Abstract
Despite extensive biochemical analyses, the biological function(s) of plant β-amylases remains unclear. The fact that β-amylases degrade starch in vitro suggests that they may play a role in starch metabolism in vivo. β-Amylases have also been suggested to prevent the accumulation of highly polymerized polysaccharides that might otherwise impede flux through phloem sieve pores. The identification and characterization of a mutant of Arabidopsis var. Columbia with greatly reduced levels of β-amylase activity is reported here. The reduced β-amylase 1 (ram1) mutation lies in the gene encoding the major form of β-amylase in Arabidopsis. Although the Arabidopsis genome contains nine known or putative β-amylase genes, the fact that the ram1 mutation results in almost complete loss of β-amylase activity in rosette leaves and inflorescences (stems) indicates that the gene affected by the ram1 mutation is responsible for most of the β-amylase activity present in these tissues. The leaves of ram1 plants accumulate wild-type levels of starch, soluble sugars, anthocyanin, and chlorophyll. Plants carrying the ram1 mutation also exhibit wild-type rates of phloem exudation and of overall growth. These results suggest that little to no β-amylase activity is required to maintain normal starch levels, rates of phloem exudation, and overall plant growth.
β-Amylases are found in vegetative tissues as well as in storage and reproductive organs, such as tubers and seeds. β-Amylases hydrolyze α-1,4-glucosidic linkages from the reducing ends of polysaccharide chains to form maltose (Beck and Ziegler, 1989). The genomes of many plant species contain several genes that encode β-amylases. For example, according to the database maintained by The Institute for Genomic Research (TIGR), the Arabidopsis genome includes nine genes that are identified as β-amylase or putative β-amylase genes (http://www.tigr.org/tdb/edb2/ath1/htmls/GeneNameSearch.html). Despite the fact that they have been well characterized in vitro, the in vivo biological function(s) of β-amylases remains under debate (Ziegler, 1999).
Several functions have been proposed for β-amylases in vivo. The fact that β-amylases hydrolyze starch in vitro (Beck and Ziegler, 1989) suggests a role for β-amylases in starch degradation. In addition, immunolocalization studies have shown an apparent association between β-amylases and starch granules (Okamoto and Akazawa, 1979; Hagenimana et al., 1992). However, these studies were conducted using polyclonal antibodies that may cross-react with starch phosphorylase. Consequently, the results of these studies should be interpreted with caution (Wang et al., 1995). In addition, starch is localized to plastids, whereas most β-amylases lack chloroplast transit peptides (Kreis et al., 1987; Monroe et al., 1991; Yoshida and Nakamura, 1991). Although chloro-plast-localized β-amylases have been identified (Lao et al., 1999), the majority of β-amylase activity is extrachloroplastic (Beck and Ziegler, 1989; Nakamura et al., 1991). For example, 80% of the amylase activity present in Arabidopsis rosette leaves is located outside of chloroplasts (Lin et al., 1988b). In addition, studies have shown that β-amylases are unable to digest starch granules unless the granules have been pretreated by either boiling to increase solubility or digestion with other enzymes (Beck and Ziegler, 1989).
A β-amylase from sweet potato (Ipomoea batatas) has been shown to inhibit starch phosphorylase activity in vitro (Pan et al., 1988). Because starch phosphorylases are involved in starch degradation, the finding that a β-amylase inhibits starch phosphorylase could imply a role for β-amylases in preventing starch breakdown. However, as stated above, most β-amylase activity is located outside of plastids. Therefore, the function of the predominant (extrachloroplastic) forms of β-amylase seems unlikely to be in starch metabolism.
Other studies have postulated that β-amylases may be localized to vacuoles (Ziegler and Beck, 1986). Vacuolar β-amylases could play a role in polysaccharide metabolism in vacuoles. However, the identity(s) of vacuolar substrates for β-amylases remains to be determined, making difficult any assessment of a possible role for β-amylases in vacuoles.
The major form of β-amylase in Arabidopsis has been shown to be localized to phloem sieve elements (Wang et al., 1995) and to be inducible by sugars (Caspar et al., 1989; Mita et al., 1995). Based on these findings, the major form of Arabidopsis β-amylase has been postulated to facilitate phloem transport by preventing accumulation of polysaccharides in sieve elements. Accumulation of polysaccharides could otherwise inhibit transport through sieve pores (Wang et al., 1995).
Several plant lines with altered levels of β-amylase activity have been identified. Studies of β-amylase-deficient lines of soybean (Glycine max; Hildebrand and Hymowitz, 1981) and rye (Secale cereale; Daussant et al., 1981) have focused primarily on characterizing the effects of reduced β-amylase activity on seed formation and germination. These studies reveal no evidence for an essential role of β-amylases in carbohydrate metabolism of developing or germinating soybean seeds (Hildebrand and Hymowitz, 1981) or in germination of rye seeds (Daussant et al., 1981). Little work has been done to determine whether the decreased β-amylase activity levels in these plant lines affect later, vegetative, stages of development. In fact, at least one of these plant lines, despite having greatly decreased levels of β-amylase activity in seeds, exhibits wild-type β-amylase levels in leaves and shoots (Daussant et al., 1991).
