Abstract
We report the identification of a nucleus-encoded minE gene, designated AtMinE1, of Arabidopsis. The encoded AtMinE1 protein possesses both N- and C-terminal extensions, relative to the eubacterial and algal chloroplast-encoded MinE proteins. The N-terminal extension functioned as a chloroplast-targeting transit peptide, as revealed by a transient expression assay using an N terminus:green fluorescent protein fusion. Histochemical β-glucuronidase staining of transgenic Arabidopsis lines harboring an AtMinE1 promoter::uidA reporter fusion unveiled specific activation of the promoter in green tissues, especially at the shoot apex, which suggests a requirement for cell division-associated AtMinE1 expression for proplastid division in green tissues. In addition, we generated transgenic plants overexpressing a full-length AtMinE1 cDNA and examined the subcellular structures of those plants. Giant heteromorphic chloroplasts were observed in transgenic plants, with a reduced number per cell, whereas mitochondrial morphology remained similar to that of wild-type plants. Taken together, these observations suggest that MinE is the third conserved component involved in chloroplast division.
Chloroplast division is one of the most critical cellular processes in plants because the plant cell is unable to synthesize this organelle de novo (Possingham and Lawrence, 1983). A series of nuclear recessive mutants of the higher plant Arabidopsis, in which the chloroplast number per cell is greater or fewer than that in the wild-type plant, have been well documented (Pyke and Leech, 1991, 1992, 1994; Pyke et al., 1994; Robertson et al., 1996; Marrison et al., 1999). Characterization of these mutants, referred to as accumulation and replication of chloroplasts (arc) mutants, indicates that chloroplast division is a complex process that involves multiple distinct steps, such as expansion, division site selection, division initiation, constriction, and scission of chloroplasts, which are controlled by different nuclear genes, although no ARC genes have yet been cloned. Intensive microscopic studies of the division process revealed that, upon chloroplast division, two concentric rings (plastid-dividing [PD] rings) appear on opposite sides of the chloroplast envelope at the constricted isthmus (Hashimoto, 1986; Kuroiwa et al., 1998). Recently, the outer cytosolic PD ring of the red alga Cyanidioschyzon merolae was observed as a bundle of novel 5-nm filaments, implying the innovation of eukaryote-specific organelle division machinery (Miyagishima et al., 2001). In addition, recent studies of transgenic land plants have shown that the same division machinery is conserved in chloroplasts and eubacterial cells. Strepp et al. (1998) and Osteryoung et al. (1998) demonstrated that a homolog(s) of the bacterial cell division protein FtsZ is required for chloroplast division, by generating knockout plants of the moss Physcomitrella patens or antisense transgenics of Arabidopsis, respectively. In bacteria, FtsZ is a cytoplasmic, tubulin-related GTPase, which assembles into a ring structure (Z-ring) surrounding the division plane, possibly serving to constrict the cell membrane (Bi and Lutkenhaus, 1991). Dysfunction of FtsZ in Escherichia coli cells leads to the formation of filamentous elongated cells (Hirota et al., 1968). Osteryoung et al. (1998) also showed that one member (AtFtsZ1-1) of the Arabidopsis FtsZ family localizes within chloroplasts, topologically consistent with that of the bacterial FtsZ. The second example of a conserved component of the division machinery is the Arabidopsis homologue of MinD, AtMinD1. Colletti et al. (2000) generated AtMinD1-antisense transgenics, which exhibited asymmetric chloroplast division and resulted in chloroplasts of variable size. These results are readily explained if the chloroplast MinD retains a function analogous to that of the bacterial MinD. In bacteria, MinD cooperates with MinC to form a complex that inhibits Z-ring formation at all potential division sites (PDSs), except for the mid-cell (RayChaudhuri et al., 2000). MinD- or MinC-deficient E. coli mutants execute Z-ring formation at all PDSs, resulting in the formation of anucleate minicells.
MinE is the third component of the eubacterial MinCDE system and serves to prevent the MinCD division inhibitor from blocking division at the proper mid-cell site, while permitting it to prevent division at other PDSs (RayChaudhuri et al., 2000). As a result, constriction is restricted to the mid-cell in the presence of MinC, MinD, and MinE, whereas a lack of MinE prohibits division at all PDSs, including the mid-cell, and results in cell filamentation similar to that of FtsZ-deficient cells (de Boer et al., 1989). Consistent with the above-mentioned ability (i.e. topological specificity), MinE is located in a cytoplasmic annular structure near the mid-cell (Raskin and de Boer, 1997; Fu et al., 2001). This MinE ring is close to, but separate from, the Z-ring. The first indication of the existence of MinE in chloroplasts came from sequencing the chloroplast genome of the green alga Chlorella vulgaris (Wakasugi et al., 1997). The C. vulgaris chloroplast genome contains minD- and minE-like open reading frames, which are tandemly arranged in the same order as in E. coli. The chloroplast genome of the cryptophyte alga Guillardia theta also encodes minD and minE in the same gene order (Douglas and Penny, 1999). Despite these examples, no recognizable minE homologs have yet been found in any chloroplast genome of land plants, including Arabidopsis (Sato et al., 1999). This suggests that gene transfer of minE has occurred, from the chloroplast to the nucleus, during the evolution of plants.
In this paper, we describe the identification of a nuclear gene of Arabidopsis, AtMinE1, which encodes a chloroplast protein homologous to MinE. This is the first report on the characterization of MinE from eukaryotes. Tissue-specific activation of the AtMinE1 promoter was observed using a β-glucuronidase (GUS) reporter assay. Furthermore, overexpression of AtMinE1 disrupted chloroplast division, suggesting a conserved function of MinE in eubacterial and chloroplast division.
