timing of expression directed by the LfKCS3 promoter (LfKCS3P) was compared with that of the L. fendleri fatty acid hydroxylase (LesqH) promoter, a strong early seed-specific promoter (Broun et al., 1998). Histochemical staining patterns of representative Arabidopsis embryos from the LfKCS3P-GUS and LesqH-GUS lines at different stages of embryo development are shown in Figure 4. For both promoters, GUS activity was evident at the earliest stages examined (4 d after flowering), and continued to be present throughout subsequent seed development.
Figure 4.
Histochemical staining of embryos from transgenic Arabidopsis expressing the GUS gene under the control of the LfKCS3 promoter (LfKCS3P) or the L. fendleri oleate 12-hydroxylase promoter (LesqH; Broun et al., 1998).
The LfKCS3 Is a Condensing Enzyme That Is Specific for Hydroxy Fatty Acids
Because the deduced amino acid sequence of LfKCS3 was highly similar to those of FAE1 and related VLCFA condensing enzymes from other plant species, we employed a transgenic approach to establish its functional identity. LfKCS3 was introduced into Arabidopsis wild-type and fad2/fae1 double-mutant plants under the control of its own promoter (LfKCS3G cassette) or LesqH promoter (LesqH-LfKCS3C cassette). Eighteen fad2/fae1 transformants were obtained containing the LfKCS3G cassette and 23 fad2/fae1 plants containing the LesqH-LfKCS3C cassette. The fatty acid composition of T2 seeds from individual plants was determined for all transformants. Even though the fad2/fae1 mutant accumulated approximately 85% (w/w) of total seed fatty acids as 18:1, none of the transgenic plants produced VLCFAs above the background level measured in untransformed double-mutant plants. Similarly, 32 independent transgenic lines obtained by transformation of Arabidopsis wild-type plants with the LfKCS3G cassette did not show any increase in VLCFAs levels compared with untransformed controls. These results indicated that 18:1 was not a substrate of the LfKCS3.
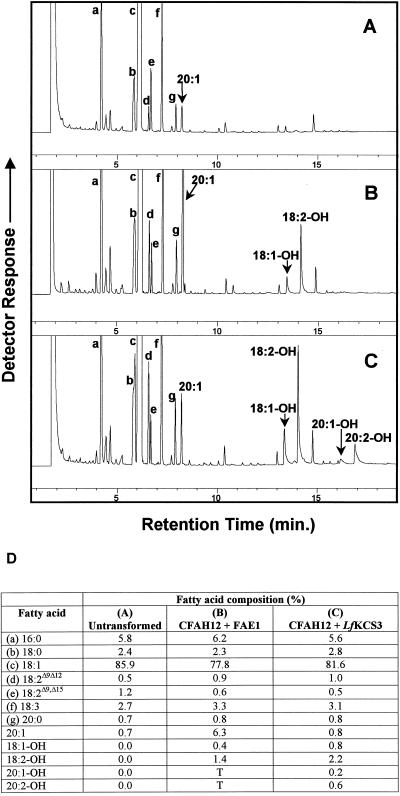
To test whether the lack of any change in fatty acid composition in the transgenic plants was due to the specificity of the LfKCS3 condensing enzyme for hydroxy fatty acids, transgenic fad2/fae1 mutant lines containing the LfKCS3G cassette or the LesqH-LfKCS3C cassette were crossed with transgenic Arabidopsis plants expressing the castor bean oleate 12-hydroxylase in the same fad2/fae1 double-mutant background (LesqH-CFAH12 lines). F1 plants were grown to maturity, and fatty acid analyses were carried out on the pools of F2 seeds harvested from individual plants. As controls, fatty acid compositions of the seeds of untransformed fad2/fae1 double-mutant (Fig. 5A) and transgenic plants expressing the Arabidopsis FAE1 and the castor hydroxylase (LesqH-FAE1 and LesqH-CFAH12 lines) in the double-mutant background (Fig. 5B) were also analyzed. Coexpression of FAE1 and the castor hydroxylase resulted in a major increase in the levels of eicosenoic acid (20:1) and the accumulation of hydroxy fatty acids that were not detected in untransformed seeds. These were identified as ricinoleic acid (18:1-OH) and densipolic acid (18:2-OH) (Fig. 5B). Densipolic acid has been shown to be a result of the Δ15 desaturation of ricinoleic acid catalyzed by the microsomal FAD3 desaturase (Engeseth and Stymne, 1996; Reed et al., 2000; Smith et al., 2000). In plants with the highest levels of hydroxy fatty acids (up to 4% of total seed fatty acids in this experiment), there were also traces of lesquerolic and (20:1-OH) and auricolic acid (20:2-OH). Previous results have shown that fad2/fae1 plants expressing CFAH12 alone accumulate ricinoleic and densipolic acid with no detectable C20 hydroxy fatty acids (Smith et al., 2000), and very low levels of 20:1 (less than 1% of total seed fatty acids). Thus, the presence of 20:1 and hydroxy fatty acids in the plants transformed with FAE1 and the CFAH12 suggests that both enzymes were active and confirmed that, as reported previously (Broun and Somerville, 1997; Smith et al., 2000), the Arabidopsis FAE1 is able to elongate hydroxy fatty acids, although this does not appear to be an efficient process.
