Abstract
Matrix metalloproteinases (MMPs) play an important role in host defense responses against pathogens in mammals where their activities lead to the production of antimicrobial peptides. We have identified a novel soybean (Glycine max) metalloproteinase gene, GmMMP2, that is transcriptionally up-regulated in infected tissues. The deduced amino acid sequence indicates that this gene belongs to the MMP family. It is a preproprotein containing an N-terminal signal peptide, a cysteine switch, a zinc-binding catalytic motif, and a C-terminal transmembrane domain. The GmMMP2 expressed in and purified from Escherichia coli exhibited an in vitro enzymatic activity in digesting myelin basic protein. All plant metalloproteinases reported so far have no known functions. However, they have been suggested to be involved in extracellular cell matrix degradation during development or senescence. Our investigations demonstrate that the GmMMP2 transcript levels were rapidly increased in compatible and incompatible interactions of soybean tissues with the oomycete pathogen Phytophthora sojae or the bacterial pathogen Pseudomonas syringae pv. glycinea. In agreement with the GmMMP2 activation, a metalloproteinase activity was gradually increased in suspension-cultured cells following the bacterial infection. GmMMP2 was also activated in response to wounding and dehydration. However, GmMMP2 activation did not correlate with the oxidative burst leading to the hypersensitive response cell death or the tissue senescence progress that involves programmed cell death. Our investigations suggest that GmMMP2 may be involved in a novel defense response of soybean against pathogenic infections.
Plants have no specific immune system, and yet they successfully fight infections with constitutive and active defense mechanisms. The active defense mechanisms against pathogenic fungi, bacteria, or viruses can be induced by a variety of biotic and abiotic elicitors. Many proteins are induced or their activities are apparently increased upon pathogen attack (Ouchi, 1983; Sequeira, 1983). These proteins can be classified into five groups on the basis of their functions. The first group includes enzymes involved in the biosynthesis of antimicrobial compounds called phytoalexins, the secondary metabolites toxic to some pathogenic microorganisms (Schmelzer et al., 1984; Schmidt et al., 1984; Welle and Grisebach, 1988; Schopfer et al., 1998). The second group consists of carbohydrolases such as chitinases and β-1,3-glucanases, which lyse cell walls of invading fungi, thereby inhibiting the growth of the invaders (Chang et al., 1992; Okinaka et al., 1995). The third group contains a number of Cys-rich proteins such as proteinase inhibitors and γ-thinion-like proteins called defensins (Broekaert et al., 1995). These Cys-rich proteins are thought to interact with the plasma membrane of the invading fungi, causing lysis and inhibition of hypha growth (Terras et al., 1995). The fourth group comprises proteinases involved in the hypersensitive response (HR)-related cell death in infected tissues (Lam et al., 1999). The fifth group includes all other proteins whose functions are unknown.
Although the actual mechanisms of those proteins involved in plant defense against adverse environment are different, they share a general response pattern. In general, the first signal is initiated as the host recognizes a pathogen. The recognition cascades a series of biochemical, physiological, and morphological responses in the host plant. These responses can be local or/and systemic. Salicylic acid (SA) and jasmonic acid (JA) are two major signaling molecules that activate many defense-related genes (for review, see Glazebrook, 1999). One of these responses is the HR, where massive protein degradation occurs around the invading site, which involves proteolytic activities (for review, see Vierstra, 1996). Most plant proteolytic enzymes examined appear to fall into the Cys and aspartic proteinase types. These two types of proteinases are involved in the metabolism of seed storage proteins and tissue senescence (Kinoshita et al., 1999). Cys proteinases were reported to be involved in programmed cell death (PCD), which is similar to apoptosis in animals (Lam et al., 1999). It was demonstrated that the cleavage of poly(ADP-Rib) polymerase by a Cys proteinase activated the HR of cowpea (Vigna unguiculata) to cowpea rust fungus (D'Silva et al., 1998). Cys proteinases have been suggested to play a regulatory role in the development of the HR in plants (Avrova et al., 1999). Development of the HR in invaded tissues presumably results in the cut-off of nutrients needed to support pathogen growth and the accumulation of antimicrobial compounds to inhibit the growth of invading pathogens. Apart from Cys proteinases, other proteolytic enzymes, including Ser, aspartic, and metalloproteinases have been speculated to play a role in the expression of active defense responses (Pak et al., 1997; Xia et al., 1999). An Arabidopsis gene CDR1 encoding an aspartic proteinase-like protein has been recently cloned by the activation T-DNA tagging approach. The activated CDR1-dominant mutant showed an enhanced resistance to Pseudomonas syringae, whereas the antisense CDR1 plants demonstrated increased susceptibility to the same pathogen. CDR1 has been shown to release a mobile signal that might induce local and systemic defense responses (Xia et al., 1999).
To date, only two plant matrix metalloproteinases (MMPs) purified from soybean (Glycine max) leaves (Graham et al., 1991) and buckwheat (Fagopyrum esculentum) seeds (Mikhail et al., 1990) have been characterized. Three full-length cDNAs encoding plant MMPs have been cloned from soybean (Pak et al., 1997), Arabidopsis (Liu et al., 1998), and cucumber (Cucumis sativus; Delorme et al., 2000). There has been no evidence showing the involvement of plant MMPs in host defense against pathogens. However, they were proposed to play a role in plant development (Graham et al., 1991) and senescence (Delorme at al., 2000). Some MMPs have been implicated to be involved in the processes such as protein turnover, the action of tetanus and botulism toxins, cancer, and arthritis in mammals (Pak et al., 1997). Recently, the regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in mouse has been elucidated in vitro and in vivo (Wilson et al., 1999). The matrilysin in vitro cleaves the pro segment from the α-defensin cryptdin precursor to activate its antibacterial properties. The matrilysin-deficient mice lack mature cryptdins, and accumulate the precursor molecules, resulting in more susceptibility to bacterial diseases. This proves that the MMP plays a defensive role in mammals.
Here, we report the identification and characterization of the Glycine max MMP 2 gene (GmMMP2) isolated from infected soybean hypocotyls in a differential display experiment. We demonstrate that the steady-state transcript levels of this single-copy gene are increased in soybean tissues following infection with a fungal or a bacterial pathogen. GmMMP2 activation by bacterial infection in suspension-cultured cells parallels the increase of a proteinase activity in digesting Azocoll, a substrate for metalloproteinase (Graham et al., 1991). In addition, GmMMP2 has exhibited a proteinase activity in vitro. Our current evidence indicates that GmMMP2 may play a role in a novel defense mechanism in plants against pathogenic infections.