RESULTS
Identification of a Nuclear-Encoded MinE Homolog Carrying a Transit Peptide
Using the TBLASTN algorithm (Altschul et al., 1990), we searched all of the Arabidopsis genomic DNA sequences available in the GenBank database using the G. theta chloroplast MinE (accession no. AAC35620) as a query sequence, and found a putative minE gene within the BAC clone F23O10 from chromosome I. This potential gene (F23O10.25) was predicted to contain an intron of 874 bp and to encode a polypeptide of 229 amino acids, whose central region (127–194) showed a 30% identity and 60% similarity with the MinE (Ssl10546) from the cyanobacterium Synechocystis sp. PCC6803 (accession no. Q55899). The predicted open reading frame and splice site of this gene were confirmed by obtaining the full-length sequence of the cDNA using a PCR-aided strategy (see “Materials and Methods”). This gene was designated AtMinE1 (accession no. AB046117), in accordance with the previously used nomenclature for Arabidopsis homologues of bacterial cell division genes, such as AtFtsZ1/2 or AtMinD1. No other sequences closely related to minE were found in the Arabidopsis genome database, even after the release of the complete genome sequence (Arabidopsis Genome Initiative, 2000), indicating that AtMinE1 is a unique gene.
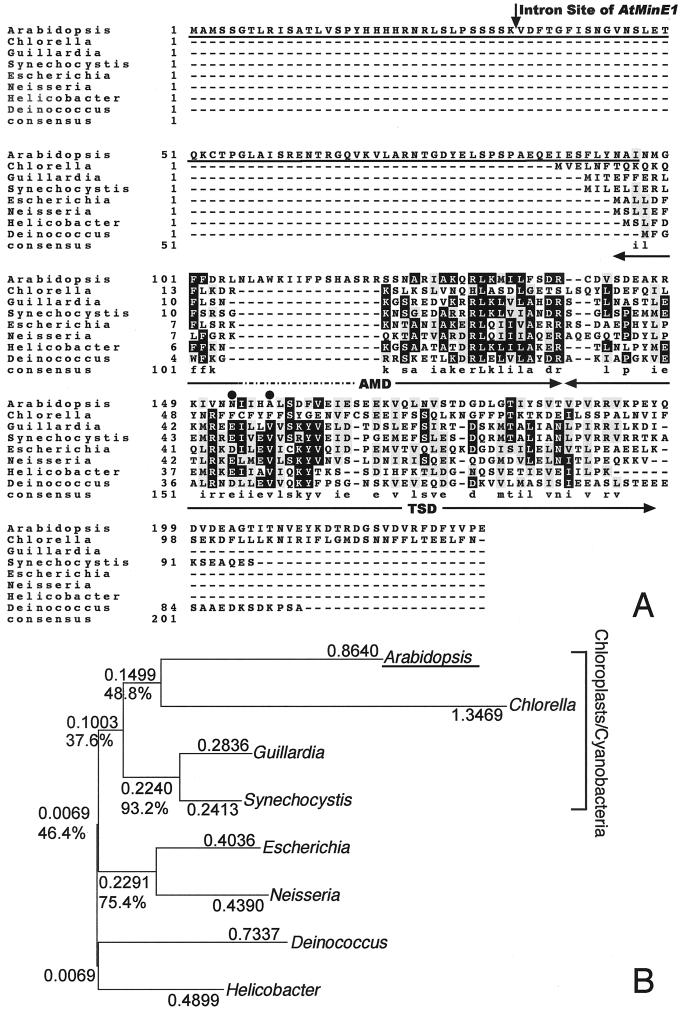
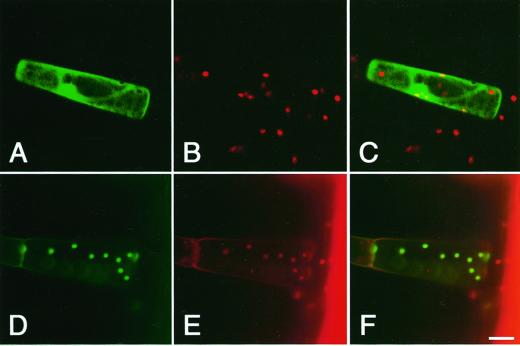
Normally, MinE from eubacteria and algal chloroplasts is a small protein composed of about 90 aa. In comparison, AtMinE1 possesses a long N-terminal extension of about 110 aa and a short C-terminal extension of about 30 aa (Fig. 1A). Construction of a phylogenetic tree of eight MinE sequences from wide-ranging eubacteria, algal chloroplasts, and Arabidopsis revealed a close relationship between AtMinE1 and the MinE protein of Synechocystis sp. and algal chloroplasts (Fig. 1B). This suggested a cyanobacterial/chloroplast origin of the nucleus-encoded AtMinE1. The very N-terminal 50-amino acids region of AtMinE1 possesses features typical of chloroplast transit peptides (Keegstra et al., 1989), such as Ala at the second residue, richness in hydroxylated residues (Ser and Thr; 30%), and a deficiency in acidic residues (Asp and Glu; 4%). In addition, computer programs that predict the subcellular localization of proteins, such as PSORT (old version; Nakai and Kanehisa, 1992), ChloroP (version 1.1; Emanuelsson et al., 1999), TargetP (version 1.01; Emanuelsson et al., 2000), and Predotar (version 0.5; http://www.inra.fr/Internet/Produits/Predotar/), unanimously concluded that there was a high probability for chloroplast targeting of AtMinE1. To determine whether the N-terminal extension of AtMinE1 could function as a transit peptide, we constructed an expression vector, pMinE-TP-GFP, consisting of 97 amino acids from the AtMinE1 N terminus fused to the N terminus of the green fluorescence protein (GFP) under the control of the cauliflower mosaic virus (CaMV) 35S promoter. This construct was introduced into tobacco (Nicotiana tabacum) leaf trichome cells by particle bombardment. As a result, transiently expressed chimeric GFP was localized exclusively in chloroplasts, whereas non-fused GFP was observed only in the cytoplasm and the nucleus (Fig. 2). Thus, the N terminus of AtMinE1 has the ability to carry this protein into chloroplasts.
Figure 1.