Figure 5.
Gas chromatographic analyses of fatty acid methyl esters prepared from mature Arabidopsis seeds. A, Untransformed fad2/fae1 mutant. B, Transformed fad2/fae1 double mutant expressing the castor oleate 12-hydroxylase (CFAH12) and Arabidopsis FAE1. C, An F2 line expressing the CFAH12 and LfKCS3. Peak identification: a, 16:0; b, 18:0; c, 18:1; d, 18:2Δ9,12; e, 18:2Δ9,15 (Reed et al., 2000); f, 18:3Δ9,12,15; g, 20:0. D, Percentage of fatty acid composition of the seed samples shown in the chromatograms. T, Trace amount, not integrated.
In the F2 lines containing the LfKCS3 condensing enzyme and CFAH12, ricinoleic and densipolic acid were again present. However, in contrast to the plants coexpressing FAE1 and CFAH12, these plants contained significant amounts of the C20 hydroxy fatty acids lesquerolic and auricolic acid (Fig. 5C). In the plants containing the highest amount of hydroxy fatty acids, just over 4% of total seed fatty acids were hydroxylated, and of these, 23% had a chain length of 20 carbons. Plants containing the LfKCS3 gene under the control of its own promoter appeared to have slightly higher levels of C20-hydroxy fatty acids than those containing the LesqH-LfKCS3C cassette. In all transgenic plants coexpressing LfKCS3 and CFAH12, levels of 20:1 remained virtually the same as the controls. Taken together, these results provided conclusive evidence that the LfKCS3 was a condensing enzyme that specifically elongated hydroxy fatty acids, not the common C18 fatty acids, in the seed of L. fendleri.
DISCUSSION
We report here the isolation and characterization of LfKCS3, a gene encoding a VLCFA condensing enzyme expressed in the seeds of L. fendleri. To date, genes for several VLCFA condensing enzymes have been isolated and characterized from higher plants. They include those with seed-specific expression that are involved in VLCFA biosynthesis for seed storage lipids in Arabidopsis (James et al., 1995), jojoba (Lassner et al., 1996), and oilseed rape (Clemens and Kunst, 1997), and a few enzymes involved in cuticular wax production such as CUT1/CER6 (Millar et al., 1999; Fiebig et al., 2000) and KCS1 (Todd et al., 1999) from Arabidopsis. The seed-specific condensing enzymes exhibit a preference for oleic acid (18:1) in vivo, whereas the CUT1/CER6 appears to catalyze the elongation of C24 fatty acids or longer in the epidermal cells of Arabidopsis (Millar et al., 1999). Arabidopsis KCS1 elongates saturated fatty acids with a chain length of C20 to C26 (Todd et al., 1999). In addition, a cDNA encoding a FAE1-related polypeptide was recently identified from a collection of Limnanthes douglasii expressed sequence tags (Cahoon et al., 2000). Expression of the L. douglasii FAE1 homolog in soybean (Glycine max) somatic embryos indicated that this enzyme utilized mainly 16:0 as a substrate, resulting in the accumulation of saturated, principally 20:0, VLCFAs. Despite differences in their substrate specificities and the types of products made, all of the plant condensing enzymes mentioned above share a high degree of sequence identity. The alignment of the predicted amino acid sequence of LfKCS3 with the known VLCFA condensing enzymes and the presence of a strictly conserved region between Gly218 and Gly224 (Fig. 1) in all condensing enzymes involved in VLCFA biosynthesis strongly suggested that LfKCS3 also encoded a VLCFA condensing enzyme.