One high β-amylase (hba) and two low β-amylase (lba) loci of Arabidopsis have also been identified (Mita et al., 1997a, 1997b). These loci do not correspond to β-amylase structural genes but instead affect regulation of β-amylase activity. Plants carrying the lba1, lba2, or hba1 loci exhibit normal levels of leaf-soluble sugars and starch. In contrast, the lba1 and lba2 loci (Mita et al., 1997b) inhibit sugar-induced anthocyanin accumulation, whereas the hba1 locus (Mita et al., 1997a) causes an increase in anthocyanin levels. Plants homozygous for the lba1 locus also exhibit decreased levels of chlorophyll when grown on media containing low (1%) concentrations of Suc but have wild-type chlorophyll levels when grown on higher (3%) Suc (Mita et al., 1997b). The lba and hba mutants, although very useful tools for studying regulation of β-amylase expression, may not be the best tools for studying β-amylase function because the mutations they carry likely affect the expression of other genes in addition to β-amylase (Mita et al., 1997a, 1997b).
Despite numerous studies, the biological function(s) of β-amylases remains unclear. Here, we report the isolation of reduced β-amylase (ram) mutants of Arabidopsis. The ram1 mutation lies in a β-amylase structural gene and results in almost complete loss of β-amylase activity, making it a useful tool for investigating β-amylase function.
RESULTS
Isolation of ram Mutants
A screen was undertaken to identify Arabidopsis mutants with decreased levels of β-amylase activity. β-Amylase activity is induced by sugars (Caspar et al., 1989; Nakamura et al., 1991; Mita et al., 1995). Mutations that affect sugar-induced β-amylase expression are expected to be useful in elucidating the molecular mechanisms by which sugar-regulated gene expression occurs in plants. Because β-amylase is believed to be regulated at both the transcriptional and post-transcriptional levels (Daussant et al., 1991; Mita et al., 1997a), the mutant screen was performed by directly measuring β-amylase activity levels in M2 plants. This screen thus allows the detection of mutations that affect post-transcriptional as well as transcriptional regulation of β-amylase. In addition, the screen allows the detection of mutants that carry defects in β-amylase structural genes. Mutants with defects in β-amylase structural genes are of interest as tools to test models regarding β-amylase function.
M2 plants derived from ethylmethane sulfonate-mutagenized Arabidopsis var. Columbia homozygous for the gl1 and pgm1 (Caspar et al., 1985) mutations were grown in soil under a 12-h photoperiod and screened for mutants with decreased levels of β-amylase activity. Plants carrying the gl1 marker were used to facilitate identification of any contaminating wild-type seeds lacking the pgm1 mutation. Plants carrying the pgm1 mutation are unable to make starch (Caspar et al., 1985). When grown under a 12-h photoperiod, pgm1 plants accumulate unusually high levels of soluble sugars at the end of the light period (Caspar et al., 1985), leading to induction of β-amylase activity (Caspar et al., 1989). Thus, pgm1 plants grown under a 12-h photoperiod have unusually high levels of β-amylase activity, facilitating identification of mutants with reduced β-amylase activity. A qualitative β-amylase activity assay (see “Materials and Methods”) was developed and used to screen approximately 6,000 M2 plants, grown as described above. These qualitative assays, as well as subsequent quantitative β-amylase assays, were conducted at pH levels of 4 to 5 to minimize α-amylase activity. In this pH range, α-amylases retain almost no activity, whereas β-amylases retain approximately 70% of maximal activity (Lin et al., 1988b). Therefore, conducting the assays at pH levels between 4 and 5 makes possible the measurement of β-amylase activity in the almost complete absence of α-amylase activity.
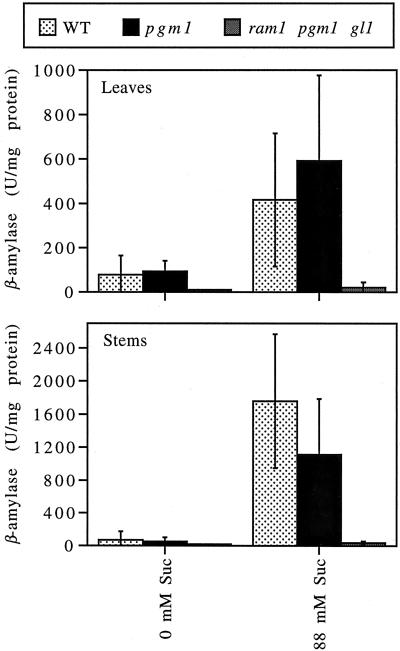
Plants that exhibited reduced levels of β-amylase activity in the qualitative assay were grown to maturity and re-screened in the next generation using a quantitative β-amylase activity assay. Eight ram mutants were found to exhibit reproducibly lower levels of β-amylase activity. Complementation analyses revealed that these mutants fall into at least four complementation groups. The ram1 mutant, which exhibits the lowest level of β-amylase activity, was chosen for further study. Quantitative β-amylase assays revealed that the ram1 mutation results in almost complete elimination of β-amylase activity in both stems (inflorescences) and rosette leaves (Fig. 1).