Alignment and phylogenetic relationship of MinE proteins. A, Sequence alignment performed with CLUSTAL W (ver. 1.8; Thompson et al., 1994) using the default parameters shown at the web site http://searchlauncher.bcm.tmc.edu/multi-align/Parameters/clustalw.html. The database accession numbers are as follows: Arabidopsis (AB046117), C. vulgaris chloroplast genome (chlorophyte; P56350), G. theta chloroplast genome (cryptophyte; AAC35620), Synechocystis sp. PCC6803 (cyanobacterium; BAA10661), E. coli (γ-proteobacterium; AAB59063), Neisseria meningitidis (β-proteobacterium; CAB83414), Helicobacter pylori (ε-proteobacterium; AAD05906), and Deinococcus radiodurans (member of Thermus/Deinococcus group; Q9RWB8). An arrow shows the position of the intron site in the AtMinE1 gene. Letters on black (gray) background show the residues identical (similar) to the consensus that is formed by four or more identical (similar) residues within the same column. The consensus residues are shown in the bottom row in lowercase except where a perfect consensus is shown in uppercase. The N-terminal region of AtMinE1, which was N terminally fused to GFP for the subsequent localization study, is underlined. The locations of the AMD and TSD in the E. coli protein are indicated by the double-headed arrows. Positions of the conserved residues, Asp-45 (Glu) and Val-49, critical for the topological specificity function in E. coli MinE (King et al., 2000) are indicated by dots. B, Unrooted phylogenetic tree of amino acid sequences of MinE proteins, constructed using the neighbor-joining method (Saitou and Nei, 1987; GENETYX-MAC version 11.0.2, Software Development, Tokyo). Relatively conserved amino acids corresponding to amino acid 121 through 193 of AtMinE1 in the above alignment (A) were utilized for tree construction. Substitutions per amino acid position and bootstrap confidence values (%) based on 500 replications are shown for each clade.
Figure 2.
Chloroplast localization of GFP fused with the N-terminal region of AtMinE1. The fusion protein was transiently expressed under the CaMV35S promoter in tobacco leaf trichome cells. GFP signals and chlorophyll autofluorescence were observed with a confocal laser scanning microscope. Fluorescence images of GFP (A and D; green), chlorophyll (B and E; red), and merged images (C and F) for non-fused GFP (A–C) or the chimeric GFP with AtMinE1 (D–F) are shown. Scale bar represents 20 μm.
The MinE protein of E. coli contains two separable functional domains: the N-terminal anti-MinCD domain (AMD), which is necessary and sufficient for counteracting MinCD activity, and the C-terminal topological specificity domain (TSD), which is essential for mid-cell localization of MinE (King et al., 2000). Sequence alignment of AtMinE1 with other MinE sequences revealed that, within the central core region of AtMinE1, an N-terminal domain corresponding to the E. coli AMD is well conserved, whereas a C-terminal domain corresponding to the E. coli TSD is poorly conserved (Fig. 1A). Asp-45 and Val-49 of the E. coli MinE TSD were recently shown to form a D2V2 tetrad at the center of an antiparallel coiled-coil within the homodimeric TSD, and to be directly involved in the topological specificity function of MinE, but not in its homodimerization (King et al., 2000). In fact, Asp-45 (or Glu) and Val-49 are strictly conserved in all currently available MinE sequences from eubacteria, cyanobacteria, and the G. theta chloroplast, and thus appear to be key functional residues for the topological specificity. Nevertheless, both Asp-45 (Glu) and Val-49 are no longer conserved in either AtMinE1 or the C. vulgaris chloroplast MinE. These results suggest functional conservation of the AMD and evolutionary divergence of the TSD function in AtMinE1.
Specific Activation of the AtMinE1 Promoter at the Shoot Apex
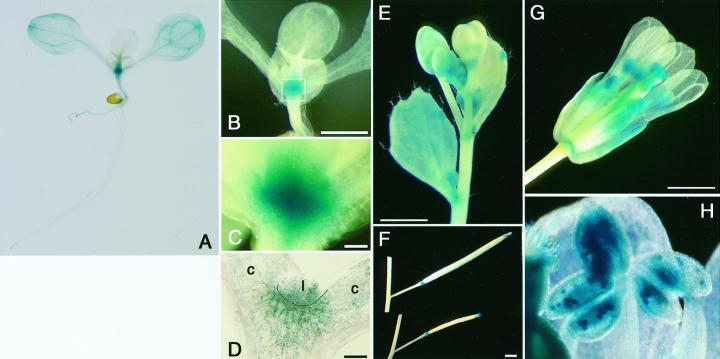
To monitor tissue-specific expression of the AtMinE1 gene in situ, a 1.6-kb genomic DNA fragment from the 5′ end of the gene was transcriptionally fused to the uidA reporter gene in the binary plasmid pBI101, and the resultant construct, pMinE-GUS, was introduced into Arabidopsis to yield stable transformants. The transgenic lines were expected to express GUS with no additional amino acids at the N terminus, under cis-acting control of the AtMinE1 upstream sequence. Of the 33 independent kanamycin-resistant plants obtained, at least 20 lines exhibited GUS activity. Seven-day-old seedlings of the GUS-positive plants were analyzed (Fig. 3, A–D). Strong GUS activity was detected in the shoot apices of these plants. The veins of the cotyledons were modestly stained, whereas very weak staining was observed at other regions of the cotyledons and tip margins of emerging leaves. Blue GUS staining was confined to green tissues in young seedlings, and was undetectable in roots (Fig. 3A), with the exception of one line (data not shown). In mature plants, blue staining was visible in leaves, sepals, siliques, and anthers (Fig. 3, E–H). The GUS activity in siliques was limited to the tip and base (Fig. 3F), and stems showed only weak activity in some areas. Staining of the stigma was not observed in some of the transgenic lines (data not shown). Strong GUS expression was observed in pollen grains inside the anthers (Fig. 3H). Because only weak activity was observed in developing anthers of the floral buds (Fig. 3E), AtMinE1 expression may only be active in the late stages of pollen development.
Figure 3.