Expression in transgenic Arabidopsis clearly demonstrated that LfKCS3 was a VLCFA condensing enzyme, and that it only catalyzed the elongation of C18 hydroxy fatty acids. When LfKCS3 and the castor hydroxylase were expressed together in the Arabidopsis fad2/fae1 double-mutant plants, C20 hydroxy fatty acids were synthesized that were not seen in plants expressing the hydroxylase alone. No changes were detected in the levels of non-hydroxy C20 fatty acids (less than 2%) in these plants, suggesting that the enzyme was only elongating hydroxy fatty acids. Further evidence for this specificity comes from the observation that the fatty acid composition of fad2/fae1 seeds transformed with LfKCS3 alone was not different from untransformed controls. Similarly, LfKCS3 expression in wild-type plants did not result in any changes in fatty acid composition. Thus, the endogenous fatty acids of Arabidopsis do not appear to be substrates for LfKCS3.
For comparison we also coexpressed FAE1, a well-characterized condensing enzyme, and the castor hydroxylase in the fad2/fae1 double-mutant plants. The introduction of FAE1 resulted in the accumulation of 20:1 in transgenic seeds, a product of 18:1 elongation. Therefore, in contrast to LfKCS3 that only utilizes hydroxy C18 fatty acids as a substrate, FAE1 is highly active with 18:1. In plants containing the highest levels of 20:1, very low levels of C20 hydroxy fatty acids were detected in the seed. This result is in agreement with earlier reports that FAE1 was capable of catalyzing the elongation of ricinoleic acid (Broun and Somerville, 1997; Smith et al., 2000). In both previous studies, the castor hydroxylase was expressed in wild-type plants that had normal levels of FAE1 activity, and as a result, accumulated around 16% 20:1. In contrast, in the work reported here, FAE1 was introduced into a mutant lacking FAE1 activity (fad2/fae1 mutant background). The highest level of 20:1 measured in fad2/fae1 transgenic lines was around 7%, indicating that FAE1 activity in these plants was likely significantly lower than in a wild-type plant. Hydroxylase activity also appeared to be low, as total levels of hydroxy fatty acids in these plants never exceeded 4%. Therefore, it is not possible to comment on the relative efficiency of elongation of hydroxy fatty acids by FAE1 in comparison with LfKCS3. However, there is a clear distinction between FAE1 and LfKCS3 in that FAE1 elongates 18:1, whereas LfKCS3 does not.
The presence of the hydroxy group at the Δ12 position of a C18 fatty acyl chain is likely to give the molecule an electrophilic profile more similar to 18:2 than 18:1. Some enzymes, for example the ER Δ12 and Δ15 desaturases, can accept substrates with electrophilic groups such as hydroxy and epoxy groups in place of a double bond (Engeseth and Stymne, 1996). However, it is not known whether the same is true for condensing enzymes. To determine if LfKCS3 can elongate 18:2 in addition to hydroxy 18:1, we transformed wild-type plants, which contain around 30% 18:2, with LfKCS3. Analysis of seed fatty acids showed no significant difference between transformed and control plants (data not shown). This result is consistent with the fatty acid profile of L. fendleri seed, which contain around 10% 18:2 but no detectable 20:2. Thus, it appears that the LfKCS3 condensing enzyme is specific for a hydroxy group at the Δ12 position of ricinoleic acid and that the hydroxy group cannot be substituted for by a double bond. If this is correct, then the mechanism of substrate recognition by the condensing enzymes is different from that of the ER Δ12 and Δ15 desaturases. The availability of condensing enzymes with specificities toward saturated, monounsaturated, or hydroxy fatty acids from plants, and polyunsaturated fatty acids from animals (Beaudoin et al., 2000), fungi (Parker-Barnes et al., 2000), and a moss (Zank et al., 2000) will allow us to further investigate the mechanism of acyl group recognition and specificity of this class of enzymes.
