Abstract
Chloroplast division is driven by a macromolecular complex containing components that are positioned on the cytosolic surface of the outer envelope, the stromal surface of the inner envelope, and in the intermembrane space. The only constituents of the division apparatus identified thus far are the tubulin-like proteins FtsZ1 and FtsZ2, which colocalize to rings at the plastid division site. However, the precise positioning of these rings relative to the envelope membranes and to each other has not been previously defined. Using newly isolated cDNAs with open reading frames longer than those reported previously, we demonstrate here that both FtsZ2 proteins in Arabidopsis, like FtsZ1 proteins, contain cleavable transit peptides that target them across the outer envelope membrane. To determine their topological arrangement, protease protection experiments designed to distinguish between stromal and intermembrane space localization were performed on both in vitro imported and endogenous forms of FtsZ1 and FtsZ2. Both proteins were shown to reside in the stromal compartment of the chloroplast, indicating that the FtsZ1- and FtsZ2-containing rings have similar topologies and may physically interact. Consistent with this hypothesis, double immunofluorescence labeling of various plastid division mutants revealed precise colocalization of FtsZ1 and FtsZ2, even when their levels and assembly patterns were perturbed. Overexpression of FtsZ2 in transgenic Arabidopsis inhibited plastid division in a dose-dependent manner, suggesting that the stoichiometry between FtsZ1 and FtsZ2 is an important aspect of their function. These studies raise new questions concerning the functional and evolutionary significance of two distinct but colocalized forms of FtsZ in plants and establish a revised framework within which to understand the molecular architecture of the plastid division apparatus in higher plants.
Evolved from prokaryotic endosymbionts (Martin and Herrmann, 1998; Gray, 1999; McFadden, 1999), plastids divide by binary fission, whereby a constriction forms at the division plane, progressively tightening to separate the new organelles. Although plastids differ structurally from their cyanobacterial ancestors (Douglas, 1998), the division processes in both share mechanistic similarities. In bacteria, the tubulin-like GTPase FtsZ assembles into a dynamic circular structure termed the Z-ring at the cell center, forming a cytoskeletal framework to which all other cell division proteins are recruited (for review, see Rothfield et al., 1999) and probably providing the force that powers the contractile machinery (Lu et al., 2000; Erickson, 2001). It is now established that homologs of FtsZ also are essential for chloroplast division in land plants (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998). In vascular plants, plastid division entails the participation of two distinct, highly conserved nuclear-encoded FtsZ protein families, FtsZ1 and FtsZ2 (Osteryoung et al., 1998; Osteryoung and McAndrew, 2001). Immunofluorescence microscopy has shown that both proteins assemble into rings encircling the division site (Vitha et al., 2001), although whether these FtsZ1 and FtsZ2 rings are components of identical or different structures has not been determined.
Ultrastructural studies of plants and algae have revealed that the plastid division apparatus contains two or three electron-dense, topologically distinct plastid-dividing (PD) rings associated with the constricted region of dividing chloroplasts (Kuroiwa et al., 1998; Miyagishima et al., 1998; Osteryoung and McAndrew, 2001). An outer PD ring is localized to the cytosolic surface of the outer envelope membrane (OEM), an inner ring is localized to the stromal surface of the inner envelope membrane (IEM), and in red algae (Cyanidioschyzon merolae) a third PD ring has been observed within the intermembrane space (IMS). The presence of a chloroplast division apparatus comprising as many as three PD rings and multiple forms of FtsZ indicates a level of complexity beyond that associated with bacterial cell division. Establishing the topological relationship among the FtsZ1 ring, the FtsZ2 ring, and the PD rings is essential for understanding the molecular architecture of the plastid division apparatus in higher plants.
Our investigations of FtsZ genes in Arabidopsis, coupled with sequence data from the Arabidopsis genome project, have shown the presence of three FtsZ genes in this organism: AtFtsZ1-1, a member of the FtsZ1 family, and AtFtsZ2-1 and AtFtsZ2-2, both members of the FtsZ2 family. All full-length FtsZ1 proteins identified to date have amino-terminal extensions that are predicted with high confidence to target them to chloroplasts (Osteryoung and McAndrew, 2001); this has been confirmed experimentally for AtFtsZ1-1 and for an FtsZ1 protein from pea (Pisum sativum) using in vitro chloroplast import assays (Osteryoung and Vierling, 1995; Gaikwad et al., 2000). Similar experiments on AtFtsZ2-1, however, which were based on the open reading frame (ORF) predicted from both cDNA and genomic AtFtsZ2-1 clones, failed to show that this protein was imported into chloroplasts, despite its demonstrated role in chloroplast division (Osteryoung et al., 1998). These data in combination with the ultrastructural studies led us to propose a model for chloroplast division whereby FtsZ1 and FtsZ2 proteins functioned as components of the inner and outer PD rings, respectively (Osteryoung et al., 1998), although the results of the import assays could not distinguish between a stromal and IMS localization for FtsZ1. However, inconsistent with the hypothesized localization of FtsZ2 in the outer PD ring, subsequent submissions of other plant FtsZ2 sequences revealed that the extreme amino termini of most FtsZ2 family members, including AtFtsZ2-2, were longer than that predicted for AtFtsZ2-1 (Osteryoung and McAndrew, 2001), suggesting that the AtFtsZ2-1 ORF used in our previous analysis (Osteryoung et al., 1998) might be incomplete, lacking the transit peptide. In addition, two recent studies demonstrated that nuclear-encoded FtsZ2-like proteins from the moss Physcomitrella patens were localized in the chloroplast when expressed as green fluorescent protein fusion proteins in vivo (Kiessling et al., 2000) and that FtsZ could not be identified immunologically in outer PD ring preparations from red algae (Miyagishima et al., 2001). Furthermore, genes encoding both FtsZ1 and FtsZ2 proteins have only been identified in higher plant genomes; thus, the relationship of the findings in moss and algae to the organization of the plastid division apparatus in higher plants remains unclear.