Histochemical GUS staining of AtMinE1 promoter::uidA-transgenic Arabidopsis plants. GUS staining patterns are shown for whole seedlings (A–C), a section of meristematic region of a seedling (D), and plant organs (E–H); cauline leaf and inflorescence (E), silique (F), open flower (G), and anther at higher magnification (H). C represents an enlargement of the boxed area in B. c, Cotyledon; l, leaf. Scale bars represent 1 mm.
Disruption of Chloroplast Division Caused by Overexpression of MinE
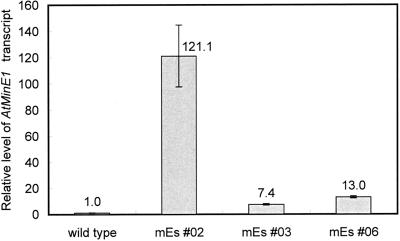
To examine the role of MinE in higher plant cells, we transformed Arabidopsis with the entire coding region of the AtMinE1 cDNA under the control of the constitutive CaMV35S promoter. Ten independent lines, named mEs (for minE-sense), that expressed AtMinE1 ectopically were obtained. Quantitative reverse transcription (RT)-PCR revealed that the mEs plants had 7- to 120-fold increases in AtMinE1 RNA levels over wild type (Fig. 4). The mEs lines appeared normal, when compared with the wild-type plants, with respect to outward appearance, growth, flowering, and fertility (data not shown). Nevertheless, microscopic observations revealed that the size, morphology, and number of chloroplasts within the cells of T1 mEs plants were abnormal (Figs. 5 and 6). The chloroplasts and their DNA (nucleoids) were simultaneously examined by fluorescence microscopy by staining resin-embedded leaf sections with 3,3′-dihexyloxacarbocyanine iodide (DiOC6) and 4′,6-diamidino-2-phenylindole (DAPI; Fujie et al., 1994; Nagata et al., 1999). In mesophyll cells, the chloroplasts were much larger and fewer in number than in wild-type plants (Fig. 5). The extent of enlargement and the decline in numbers of chloroplasts varied between cells, even within the same tissue. In the most extreme case, a single giant chloroplast occupied the majority of the cell volume. Even less affected cells contained five or fewer chloroplasts. Chloroplast shape was also aberrant, and varied from the normal lens shape to snaky, cup shaped, or multilobed. Despite the aberrant morphology of the chloroplasts, their nucleoids were small and uniformly dispersed throughout the stromal region (Fig. 5A), which is characteristic of photosynthetically competent plastids of higher plants (Kuroiwa, 1991). Chloroplasts of cotyledons, hypocotyls, and petioles were also visualized by chlorophyll autofluorescence and were similarly affected (i.e. enlarged and reduced) by overexpression of AtMinE1 (data not shown). These phenotypes were inherited in the T2 progeny.
Figure 4.
Relative levels of AtMinE1 transcript in wild-type and mEs plants (with wild type = 1) determined by quantitative RT-PCR. Fluorescence for AtMinE1 in each sample was normalized to the 18S control.
Figure 5.
Subcellular phenotypes of mEs plants. Thin sections of the foliage leaves of Arabidopsis, simultaneously stained with DAPI (A and B) and DiOC6 (C and D), were observed by epifluorescence microscopy. The fluorochrome DiOC6 stains membranous organelles, including chloroplasts, in fixed tissues. Cells from mEs (A and C) and wild-type (B and D) seedlings are shown. Scale bar represents 20 μm.
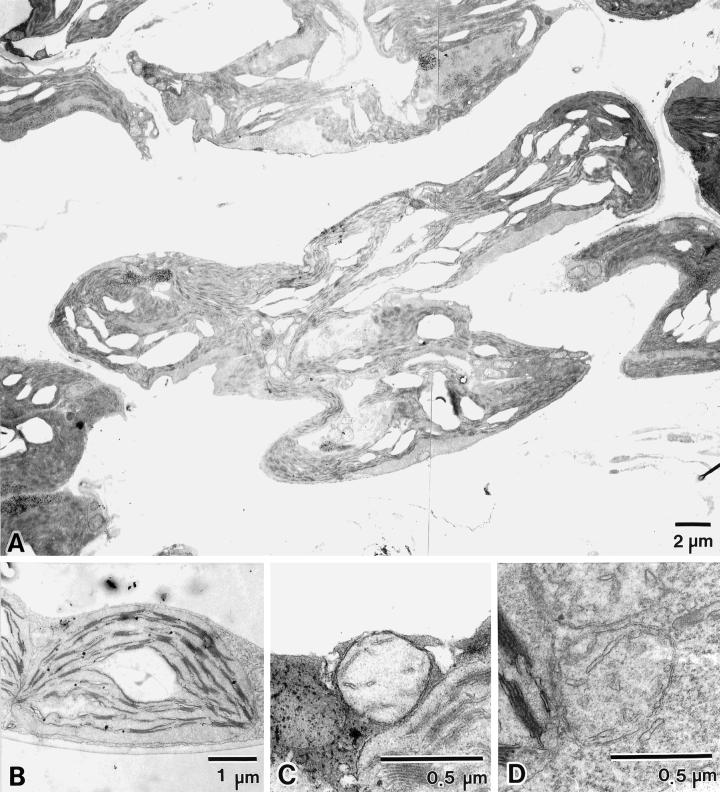
Figure 6.
Ultrastructure of mEs macro-chloroplasts. Electron micrographs of leaf mesophyll chloroplasts (A and B) and mitochondria (C and D) from mEs (A and C) and wild-type (B and D) plants are shown.
The chloroplast ultrastructure of mEs plants was examined by transmission electron microscopy (Fig. 6A). The profiles of the mEs chloroplasts appeared extremely elongated and distorted, meandering throughout a large part of the cytoplasm, confirming the results of fluorescent microscopy. Despite such extraordinary profiles, mEs chloroplasts still retained a well-developed thylakoid membrane system, which was arranged parallel to the long axis of the chloroplast and had an undulating structure that mirrored the undulations in the chloroplast surface. The arrangement of membranes into grana and inter-granal lamellae resembled that of wild-type chloroplasts, suggestive of normal photosynthetic ability. The morphology of mitochondria was also observed in these samples by transmission electron microscopy; it appeared normal with respect to shape, size, and internal membrane structure (i.e. cristae), and was indistinguishable from that of wild-type mitochondria (Fig. 6, C and D). This implies that mitochondrial division was not affected by overexpression of AtMinE1, although an accurate count of mitochondrial numbers was not obtained.