The predominant hydroxy fatty acids in plants expressing LfKCS3 and the castor hydroxylase are the diunsaturated fatty acids, densipolic and auricolic acid. Several reports (Engeseth and Stymne, 1996; Reed et al., 2000; Smith et al., 2000) have shown that densipolic acid is formed by the Δ15 (n-3) desaturation of ricinoleic acid catalyzed by the FAD3 desaturase. The mechanism of synthesis of auricolic acid is less clear, and it is still uncertain whether 20:2-OH is formed by the elongation of 18:2-OH, as suggested by Engeseth and Stymne (1996), or by the FAD3 catalyzed desaturation of 20:1-OH, as proposed by Reed et al. (1997). Isolation of a cDNA encoding LfKCS3 and in vitro analysis, for example by expression in yeast, may help answer these questions.
In the transgenic Arabidopsis plants reported here, levels of hydroxy fatty acids were generally low, reaching up to 4% of the total seed fatty acids in the best lines. There could be a number of reasons for this, including low levels of gene expression, poor incorporation of hydroxy fatty acids into triacylglycerol, or degradation of hydroxy fatty acids. However, it is interesting to note that in this and previous studies, hydroxy fatty acids of C18 and C20 chain lengths accumulated in transgenic Arabidopsis. This suggests that although enzymes of triacylglycerol synthesis such as acyltransferases tend to show specificity and selectivity biased toward the fatty acids normally present in the plant (Wiberg et al., 1994; Frentzen, 1998), they do not entirely exclude hydroxy fatty acids from storage lipids.
Analysis of fatty acid composition of developing L. fendleri embryos revealed that hydroxy fatty acids are not readily detectable until developmental stage 4, which seems to coincide with the beginning of seed oil deposition. C18 and C20 hydroxy fatty acids appeared at the same time, indicating that LfKCS3 condensing enzyme must be expressed at the same time as, or earlier than, LFAH12 hydroxylase in the L. fendleri embryos. We compared the timing of both promoter activities using the GUS reporter gene in transgenic Arabidopsis plants. Histochemical staining showed that with the LfKCS3 or the LesqH promoter (Broun et al., 1998), GUS expression was first observed in very young torpedo stage embryos 4 d after flowering, and continued throughout subsequent embryo development. Because the LfKCS3 promoter directs gene expression only in developing embryos, it will benefit a variety of seed-specific applications, especially those aimed at genetic engineering of storage lipid composition in oilseed crops.
MATERIALS AND METHODS
Screening of the Genomic DNA Library
A Lesquerella fendleri genomic DNA library was obtained from Dr. Chris Somerville (Carnegie Institution of Washington, Stanford, CA). The genomic library was plated on Escherichia coli LE392 (Promega, Madison, WI), and about 150,000 clones were screened using the entire deduced coding region of Arabidopsis FAE1 as a probe. The probe was amplified by PCR from the pGEM7-FAE1 template (Millar and Kunst, 1997) using FAE1 upstream primer, 5′-CCGAGCTCAAAGAGGATACATAC-3′ and FAE1 downstream primer, 5′-GATACTCGAGAACGTTGGCACTCAGATAC-3′. PCR was performed in a 10-μL reaction containing 10 ng of the template, 2 mm MgCl2, 1.1 μm of each primer, 100 μm of dCTP + dGTP + dTTP mix, 50 μCi of [α-32P]dATP, 1× PCR buffer, and 2.5 units of Taq DNA polymerase (Invitrogen, Carlsbad, CA). Amplification conditions were 2 min of initial denaturation at 94°C, 30 cycles of 94°C for 15 s, 55°C for 30 s, 72°C for 1 min and 40 s, followed by a final extension at 72°C for 7 min. The amplified radioactive probe was purified using QIAquick PCR purification kit (Qiagen, Valencia, CA) and was denatured by boiling before addition to the hybridization solution. Hybridization was carried out overnight at 65°C in a solution containing 6× standard saline citrate (SSC), 20 mm NaH2PO4, 0.4% (w/v) SDS, 5× Denhardt's solution, 0.1% [w/v] of Ficoll [Type 400, Pharmacia, Buckinghamshire, UK], 0.1% [w/v] polyvinylpyrrolidone, 0.1% [w/v] bovine serum albumin [Fraction V; Sigma, St. Louis]), and 50 μg mL−1 sonicated, denatured salmon sperm DNA (Sigma), followed by three washes for 20 min each in 2× SSC and 0.5% (w/v) SDS at 65°C.