Figure 1.
The ram1 mutant exhibits severely reduced levels of β-amylase activity. Wild-type (WT), pgm1, and ram1 pgm1 gl1 plants were grown in soil under continuous light for 3 to 5 weeks. Cuttings (leaves or stem sections) were then removed and placed in water (0 mm Suc) or 88 mm Suc. After approximately 3 d, total proteins were prepared from the leaf or stem sections and assayed for β-amylase activity (1 unit [U] = the amount of β-amylase required to generate 1 nmol of reducing sugar per min from amylopectin). Note that, although pgm1 mutants grown under a 12-h photoperiod accumulate high levels of soluble sugars leading to induction of β-amylase activity, pgm1 mutants grown under continuous light exhibit wild-type levels of soluble sugars and of β-amylase activity (Caspar et al., 1985, 1989). The data presented represent the combined results of multiple independent experiments. Values indicate the means ± sd (n = 2–6).
The ram1 Mutation Lies in a β-Amylase Structural Gene
The ram1 locus was mapped and found to lie approximately 7 cM from marker g4539 (Nam et al., 1989) on chromosome 4. A search of the GenBank database and genomic maps available through The Arabidopsis Information Resource (TAIR) website (http://www.Arabidopsis.org/) revealed that a β-amylase structural gene with GenBank accession no. S77076 (Monroe et al., 1991; Mita et al., 1995) maps to the same region of the genome. It is interesting that the β-amylase encoded by this gene has been shown to be localized to phloem sieve elements (Wang et al., 1995). The similar map positions of this β-amylase gene and the ram1 locus raised the possibility that the ram1 mutation lies in the gene encoding the phloem-specific β-amylase.
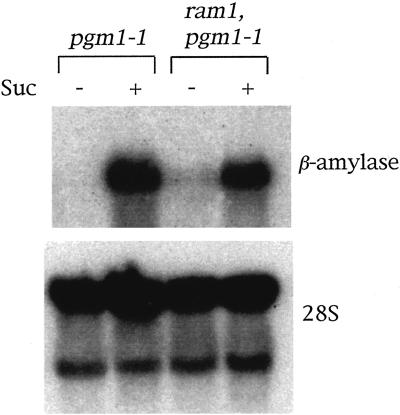
To determine whether the ram1 mutation lies in the β-amylase gene, PCR was used to amplify DNA fragments containing the β-amylase gene from the ram1 mutant. Sequencing of the PCR products revealed that ram1 carries a defective β-amylase gene. The ram1 mutation consists of a G to A transition that alters the sequence of nucleotide 1,386 (counting the A of the translational initiation codon as 1) of the β-amylase genomic DNA (TTTTAGTGTTATGACAAGTAC to TTTTAATGTTATGACAAGTA). The altered G residue is predicted to lie at the −1 position of a 3′ RNA splice site (Monroe et al., 1991; Mita et al., 1995). As shown in Figure 2, northern blot analysis revealed that ram1 has approximately wild-type steady-state levels of β-amylase mRNA. In addition, the ram1 and wild-type β-amylase transcripts appear similar in size. These results suggest that a cryptic 3′-splice site near the mutated splice site is used in processing of β-amylase mRNA from ram1. However, the intron affected by the ram1 mutation is only 82 bp in size (Mita et al., 1995), raising the possibility that this intron is not spliced in the ram1 mutant but that the resulting difference in transcript size is difficult to detect by northern blot analysis. Western blot analyses failed to detect β-amylase protein in extracts made from ram1 (data not shown). This result suggests that β-amylase mRNA from ram1 is inefficiently translated or results in production of an unstable protein.
Figure 2.
The ram1 mutant has wild-type levels of β-amylase mRNA. Wild-type and ram1 pgm1 seedlings were grown on solid minimal Arabidopsis media for approximately 2 weeks and then for an additional 2 weeks in soil under continuous light conditions. Plants were then removed from the soil, rinsed with water, and placed with their root systems in either water (Suc −) or an 88 mm Suc solution (Suc +). After 3 d under 66 to 80 μmol photons m−2 s−1 continuous light, rosette leaves were harvested and used to prepare total RNA. A northern blot prepared using this RNA was probed with labeled DNA corresponding to an Arabidopsis β-amylase cDNA. The blot was then stripped and reprobed with labeled DNA corresponding to the 28S rDNA.