RESULTS
Identification of a Soybean MMP Transcript
To isolate defense-related genes, differential display experiments were carried out using RNA samples prepared from etiolated soybean hypocotyls infected with the fungus Phytophthora sojae or mock inoculated with water. We identified a transcript that accumulated in compatible and incompatible interactions, but not in the water-treated hypocotyls. The PCR-amplified DNA molecules specific to this transcript were cloned and sequenced. Using this DNA fragment as a probe, a full-length cDNA clone was isolated by screening a soybean suspension-cultured cell cDNA library. The deduced protein structure is similar to that of the soybean metalloproteinase SMEP1 and to that of members of the mammalian MMP family. It carries a signal peptide at the N terminus, followed by a propeptide where a Cys switch motif exists (Fig. 1). The mature protein contains a zinc-binding motif and a transmembrane domain at the C terminus that SMEP1 lacks (Fig. 1). The computer analyses of the deduced peptide reveal that this metalloproteinase is synthesized as a zymogen, an inactive form, and is possibly secreted into an extracellular location because of the N-terminal signal peptide (von Heijne, 1986). The proprotein comprises eight potential N-linked glycosylation sites. Based on the presence of all conserved hallmarks of MMP, specifically the existence of the zinc-binding (HEXXHXXGXXHS) motif (Bode et al., 1993; Hooper, 1994), we considered that this is a MMP and named this novel protein GmMMP2. Southern hybridization indicated that GmMMP2 is a single-copy gene (data not shown). cDNA cloning experiments indicated that there was only one transcript species representing GmMMP2 in soybean cell suspensions. Sequencing of genomic DNA revealed that the gene does not carry any introns. Northern-blot analysis conducted using RNA samples from various organs of mature plants showed a detectable level of GmMMP2 transcript only in leaves (data not shown).
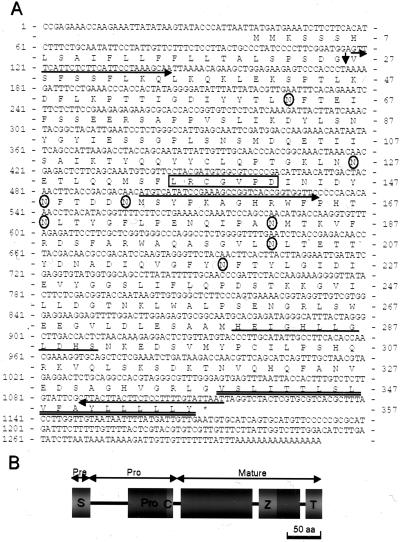
Figure 1.
Sequence analyses of the GmMMP2 cDNA and the deduced peptide. A, The deduced amino acid sequence is shown below the coding sequence, and the asterisk indicates the termination codon. The numbers at the left are for base pairs, whereas those at the right are for amino acids. A predicted signal peptide is located at amino acid position 1 through 26. The cleavage site is indicated by a vertical arrow. A forward primer denoted by the first rightward arrow right after the signal peptide below the sequence is used together with the reverse primer denoted by a leftward arrow below the sequence to generate pro-GmMMP2 sequence. Another forward primer denoted by the second rightward arrow is used together with the same reverse primer to generate the mature GmMMP2 sequence. A putative Cys switch sequence LRCGVPD is framed, whereas the zinc-binding motif is underlined. A putative C-terminal transmembrane domain is indicated by a double underline. All N residues circled by an oval are potential glycosylation sites. B, A schematic representation of the putative domains and motifs of GmMMP2. The cylinders represent those conserved domains and motifs, whereas the black lines between these domains denote the variable regions among the MMP members. S and Pre, Signal peptide; Pro, propeptide; C, Cys switch motif; Z, zinc-binding motif; T, transmembrane domain. The scale bar is equal to 50 amino acids.
Rapid Accumulation of GmMMP2 Transcripts following Infection
Northern-blot hybridization experiments were carried out for soybean hypocotyls and leaves that were inoculated with zoospores of the oomycete pathogen P. sojae, race 1, or treated with water droplets (Fig. 2). The steady-state GmMMP2 transcript levels in hypocotyls increased rapidly in incompatible and compatible interactions (Fig. 2A). The GmMMP2 transcript accumulated more slowly in infected leaves (Fig. 2B) compared with that in hypocotyls infected with P. sojae. In both cases, the GmMMP2 transcript accumulation occurred before the appearance of disease symptoms, i.e., 8 or 36 h post-inoculation (hpi) for the infected hypocotyls and leaves, respectively. Unexpectedly, 0-h leaf samples showed a higher expression of GmMMP2. To investigate whether abiotic stresses induce the accumulation of GmMMP2 when leaves are detached from plants, we examined the impact of dehydration and wounding on activation of GmMMP2. An apparent accumulation of GmMMP2 transcripts was observed in dehydrated and wounded leaves (Fig. 2C).
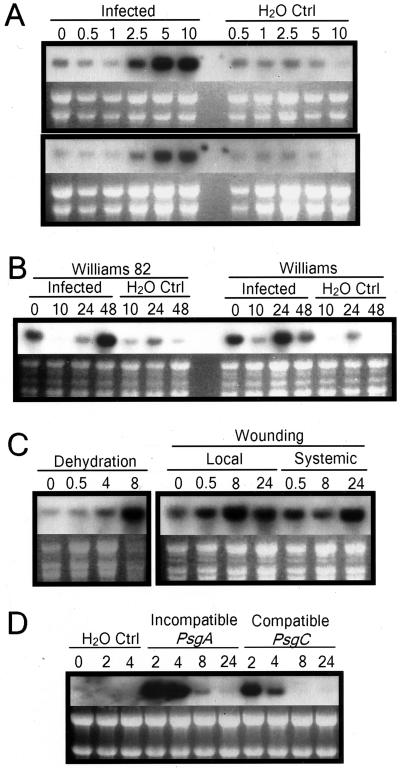
Figure 2.