Plasmid Construction and Plant Transformation
After tertiary screening, nine positive clones were purified from the L. fendleri genomic library. The phage DNA from those nine clones was extracted using the Lambda mini kit (Qiagen) according to the manufacturer's protocol. One of them was digested with EcoRI, and a 4.3-kb fragment, designated LfKCS3G, was subcloned into the pGEM-7Zf(+) vector (Promega) cut with EcoRI, resulting in the vector pMHS15. The whole insert was sequenced in both directions with a Prism 377 automated DNA sequencer (Applied Biosystems, Foster City, CA) using di-deoxy Big Dye chemistry (Applied Biosystems) according to the Terminator Cycle Sequencing protocol.
To prepare the promoter-GUS fusion construct, the fragment directly upstream of the LfKCS3 coding region was amplified using the high-fidelity Pfu polymerase (Stratagene, La Jolla, CA) with a forward primer 5′-CGCA-AGCTTGAATTCGGAAATGGGCCAAG-3′ and a reverse primer 5′-CGCGTCGACTGTTTTGAGTTTGTGTCGGG-3′. The amplified 573-bp promoter fragment (designated LfKCS3P) was inserted upstream of the uidA gene in pBI101 (CLONTECH, Palo Alto, CA) cut with HindIII and SalI, resulting in the vector pLfKCS3P-GUS.
Two constructs were generated for LfKCS3 expression in Arabidopsis. In the first, a 3.1-kb genomic fragment containing the promoter and the coding sequence was excised from pMHS15 with EcoRI and HpaI and was ligated into a binary plant transformation vector, pRD400 (Datla et al., 1992) cut with EcoRI and SmaI, resulting in the vector pLfKCS3G. For comparison, the coding region of LfKCS3 (designated LfKCS3C) was also placed under the control of the L. fendleri oleate 12-hydroxylase promoter (designated LesqH) to produce the LesqH-LfKCS3C cassette. This promoter was isolated from L. fendleri and was reported to be highly active in developing seeds of transgenic Arabidopsis starting early in embryogenesis (Broun et al., 1998). To generate the LesqH-LfKCS3C cassette, the coding region of LfKCS3 was amplified from pMHS15 using the high-fidelity Platinum Pfx DNA polymerase (Invitrogen), forward primer 3ApaI (5′-TTAGGGCCCATCTCATCCC-TTAGTACCCTC-3′), and the reverse primer 3NotI (5′-TATTGCGGCCGCTTCGATCAAGCGTGCTAC-3′). The resulting fragment was cut with ApaI and NotI and was ligated in sense orientation between the LesqH promoter and nopaline synthase terminator to give the vector pMHS23. The entire cassette was then cut from pMHS23 with EcoRI and SalI and was ligated into the binary vector pRD400, cut with EcoRI and SalI, to produce the vector pLesqH-LfKCS3C.