Because the previous import experiments and the cytological data demonstrating colocalization of the FtsZ1 and FtsZ2 rings in Arabidopsis did not indicate whether these proteins were positioned outside the chloroplast, inside the chloroplast, or in the IMS, we used a biochemical approach to re-evaluate and more definitively establish the localization of FtsZ1 and FtsZ2 in higher plants. We have isolated a new cDNA for AtFtsZ2-1 as well as a cDNA for AtFtsZ2-2, with ORFs longer than those previously predicted, and used these clones to conduct a new series of localization experiments. The results indicate that both Arabidopsis FtsZ2 proteins, like FtsZ1, are imported into isolated chloroplasts, processed to mature form, and protected from proteolytic challenge following import. Moreover, protease protection assays analyzing the suborganellar localization of both in vitro imported and endogenous FtsZ1 and FtsZ2 provide strong evidence that both proteins are localized in the stromal compartment of the chloroplast. Results indicating precise colocalization of FtsZ1 and FtsZ2 in various transgenic plants and mutants even under conditions in which FtsZ filament assembly is abnormal provide evidence that these two forms of FtsZ interact in vivo. These findings compel us to consider new models for the roles of FtsZ1 and FtsZ2 in plastid division.
RESULTS
Longer ORFs for AtFtsZ2-1 and AtFtsZ2-2 Are Confirmed by Isolation of New cDNAs
Evaluation of the genomic sequences predicted to encode the FtsZ2 proteins of Arabidopsis indicated that in-frame Met codons corresponding to potential alternative start sites were present 243 and 258 nucleotides upstream from those previously predicted for AtFtsZ2-1 and AtFtsZ2-2, respectively (Osteryoung et al., 1998). Because cDNA clones corresponding to these longer ORFs had not previously been identified, we wished to ascertain whether the longer ORFs were transcribed. Therefore, we used gene-specific primers for AtFtsZ2-1 and AtFtsZ2-2 to determine whether cDNAs corresponding to the longer ORFs could be amplified by reverse transcription-PCR. Products were obtained in both cases, all introns removed, indicating that both cDNAs represented mature transcripts and that both ORFs were expressed.
Both AtFtsZ2-1 and AtFtsZ2-2 Are Imported into Chloroplasts in Vitro
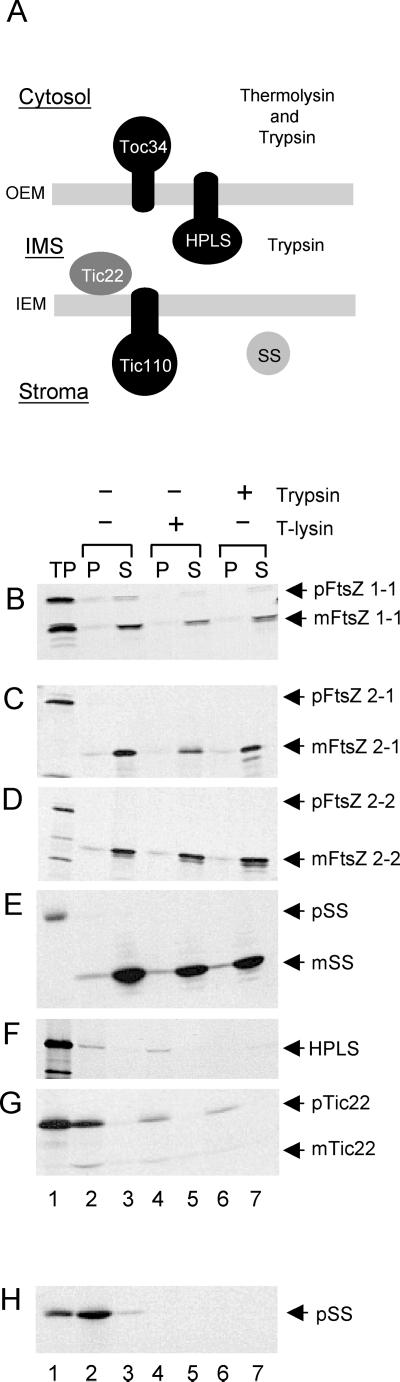
We used the subcellular targeting prediction program TargetP (Emanuelsson et al., 2000) to determine whether N-terminal chloroplast transit peptides could be identified in the full-length form of AtFtsZ2-1 or AtFtsZ2-2 (Table I). Although the TargetP score for AtFtsZ1-1 predicted both the presence of a transit peptide and targeting to chloroplasts with high confidence, as was confirmed experimentally (Osteryoung and McAndrew, 2001), the targeting predictions for the FtsZ2 sequences were unclear (Table I). To test this experimentally, we performed in vitro import assays using isolated pea chloroplasts. Because PD rings have been visualized associated with both the stromal and cytosolic surfaces of the envelopes in plants (Kuroiwa, 1998) as well as in the IMS in red algae (Miyagishima et al., 1998), we incorporated into these assays a series of protease protection experiments to identify the suborganellar localization of AtFtsZ2-1 and AtFtsZ2-2 import products. For this, we used two proteases with established differences in their abilities to penetrate the OEM and IEM (Fig. 1A): thermolysin, which cannot penetrate the OEM and therefore selectively degrades proteins localized on the cytosolic surface of the chloroplast (Cline et al., 1984), and trypsin, which penetrates the OEM but not the IEM and, therefore, degrades proteins localized either on the cytosolic surface or in the IMS, but not in the stromal compartment (Jackson et al., 1998). Parallel assays with marker proteins whose topologies with respect to the OEM and IEM are well established (Fig. 1A) were performed as controls.
Table I.
Comparison of the predictions made by TargetP for the presence of a chloroplast transit peptide at the amino terminus of full-length FtsZ sequences
| Organism | Accession No. | Family Grouping | TargetPa Scoreb (+/−)c | Import Assay Results |
|---|---|---|---|---|
| Arabidopsis (AtFtsZ2-1) | AF089738 | FtsZ2 | 0.555 (+) | Importedd |
| Arabidopsis (AtFtsZ2-2) | AF384167 | FtsZ2 | 0.374 (−) | Importedd |
| Gentiana lutea | AAF23771 | FtsZ2 | 0.555 (−) | NPDe |
| Nicotiana tabacum | AJ271750 | FtsZ2 | 0.131 (−) | NPD |
| Lilium longiflorum | AB042101 | FtsZ2 | 0.712 (+) | NPD |
| P. patens | AJ249140 | FtsZ2 | 0.784 (+) | Importedf |
| P. patens | AJ249138 | FtsZ2 | 0.871 (+) | Importedf |
| Arabidopsis (AtFtsZ1-1) | Q43545 | FtsZ1 | 0.974 (+) | Importedg |
Scores were calculated using the N-terminal 130 amino acids of the predicted ORF.
Positive or negative chloroplast-targeting prediction is indicated by + or −, respectively.
This study.
No published data are available.