Northern-blot analyses of the GmMMP2 expression. A, Accumulation of the GmMMP2 transcripts in soybean hypocotyls following infection with the oomycete pathogen P. sojae race 1. The etiolated hypocotyls of 7-d-old seedlings of the resistant cv Williams 82 (top) and susceptible cv Williams (bottom) were inoculated with the zoospores of P. sojae race 1 (infected) or water droplets (H2O Ctrl), and were sampled at the indicated hours after inoculation. B, Accumulation of the GmMMP2 transcripts in soybean leaves following infection with the oomycete pathogen P. sojae race 1. The first trifoliate leaves of 4-week-old plants of the cv Williams 82 and cv Williams were inoculated with the zoospores of P. sojae race 1 (infected) or water droplets (H2O Ctrl), and were sampled at the indicated hours after inoculation. The GmMMP2 activation for the 0-h samples is due to abiotic stresses when the leaves were detached from plants. C, Accumulation of the GmMMP2 transcripts in soybean leaves following dehydration or wounding treatment. At 8 h post-dehydration, the transcription of GmMMP2 reaches the highest level even though the total RNA degraded significantly by that time. The wounding causes a moderate increase in the GmMMP2 transcript accumulation locally and systemically. D, Accumulation of the GmMMP2 transcripts in the suspension-cultured cells of cv Williams 82 following infection with the bacterial pathogen PsgA (incompatible) or PsgC (compatible). The amounts of RNA loaded were visualized by ethidium bromide staining under UV light.
To investigate if the GmMMP2 transcript levels increase following infection of soybean with a different pathogen, cell suspensions prepared from the cultivar Williams 82 were inoculated with the bacterial pathogen P. syringae pv. glycinea (Psg) harboring the avrA (incompatible) or avrC (compatible) gene (Levine et al., 1994). A rapid but transient accumulation of GmMMP2 was observed for compatible and incompatible interactions (Fig. 2D).
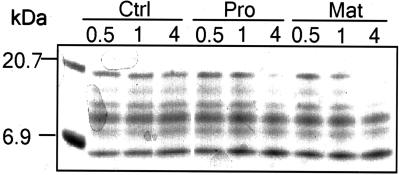
Induction of many pathogenesis-related or wound-inducible genes is triggered by SA and JA, respectively. Therefore, we studied the effect of these two signaling components on the activation of GmMMP2. We also investigated if biotic elicitor such as crude elicitors derived from yeast extract can activate GmMMP2 transcription. As with the bacterial infection, a rapid and transient accumulation of GmMMP2 transcripts was observed following elicitation of soybean cell suspensions with the yeast elicitor (YE; Fig. 3). However, SA and JA did not provoke the accumulation of GmMMP2 transcripts (Fig. 3).
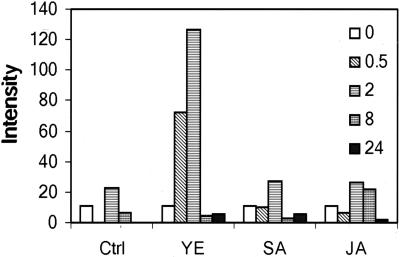
Figure 3.
GmMMP2 expression profiles in suspension-cultured cells following YE, SA, JA, and water control (Ctrl) treatment at the final concentrations of 50 μL mL−1, 50 μm, and 20 μm, respectively. Data, in intensity of the bands from an autoradiogram, normalized on the basis of 28S rRNA intensity, are from a northern-blot analysis of the cells that were treated with the indicated substance for 0, 0.5, 2, 8, and 24 h. The experiment was repeated twice and the average values are presented.
GmMMP2 Activation Is Not Correlated with the HR or PCD
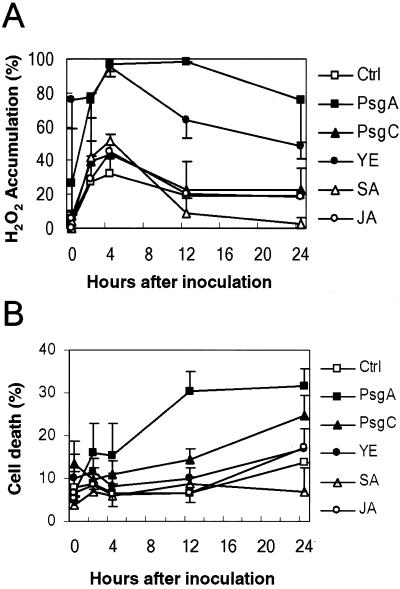
The generation of hydrogen peroxide has emerged as a key event in plant defense. Hydrogen peroxide can induce certain defense-related genes. To address questions such as whether hydrogen peroxide is a possible cause for the GmMMP2 activation and whether the GmMMP2 activation leads to plant cell death or vice versa, we investigated their relationships by challenging soybean cell suspensions with various elicitors. Upon inoculation with Psg carrying avrA (PsgA) or yeast elicitor, the levels of GmMMP2 transcript and oxidative burst immediately increased (Figs. 2D and 4A). The GmMMP2 transcription declined following a peak at 2 h after infection or elicitation (Fig. 2D) and so did the oxidative burst following a peak at 4 h (Fig. 4A). A high level of cell death was recorded for cells infected with PsgA, but not with the YE treatment (Fig. 4B). However, PsgA and the YE caused the GmMMP2 accumulation (Figs. 2D and 3). This demonstrates that GmMMP2 expression does not correlate with the onset of the cell death following infection or elicitation. Unlike PsgA and YE treatments, Psg carrying avrC (PsgC) did not cause significant increase in the oxidative burst, but it did cause a higher cell death rate in comparison with water control (Fig. 4).
Figure 4.
Oxidative burst and cell death in soybean cell suspensions following infection and elicitation. A, Data were normalized and presented as a percentage of untreated cells. B, For each replicate, three observation fields were counted for an average value. All oxidative burst and cell death data are presented as means with standard deviations of three replicates. Ctrl, H2O control.