To further investigate the role of MinE in chloroplast division, we constructed a transformation vector containing the coding region of AtMinE1 cDNA in the antisense orientation under the control of the CaMV35S promoter. This vector was introduced into Arabidopsis plants, resulting in the generation of 11 kanamycin-resistant T1 plants. Ten of these T1 plants and their T2 progenies were no different from wild-type plants with respect to size, shape, and number of chloroplasts (data not shown). Only one remaining T1 plant produced T2 progeny, which displayed a striking reduction in chloroplast number (data not shown). However, owing to the paucity of AtMinE1 antisense lines showing this phenotype, we could not definitively conclude that repressed expression of AtMinE1 disrupts chloroplast division.
DISCUSSION
ftsZ and minD are the most well conserved among prokaryotic cell division genes, and are present in most species of prokaryotes (Margolin, 2000). Given the eubacterial origins of chloroplasts and mitochondria, it is not unexpected that FtsZ homologs are also utilized for division of chloroplasts (Osteryoung et al., 1998; Strepp et al., 1998) and mitochondria (Beech et al., 2000; Takahara et al., 2000), or that MinD regulates chloroplast division (Colletti et al., 2000; Kanamaru et al., 2000). In contrast to FtsZ and MinD, MinE is not present in the majority of characterized species (Margolin, 2000). The present study demonstrated for the first time that MinE is also utilized for chloroplast division, despite its dispensable nature among prokaryotes.
Transient expression of the AtMinE1 N terminus:GFP fusion indicates that AtMinE1 is targeted to chloroplasts (Fig. 2). Because AtMinE1 is predicted to be soluble, lacking both a membrane-spanning hydrophobic region and a lumenal targeting domain, this protein is likely a stromal one. In E. coli, it was previously shown that a biologically active MinE:GFP fusion protein accumulates near the middle of the cells in a ring-like structure that appears to be associated with the plasma membrane (Raskin and de Boer, 1997). Recent time-lapse studies of MinE:GFP localization unveiled a more dynamic nature of the MinE ring structure, which undergoes a repetitive cycle of movement first to one cell pole and then to the opposite pole (Fu et al., 2001; Hale et al., 2001). Therefore, it is likely that AtMinE1 localizes in a confined subregion(s) inside the chloroplast inner envelope, rather than distributing uniformly throughout the stromal region. To complicate matters, Kiessling et al. (2000) reported that in chloroplasts of the moss P. patens, transiently expressed FtsZ:GFP assembles into a basket-like framework, appearing to scaffold the entire chloroplast envelope from the stromal side. This FtsZ network, referred to as a plastoskeleton, may be involved in the maintenance of the structural integrity of chloroplasts, rather than in chloroplast division per se. Complementing the P. patens study, Vitha et al. (2001) observed that chloroplast Z-rings formed at the mid-plastid division site in Arabidopsis. E. coli MinE localizes in an annular structure near the Z-ring and excludes the FtsZ-inhibitory MinCD complex from the mid-cell to permit Z-ring assembly. Therefore, it is possible that the chloroplast MinE also assembles into an intra-organellar network near the FtsZ plastoskeleton, or into a mid-plastid ring juxtaposed with the stromal Z-ring, to exclude chloroplast MinD from the plastoskeleton/Z-ring. To explore this hypothesis, we are currently preparing a full-length version of the AtMinE1:GFP fusion vector and anti-AtMinE1-peptide antibodies.
GUS reporter assay is a convenient way to obtain spatial information on promoter activity qualitatively, although the intensity of GUS staining does not necessarily correlate with the actual product level. To unambiguously define the AtMinE1 protein levels in distinct tissues, anti-AtMinE1 antibodies would be used for quantitative immunoblotting in our future study. The currently available data from the GUS assay suggest that the AtMinE1 promoter is mainly activated at the shoot apex in young seedlings (Fig. 3, A–D), where active cell division occurs. This high expression of AtMinE1 at the shoot apex may be required for the coordination of proplastid division and cell division, because the rate of proplastid division must be at least the same as the rate of cell division to maintain plastid continuity. Detailed studies of the AtMinE1 promoter, utilizing cultured cells, may pave the way for a new understanding of the mechanism that couples chloroplast division to cell division. In contrast to the specific activation of the AtMinE1 promoter at the shoot apex, no promoter activity was detected in root apical meristems (Fig. 3A). This implies that root plastids (amyloplasts and leucoplasts) require another set of molecular components for their effective division, which differs from (but may partially overlap) that required for chloroplast division. Further analysis of AtMinE1 may also elucidate this variation in plastid division machinery that depends on the plastid type. In mature plants, the AtMinE1 promoter was activated in green tissues of the leaves, sepals, and siliques, and inside the anthers (Fig. 3, E–H). These observations indicate that AtMinE1 primarily functions within the chloroplasts of green tissues. The biological significance of the specific expression of the AtMinE1 promoter::uidA in anthers is currently difficult to explain. However, Mori and Tanaka (2000) reported that high expression of a lily (Lilium longiflorum) ftsZ occurs in plastid-deficient generative cells. This suggests that the eubacteria-derived system has unknown functions besides chloroplast division in these cells.