Figure 1.
FtsZ proteins are imported into isolated pea chloroplasts. A, Model illustrating the localization and orientation of control proteins used in protease protection assays. B to G, Radiolabeled AtFtsZ1-1 (B), AtFtsZ2-1(C), AtFtsZ2-2 (D), or the control proteins, SS (E), HPLS (F), and Tic22 (G), were imported into isolated pea chloroplasts. In H, pSS was bound to the import machinery of the chloroplast outer membrane but not imported. Both import and binding reactions were incubated with (+) or without (−) trypsin or thermolysin (T-lysin) for 30 min on ice and then quenched. Intact chloroplasts were recovered by sedimentation through a 40% (v/v) Percoll cushion, lysed, and separated into a total membrane (P) or soluble (S) fraction. All samples were separated by SDS-PAGE and analyzed by fluorography. TP, 10% of the radiolabeled translation product added to the reaction, precursor protein (p), and mature protein (m) are indicated by arrows.
The import assays indicated that both FtsZ2 proteins as well as FtsZ1 were imported into pea chloroplasts (Fig. 1, B–D), processed to mature proteins (compare lanes 1 and 3 in B–D), indicating removal of the transit peptide, and associated with the soluble fractions following import (Fig. 1, B–D, lanes 3, 5, and 7). In addition, in all three cases the processed import products were protected from proteolytic degradation by both thermolysin (Fig. 1, B–D, lane 5) and trypsin (Fig. 1, B–D, lane 7). This behavior was identical to that of the small subunit of Rubisco (SS), a stromal marker protein (Fig. 1E). In contrast, the IMS marker proteins hydroperoxide lyase (HPLS), an integral protein of the OEM that remains unprocessed following import and is predominantly orientated toward the IMS (Froehlich et al., 2001), and Tic22 (Translocon at the inner membrane of chloroplasts), a peripheral IEM protein likewise orientated toward the IMS (Kouranov et al., 1998), were both susceptible to degradation by trypsin but not thermolysin (Fig. 1, F–G, compare lanes 4–6). In a parallel reaction, the precursor of SS (pSS) bound to the OEM surface of chloroplasts was completely degraded by trypsin and thermolysin (Fig. 1H), confirming the activity of both proteases.
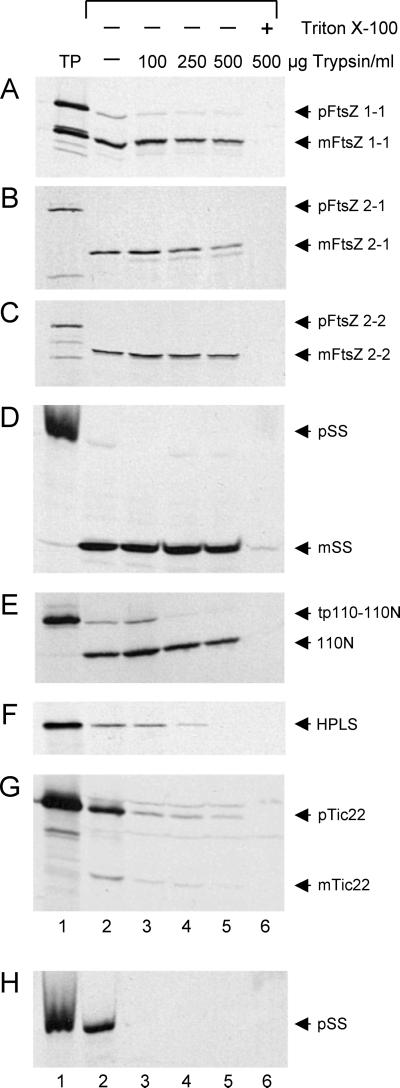
Although trypsin has been a valuable tool in defining the topology of outer and inner envelope proteins and in localizing soluble proteins to the chloroplast IMS, the susceptibility of IMS proteins to trypsin is often dependent on protease concentration (Jackson et al., 1998; Kouranov et al., 1998, 1999; Froehlich et al., 2001). To rule out the possibility that the protection of the FtsZs from trypsin was a function of protease concentration in the IMS rather than localization of the proteins in the stroma, a trypsin titration experiment was performed (Fig. 2). The results show that after import all mature FtsZ proteins as well as mSS were protected from trypsin at concentrations up to 500 μg mL−1 (Fig. 2, A–D, lanes 2–5). As an additional control, we demonstrated that a truncated version of Tic110, tp110-110N, which is orientated toward the stroma (Fig. 1A), was not degraded (Fig. 2E, lanes 2–5), confirming that trypsin did not gain access to the stromal compartment. Only solubilization of the chloroplast membranes by detergent rendered these proteins susceptible to proteolysis (Fig. 2, A–E, lane 6). In contrast, the IMS marker proteins HPLS and mTic22 became vulnerable to trypsin at much lower concentrations (Fig. 2, F and G, lanes 3–5), indicating that trypsin had gained access to the IMS. Trypsin activity also was confirmed by the complete digestion of OEM-bound pSS (Fig. 2H, lanes 3–6). The import and protease susceptibilities of the FtsZ proteins relative to those of the control proteins leads us to conclude that AtFtsZ1-1, AtFtsZ2-1, and AtFtsZ2-2 are targeted to the chloroplast by cleavable transit peptides, imported, and localized to the chloroplast stroma. Because these results are inconsistent with the predictions of TargetP (Table I), they underscore the limitations of bioinformatics resources in revealing subcellular localization. In this context, it is worth noting that neither AtFtsZ2-1 nor AtFtsZ2-2 is represented in the list of chloroplast-targeted proteins (http://mips.gsf.de/proj/thal/db/tables/tables_gen_frame.html) predicted to be encoded in the Arabidopsis genome.
Figure 2.
The protection of FtsZ proteins from trypsin digestion suggests that these proteins are localized to the chloroplast stroma following import in vitro. A to G, Large-scale import reactions using either radiolabeled AtFtsZ1-1 (A), AtFtsZ2-1 (B), AtFtsZ2-2 (C), or the control proteins, SS (D), tp110-110N, truncated Tic110 (E), HPLS (F), and Tic22 (G), were incubated for 30 min at room temperature. Each import reaction was then divided into five equal fractions. H, Likewise, a large-scale binding reaction with pSS was performed and divided into five equal fractions. Individual import and binding reactions were incubated either without (−) or with 100, 250, or 500 μg trypsin mL−1 in the absence (−) or presence (+) of Triton X-100. Protease digestion reactions were incubated on ice for 30 min and then quenched. Intact chloroplasts recovered by sedimentation were solubilized in sample buffer, separated by SDS-PAGE, and analyzed by fluorography. TP represents 10% of radiolabeled translation product added to the reaction; arrows indicate precursor protein (p) and mature protein (m).