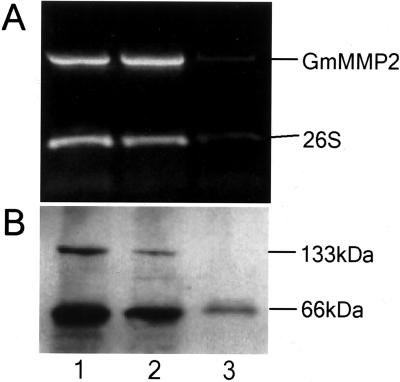
To see if the expression of GmMMP2 is correlated with tissue senescence process, we conducted reverse transcriptase (RT)-PCR and western analyses with leaves at three distinct developmental stages, i.e., young (light green), mature (dark green), and senescent (completely yellow but healthy) leaves. No differences in the expression of GmMMP2 were detected in comparison with 26S rRNA expression pattern, although the amounts of the transcripts decreased when senescence occurred (Fig. 5). Correspondingly, the western profiles showed that the amounts of both GmMMP2 forms, i.e., 66 and 133 kD, respectively, decreased in agreement with the transcript levels (Fig. 5).
Figure 5.
Analyses of quantitative RT-PCR and immunodetection. A, Equal amount of total RNA samples isolated from three different developmental stages of cv Williams 82 soybean leaves were used to quantitatively amplify GmMMP2 fragment. A 26S ribosome fragment was amplified as an internal control. One forward gene-specific primer was used in the amplifications together with a reverse polyT-adaptor primer for the GmMMP2 and 26S ribosome genes. B, Ten micrograms of total soluble protein extract was separated with a 4% to 20% (w/v) gradient SDS-PAGE gel, and immunodetection was performed using rabbit antisera against GmMMP2. Two different-size GmMMP2 forms are presented. Lanes 1 through 3 represent young, mature, and senescent leaves, respectively.
Proteolytic Activity
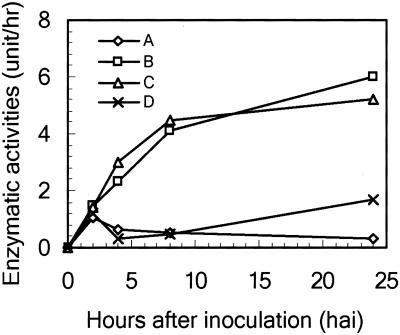
Based on computer analyses, we generated two fragments for putative pro- and mature GmMMP2 with the designed primers (Fig. 1) by RT-PCR using hypocotyl tissues infected with P. sojae. Both peptides were expressed in and purified from Escherichia coli. The purified proteins were subjected to activity assay in vitro upon refolding. Pro- and mature GmMMP2 showed a proteolytic activity in degrading myelin basic protein (MBP) although mature GmMMP2 had higher activity than pro-GmMMP2 (Fig. 6). In addition, we found that the soybean suspension-cultured cells infected with PsgA exhibited a higher proteolytic activity in digesting Azocoll, a substrate for metalloproteinase activity assay (Graham et al., 1991). The metal-chelator EDTA could eliminate this activity, whereas a proteinase inhibitor mixture without EDTA had little impact on the proteinase activity (Fig. 7). As a consequence, this EDTA-sensitive proteinase activity is in agreement with the GmMMP2 activation following the bacterial infection (Fig. 2D).
Figure 6.
Degradation of bovine MBP by pro- and mature GmMMP2. The proteolytic activity of pro- and mature GmMMP2 purified from E. coli was determined with Tris-HCl, pH 7.5, buffer containing bovine MBP. The reaction mixtures were incubated at 37°C for 0.5, 1, and 4 h. The degrading products were separated with a 4% to 20% (w/v) gradient SDS-PAGE gel and were visualized by Coomassie Brilliant Blue staining. Ctrl, H2O control; Pro, pro-GmMMP2; Mat, mature GmMMP2.
Figure 7.
Increased protease activity of the suspension-cultured cells upon infection with PsgA. Azocoll-digesting specific activity in the presence and absence of EDTA or a protease inhibitor mixture without EDTA was determined in total soluble extracts from suspension-cultured cells treated with water or infected with PsgA. A, Cells treated with water. B, Cells infected with PsgA in the presence of the protease inhibitor mixture without EDTA. C, Cells infected with PsgA. D, Cells infected with P. syringae in the presence of the proteinase inhibitor mixture and 1 mm EDTA. Two hundred micrograms of total soluble protein extract was used for each protease activity assay.
DISCUSSION
MMP is a family determined by the consensus HEXXHXXGXXHS (Jiang and Bond, 1992; Bode et al., 1993). As the sequence HEIGHLLGLDHS of GmMMP2 matches the consensus, we defined GmMMP2 as a member of this family. The consensus HEXXHXXGXXHS comprises three histidines that bind to the zinc, forming a catalytic center of this enzyme (Bode et al., 1993). Pro-GmMMP2 contains eight potential N-linked glycosylation sites, indicating that the actual GmMMP2 may have much larger size in vivo than theoretically estimated. Theoretical calculation shows that pro-GmMMP2 should be around 36.6 kD. However, the smallest peptide in healthy soybean leaves detected by the antisera raised against putative pro-GmMMP2 is about 66 kD. Therefore, we speculated that the peptide of 66 kD was glycosylated. Using the N-glycosidase F, also known as PNGase F, we have proved our speculation that the peptide of 66 kD is heavily glycosylated (Y. Liu and M.K. Bhattacharyya, unpublished data). We also speculate that the larger peptide of 133 kD is a dimer form because its size doubles that of the smaller peptide of 66 kD. However, this speculation needs further investigation.
Plant metalloproteinase activity was first detected in soybean leaves as Azocollase (Ragster and Chrispeels, 1979). This protein could not be purified until 1991 when Graham et al. (1991) finally obtained and characterized the first plant metalloproteinase SMEP1 from soybean leaves. However, the roles of MMPs in higher plants remain unclear. It has been speculated that MMPs are involved in remodeling of plant extracellular matrix in association with plant growth, development, and possibly defense processes (Pak et al., 1997; Maidment et al., 1999). SMEP1 is a single-copy gene and its transcription is under a developmental control in leaf tissue in a temporal fashion (Pak et al., 1997). GmMMP2 was also found to be a single-copy gene and has 28% amino acid sequence identity with SMEP1. GmMMP2 has all putative motifs of SMEP1 and an additional C-terminal transmembrane domain. Thus, we assume that they are different isoforms of zinc metalloproteinases in soybean, probably carrying out similar functions under various internal and external conditions.