All three plant transformation vectors, pLfKCS3P-GUS, pLfKCS3G, and pLesqH-LfKCS3C were introduced into Agrobacterium tumefaciens strain GV3101 (pMP90; Koncz and Schell, 1986) by heat shock and were selected for resistance to kanamycin (50 μg mL−1). They were then used to transform Arabidopsis Columbia-2 wild type and/or fad2/fae1 double mutant. The fad2/fae1 double mutant is characterized by a very high level (>80%) of oleic acid (18:1) in its seed oil due to deficiency in the activities of cytoplasmic oleate Δ12 desaturase (FAD2) and the FAE1 condensing enzyme. Plants were grown in a growth chamber at 20°C under continuous light. Transformation of Arabidopsis was performed by the floral dip method (Clough and Bent, 1998). Screening for transformed seed was done on 50 μg mL−1 kanamycin as previously described (Katavic et al., 1994).
To test the activity of the LfKCS3 with hydroxy fatty acids, fad2/fae1 double-mutant lines transformed with the LfKCS3G or LesqH-LfKCS3C cassettes were crossed with the transgenic fad2/fae1 Arabidopsis plants expressing the castor oleate 12-hydroxylase under the control of the LesqH promoter (LesqH-CFAH12 lines; Smith et al., 2000). For comparison, transgenic fad2/fae1 double-mutant plants were also generated that expressed the castor oleate 12-hydroxylase and the Arabidopsis FAE1 condensing enzymes. These expression cassettes (LesqH-CFAH12 and LesqH-FAE1) contained the coding regions of each gene, under the control the LesqH promoter and nopaline synthase terminator.
GUS Assay
GUS assay was performed by immersing tissues in GUS histochemical staining solution (Jefferson, 1987) for 4 to 7 h at 37°C. The assay solution was composed of 50 mm sodium phosphate, pH 7.0, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 10 mm EDTA, 0.05% (w/v) Triton X-100, and 0.35 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. Following staining, the samples were fixed in 70% (v/v) ethanol.
RNA Gel-Blot Analysis
For RNA gel-blot analyses, total RNA from various tissues of L. fendleri was extracted using the RNeasy Plant Mini Kit (Qiagen) or a conventional phenol-chloroform method. For the phenol-chloroform method, frozen tissue was pulverized in a precooled mortar using liquid nitrogen and was transferred to a centrifuge tube containing 9 mL of NTES buffer (100 mm NaCl, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1% [w/v] SDS) and 6 mL of phenol:chloroform:isoamyl alcohol (25:24:1, v/v). After shaking for a minimum of 10 min, the tube was centrifuged at 5,000g for 10 min at 4°C to separate the two phases. Nucleic acids were precipitated by the addition of 0.1 volume of 3 m sodium acetate, pH 5.2, and 2 volumes of ethanol to the aqueous layer. Following centrifugation, the precipitated nucleic acids were resuspended in water. Lithium chloride was then added to a final concentration of 2 m. Following precipitation on ice for at least 3 h, total RNA was collected by centrifugation at 10,000g for 10 min at 4°C and was resuspended in water.
Ten micrograms of total RNA lane−1 was separated in an 1.2% (w/v) agarose gel containing formaldehyde, blotted onto a Hybond-XL membrane (Amersham Pharmacia Biotech, Piscataway, NJ) in 20× SSC, and baked for 2 h at 80°C. The membrane was then hybridized to a 461-bp probe from the 5′ region of the genomic clone that had been labeled with [α-32P]dATP by PCR using pMHS15 as a template with the oligonucleotides 5′-ATGAAAGCAACGCACCACAAAACGAAGAC-3′ (forward primer) and 5′-ACTCAAGAGAAGAATCATCACAACCCACC-3′ (reverse primer). The following PCR program was used: 2 min of initial denaturation at 94°C, 36 cycles of 94°C for 15 s, 60°C for 30 s, 72°C for 30 s, followed by a final extension at 72°C for 5 min. The amplified radioactive probe was purified by QIAquick PCR purification kit (Qiagen), denatured by boiling, and was added to the hybridization solution. Hybridization was performed overnight at 65°C in 0.5 m phosphate buffer, pH 7.2, 7% (w/v) SDS, and 10 mm EDTA. The blots were washed three times in 2× SSC and 0.1% (w/v) SDS at 65°C for 30 min each, before exposing the x-ray film (Eastman-Kodak, Rochester, NY). To show equal loading of the RNA samples, Arabidopsis 18S rRNA (Unfried et al., 1989) was used as a probe. A 32P-labeled rRNA probe was prepared by PCR using Taq DNA polymerase (Invitrogen) and the oligonucleotides 5′-CTGCCAGTAGTCATATGC-3′ and 5′-ATGGATCCTCGTTAAGGG-3′ with 10 ng of Arabidopsis (ecotype Columbia) DNA as a template. Amplification conditions were: 2 min of initial denaturation at 94°C, 30 cycles of 94°C for 15 s, 50°C for 30 s, 72°C for 30 s, followed by a final extension at 72°C for 7 min. After overnight hybridization at 65°C, the blot was washed three times in 0.1× SSC and 0.1% (w/v) SDS at 65°C before exposure of the x-ray film.