Overexpression of minE within E. coli led to the formation of anucleate minicells, consistent with the MinCDE division site selection model (de Boer et al., 1989). In contrast, overexpression of AtMinE1 in Arabidopsis produced enlarged chloroplasts, possibly caused by the inhibition of chloroplast division (Figs. 5 and 6). In addition, we did not find any chloroplast heterogeneity within a single mEs cell, as was seen with AtMinD1 antisense plants (Colletti et al., 2000). The eubacterial MinCDE model predicts that overexpression of minE should have the same effect on chloroplast division as does repression of minD, and vice versa. The apparent contradiction between this prediction and our observations of the mEs lines may reflect only a superficial, rather than a crucial, difference. High-level expression of ftsZ in E. coli inhibits, rather than accelerates, cell division (Ward and Lutkenhaus, 1985). Similarly, Dai and Lutkenhaus (1992) and Dewar et al. (1992) stressed the importance of the proper balance between FtsZ and its interacting partner FtsA for normal cell division, because inhibition of cell division due to increased FtsZ could be suppressed by increased FtsA and vice versa. In addition, overexpression of FtsZ1 or FtsZ1:GFP in Arabidopsis inhibited chloroplast division, in proportion to its protein level (Stokes et al., 2000; Vitha et al., 2001), rather than the formation of increased minichloroplasts, as predicted by the eubacterial model. Combined, it appears that a proper balance is required between Fts, Min, and other division proteins to facilitate cell or chloroplast division. An alternative explanation is that the function of the chloroplast MinE is different from that of the E. coli (or eubacterial) MinE. Complete genome sequencing of Synechocystis sp. (Kaneko et al., 1996) and Arabidopsis (Arabidopsis Genome Initiative, 2000) has revealed the presence of a MinC homolog in Synechocystis sp. and its absence in Arabidopsis. Consequent to its absence, chloroplast MinD or MinE in plants may have acquired a novel function(s).
Finally, chloroplast targeting of AtMinD1 (Colletti et al., 2000; Kanamaru et al., 2000) and AtMinE1 (this study; Fig. 2) raises a crucial problem: If these proteins define the site of assembly of the outer cytosolic PD ring, then there must be a means of transmitting topological information across the double membranes of the plastid envelope. In the alga C. merolae, a middle PD ring has been observed in the intermembrane space, in addition to the inner stromal and outer cytosolic PD rings (Miyagishima et al., 1998). Although this has not been observed in other organisms, such a structure could have a role in transmitting topological information from the stromal MinDE system to the cytosolic PD ring.
MATERIALS AND METHODS
Plant Materials
Seeds of Arabidopsis ecotype Columbia were purchased from Lehle Seeds (Round Rock, TX) and used throughout in this work.
Isolation of the AtMinE1 cDNA Clone
The oligonucleotides mE-F 5′-GTCGACCCGGGCGAG-CAATTTCAAGTTTCTCGG-3′ and mE-R 5′-CCCGGGC-TGTCTTTGTTCCCTCCAGACTAAAC-3′, with an SmaI restriction site (underlined), were used to amplify AtMinE1 cDNA from an Arabidopsis MATCHMAKER cDNA Library (CLONTECH, Palo Alto, CA) using the Expand High Fidelity PCR System (Roche Diagnostics, Mannheim, Germany). The PCR product was subcloned into a pCRII vector (Invitrogen, Carlsbad, CA) to produce pCRIImE and then sequenced with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). To determine both the 5′ and 3′ ends of the AtMinE1 cDNA, the first PCR was carried out with the vector primer GAD10F1 5′-GGACGGACCAAACTGCGTATAACGCG-3′ and the cDNA-specific primer mE-R2 5′-CCCGGGCCAGCAAATAAATCTAAGACTGTGCC-3′ for the 5′ end, or with the vector primer GAD10R1 5′-CAAACCTCTGGCGAAGAAGTCCAAAGC-3′ and the cDNA-specific primer mE-F for the 3′ end, using the Arabidopsis cDNA library as template. The PCR products were size fractionated by agarose gel electrophoresis and DNA fragments of the appropriate length were purified with a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). Using the purified DNA fragments as a template, a second PCR was performed with the vector primer GAD10F2 5′-CGATGATGAAGATACCCCACCAAACCC-3′ and the cDNA-specific primer mE-5R 5′-GAGAGTTCCAGAAGACATCGCC-3′, for the 5′ end, or with the vector primer GAD10R2 5′-CGATGCACAGTTGAAGTGAACTTGCGG-3′ and the cDNA-specific primer mE-3F 5′-CCAGAGTGATGAAAAGGCACAGTC-3′, for the 3′ end. The products were subcloned and the clones containing the longest insert of five arbitrarily chosen clones were subjected to sequencing for each end.
Stable Plant Transformation and Selection
Plasmids were transferred from Escherichia coli to Agrobacterium tumefaciens strain C58C1 using the triparental mating procedure (Walkerpeach and Velten, 1994). Arabidopsis was infected with A. tumefaciens containing the plasmid using the floral dip method (Clough and Bent, 1998). Transformed seeds were selected on one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) containing 50 μg mL−1 of kanamycin and 100 μg mL−1 of cefotaxime.
Expression and Visualization of an AtMinE1:GFP Fusion in Living Plant Cells
GFP was used as a reporter to examine the subcellular localization of AtMinE1 (Chiu et al., 1996). A cDNA fragment encoding the N-terminal 97-aa sequence of AtMinE1 was amplified by PCR with an oligonucleotide primer set with restriction sites (underlined) ME-SalI 5′-AAGGTC-GACTTCTCCGGCGAGCAATT-3′ and ME-NcoI 5′-AAC-CCATGGTGATGGCATTATAGAGAAAG-3′. The PCR product (0.3 kb) was digested with SalI and NcoI, and ligated into SalI-NcoI-digested CaMV35S-sGFP(S65T)-nos vector (Isono et al., 1997; provided by Dr. Yasuo Niwa, University of Shizuoka, Japan) to yield the construct pMinE-TP-GFP. This construct was introduced into young leaf cells of tobacco (Nicotiana tabacum cv Xanthi) with a particle bombardment device (Biolistic PDS-1000/He; Bio-Rad, Hercules, CA) with DNA-coated, 1.0-μm gold particles (Bio-Rad). Gold particles were coated by precipitating 20 μg of purified plasmid DNA (10 μL) onto 50 μL of suspended gold particles with 50 μL of 2.5 m CaCl2 and 20 μL of 0.1 m spermidine, followed by washing with ethanol. Bombardment was performed at a helium pressure of 1,100 p.s.i. (pound-force per square inch) with a vacuum of 27 inches of Hg in the chamber, and a distance to the target tissues of 6 cm. After bombardment, leaves were incubated for 2 d at 28°C before observation with a confocal laser scanning microscope (TCS-NT, Leica Microsystems, Heidelberg) or a fluorescence microscope (IX70, Olympus, Tokyo) with a color CCD camera (DP50-C, Olympus) attachment. For confocal microscopy, GFP was excited at 488 nm with an argon/krypton laser. Recorded images of GFP and chlorophyll were imported into the RGB channels of Adobe Photoshop (Adobe Systems, San Jose, CA), and merged images were processed.