ram1 Leaves Contain Wild-Type Levels of Starch and Soluble Sugars
Because the ram1 mutation results in the almost complete elimination of β-amylase activity, the ram1 mutant provides a valuable tool for investigating the biological role of β-amylases. Findings that the majority of β-amylase activity is extrachloroplastic (Beck and Ziegler, 1989; Nakamura et al., 1991), whereas starch is localized to the plastids, suggest that β-amylases are not involved in starch metabolism. However, β-amylases are able to degrade starch in vitro (Beck and Ziegler, 1989). In addition, the possibility remains that a small percentage of β-amylase activity could be localized to plastids. As a result, a role for β-amylases in starch metabolism cannot be ruled out. To test this possibility, it was first necessary to obtain a plant line that is homozygous for the ram1 mutation but that lacks the pgm1 and gl1 mutations. It was particularly important to obtain a ram1 line lacking the pgm1 mutation because the phosphoglucomutase encoded by PGM1 is needed for starch biosynthesis (Caspar et al., 1985). Therefore, the original ram1 pgm1 gl1 line was back-crossed with wild-type plants and a line that is homozygous for the ram1 mutation, but that lacks the pgm1 and gl1 mutations, was isolated.
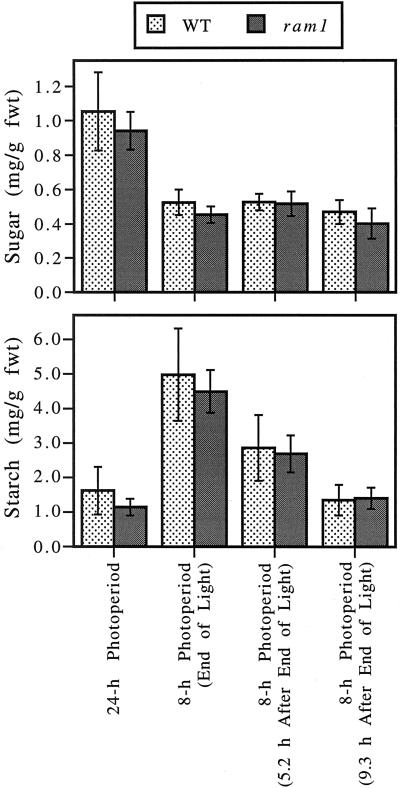
Starch and soluble sugar levels were measured in ram1 and wild-type plants grown under different light regimes. As shown in Figure 3, ram1 leaves contain wild-type levels of soluble sugars (a combination of Glc, Suc, and Fru) under all growth conditions tested. Leaves of ram1 plants grown under continuous light conditions also have wild-type starch levels, indicating that the ram1 mutation does not affect steady-state starch levels. In addition, leaves of ram1 plants contain wild-type starch levels when plants are grown under an 8-h photoperiod and leaves are harvested at the end of the light period and at different times during the dark period. These results indicate that severe reductions in β-amylase activity have no significant effect on starch accumulation during the light period or on starch degradation rates during the dark period (Fig. 3).
Figure 3.
Severe decreases in β-amylase activity do not affect leaf starch or soluble sugar levels. Wild-type (WT) and ram1 plants were grown under continuous light for 44 d (= 24-h photoperiod plants); the final 5 d was at a light intensity of 140 to 150 μmol photons m−2 s−1. Additional plants were grown for 33 d under continuous light and then for an additional 11 d under an 8-h photoperiod (= 8-h photoperiod plants); the final 5 d was at a light intensity of 220 to 230 μmol photons m−2 s−1. Leaves were harvested from the plants growing in the 8-h photoperiod at the end of the light period, 5.2 h after the end of the light period and 9.3 h after the end of the light period. Two rosette leaves, each from a different plant, were used for each sugar/starch assay. The sugar assays measured the combined amounts of Glc, Suc, and Fru present in a particular sample. The ram1 plants used in these experiments lack the pgm1 mutation and are from a line that has been back-crossed four times to wild-type plants. Values indicate the means ± sd (n = 8). This experiment was repeated, with similar results. fwt, Fresh weight.
Phloem Exudates of ram1 Plants Contain Wild-Type Sugar Levels
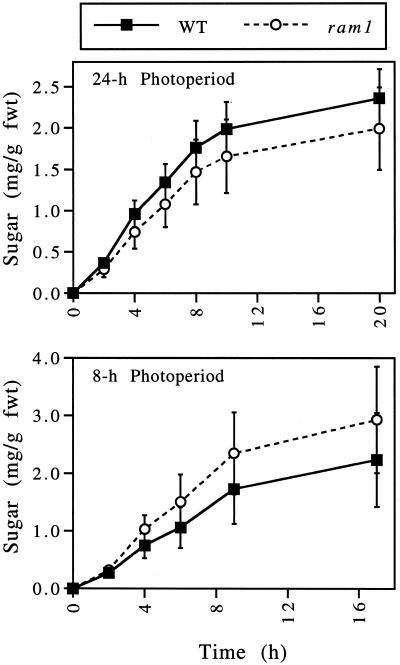
The β-amylase encoded by the gene disrupted in the ram1 mutant is localized to sieve elements (Wang et al., 1995). Based on this and other work, β-amylases have been postulated to play a role in preventing the accumulation of polysaccharides that might otherwise impede flux through the sieve elements (Wang et al., 1995). To test this hypothesis, phloem exudation rates were measured in wild-type and ram1 plants. As shown in Figure 4, the ram1 mutation does not affect the rate at which sugars (a combination of Glc, Suc, and Fru) are exuded from the cut ends of leaf petioles harvested from plants grown under continuous light conditions or an 8-h photoperiod.