Analysis of Fatty Acid Composition
To determine the fatty acid composition of the seed oil, fatty acid methyl esters were prepared by refluxing the seed samples in 2 mL of 1 n methanolic-HCl for 90 min at 80°C. After cooling, 2 mL of 0.9% (w/v) NaCl solution and 150 μL of hexane were added and the mixture was vortexed vigorously. The fatty acid methyl esters in the hexane phase were analyzed by gas-liquid chromatography as described previously (Kunst et al., 1992). The identity of fatty acids in the samples was determined by comparing retention times with those of standards (Sigma). Lipid extracts from castor bean and L. fendleri were used as sources of ricinoleic (18:1-OH), densipolic (18:2-OH), lesquerolic (20:1-OH), and auricolic (20:2-OH) acid.
ACKNOWLEDGMENTS
We thank Chris Somerville and Pierre Broun (Carnegie Institution of Washington, Stanford, CA) for L. fendleri genomic DNA library and LesqH-GUS transgenic seeds, and David Dierig (U.S. Department of Agriculture-Agricultural Research Service Water Conservation Laboratory, Phoenix) for the L. fendleri plants. We also thank Tanya Hooker (University of British Columbia, Vancouver) for taking pictures of the L. fendleri developing embryos and Sabine Clemens (Simon Fraser University, Vancouver) for providing the developing embryos from the LesqH-GUS transgenic Arabidopsis plants.
Footnotes
This work was supported by a grant from the Science Council of British Columbia, Canada, by a Natural Sciences and Engineering Research Council of Canada Strategic grant, and by Linnaeus Plant Sciences, Inc.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010544.
LITERATURE CITED
- Badami RC, Patil KB. Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res. 1981;19:119–153. doi: 10.1016/0163-7827(80)90002-8. [DOI] [PubMed] [Google Scholar]
- Bafor M, Smith M, Jonsson L, Stobart K, Stymne S. Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor bean (Ricinus communis) endosperm. Biochem J. 1991;280:507–514. doi: 10.1042/bj2800507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Michaelson LV, Hey SJ, Lewis MJ, Shewry PR, Sayanova O, Napier JA. Heterologous reconstitution in yeast of the polyunsaturated fatty acid biosynthetic pathway. Proc Natl Acad Sci USA. 2000;97:6421–6426. doi: 10.1073/pnas.110140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Boddupalli S, Somerville C. A bifunctional oleate 12-hydroxylase:desaturase from Lesquerella fendleri. Plant J. 1998;13:201–210. doi: 10.1046/j.1365-313x.1998.00023.x. [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C. Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol. 1997;113:933–942. doi: 10.1104/pp.113.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Marillia E-F, Stecca KL, Hall SE, Taylor DC, Kinney AJ. Production of fatty acid components of meadowfoam oil in somatic soybean embryos. Plant Physiol. 2000;124:243–251. doi: 10.1104/pp.124.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Kunst L. Isolation of a Brassica napus cDNA (accession no. AF009563) encoding 3-ketoacyl-CoA synthase, a condensing enzyme involved in the biosynthesis of very long chain fatty acids in seeds (PGR97-125) Plant Physiol. 1997;115:313–314. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Datla RSS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W. Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene. 1992;211:383–384. doi: 10.1016/0378-1119(92)90232-e. [DOI] [PubMed] [Google Scholar]
- Edelman I, Olson S, Devereux J. Wisconsin Sequence Analysis Package, version 8.0. Genetic Computer Group, Inc; 1994. [Google Scholar]
- Engeseth N, Stymne S. Desaturation of oxygenated fatty acids in Lesquerella and other oil seeds. Planta. 1996;198:238–245. [Google Scholar]