Analysis of AtMinE1 Promoter Activity Using a uidA (GUS) Reporter
GUS was used as a reporter to study the tissue-specific expression of AtMinE1. A 1.6-kb Arabidopsis genomic DNA sequence upstream from the AtMinE1-coding region was fused to the GUS gene (uidA) as a transcriptional fusion. The 1.6-kb genomic DNA fragment was amplified by PCR using oligonucleotides containing SalI and BamHI restriction sites, digested with BamHI and SalI, and ligated into these sites in the vector pBI101 to yield pMinE-GUS. pMinE-GUS was used for stable transformation of Arabidopsis using A. tumefaciens as described above. GUS staining was performed with intact seedlings or excised plant organs, essentially as described (Jefferson, 1987). Plant tissues were soaked in staining buffer (50 mm sodium phosphate [pH 7.0], 10 mm EDTA, 0.5 mm potassium ferrocyanide, 0.1% [v/v] Triton X-100, 2% [v/v] dimethyl sulfoxide, and 0.7 mm 5-bromo-4-chloro-3-indoryl-β-d-glucuronide [X-Gluc; Nacalai tesque, Kyoto]) and incubated at 37°C for 24 h. These were cleared of chlorophyll in a graded ethanol series, and observed under a stereomicroscope (SZH10, Olympus) with a digital camera (HC-300, Fujifilm, Tokyo) attachment. For detailed observation, stained materials were embedded in 5% (w/v) agar, sectioned at 80 μm with a microslicer (DTK-1000, DOSAKA, Kyoto), and observed under a microscope (IX70, Olympus).
Construction and Microscopic Observations of AtMinE1-Overexpressing Transgenic Arabidopsis
To overexpress the entire coding region of AtMinE1 under the control of the CaMV35S promoter in Arabidopsis, a cDNA fragment was PCR amplified from pCRIImE using oligonucleotides mE-F and mE-SacR 5′-AGACAAGATGAGCTCCAGCAAATAAATCTAAGACTGTG-3′. This fragment was digested with SmaI and SacI and ligated into the same restriction sites of pBI121 (TOYOBO, Tokyo) to yield pBImEs. pBImEs was used for stable transformation of Arabidopsis with A. tumefaciens as described above. Kanamycin-resistant T1 seedlings of mEs lines were grown on selection plates for 2 weeks, and then transplanted to soil (Golden Peatban, Sakata Seed, Yokohama, Japan). Using a razor blade, leaf sections were cut from plants that had been grown for 1 week in soil and were processed for microscopy as follows.
Fluorescence Microscopy
Samples were fixed in 4% (w/v) paraformaldehyde, buffered with 20 mm sodium cacodylate (pH 7.2) for 20 h at 4°C, dehydrated through an ethanol series, and then embedded in Technovit 7100 resin (Kulzer and Co., Wehrheim, Germany). Thin sections (0.6 μm thick) were cut with a glass knife on an ULTRACUT UCT ultramicrotome (Leica, Wien, Austria), placed on cover slips, and dried. Sections were stained with 100 μg mL−1 DiOC6 in ethanol for 30 s, washed in 70% (v/v) ethanol for 10 s, and in distilled water for 10 s, before further staining with 1 μg mL−1 DAPI, as described previously (Fujie et al., 1994; Nagata et al., 1999). The samples were observed with an Olympus IX70 microscope.
Electron Microscopy
Samples were fixed in 4% (w/v) glutaraldehyde, buffered with 20 mm sodium cacodylate (pH 7.0) for 20 h at 4°C, and then washed with the same buffer for 4 h at 4°C. Samples were post-fixed in 2% (w/v) osmium tetroxide in 20 mm cacodylate buffer (pH 7.0) for 20 h at 4°C. The fixed samples were run through an alcohol series and embedded in Spurr's resin (Spurr, 1969). Ultrathin sections were cut with a diamond knife on an ultramicrotome and transferred to Formvar-coated grids. They were double stained with 1% (w/v) uranyl acetate for 15 min at 37°C and with lead citrate solution for 10 min at room temperature. After washing with distilled water, the samples were observed with a JEM-2000 FX II electron microscope (JEOL, Tokyo).
Measurement of AtMinE1 RNA Levels by Quantitative RT-PCR
Total RNA was isolated from the wild type or from independent mEs lines (T2) grown on kanamycin-free Murashige and Skoog medium for 2 weeks, using SV Total RNA Isolation System (Promega, Madison, WI). A SUPERSCRIPT First-Strand Synthesis System for RT-PCR (Life Technologies, Rockville, MD) was used for RT reaction. PCR primers and TaqMan Probes were designed by using the program Primer Express ver.1.5 (Applied Biosystems) as follows. Primers for AtMinE1 were 5′-CGCATCAA-GAAGAAGCTCCAA-3′ (forward) and 5′-CCTGTCGGAG-AAGAGGATCATC-3′ (reverse), and the TaqMan Probe was 5′-FAM-CAAGAATTGCAAAGCAGCGGCTCA-TAMRA-3′. Primers for 18S ribosomal RNA (used as a control) were 5′-CGGCTACCACATCCAAGGA-3′ (forward) and 5′-GCTGGAATTACCGCGGCT-3′ (reverse), and the TaqMan Probe was 5′-VIC-CAGCAGGCGCGCAAATTACCCA-TAMRA-3′. The RT products were added to the recommended PCR master mix, 10 μm primers, and 5 μm TaqMan probe. PCR was performed in sextuplicate on the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The amplification conditions were 1 cycle for 2 min at 50°C and 10 min at 95°C, followed by 40 cycles for 15 s at 95°C and 1 min at 60°C. A dilution series of the pCRIImE DNA was used to generate a standard curve. AtMinE1 product was normalized by using 18S as an endogenous control and calculated relative to wild-type message levels.