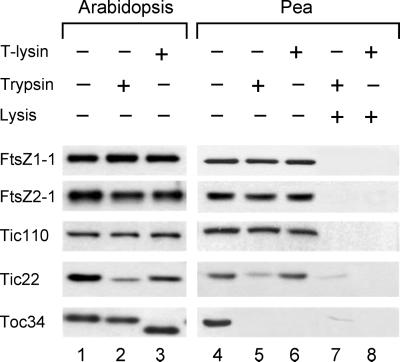
Endogenous FtsZ Proteins of Arabidopsis and Pea Behave Similarly to the in Vitro FtsZ Chloroplast Import Products
To complement the in vitro experiments (Figs. 1 and 2), we subjected chloroplasts isolated from Arabidopsis and pea to treatments with either thermolysin or trypsin and monitored the sensitivity of endogenous FtsZ proteins to the proteases by immunoblotting (Fig. 3). The immunoreactive signals obtained following chloroplast incubation in the absence of added protease are shown in Figure 3 (lanes 1 and 4). Consistent with the in vitro results (Figs. 1 and 2), endogenous FtsZ1, FtsZ2, and Tic110 proteins were protected from degradation by either thermolysin (lanes 3 and 6) or trypsin (lanes 2 and 5), whereas endogenous Tic22, an IMS control protein, was vulnerable to trypsin digestion in chloroplasts from both plants (Fig. 3, lanes 2 and 5). To further confirm that the proteases were active, we monitored the protease susceptibility of Translocon at the outer membrane of chloroplasts (Toc) 34, a protein of the OEM oriented toward the cytosol (Fig. 1A). Consistent with its topology, Toc34 was completely degraded by both proteases in pea (Fig. 3, lanes 5 and 6) and partially digested in Arabidopsis (Fig. 3, lanes 2 and 3). Variability in the susceptibility of Toc34 to protease digestion has previously been reported (Chen and Schnell, 1997). Finally, endogenous FtsZ1, FtsZ2, and control proteins were fully susceptible to protease digestion when chloroplasts were osmotically lysed (Fig. 3, lanes 7 and 8), indicating that the invulnerability of these proteins to the proteases was not a consequence of protein folding. These data provide strong evidence that FtsZ1 and FtsZ2 proteins in plants are localized in the stromal compartment.
Figure 3.
The protection of endogenous FtsZ proteins from both thermolysin and trypsin digestion in chloroplasts isolated from Arabidopsis or pea indicates that both FtsZ1 and FtsZ2 are localized to the stroma of chloroplasts in vivo. Intact chloroplasts equivalent to 50 μg of chlorophyll were treated with (+) or without (−) thermolysin (T-lysin) or trypsin either before (−) or after (+) lysis. Following protease treatments at 500 μg mL−1 (lanes 2 and 3) or 800 μg mL−1 (lanes 5–8) and solubilization in sample buffer, SDS-PAGE-resolved polypeptides remaining in Arabidopsis (lanes 1–3) or pea (lanes 4–8) chloroplasts were analyzed by immunoblotting with antigen-specific antibodies, as indicated, using chemiluminescence detection. Equal sample volumes, equivalent to 2 μg of chlorophyll, for analysis of FtsZ1, FtsZ2, and Tic22, or 3 μg of chlorophyll, for analysis of Toc34 and Tic110, were loaded onto gels.
Overexpression of AtFtsZ2-1 Inhibits Chloroplast Division
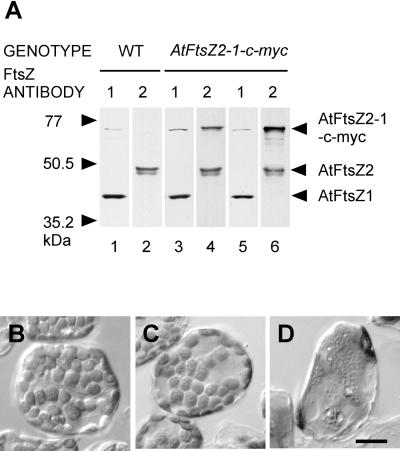
The isolation of a new cDNA for AtFtsZ2-1 with a longer ORF indicates that the cDNA sequence identified previously was not full length and explains the failure of Arabidopsis plants transformed with the shorter ORF to accumulate AtFtsZ2-1 above wild-type levels (Stokes et al., 2000). To revisit the effect of AtFtsZ2-1 overexpression on plastid division, we introduced into transgenic Arabidopsis an AtFtsZ2-1 cDNA to which a c-myc tag had been fused two codons upstream from the AtFtsZ2-1 stop codon. The transgene was expressed under control of the native AtFtsZ2-1 promoter, and the phenotypes of mesophyll cell chloroplasts in numerous independent transgenic lines were investigated by light microscopy. In contrast to wild-type cells containing normal levels of AtFtsZ1-1 and AtFtsZ2-1 proteins (Fig. 4A, lanes 1 and 2) and normal chloroplast numbers (Fig. 4B), severe plastid division defects were frequently observed in the AtFtsZ2-1-c-myc transgenic plants as indicated by the presence of single enlarged chloroplasts (Fig. 4D). Higher levels of the fusion protein were detected in these plants (Fig. 4A, lane 6) compared with transgenic plants without plastid division defects (Fig. 4A, lane 4, and 4C). These results are similar to the dosage-dependent chloroplast division defects observed in plants overexpressing AtFtsZ1-1 (Stokes et al., 2000) and to the cell division defects resulting from high levels of FtsZ overexpression in bacteria (Ward and Lutkenhaus, 1985).
Figure 4.