The study of PCD has been intensified in the past decade. The central enzymes are caspases, a growing family of Cys proteinase in mammals. They are synthesized as zymogens that require specific cleavage and subsequent oligomerization to reveal their enzymatic activities (Lam et al., 1999). Caspase-like proteolytic activity has been observed to be transiently activated in plants synchronized to undergo the HR (del Pozo and Lam, 1998). In addition to caspases, other Cys proteinases may also be involved in the HR cell death (D'Silva et al., 1998). In our investigations, GmMMP2 was highly expressed following incompatible or compatible interaction with the pathogens. However, it is well known that the HR cell death occurs only when the interaction between host and pathogen is incompatible. Furthermore, the YE did not cause significant cell death as compared with the control, but it did induce GmMMP2 expression. Therefore, it appears unlikely that GmMMP2 is involved in plant HR cell death. It has been reported recently that a cucumber MMP gene Cs1-MMP is expressed at late stages of developmental senescence prior to the appearance of DNA fragmentation, indicating that Cs1-MMP may be involved in PCD (Delorme et al., 2000). However, as with a similar approach, we could not see a positive correlation between tissue senescence progress and the GmMMP2 expression in soybean leaves (Fig. 5). On the contrary, the amounts of GmMMP2 mRNA and protein declined in the senescent leaves. Again, it appears unlikely that GmMMP2 is involved in PCD.
It has long been known that soybean seedlings infected with P. sojae or soybean suspension-cultured cells treated with a glucan elicitor from this oomycete accumulate glyceollin, the isoflavanoid phytoalexin of this plant species (Zähringer et al., 1979). Phytoalexin accumulation in soybean is associated with rapid transient increases in the activities of phytoalexin biosynthetic enzymes. These changes are implicated by increase in enzymatic activities and amounts of the transcripts encoding the enzymes (Habereder et al., 1989). Apparently, phytoalexin synthesis in soybean, as in other systems, is regulated by the transient gene activation. The mechanisms by which pathogens activate those genes involved in phytoalexin biosynthesis are not known. However, a number of investigations have shown that two agents, i.e., active oxygen species, usually measured as hydrogen peroxide, and calcium ions accumulated immediately upon a pathogen attack, might be among the first signals causing a cascade of responses of plants against pathogenic infections (Stäb and Ebel, 1987; Levine et al., 1994; Shirasu et al., 1997). In soybean suspension-cultured cells infected with P. syringae pv. glycinea, the release of reactive oxygen radicals paralleled the induction of GmMMP2, although the generation of hydrogen peroxide lasted longer than the activation of GmMMP2 (Fig. 4).
GmMMP2 may not directly regulate the activities of those crucial enzymes involved in phytoalexin synthesis because the speculation of the extracellular existence of GmMMP2 contradicts the possibility that the GmMMP2 may regulate those enzymes in the cytosol. However, GmMMP2 may still play a role in the induction of glyceollin biosynthesis in an indirect way. For instance, it may regulate the activities of β-1,3-glucanase and chitinase that digest the fungal cell walls, thus releasing the glucan elicitors. Recently, a mechanism by which the metalloproteinase matrilysin in mouse activates the α-defensin cryptdin has been elucidated in detail. The matrilysin cleaves the pro peptide from the cryptdin precursors, thus activating its antibacterial activities (Wilson et al., 1999). In addition, matrilysin-deficient mice that lack mature cryptdins and accumulate precursor molecules are more susceptible to bacteria or cholinergic agents. The matrilysin that mediates removal of the pro domain of cryptdin, thereby regulating the level of the functional peptides, has been identified. Because mammalian matrilysin and plant metalloproteinases belong to the same MMP family, these two types of metalloproteinases may act in a similar way. Many plant defensins are produced in a precursor form that must be processed for activation (Chiang and Hadwiger, 1991). To test if GmMMP2 is directly involved in inhibition of pathogen growth, we cocultured PsgA or P. sojae with the putative mature GmMMP2 that was expressed and purified from E. coli, in combination with or without total soluble proteins extracted from soybean suspension cells (Williams 82). We found that the mature GmMMP2, together with the total soybean soluble protein extract, strongly inhibited the growth of these two pathogens (Y. Liu and M.K. Bhattacharyya, unpublished data). These results suggest that most likely GmMMP2 release an antimicrobial substance(s) in retarding the growth of invading pathogens. We are currently pursuing our work to identify possible natural substrates for GmMMP2.
Many defense-related genes are activated via a SA-dependent or JA-dependent pathway (for review, see Glazebrook, 1999). However, the application of these two substances had no effects on the accumulation of GmMMP2 transcript in soybean suspension-cultured cells (Fig. 3), indicating that the activation of GmMMP2 is probably independent of both components. Nevertheless, we found that mechanical wounding could stimulate GmMMP2 transcription locally and systemically (Fig. 2C). This suggests that there might be another signaling pathway where SA and JA are not the key molecules.
MATERIALS AND METHODS
Plant Materials, Inoculation, Elicitation, Wounding, and Dehydration
Soybean (Glycine max) seeds of near-isogenic lines cv Williams (Phytophthora sojae race 1 susceptible) and cv Williams 82 (P. sojae race 1 resistant) were grown in the dark for 7 d under conditions described before (Ward et al., 1989). P. sojae race 1 was grown on plates of V8 medium, whereas zoospores were produced and etiolated hypocotyls were inoculated according to Ward et al. (1989). The disease symptoms started to show around 8 to 10 hpi. One-centimeter-long segments carrying the inoculated site were harvested and immediately frozen in liquid nitrogen after quick blotting on a dry paper towel. The collected materials were stored at −80°C for late use or were immediately used for total RNA extraction.
Leaf inoculation was performed using 4-week-old plants of cv Williams and cv Williams 82 grown in a growth chamber under conditions reported earlier (Bhattacharyya and Ward, 1986). Two opposite leaves of the first completely open trifoliate were detached and placed on a layer of moistened 3M filter paper (3M, St. Paul) in petri dishes. The leaves were inoculated with the zoospores of P. sojae race 1 or were treated with water droplets (Bhattacharyya and Ward, 1986). The disease symptoms started to appear at about 36 hpi.
Leaf wounding was done on one leaf blade of the first and second trifoliate leaves of 4-week-old cv Williams 82 plants. Leaves were scratched with a sawtooth wheel and collected as wounded samples, whereas the opposite intact leaf blades of individual trifoliate leaves were collected to study the distance effect of wounding on GmMMP2 expression. The wounded sites started to become brown at 4 h following wounding.