ACKNOWLEDGMENTS
We thank Dr. Yasuo Niwa for providing the GFP vector and Dr. Yukihisa Shimada and Mr. Narumasa Miyauchi (Plant Science Center, RIKEN) for help with the real-time PCR system.
Footnotes
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid no. 12740452 to R.I.), by the President's Special Research Grant from RIKEN (grant to R.I.), and by the Special Postdoctoral Researcher's Program of RIKEN (R.I., M.F., and N.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010386.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Beech PL, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson PR, McFadden GI. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol. 2000;10:507–516. doi: 10.1016/s0960-9822(00)00466-8. [DOI] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: Complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48:236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Shih Y-L, Zhang Y, Rothfield LI. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia colidivision cycle. Proc Natl Acad Sci USA. 2001;98:980–985. doi: 10.1073/pnas.031549298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie M, Kuroiwa H, Kawano S, Mutoh S, Kuroiwa T. Behavior of organelles and their nucleoids in the shoot apical meristem during leaf development in Arabidopsis thalianaL. Planta. 1994;194:395–405. doi: 10.1007/BF00194444. [DOI] [PubMed] [Google Scholar]
- Hale CA, Meinhardt H, de Boer PAJ. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 2001;20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H. Double ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma. 1986;135:166–172. [Google Scholar]
- Hirota Y, Ryter A, Jacob F. Thermosensitive mutants of E. coliaffected in the process of DNA synthesis and cellular division. Cold Spring Harbor Symp Quant Biol. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H. Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of ς70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:14948–14953. doi: 10.1073/pnas.94.26.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Kanamaru K, Fujiwara M, Kim M, Nagashima A, Nakazato E, Tanaka K, Takahashi H. Chloroplast targeting, distribution and transcriptional fluctuation of AtMinD1, a eubacteria-type factor critical for chloroplast division. Plant Cell Physiol. 2000;41:1119–1128. doi: 10.1093/pcp/pcd037. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystissp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- Kiessling J, Kruse S, Rensing SA, Harter K, Decker EL, Reski R. Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J Cell Biol. 2000;151:945–950. doi: 10.1083/jcb.151.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GF, Shih Y-L, Maciejewski MW, Bains NPS, Pan B, Rowland SL, Mullen GP, Rothfield LI. Structural basis for the topological specificity function of MinE. Nat Struct Biol. 2000;7:1013–1017. doi: 10.1038/80917. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T. The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytol. 1991;128:1–62. [Google Scholar]
- Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. The division apparatus of plastids and mitochondria. Int Rev Cytol. 1998;181:1–41. doi: 10.1016/s0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- Marrison JL, Rutherford SM, Robertson EJ, Lister C, Dean C, Leech RM. The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 1999;18:651–662. doi: 10.1046/j.1365-313x.1999.00500.x. [DOI] [PubMed] [Google Scholar]
- Miyagishima S, Itoh R, Toda K, Takahashi H, Kuroiwa H, Kuroiwa T. Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J Electron Microsc. 1998;47:269–272. [Google Scholar]
- Miyagishima S, Takahara M, Kuroiwa T. Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell. 2001;13:707–721. doi: 10.1105/tpc.13.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Tanaka I. Isolation of the ftsZ gene from plastid-deficient generative cells of Lilium longiflorum. Protoplasma. 2000;214:57–64. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;103:473–497. [Google Scholar]
- Nagata N, Saito C, Sakai A, Kuroiwa H, Kuroiwa T. Decrease in mitochondrial DNA and concurrent increase in plastid DNA in generative cells of Pharbitis nilduring pollen development. Eur J Cell Biol. 1999;78:241–248. doi: 10.1016/s0171-9335(99)80057-0. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possingham JV, Lawrence ME. Controls to plastid division. Int Rev Cytol. 1983;84:1–56. [Google Scholar]
- Pyke KA, Leech RM. Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana(L.) Heynh. Plant Physiol. 1991;96:1193–1195. doi: 10.1104/pp.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 1992;99:1005–1008. doi: 10.1104/pp.99.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 1994;104:201–207. doi: 10.1104/pp.104.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Rutherford SM, Robertson EJ, Leech RM. arc6, a fertile Arabidopsismutant with only two mesophyll cell chloroplasts. Plant Physiol. 1994;106:1169–1177. doi: 10.1104/pp.106.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PAJ. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D, Gordon GS, Wright A. How does a bacterium find its middle? Nat Struct Biol. 2000;7:997–999. doi: 10.1038/81014. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Rutherford SM, Leech RM. Characterization of chloroplast division using the Arabidopsis mutant arc5. Plant Physiol. 1996;112:149–159. doi: 10.1104/pp.112.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2genes in Arabidopsis. Plant Physiol. 2000;124:1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Takano H, Sakai A, Kawano S, Kuroiwa T. A putative mitochondrial ftsZ gene is present in the unicellular primitive red alga Cyanidioschyzon merolae. Mol Gen Genet. 2000;264:452–460. doi: 10.1007/s004380000307. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW. FtsZ ring formation at the chloroplast division site in plants. J Cell Biol. 2001;153:111–119. doi: 10.1083/jcb.153.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, Nagai T, Kapoor M, Sugita M, Ito M, Ito S, Tsudzuki J, Nakashima K, Tsudzuki T, Suzuki Y. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc Natl Acad Sci USA. 1997;94:5967–5972. doi: 10.1073/pnas.94.11.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkerpeach CR, Velten J. Agrobacterium-mediated gene transfer to plant cells: cointegrate and binary vector systems. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–19. [Google Scholar]
- Ward JE, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]