Figure 4.
The ram1 mutant exhibits wild-type (WT) rates of phloem exudation. Wild-type and ram1 (lacking the pgm1 mutation) plants were grown under continuous light for 46 d; the final 5 d was at a light intensity of 200 to 210 μmol photons m−2 s−1 (= 24-h photoperiod plants). Additional plants were grown for 35 d under continuous light and then for an additional 11 d under an 8-h photoperiod; the final 5 d was at a light intensity of 220 to 230 μmol photons m−2 s−1 (= 8-h photoperiod plants). Phloem exudates were collected over different time intervals from groups of three leaves. The phloem exudates were analyzed to determine the amount of soluble sugar (Suc, Glc, and Fru) exuded from each group of leaves within a particular time period. The amount of soluble sugar in each sample was then divided by the combined fresh weights (fwt) of the three leaves from which that sample was collected. Values indicate the means ± sd (n = 8). This experiment was repeated, with similar results.
ram1 Accumulates Wild-Type Levels of Anthocyanin and Chlorophyll
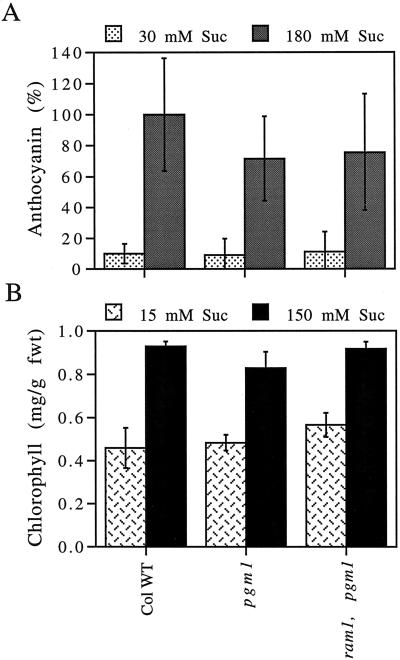
Mutations that affect regulation of β-amylase activity in Arabidopsis have also been shown to affect anthocyanin and chlorophyll levels (Mita et al., 1997a, 1997b). Therefore, it was of interest to determine whether ram1 exhibits altered anthocyanin or chlorophyll levels. As shown in Figure 5, ram1 accumulates wild-type anthocyanin and chlorophyll levels, when grown both in the presence of low sugar concentrations and in the presence of high sugar concentrations.
Figure 5.
Anthocyanin and chlorophyll accumulation are unaffected by the ram1 mutation. A, For anthocyanin determinations, plants were grown under continuous light for 2 weeks on minimal Arabidopsis medium supplemented with 30 mm Suc and then for an additional week on medium containing either 30 or 180 mm Suc. Anthocyanin levels were divided by sample fresh weight (fwt) and scaled relative to values obtained for wild-type (WT) seedlings on 180 mm Suc. B, For chlorophyll determinations, plants were grown under continuous light for 2 weeks on minimal Arabidopsis medium supplemented with either 15 or 150 mm Suc. Total shoot systems of three seedlings were used for each anthocyanin and chlorophyll assay. Values indicate the means ± sd (n = 3). These experiments were repeated, with similar results.
ram1 Exhibits Wild-Type Growth Rates
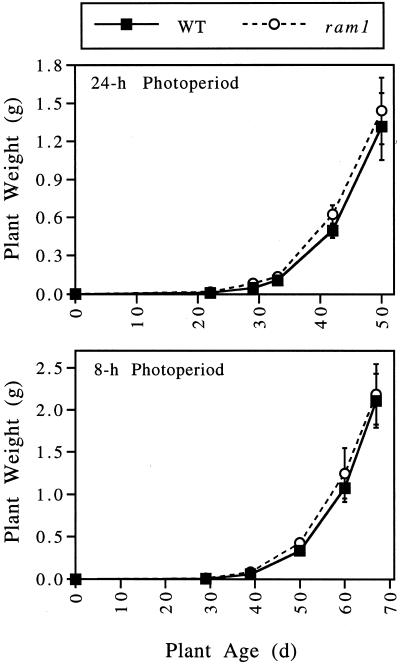
As a more generalized test for deleterious effects due to loss of β-amylase activity, the growth rates of wild-type and ram1 plants were determined. Growth rates were measured by harvesting and weighing shoot systems (all above ground parts) at regular intervals. As shown in Figure 6, ram1 and wild-type plants have similar growth rates when grown under continuous light conditions or an 8-h photoperiod.