For dehydration treatments, the entire 4-week-old plants in the growth chamber were carefully removed from soil without any obvious mechanical injury on roots and were placed in a glass beaker with water for 24 h at room temperature with 45% relative humidity. Thereafter, water was removed. The dehydrated plants started to wilt at 4 h post-treatment.
The suspension-cultured cells of the soybean cv Williams 82 were grown in Murashige and Skoog medium (Murashige and Skoog, 1962) containing 2.22 μm 6-benzylaminopurine and 0.3 mg L−1 2,4-dichlorophenoxyacetic acid in the dark at 25°C on an orbital shaker (130 rpm). The culture was subcultured every 7 d by diluting it 5-fold with fresh medium. Five days after subculturing, cells were inoculated with PsgA or PsgC (Keen and Buzell, 1991). The bacterial pathogen was grown and inoculum was prepared according to Keen and Buzell (1991). YE was prepared following the protocol described by Schumacher et al. (1987). The suspension cells were inoculated with PsgA and PsgC at a final concentration of 1 × 108 colony-forming units mL−1, or they were treated with the YE, SA, and JA at final concentrations of 50 μg mL−1, 50 μm, and 20 μm, respectively. All chemicals were obtained from Sigma (St. Louis) unless otherwise indicated.
RNA Isolation, Differential Display, RT-PCR, and Northern Analysis
Differential display experiments were performed according to the instructions from GenHunter, Inc. (Nashville, TN). Each PCR reaction was repeated independently at least two times. Fragments of interest were isolated from the differential display gels and were subcloned into a T-vector (Bhattacharyya et al., 1997). Four clones from each ligation reaction were randomly selected for sequencing. A clone-specific primer for each of these clones was synthesized. RT-PCR was carried out using these clone-specific primers and a touchdown program with annealing temperature dropping from 62°C to 58°C. Based on the results of these RT-PCR experiments, fragments that showed infection-associated increase in transcript accumulation were selected and applied in cloning full-length cDNAs and northern-blot analyses.
For northern analysis, total RNA was extracted using a RNAwiz reagent kit (Ambion, Austin, TX). Ten micrograms of denatured RNA was electrophoresed on a 1.5% (w/v) agarose gel in 16% (v/v) formaldehyde and MEN buffer (200 mm MOPS [3-(N-morpholino)propanesulfonic acid], 50 mm NaAc, and 20 mm EDTA, pH 7.0). The separated mRNAs were transferred onto a GeneScreen Plus nylon membrane (NEN Life Science Products, Boston) using 10× saline-sodium citrate (SSC) and crosslinked with UV light. Filters were prehybridized at 42°C in buffer containing 50% (v/v) formamide for at least 4 h. Radiolabeled DNA probes were prepared using a random primer kit from Amersham Pharmacia Biotech (Buckinghamshire, UK). Blots were hybridized at 42°C overnight and were washed at 65°C in 2× SSC once for 60 min and in 0.1× SSC containing 0.1% (w/v) SDS twice for 15 min each. The intensity of each band in the autoradiograms was quantified and normalized on the basis of 28S rRNA intensity using the software ImageQuaNT (Molecular Dynamics, Sunnyvale, CA).
Full-Length cDNA Cloning and Sequence Analyses
A soybean cDNA library prepared from suspension-cultured cells was kindly provided by Dr. Toshiro Shigaki (Baylor College of Medicine, Houston; unpublished data). A fragment of 598-bp homology to MMPs, identified originally in a differential display experiment, served as a probe in screening the cDNA library. Approximately 3 × 106 plaque-forming units were plated and screened by Southern hybridization (Sambrook et al., 1989). Six putative clones were purified and sequenced. They were all found identical in sequence, containing one open reading frame. Deduced amino acid sequence was then analyzed by the PredictProtein program of Columbia University (http://dodo.cpmc.columbia.edu/predictprotein) to determine the structure (Rost and Sander, 1993).
Genomic DNA Extraction, PCR, and Southern Analysis
Genomic DNA was extracted from mature leaves of cv Williams and cv Williams 82 according to Kasuga et al. (1997). A touchdown PCR program from 63°C to 58°C was performed to amplify the corresponding fragments using Taq:Pfu DNA polymerase cocktail mixture in a ratio of 16:1. Three restriction enzymes DraI, EcoRI, and EcoRV (Life Technologies, Rockville, MD) were chosen for complete DNA digestion because they do not cut within the cDNA. The digested DNA was separated on a 0.8% (w/v) agarose gel in 0.5× Tris-boric acid-EDTA buffer overnight and was blotted onto a nylon membrane through alkaline transfer. The membrane was neutralized in 100 mm Tris-HCl, pH 7.5, for 2 min followed by treatment with 2× SSC for 5 min. The filter was oven dried at 80°C under vacuum for at least 2 h. The Southern hybridization and washing procedures were the same as that for northern hybridization with the exception that the final washing step was continued twice for 30 min.
Hydrogen Peroxide Accumulation and Cell Death Determination
Hydrogen peroxide accumulation in cell suspensions was assayed by scopoletin destruction indicated by the loss of fluorescence at 460 nm after excitation at 350 nm (Levine et al., 1994). Four-day-old suspension-cultured cells were used for inoculation. At the indicated time point, scopoletin was added into the cell suspensions at final concentration of 20 μm. The incubation was conducted with constant shaking on a horizontal shaker at 120 rpm at room temperature for 5 min. For each sample, 200 μL of supernatant upon centrifugation at 1,000g for 2 min was loaded into a microtiter plate for reading. The values were normalized and presented as a percentage of the untreated cells. Cell death was determined with 0.05% (w/v) Evans blue according to Shigaki and Bhattacharyya (1999). For each replicate, three observation fields were evaluated for an average value. All data are presented as means with the standard deviations of three replicates.