Correlation of protein levels with chloroplast phenotypes of plants expressing the AtFtsZ2-1-c-myc transgene. A, Immunoblots of leaf extracts from wild-type (WT) plants (lanes 1 and 2) and transgenic plants expressing AtFtsZ2-1-c-myc at moderate (lanes 3 and 4) or high levels (lanes 5 and 6) were probed with affinity-purified anti-AtFtsZ1-1 (lanes 1, 3, and 5) or anti-AtFtsZ2-1 (lanes 2, 4, and 6). The identities of the immunoreactive polypeptides and the migration of molecular mass markers are indicated on the right and left, respectively. Immunoreactivity of anti-FtsZ2 with AtFtsZ2-1-cmyc is shown in lanes 4 and 6. The weak high molecular mass band in lanes 1, 3, and 5 probably represents a dimeric form of AtFtsZ1-1. B, Chloroplasts in a wild-type leaf mesophyll cell. C and D, Leaf mesophyll cells from transgenic plants expressing AtFtsZ2-1-c-myc at moderate (C) or high (D) levels. The plants shown in C and D are identical to those represented in the immunoblot in A. Scale bar, 20 μm.
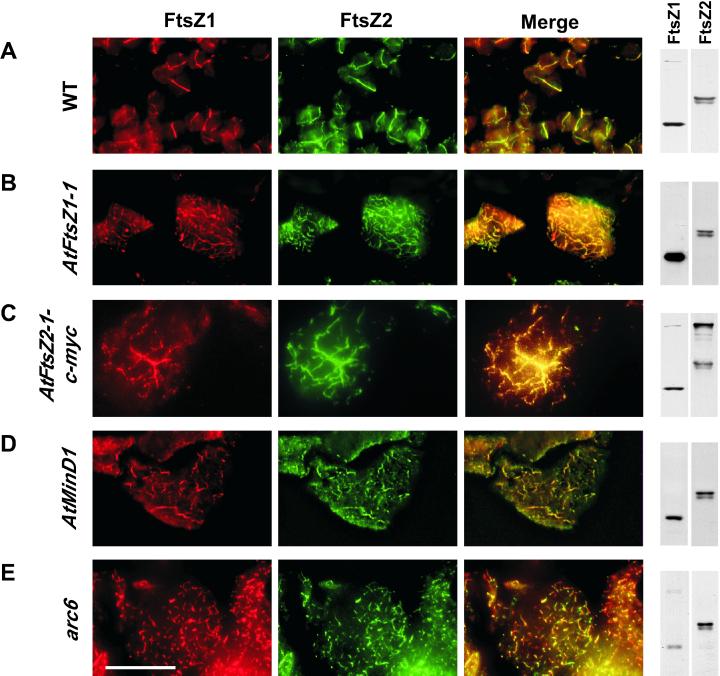
Aberrations in FtsZ Filament Morphology Do Not Disturb Colocalization of FtsZ1 and FtsZ2
We previously demonstrated that FtsZ1 and FtsZ2 colocalized to midplastid rings in wild-type Arabidopsis (Fig. 5A) and in other plants (Vitha et al., 2001). Combined with our present findings, that these proteins are localized in the same subcompartment of the chloroplast, these data suggest that FtsZ1 and FtsZ2 could be components of the same ring structure. To investigate whether uncoupling of FtsZ1 and FtsZ2 localization could be observed, we used double immunofluorescence labeling to analyze their localization patterns in several transgenic lines and mutants with plastid division defects and abnormal FtsZ filaments. These included plants that overexpress AtFtsZ1-1 (Stokes et al., 2000), AtFtsZ2-1-c-myc (this report), AtMinD1 (Colletti et al., 2000), and the plastid division mutant arc6 (Pyke et al., 1994). Chloroplasts from plants with high AtFtsZ1-1 or AtFtsZ2-1-c-myc levels contained relatively long, disorganized filaments (Fig. 5, B and C), whereas the AtMinD1 overexpression lines and chloroplasts from the arc6 mutant plants contained shorter FtsZ fragments (Fig. 5, D and E, respectively). Despite variations in filament morphology and FtsZ1-to-FtsZ2 ratios among these plants (see corresponding immunoblots in Fig. 5), in every case FtsZ1 and FtsZ2 were tightly colocalized (see Fig. 5, yellow overlay). Because of the differences in FtsZ1 and FtsZ2 immunofluorescence signal intensities, particularly in plants overexpressing AtFtsZ1-1 or AtFtsZ2-1-c-myc (Fig. 5, B and C), some of the areas of the overlay images show predominantly green or red color. Nevertheless, the anti-FtsZ1 and anti-FtsZ2 antibodies consistently revealed identical filament patterns for both proteins within a given specimen. A series of controls for specificity and order of labeling (Vitha et al., 2001) confirmed that the fluorescence signals detected in these experiments represented distinct proteins (not shown).
Figure 5.
Colocalization of AtFtsZ1-1 and AtFtsZ2-1 is unaltered by various plastid division defects. Leaf sections from Arabidopsis wild-type (WT) plants (A), transgenic plants highly overexpressing AtFtsZ1-1, AtFtsZ2-1-c-myc, or AtMinD1 (B–D, respectively), or arc6 mutant plants (E) were subjected to sequential, double immunofluorescence labeling of AtFtsZ1-1 and AtFtsZ2-1. Localization of AtFtsZ2-1-c-myc fusion-proteins (C) was achieved using anti-c-myc antibodies. Immunoblots of proteins in leaf extracts from the same plants, probed with anti-AtFtsZ1-1 or anti-AtFtsZ2-1 antibodies, are shown on the right. The yellow color in the overlay of the red and green signals indicates colocalization of AtFtsZ1-1 and AtFtsZ2-1. Scale bar, 20 μm.
DISCUSSION
Our previous hypothesis that FtsZ1 and FtsZ2 proteins were components of the inner and outer PD rings, respectively, was compatible with the existing cytological data and provided an attractive explanation for the presence of two distinct FtsZ families in plants (Osteryoung et al., 1998). In an initial test of this model, we demonstrated that both FtsZ1 and FsZ2 localize to rings at the plastid division site (Vitha et al., 2001). However, their topologies could not be interpreted from the methods used. By defining the suborganellar localizations of FtsZ1 and FtsZ2 in the present study, we have now established that both rings are localized in the stromal compartment of the chloroplast. These findings are significant for two reasons. First, they indicate that FtsZ2 is not a component of the outer PD ring. This is consistent with a recent report by Miyagishima et al. (2001) indicating that FtsZ could not be detected in outer PD ring extracts from red algae. Based on these and other data, these investigators hypothesized that the outer PD ring is not derived from the endosymbiotic progenitor of chloroplasts, as are the FtsZ rings, but rather from the host cell. Furthermore, neither FtsZ1 nor FtsZ2 proteins are indicated as constituents of the PD ring observed in the IMS. Second, the colocalization of FtsZ1 and FtsZ2 has important implications for the functional relationship between these two proteins. In most prokaryotes, a single FtsZ is sufficient to orchestrate cell division (Wang and Lutkenhaus, 1996; Erickson, 2000; Margolin, 2000; Gilson and Beech, 2001). Why have two forms of FtsZ evolved to function in the same suborganellar compartment during plastid division in higher plants?