Figure 6.
The ram1 mutant exhibits wild-type (WT) growth rates. Wild-type and ram1 plants were grown in soil at 19°C to 20°C under either continuous light (24-h photoperiod) or an 8-h photoperiod. At regular intervals, shoot systems (corresponding to all above ground parts) of groups of five plants were harvested and weighed. The ram1 plants used in these experiments lack the pgm1 mutation and are from a line that has been back-crossed four times to wild-type plants. The values presented represent the weights of individual shoot systems. Values indicate the means ± sd (n = 5–6). This experiment was repeated, with similar results.
DISCUSSION
A qualitative assay for β-amylase activity was developed (see “Materials and Methods”). This assay is technically simple and quick to perform, allowing 200 to 300 assays to be conducted within a single day. This assay allows mutants with decreased β-amylase activity levels to be identified by measuring β-amylase activity levels directly rather than by measuring the activity of a reporter gene in transgenic lines carrying a β-amylase promoter/reporter fusion construct, for example. This screen thus allows for detection of mutations that affect post-transcriptional as well as transcriptional gene regulation. Mutations that affect β-amylase structural genes can also be identified.
The qualitative β-amylase activity assay was used to identify eight ram mutants of Arabidopsis that fall into at least four complementation groups. One of these mutants, the ram1 mutant, retains almost no β-amylase activity. Characterization of the ram1 mutation reveals that it lies within a previously identified β-amylase structural gene with GenBank accession no. S77076 (Monroe et al., 1991; Mita et al., 1995). The ram1 mutation converts a G residue at the −1 position of a 3′ RNA splice site to an A. All 998 Arabidopsis intron sequences compiled by TAIR (http://www.Arabidopsis.org/splice_site.html) from published (Brown et al., 1996) and unpublished studies have a G residue at the −1 position of their 3′ RNA splice sites. Therefore, the ram1 mutation is expected to result in a nonfunctional 3′-splice site. Such mutations frequently cause activation of a cryptic 3′-splice site downstream of the mutated splice site (Brown, 1996). In the case of the ram1 mutation, the nearest AG dinucleotide that may function as a cryptic 3′-splice site lies 12 bp downstream of the mutated splice site. Northern blot analysis reveals that the ram1 mutant produces a β-amylase transcript that is similar in size and abundance to the transcript produced by wild-type plants. These results suggest that a cryptic 3′-splice site near the mutated splice site is used in processing of β-amylase mRNA from ram1 and that use of this cryptic 3′-splice site does not result in a significant decrease in transcript stability.
If the AG dinucleotide located 12 bp downstream of the mutated 3′ RNA splice site acts as a cryptic splice site, a transcript missing four codons will result. These four codons encode the amino acids CYDK. Although these four amino acids are not believed to interact directly with the β-amylase substrate (Mikami et al., 1994), a search of the literature (Totsuka et al., 1994) and of the GenBank database reveals that these amino acid residues are highly conserved among β-amylases from a variety of plant species. The Y residue is particularly well conserved, because it is present not only in β-amylases from plants but also in several species of bacteria (Totsuka et al., 1994). Mutating the C residue present at the equivalent position in a β-amylase from soybean did not result in a significant reduction in β-amylase activity (Totsuka et al., 1994). However, because the four amino acids predicted to be missing from the β-amylase produced by ram1 are highly conserved and include two that are charged, the removal of all four amino acids is likely to result in severe alterations in protein structure and activity.
Although the β-amylase transcripts produced by ram1 and wild-type plants appear similar in size, the fact that the intron affected by the ram1 mutation is only 82 bp long (Mita et al., 1995) leaves open the possibility that this intron is not removed in ram1 plants but that the resulting alteration in transcript size is difficult to detect by northern analysis. Failure to remove this intron would cause a frameshift in the middle of the coding region. Site-directed mutagenesis experiments indicate that at least one amino acid within the region that would be affected by this frameshift plays a critical role in β-amylase activity (Totsuka et al., 1994). In addition, x-ray crystallographic data reveal that amino acids that would be affected by this frameshift form part of the region of the protein comprising the catalytic center (Mikami et al., 1994). Therefore, a frameshift caused by failure to remove the intron affected by the ram1 mutation would be expected to result in a severe decrease in enzyme activity and possibly also in protein stability.
β-Amylase assays reveal that ram1 rosette leaves and stems retain almost no β-amylase activity. According to the database (http://www.tigr.org/tdb/edb2/ath1/htmls/GeneNameSearch.html) maintained by TIGR, the Arabidopsis genome contains nine genes that are identified as β-amylase or putative β-amylase genes. The results presented here indicate that one of those genes (GenBank accession no. S77076), the gene affected by the ram1 mutation, is responsible for at least most of the β-amylase activity present in rosette leaves and stems of wild-type plants. The very slight amount of amylase activity detected in ram1 mutants may be the result of the gene affected by the ram1 mutation producing a small amount of correctly processed transcript or of the protein produced by an altered transcript retaining a small amount of activity. Alternatively, despite the fact that the β-amylase assays are conducted at low pH to minimize α-amylase activity (Lin et al., 1988b), a small amount of α-amylase activity may still be detectable. Finally, residual amylase activity in ram1 plants may be due to expression of other β-amylase structural genes.