GmMMP2 Expression, Purification, and Refolding
Total RNA was extracted from hypocotyls of soybean cv Williams 82 infected with P. sojae using RNAwiz reagent kit (Ambion). Putative pro- and mature GmMMP2 DNA fragments were generated by RT-PCR using primers stated in Figure 1. The amplified fragments were purified with a Gel Extraction kit (Qiagen, Hilden, Germany). The purified fragments were subcloned into the protein expression vector pET28a containing a His-tag at the N terminus of the expressed protein (Novagen, Madison, WI). In-frame sequences were verified by sequencing. The cells of E. coli strain BL21 were transformed with the cloned vector containing GmMMP2 fragment or the original “blank” vector pET28a as a control. Protein expression was induced in the transformed E. coli strain BL21 with 1 mm isopropyl-1-thio-β-d-galactopyranoside at 37°C for 3 h. Cells were harvested by centrifugation, the pellet was resuspended in 20 mm Tris-HCl, pH 8.0, buffer containing a protease inhibitor mix without the metal-chelator EDTA (Sigma), and was it lysed by sonication. Insoluble material was pelleted by a centrifugation at 10,000g at 4°C for 15 min and was washed with the same buffer. The final pellet was resolubilized in 20 mm Tris-HCl, pH 7.5, buffer plus 6 m urea because most recombinant protein was expressed in inclusion bodies. The expressed protein was clarified by a centrifugation at 10,000g at 4°C for 30 min and was visualized with Coomassie Brilliant Blue after separation by SDS-PAGE. The recombinant protein was purified on a Ni-NTA column (Qiagen) and was further purified by SDS-PAGE separation and gel slicing followed by electric elution. The purified protein was used to raise antibodies in rabbit. Fifty micrograms of the purified GmMMP2 was diluted into 200 μL of refolding buffer (100 mm Tris-HCl, pH 7.5, 10% (v/v) glycerol, 10 mm CaCl2, 100 mm NaCl, 1 μm ZnCl2, and 0.05% [w/v] Brij 35) and was incubated at 4°C overnight.
Total Plant Protein Extraction and Western Analysis
Soybean leaves of the cv Williams 82 at different developmental stages were frozen in liquid nitrogen and ground into a fine powder. Total proteins were extracted with Tris-HCl buffer (50 mm Tris-HCl, pH 8.0, 100 mm KCl, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 5% [v/v] glycerol). Soluble protein fraction was collected upon centrifugation and was stored at −20°C. Protein concentrations were determined by Bradford's method using bovine serum albumin as a standard. Ten micrograms of total proteins was separated with a 4% to 20% (w/v) gradient SDS-PAGE. Western blotting was conducted with Nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). Western immunodetection was performed using the antibodies against GmMMP2 according to the manufacturer's instruction using an enhanced chemiluminescence system (Amersham Pharmacia Biotech).
GmMMP2 Enzymatic Activity Assay
The catalytic activity of pro- and mature GmMMP2 was determined by incubating 0.2 μg of refolded GmMMP2 in 10 μL of 20 mm Tris-HCl, pH 7.5, buffer containing 5 μg of bovine MBP. The degradation of MBP was visualized with the 4% to 20% (w/v) SDS-PAGE followed by Coomassie Brilliant Blue staining. The proteinase activity of the suspension-cultured cells was determined by incubation of the total soluble protein extract with 0.001% (w/v) Azocoll, a metalloproteinase substrate in 10 μL of 20 mm Tris-HCl, pH 7.5, buffer. The reaction mixtures were clarified upon a brief centrifugation, and were then measured with a photospectrometer at 520 nm. A unit is defined as 0.01 Δ optical density h−1 (Graham et al., 1991).
ACKNOWLEDGMENTS
We thank Drs. Elison B. Blancaflor and Richard A. Dixon (Noble Foundation, Ardmore, OK) for critically reading the manuscript, Dr. Salvatore Sparace (McGill University, Toronto, Canada) for the help in oxidative burst assay, Ann Harris (Noble Foundation) for DNA sequencing and oligo primer synthesis, and Dr. Toshiro Shigaki for providing the soybean suspension cell cDNA library.
Footnotes
This work was supported by the Samuel Roberts Noble Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010593.
LITERATURE CITED
- Avrova AO, Stewart HE, De Jong WD, Heilbronn J, Lyon GD, Birch PR. A cysteine protease gene is expressed early in resistant potato interaction with Phytophthora infestans. Mol Plant-Microbe Interact. 1999;12:1114–1119. doi: 10.1094/MPMI.1999.12.12.1114. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya MK, Gonzales RA, Kraft M, Buzzell RI. A copia-like retrotransposon Tgmr closely linked to the Rps1-k allele that confers race-specific resistance of soybean to Phytophthora sojae. Plant Mol Biol. 1997;34:255–264. doi: 10.1023/a:1005851623493. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya MK, Ward EWB. Resistance, susceptibility and accumulation of glyceollins I-III in soybean organs inoculated with Phytophthora megasperma f. sp. glycinea. Physiol Mol Plant Pathol. 1986;29:227–273. [Google Scholar]
- Bode W, Gomis-Rüth F-X, Stöchler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the “metzincins.”. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FR, Cammue BP, Osborn RW. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M-M, Hadwiger LA, Horovitz D. Molecular characterization of a pea β-1,3-endoglucanase induced by Fusarium solani and chitosan challenge. Plant Mol Biol. 1992;20:609–618. doi: 10.1007/BF00046446. [DOI] [PubMed] [Google Scholar]
- Chiang CC, Hadwiger LA. The Fusarium solani-induced expression of a pea gene family encoding high cysteine content proteins. Mol Plant-Microbe Interact. 1991;4:324–331. doi: 10.1094/mpmi-4-324. [DOI] [PubMed] [Google Scholar]
- del Pozo O, Lam E. Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol. 1998;8:1129–1132. doi: 10.1016/s0960-9822(98)70469-5. [DOI] [PubMed] [Google Scholar]