That FtsZ1 and FtsZ2 are, in fact, functionally distinct is suggested by several lines of evidence. These include the presence of both families in dicots and monocots (Mori and Tanaka, 2000; Beech and Gilson, 2000; Gilson and Beech, 2001; Osteryoung and McAndrew, 2001), experiments showing that depletion of either protein in Arabidopsis disrupts plastid division (Osteryoung et al., 1998; Stokes et al., 2000), and analyses revealing conserved differences in their primary and secondary structures (Osteryoung and McAndrew, 2001). A particularly noteworthy difference is that only FtsZ2 proteins contain a short, conserved peptide sequence at the extreme carboxy terminus that in prokaryotes binds the integral membrane protein ZipA (Erickson, 2001; Osteryoung and McAndrew, 2001). In bacteria, ZipA is thought to stabilize the Z-ring in vivo by cross-linking FtsZ polymers (RayChaudhuri, 1999; Hale et al., 2000; Mosyak et al., 2000). Although ZipA homologs have not been identified in plants, it is possible that an undiscovered plastid division protein interacts specifically with FtsZ2. The absence of the ZipA-binding peptide in FtsZ1 and its presence in FtsZ2 could, therefore, endow these two proteins with unique functions. However, recent studies, including the results shown in Figure 4, showing that the severity of the plastid division defects in plants with altered FtsZ1 or FtsZ2 levels is strongly dose dependent (Kiessling et al., 2000; Stokes et al., 2000; Vitha et al., 2001), raise the question of whether FtsZ1 and FtsZ2 could substitute functionally for one another if total FtsZ levels were unchanged. We are currently addressing this issue experimentally.
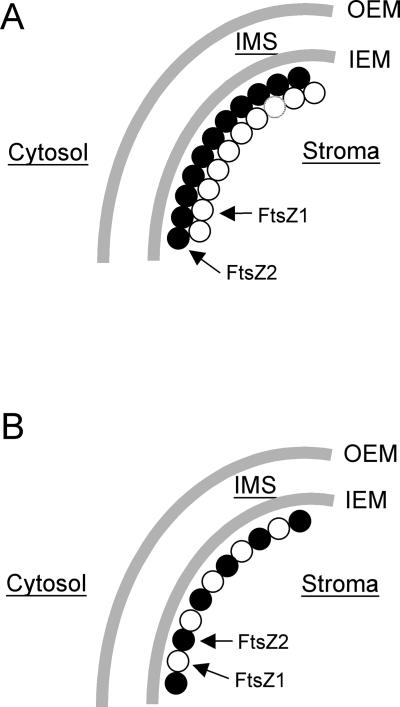
If FtsZ1 and FtsZ2 are functionally distinct, as we believe, our results collectively suggest two alternative explanations for their functional relationship. In the first model (Fig. 6A), FtsZ1 and FtsZ2 could form separate, homopolymeric protofilaments that associate laterally to form the plastid Z-ring just inside the IEM. In the second model (Fig. 6B), FtsZ1 and FtsZ2 could co-assemble as heteropolymeric filaments, perhaps analogous to the association between α- and β-tubulin in microtubules (Nogales et al., 1998). Numerous stoichiometric variations on both scenarios can be imagined, any of which could be supported by the tight and invariant colocalization of FtsZ1 and FtsZ2 shown in Figure 5. Investigating the interactions between FtsZ1 and FtsZ2 will be important for determining their functional relationship, and for distinguishing between and further refining these two models.
Figure 6.
Possible configurations of FtsZ1- and FtsZ2-containing rings at the chloroplast division site in the stromal compartment of chloroplasts. In A, FtsZ1 (○) and FtsZ2 (●) proteins could be assembled into colocalized homopolymeric rings that may be laterally associated at the division site. In B, FtsZ1 and FtsZ2 proteins could be co-assembled into heteropolymeric ring structures at the division site.
MATERIALS AND METHODS
Isolation of Full-Length AtFtsZ2-1 and AtFtsZ2-2 cDNAs
Arabidopsis, ecotype Columbia (Col-0) plants were grown for 21 d as described previously (Osteryoung et al., 1998). Total RNA was extracted from 1 g of leaf tissue by a modification of the procedure described by Chomczynski and Sacchi (1987) using the reagent TRIZOL (Gibco-BRL, Grand Island, NY). Gene-specific oligonucleotide primers for either AtFtsZ2-1 (5′-TATGGCAACTTACGTTTCACCGTGT-3′, forward, and 5′-CTCACATCAGCAAAATCCAC-3′, reverse) or AtFtsZ2-2 (5′-CAGAATGGCAGCTTATGTTTCTCC-3′, forward, and 5′-AGTGGGTCTAGAGGCGAGGA-3′, reverse) were used to amplify their corresponding cDNAs by reverse transcription-PCR using an M.J. Research thermocycler (Waltham, MA). Superscript II reverse transcriptase (Gibco-BRL) was used to synthesize first-strand cDNAs from 10 μg of total RNA. PCR amplification was performed using Pfu DNA polymerase (Stratagene, La Jolla, CA) or Deep Vent DNA polymerase (New England Biolabs, Beverly, MA) for AtFtsZ2-1 or AtFtsZ2-2, respectively, and was carried out using the corresponding forward and reverse gene-specific primers. Amplified cDNAs were purified, subcloned into pBluescript II KS (Stratagene), and sequenced to confirm the fidelity of the PCR reaction.
Chloroplast Isolation
Intact chloroplasts used for in vitro import or protease protection assays were purified from 8- to 12-d-old pea (Pisum sativum var. Little Marvel) seedlings or 4-week-old Arabidopsis ecotype Columbia (Col-0) plants over a Percoll gradient as previously described (Bruce et al., 1994), except that sodium ascorbate and glutathione were omitted from the buffers used for pea chloroplast isolation.