Mutants with decreased levels of β-amylase activity have previously been identified. However, the effects of the available soybean (Hildebrand and Hymowitz, 1981) and rye (Daussant et al., 1981) mutations on vegetative development have not been well characterized. In addition, previously identified Arabidopsis mutants with alterations in β-amylase activity carry defects in genes involved in β-amylase regulation rather than in β-amylase structural genes. These mutants are expected to be defective in the expression of other genes in addition to β-amylase (Mita et al., 1997a, 1997b). Consequently, distinguishing between the phenotypes of these mutants that are the result of altered β-amylase levels and those that are the result of altered expression of other genes may be problematic.
Several models have been proposed to explain the biological function(s) of β-amylases. The identification of a mutation that lies in a β-amylase structural gene and that results in severely reduced β-amylase activity levels provides a useful tool for testing these models. The fact that β-amylases degrade starch in vitro (Beck and Ziegler, 1989) suggests that β-amylases may play a role in carbohydrate metabolism in vivo. However, leaves of light-grown ram1 plants accumulate wild-type levels of soluble sugars and of starch, indicating that the ram1 mutation does not affect starch synthesis or accumulation of soluble sugars. The ram1 mutation also has no apparent effect on starch degradation. When ram1 and wild-type plants are grown under an 8-h photoperiod, ram1 starch levels decline during the dark period at the same rate as wild-type starch levels.
The β-amylase gene in which the ram1 mutation lies has been shown to encode a β-amylase that is localized to phloem sieve elements (Wang et al., 1995). This β-amylase is also induced by soluble sugars, such as Suc (Caspar et al., 1989; Mita et al., 1995). Based on these findings, this β-amylase has been postulated to prevent the buildup of highly polymerized polysaccharides that might otherwise accumulate in sieve tubes during times of increased sugar availability (Wang et al., 1995). Such a buildup of polysaccharide particles could impede flux of materials through sieve pores. To test this model, rates of phloem exudation were measured in ram1 and wild-type Arabidopsis. These experiments indicate that the ram1 mutation does not result in decreased flux of soluble sugars in phloem exudates. Additional experiments indicate that the ram1 mutation does not inhibit plant growth rates under either continuous light conditions or an 8-h photoperiod. These results suggest that little to no β-amylase activity is required to maintain normal rates of phloem transport.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Media
All plant lines used in these experiments are from the Arabidopsis var. Columbia ecotype. Wild-type seeds were originally obtained from Dr. Chris Somerville (Carnegie Institute of Washington, Palo Alto, CA). The M2 seeds used to screen for plants with reduced β-amylase activity and seeds of the pgm1 mutant were obtained from Dr. Tim Caspar (DuPont Co., Wilmington, DE). The M2 seeds were generated by first crossing a pgm1 mutant line, originally designated as line TC7 (Caspar et al., 1985), with a plant line carrying the gl1 mutation. Plants homozygous for both mutations were isolated, and seeds from these plants were treated with ethyl methanesulfonate to generate M1 seeds. These M1 seeds were sown, the resulting plants allowed to self fertilize, and the M2 seeds harvested. The original plant line carrying the ram1 mutation is thus also homozygous for the pgm1 and gl1 mutations. This line was back-crossed four times to wild-type Arabidopsis of the Columbia ecotype to generate a line that is homozygous for the ram1 mutation but that lacks the gl1 and pgm1 mutations. Unless otherwise noted, plants were grown under continuous cool-white fluorescent light at approximately 21°C. Minimal Arabidopsis media was prepared according to a previously published protocol (Kranz and Kirchheim, 1987).
Mutant Screen and Qualitative β-Amylase Activity Assay
Approximately 6,000 M2 plants (derived from ethyl methanesulfonate-mutagenized populations of plants homozygous for the pgm1 and gl1 mutations) were grown under a 12-h photoperiod. A hole punch was used to remove two leaf discs from each M2 plant. These leaf discs were submerged in the wells of 96-well microtiter plates containing 50 μL of 0.2 m Na acetate (pH 4.3) and 0.1% (w/v) Triton X-100. The 96-well microtiter plates were then placed under a partial vacuum for several minutes. To aid in cell/tissue disruption, the microtiter plates and accompanying leaf samples were subjected to three freeze/thaw cycles by moving the microtiter plates back and forth between liquid N and a container of warm water. The microtiter plates were then placed in a tray of warm (approximately 40°C) water and 50 μL of 0.2 m Na acetate (pH 4.3), 1.2% (w/v) molten low-melt agarose, and 0.5% (w/v) soluble starch, maintained at a temperature of approximately 45°C, was added to each well of the microtiter plate. The microtiter plates were sealed with plastic wrap and incubated at 37°C for approximately 1 h. To determine which wells of the microtiter plates still contained starch, 50 μL of 0.2 n HCl, 0.36 mm iodine, and 43.4 mm KI were added to each well. Upon addition of the iodine solution, wells that still contained undigested starch were stained blue, whereas wells that no longer contained starch became yellow. Putative mutants, corresponding to M2 plants whose leaf discs failed to digest all the starch in the wells in which they were placed, were rescreened using the same assay.