- Delorme VG, McCabe PF, Kim D-J, Leaver CJ. A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiol. 2000;123:917–927. doi: 10.1104/pp.123.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva I, Poirier GG, Heath MC. Activation of cysteine proteases in cowpea plants during the hypersensitive response: a form of programmed cell death. Exp Cell Res. 1998;245:389–399. doi: 10.1006/excr.1998.4256. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis. Curr Opin Plant Biol. 1999;2:280–286. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- Graham JS, Xiong J, Gillikin JW. Purification and developmental analysis of a metalloendoproteinase from leaves of Glycine max. Plant Physiol. 1991;97:786–792. doi: 10.1104/pp.97.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habereder H, Schröder G, Ebel J. Rapid induction of phenylalanine ammonia-lyase and chalcone synthase mRNAs during fungus infection of soybean (Glycine max L.) roots or elicitor treatment of soybean cell cultures at the onset of phytoalexin synthesis. Planta. 1989;177:58–65. doi: 10.1007/BF00392154. [DOI] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloprotease. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bond J. Families of metalloproendopeptidases and their relationships. FEBS Lett. 1992;312:110–114. doi: 10.1016/0014-5793(92)80916-5. [DOI] [PubMed] [Google Scholar]
- Kasuga T, Salimath SS, Shi J, Gijzen M, Buzzell RI, Battacharyya MK. High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol Plant-Microbe Interact. 1997;10:1035–1044. [Google Scholar]
- Keen NT, Buzell RI. New resistance genes in soybean against Pseudomonas syringy pv. glycinea: evidence that one of them interacts with a bacterial elicitor. Theor Appl Genet. 1991;81:133–138. doi: 10.1007/BF00226123. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissue during senescence and under various stressed conditions. Plant J. 1999;19:43–53. doi: 10.1046/j.1365-313x.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- Lam E, Pontier D, del Pozo O. Die and let live: programmed cell death in plants. Curr Opin Plant Biol. 1999;2:502–507. doi: 10.1016/s1369-5266(99)00026-6. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenkaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Liu CY, Xu H, Graham JS. Cloning and characterization of an Arabidopsis cDNA homologous to the matrix metalloproteinases. Plant Physiol. 1998;117:1127. [Google Scholar]
- Maidment JM, Moore D, Murphy GP, Murphy G, Clark IM. Matrix metalloproteinase homologues from Arabidopsis thaliana: expression and activity. J Biol Chem. 1999;274:34706–34710. doi: 10.1074/jbc.274.49.34706. [DOI] [PubMed] [Google Scholar]
- Mikhail A, Yakov ED, Natalia EV. Isolation and properties of a metalloproteinase from buckwheat (Fagopyrum esculentum) seeds. Biochem J. 1990;272:677–682. doi: 10.1042/bj2720677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Okinaka Y, Mimori K, Takeo K, Kitamura S, Takeuchi Y, Yamaoka N, Yoshikawa M. A structural model for the mechanisms of elicitor release from fungal cell walls by plant β-1,3-endoglucanase. Plant Physiol. 1995;109:839–845. doi: 10.1104/pp.109.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi S. Induction of resistance or susceptibility. Annu Rev Phytopathol. 1983;21:289–315. [Google Scholar]
- Pak JH, Liu CY, Huangpu J, Graham JS. Construction and characterization of the soybean leaf metalloproteinase cDNA. FEBS Lett. 1997;404:283–288. doi: 10.1016/s0014-5793(97)00141-5. [DOI] [PubMed] [Google Scholar]
- Ragster L, Chrispeels MJ. Azocoll-digesting proteinases in soybean leaves: characteristics and changes during leaf maturation and senescence. Plant Physiol. 1979;64:857–861. doi: 10.1104/pp.64.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neutral networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 2.108–2.117. [Google Scholar]
- Schmelzer E, Börner H, Grisebach H, Ebel J, Hahlbrock K. Phytoalexin synthesis in soybean (Glycine max): similar time course of mRNA induction in hypocotyls infested with a fungal pathogen and in cell culture treated with fungal elicitor. FEBS Lett. 1984;172:59–63. [Google Scholar]
- Schmidt WE, Ebel J, Loyal R. Phytoalexin synthesis in soybean cells: elicitor induction of phenylalanine ammonia-lyase and chalcone synthase mRNA and correlation with phytoalexin accumulation. Arch Biochem Biophys. 1984;232:240–248. doi: 10.1016/0003-9861(84)90540-x. [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Kochs G, Lottspeich F, Ebel J. Molecular characterization and functional expression of dihydroxypterocarpan 6a-hydroxylase, an enzyme specific for pterocarpanoid phytoalexin biosynthesis in soybean (Glycine max L.) FEBS Lett. 1998;432:182–186. doi: 10.1016/s0014-5793(98)00866-7. [DOI] [PubMed] [Google Scholar]
- Schumacher HM, Gundlach H, Fiedlar F, Genk MH. Elicitation of benzophenanthridine alkaloid synthesis in Escholtzia cell cultures. Plant Cell Rep. 1987;6:410–413. doi: 10.1007/BF00272770. [DOI] [PubMed] [Google Scholar]
- Sequeira L. Mechanisms of induced resistance in plants. Annu Rev Microbiol. 1983;37:51–79. doi: 10.1146/annurev.mi.37.100183.000411. [DOI] [PubMed] [Google Scholar]
- Shigaki T, Bhattacharyya MK. Color coding the cell death status of plant suspension cells. BioTechniques. 1999;26:1060–1062. doi: 10.2144/99266bm12. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stäb MR, Ebel J. Effects of Ca2+ on phytoalexin induction by fungal elicitor in soybean cells. Arch Biochem Biophys. 1987;257:416–423. doi: 10.1016/0003-9861(87)90585-6. [DOI] [PubMed] [Google Scholar]
- Terras FR, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EWB, Cahill DM, Bhattacharyya MK. Early cytological differences between compatible and incompatible interaction of soybean with Phytophthora megasperma f. sp. glycinea. Physiol Mol Plant Pathol. 1989;34:267–283. [Google Scholar]
- Welle R, Grisebach H. Induction of phytoalexin synthesis in soybean: enzymatic cyclization of prenylated pterocarpans to glyceollin isomers. Arch Biochem Biophys. 1988;263:191–198. doi: 10.1016/0003-9861(88)90627-3. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Xia Y, Blount J, Dixon RA, Borevitz J, Guo Z, Lamb C. Activation tagging approach for identification of genes involved in plant disease resistance responses. The Abstracts of ASPP Annual Meeting, Plant Biology ‘99, Baltimore, MD. 1999. p. 178. [Google Scholar]
- Zähringer U, Ebel J, Mulheirn LJ, Lyne RL, Grisebach H. Induction of phytoalexin synthesis: dimethylallylpyrophosphate:trihydroxypterocarpan dimethylallyl transferase from elicitor-induced cotyledons. FEBS Lett. 1979;101:90–92. doi: 10.1016/0014-5793(79)81301-0. [DOI] [PubMed] [Google Scholar]