In Vitro Translation and Import Reactions
All in vitro synthesized, radiolabeled proteins used in import assays were generated using a coupled transcription-translation system containing nuclease-treated rabbit reticulocyte lysate (Promega, Madison, WI), and [35S] Met (DuPont/NEN, Wilmington, DE), according to the manufacturers' recommended protocol. Plasmids containing the AtFtsZ1-1 (Osteryoung and Vierling, 1995), AtFtsZ2-1, AtFtsZ2-2, pTic22 (Kouranov et al., 1998), or tp110-110N (Lübeck et al., 1997) cDNA required T7 RNA Polymerase (Promega) for transcription. Plasmids containing pSS (Olsen and Keegstra, 1992) or HPLS (Froehlich et al., 2001) cDNA required SP6 RNA polymerase (Promega) or T3 RNA polymerase (Promega), respectively.
Binding or import reactions (adapted from Bruce et al., 1994) received 500,000 dpm of radiolabeled translation product Mg-ATP (Sigma, St. Louis), either 0.1 or 4 mm, to promote binding or translocation, respectively, and intact chloroplasts corresponding to 25 μg of chlorophyll in a final volume of 150 μL. Treatment of binding or import reactions with thermolysin was performed as described by Cline et al. (1984). Treatment with trypsin was performed as described by Jackson et al. (1998). After protease digestions were quenched, intact chloroplasts were re-isolated through 40% (v/v) Percoll and washed in import buffer (50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-KOH, pH 8.0, 0.33 m sorbitol). Pelleted chloroplasts were either solubilized directly in sample buffer for analysis by SDS-PAGE (Laemmli, 1970) and fluorography or resuspended in lysis buffer (25 mm HEPES-KOH, pH 8.0, 4 mm MgCl2), vigorously vortexed, and incubated on ice for 15 min. Lysed chloroplasts were centrifuged (100,000g) to separate total membranes from soluble fractions.
Protease Protection Assays
Protease treatments of intact chloroplasts isolated from Arabidopsis or pea were performed as described by Cline et al. (1984) and Kouranov et al. (1999), with some modifications. Briefly, isolated intact chloroplasts (equivalent to 50 μg of chlorophyll) were incubated for 30 min in ice-cold reaction buffer (50 mm HEPES-KOH, pH 7.9, 0.33 m sorbitol, 0.8 mm CaCl2) in the absence or presence of either trypsin (bovine pancreatic, 13,000 units mg−1 protein, Sigma) or thermolysin (Sigma) at final concentrations of 500 or 800 μg mL−1 for each protease in a total volume of 250 μL. Proteolysis was arrested with an equal volume of quenching buffer (50 mm HEPES-KOH, pH 7.9, 0.33 m sorbitol, 10 mm EDTA, 0.05 mg mL−1 1-chloro-3-tosylamido-7-amino-2-heptanone, 1.5 μg of soybean trypsin inhibitor per μg of trypsin, 1 mm benzamide, 1 mm benzamidine HCl, 5 mm ε-amino-N-caproic acid, 1 μm leupeptin, and 1 μm pepstatin A). After 15 min on ice, chloroplasts were re-isolated over 40% (v/v) Percoll containing all inhibitors, washed, and resuspended in quenching buffer to a final volume of 50 μL. Chlorophyll content was determined by the method of Arnon (1949). Alternatively, intact chloroplasts (50 μg of chlorophyll) were lysed under hypotonic conditions by vigorous mixing in ice-cold lysis buffer (200 μL of 10 mm HEPES-KOH, pH 7.9, 0.25 m KCl) and subjected to the same proteolytic treatments described above.
Antibodies and Immunoblotting
Anti-peptide antibodies specific for FtsZ1 or FtsZ2 proteins were generated as previously described (Stokes et al., 2000). Tic110 antibodies were the generous gift of F. Kessler (Institute of Plant Sciences, Zurich). Toc34 (Arabidopsis specific) and Tic22 antibodies were the generous gift of D. Schnell (Rutgers University, Newark, NJ). Antibodies for Toc34 (pea specific) were a generous gift of the laboratory of K. Keegstra (Michigan State University, East Lansing). Electrophoresis and immunoblotting were performed as previously described (Stokes et al., 2000). Proteins were transferred either to nitrocellulose (0.45 μm, NitroBind, Osmonics Inc., Westborough, MA) for immunoblot analysis of Tic22, FtsZ1, or FtsZ2 proteins or to polyvinylidene difluoride membranes (0.2 μm, Immun-Blot, Bio-Rad, Hercules, CA) for analysis of Tic110, Toc75, or Toc34. Protein transfer was assessed as uniform and complete by blot and gel staining. Nitrocellulose membranes were blocked for 30 min in TBST (50 mm Tris-HCl, pH 7.4, 200 mm NaCl, 0.2% [v/v] Tween 20) containing 2% or 3% (w/v) nonfat dry milk (Meijer Distribution, Inc., Grand Rapids, MI) prior to overnight incubation with antibodies FtsZ (diluted 1:3,000) or Tic22 (1:2,000), respectively. Polyvinylidene difluoride membranes were blocked for 45 min in TBST containing 3% (w/v) nonfat dry milk for Tic110 (1:800) or 5% (w/v) nonfat dry milk for Toc75 (1:5,000) and Toc34 (1:1,000), respectively. Immunoreactivity was visualized by chemiluminescent detection as previously described (Stokes et al., 2000).
Preparation of AtFtsZ2-1-c-myc Fusion Construct
AtFtsZ2-1 cDNA (accession no. AF089738) in a pUC19-based cloning vector was prepared for C-terminal epitope tagging by digestion at a unique AvaI site in front of the penultimate triplet of the coding sequence, and the overhangs were filled in with the Klenow fragment of DNA polymerase (Gibco-BRL). The 6× c-myc epitope tag was excised as a ClaI fragment from CD3-128 (clone obtained from Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH), treated with the Klenow fragment of DNA polymerase, and cloned into the AtFtsZ2-1 cDNA described above. Clones with correct insert orientation were selected and confirmed by sequencing. The initial portion of AtFtsZ2-1 cDNA (up to the SpeI restriction site) was replaced with a corresponding region from the genomic sequence, which was amplified by PCR from Arabidopsis BAC clone F2H17 (accession no. AC006921). This included 1.8-kb 5′ from the coding region (promoter region) and the first 257 nucleotides of the AtFtsZ2-1-coding region (up to the SpeI site). The fusion construct containing the full N-terminal portion of AtFtsZ2-1, driven by the native promoter, was then transferred to the pART27 (Gleave, 1992) plant transformation vector.