- Fehling E, Mukherjee KD. Acyl-CoA elongase from a higher plant (Lunaria annua): metabolic intermediates of very-long-chain acyl-CoA products and substrate specificity. Biochim Biophys Acta. 1991;1082:239–246. doi: 10.1016/0005-2760(91)90198-q. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell. 2000;12:2001–2008. doi: 10.1105/tpc.12.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen M. Acyltransferases from basic science to modified seed oils. Fett/Lipid. 1998;100:161–166. [Google Scholar]
- Ghanevati M, Jaworski JG. Active site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA sythase, FAE1 KC5. Biochim Biophys Acta. 2001;1530:77–85. doi: 10.1016/s1388-1981(00)00168-2. [DOI] [PubMed] [Google Scholar]
- Hayes DG, Kleiman R, Phillips BS. The triglyceride composition, structure, and presence of estolides in the oils of Lesquerella and related species. J Am Oil Chem Soc. 1995;72:559–569. [Google Scholar]
- James DW, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Bol Rep. 1987;5:387–405. [Google Scholar]
- Katavic V, Haughn GW, Reed D, Martin M, Kunst L. In planta transformation of Arabidopsis thaliana. Mol Gen Genet. 1994;245:363–370. doi: 10.1007/BF00290117. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Kunst L, Taylor DC, Underhill EW. Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem. 1992;30:425–434. [Google Scholar]
- Lassner MW, Lardizabal K, Metz JG. A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell. 1996;8:281–292. doi: 10.1105/tpc.8.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- Parker-Barnes JM, Das T, Bobik E, Leonard AE, Thurmond JM, Chaung L-T, Huang Y-S, Mukerji P. Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc Natl Acad Sci USA. 2000;97:8284–8289. doi: 10.1073/pnas.97.15.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DW, Schafer UA, Covello PS. Characterization of the Brassica napus extraplastidial linoleate desaturase by expression in Saccharomyces cerevisiae. Plant Physiol. 2000;122:715–720. doi: 10.1104/pp.122.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DW, Taylor DC, Covello PS. Metabolism of hydroxy fatty acids in developing seeds in the genera Lesquerella (Brassicaceae) and Linum (Linaceae) Plant Physiol. 1997;114:63–68. doi: 10.1104/pp.114.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Moon H, Kunst L. Production of hydroxy fatty acids in the seeds of Arabidopsis thaliana. Biochem Soc Trans. 2000;28:947–950. [PubMed] [Google Scholar]
- Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC. BCM Search Launcher: an integrated interface to molecular biology data base search and analysis services available on the world wide web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- Unfried I, Stocker U, Gruendler P. Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Col0. Nucleic Acids Res. 1989;17:7513. doi: 10.1093/nar/17.18.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C. An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA. 1995;92:6743–6747. doi: 10.1073/pnas.92.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo FJ, Fox BG, Somerville C. Unusual fatty acids. In: Moore TS, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 91–126. [Google Scholar]
- von Wettstein-Knowles PM. Elongase and epicuticular wax biosynthesis. Physiol Veg. 1982;20:797–809. [Google Scholar]
- Wiberg E, Tillberg E, Stymne S. Substrates of diacylglycerol acyltransferase in microsomes from developing oil seeds. Phytochemistry. 1994;36:573–577. [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zank TK, Zähringer U, Lerchl J, Heinz E. Cloning and expression of the first plant fatty acid elongase specific for Δ-6-polyunsaturated fatty acids. Biochem Soc Trans. 2000;28:654–658. [PubMed] [Google Scholar]