Plant Transformation
Agrobacterium tumefaciens-mediated transformation of wild-type Arabidopsis, ecotype Columbia (Col-0), and growth of the T1 generation of transgenics were performed as previously described (Osteryoung et al., 1998).
Immunofluorescence Labeling
Tissue was taken from the base of expanding Arabidopsis leaves (15 mm long, 5 weeks old). Fixation, embedding, and double immunofluorescence labeling with anti-AtFtsZ1 and anti-AtFtsZ2 antibodies was performed as described previously (Vitha et al., 2001). The protocol used for double immunofluorescence labeling of plants carrying AtFtsZ2-1-c-myc constructs was similar to that described for single immunofluorescence labeling (Vitha et al., 2001). Anti-AtFtsZ1 antibodies (1:200) were combined with mouse monoclonal anti-c-myc antibodies (1:200; Zymed Laboratories, San Francisco), followed by a mixture of secondary antibodies, including anti-rabbit Oregon Green 488 and anti-mouse Alexa 598 (both from Molecular Probes, Eugene, OR) at 1:100 and 1:400 dilutions, respectively. Specimens were viewed and images from the green and red fluorescence channels captured as described before (Vitha et al., 2001). Grayscale images from the separate channels were then pseudocolored and prepared for publication using Adobe Photoshop 5.0 (Adobe Systems Inc., San Jose, CA) and Canvas 6.0 (Deneba Software, Miami) software.
ACKNOWLEDGMENTS
We thank Brandon Castiglione for preparing intact pea chloroplasts and Dr. Ken Keegstra for sharing protocols and enlightening discussions.
Footnotes
This work was supported, in part, by the National Science Foundation (grants MCB–9604412 and MCB–9904524) and the Division of Energy Biosciences at the U.S. Department of Energy.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010542.
LITERATURE CITED
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech PL, Gilson PR. FtsZ and organelle division in Protists. Protist. 2000;151:11–16. doi: 10.1078/1434-4610-00003. [DOI] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K. In vitroimport of proteins into chloroplasts. In: Gelvin SB, Schilperoort RB, editors. Plant Molecular Biology Manual J1. Boston: Kluwer Academic Publishers; 1994. pp. 1–15. [Google Scholar]
- Chen D, Schnell DJ. Insertion of the 34-kDa chloroplast protein import component, IAP34, into the chloroplast outer membrane is dependent on its intrinsic GTP-binding capacity. J Biol Chem. 1997;272:6614–6620. doi: 10.1074/jbc.272.10.6614. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Andrews J, Keegstra K. Thermolysin is a suitable protease for probing the surface on intact pea chloroplasts. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol. 2000;10:507–516. doi: 10.1016/s0960-9822(00)00466-8. [DOI] [PubMed] [Google Scholar]
- Douglas SE. Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev. 1998;8:655–661. doi: 10.1016/s0959-437x(98)80033-6. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Dynamin and FtsZ: missing links in mitochondrial and bacterial division. J Cell Biol. 2000;148:1103–1105. doi: 10.1083/jcb.148.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. The FtsZ protofilament and attachment of ZipA: structural constraints on the FtsZ power stroke. Curr Opin Cell Biol. 2001;13:55–60. doi: 10.1016/s0955-0674(00)00174-5. [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Itoh A, Howe GA. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 2001;75:675–678. doi: 10.1104/pp.125.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaikwad A, Babbarwal V, Pant V, Mukherjee SK. Pea chloroplast FtsZ can form multimers and correct the thermosensitive defect of an Escherichia coli ftsZmutant. Mol Gen Genet. 2000;263:213–221. doi: 10.1007/s004380051162. [DOI] [PubMed] [Google Scholar]
- Gilson PR, Beech PL. Cell division protein FtsZ: running rings around bacteria, chloroplasts and mitochondria. Res Microbiol. 2001;152:3–10. doi: 10.1016/s0923-2508(00)01162-1. [DOI] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Gray MW. Evolution of organellar genomes. Curr Opin Genet Dev. 1999;9:678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem. 1998;273:16583–16588. doi: 10.1074/jbc.273.26.16583. [DOI] [PubMed] [Google Scholar]
- Kiessling J, Kruse S, Rensing SA, Harter K, Decker EL, Reski R. Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J Cell Biol. 2000;151:945–950. doi: 10.1083/jcb.151.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Wang H, Schnell DJ. Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J Biol Chem. 1999;274:25181–25186. doi: 10.1074/jbc.274.35.25181. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T. The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolaeas model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays. 1998;20:344–354. [Google Scholar]
- Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. The division apparatus of plastids and mitochondria. Int Rev Cytol. 1998;181:1–41. doi: 10.1016/s0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübeck J, Heins L, Soll J. A nuclear-encoded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J Cell Biol. 1997;137:1279–1286. doi: 10.1083/jcb.137.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Endosymbiosis and evolution of the plant cell. Curr Opin Plant Biol. 1999;2:513–519. doi: 10.1016/s1369-5266(99)00025-4. [DOI] [PubMed] [Google Scholar]
- Miyagishima S, Itoh R, Toda K, Takahashi H, Kuroiwa H, Kuroiwa T. Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J Electron Micros. 1998;47:269–272. [Google Scholar]
- Miyagishima S, Takahara M, Kuroiwa T. Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell. 2001;13:707–721. doi: 10.1105/tpc.13.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Tanaka I. Isolation of the ftsZ gene from plastid-deficient generative cells of Lilium longiflorum. Protoplasma. 2000;214:57–64. [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Olsen LJ, Keegstra K. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J Biol Chem. 1992;267:433–439. [PubMed] [Google Scholar]
- Osteryoung KW, McAndrew RS. The plastid division machine. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:315–333. doi: 10.1146/annurev.arplant.52.1.315. [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Pyke KA, Rutherford SM, Robertson EJ, Leech RM. arc6, a fertile Arabidopsismutant with only two mesophyll cell chloroplasts. Plant Physiol. 1994;106:1169–1177. doi: 10.1104/pp.106.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Justice S, García-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–428. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2genes in Arabidopsis. Plant Physiol. 2000;124:1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW. FtsZ ring formation at the chloroplast division site in plants. J Cell Biol. 2001;153:111–119. doi: 10.1083/jcb.153.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria. Mol Microbiol. 1996;21:313–319. doi: 10.1046/j.1365-2958.1996.6421360.x. [DOI] [PubMed] [Google Scholar]
- Ward JE, Jr, Lutkenhaus J. Overproduction of Ftsz induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]