Quantitative β-Amylase Activity Assays
Tissue for β-amylase assays was prepared by cutting rosette leaves and inflorescences (stems) from 3- to 6-week-old plants. The cut ends of the leaf petioles and inflorescences were placed in either water or 88 mm Suc for 3 d prior to harvesting. Quantitative β-amylase activity assays were performed on these tissues using a modified version of a published procedure (Lin et al., 1988b). In brief, β-amylase activity was measured by determining the amount of amylopectin digested to sugar (maltose) by a specific amount of total protein within a specific time interval. Total protein extracts for use in β-amylase assays were prepared from approximately 0.01 g of leaf or stem material. The plant tissue was ground on ice in a 1.5-mL microtube containing 150 μL of 50 mm Tris-HCl (pH 8). Samples were spun for 20 min at maximum speed in a microfuge kept in a 4°C room. Supernatants were transferred to fresh microtubes and assayed for total protein levels using the Coomassie Protein Assay Reagent (Pierce Chemical Co., Rockford, IL), according to the manufacturer's instructions.
β-Amylase reaction mixtures were prepared by mixing 10 μg of total protein, diluted with deionized water to a total volume of 85 μL, with 75 μL of 20 mg mL−1 amylopectin and 150 μL of 0.1 n Na acetate (pH 4.6). To prepare the amylopectin stock solution, amylopectin was boiled in 0.2 n KOH for 10 min and then stored in aliquots at −20°C prior to use. Immediately after mixing, 95-μL aliquots of each β-amylase reaction mixture were transferred to new microtubes, and the reactions were stopped by placing the microtubes in boiling water for 2 to 3 min (= zero time point). The remainders of each reaction mixture were incubated at 37°C for 60 min before removing and boiling additional 95-μL aliquots. To quantify the amount of sugar present in each sample, 20-μL aliquots of each sample were mixed with 230 μL of deionized water and 750 μL of p-hydroxybenzoic acid hydrazide (PAHBAH)/NaOH solution (Lever, 1977). The PAHBAH/NaOH solution was prepared immediately prior to use by mixing one part 5% (w/v) PAHBAH in 0.5 n HCl with 4 parts 0.5 n NaOH. The sugar assay reaction mixtures were incubated at 100°C for 5 min and allowed to cool, and then A410 values were determined and compared with the values obtained using a maltose standard curve.
One unit of β-amylase activity is defined as the amount of β-amylase required to generate 1 nmol of reducing sugar (maltose) per min by digestion of amylopectin at 37°C.
Identifying the ram1 Mutation
Oligonucleotides complementary to an Arabidopsis β-amylase genomic DNA clone (GenBank accession no. S77076) were used to amplify DNA from the ram1 mutant via PCR. The PCR products were separated on an agarose gel and purified using the Qiaex 2 gel purification kit (Qiagen Inc., Santa Clarita, CA). DNA samples were submitted to a commercial facility for sequencing (Lone Star Labs Inc., Houston). DNA spanning the region from approximately 900 bp upstream of the start codon to 300 bp downstream of the stop codon was sequenced on at least one strand. The region containing the ram1 mutation was sequenced on both DNA strands.
RNA Preparation and Analysis
Rosette leaves were harvested from approximately 4-week-old plants, grown for the last 3 d with their root systems in either water or 88 mm Suc. The leaves were frozen in liquid N and ground to a powder, and total RNA was extracted using the TRIzol Reagent (Invitrogen, Rockville, MD), according to the manufacturer's instructions. Ten-μg aliquots of total RNA were then separated on an agarose gel and transferred to a BrightStar membrane (Ambion Inc., Austin, TX) using the NorthernMax kit (Ambion), according to the manufacturer's instructions. A DNA fragment containing almost the entire coding region of an Arabidopsis β-amylase cDNA clone (EMBL accession no. M73467) was 32P-labeled and hybridized to the northern blot using the NorthernMax kit (Ambion), according to the manufacturer's instructions. Note that this β-amylase cDNA represents the same gene identified by the genomic β-amylase DNA sequence with GenBank accession no. S77076. The northern blot was exposed to x-ray film, stripped, and reprobed with labeled 28S rDNA.
Immunoblots
Total proteins were separated by SDS-PAGE and transferred to a 0.22-μm nitrocellulose transfer membrane (NitroME from Micron Separations Inc., Westboro, MA). The membrane was probed using anti-β-amylase antibodies obtained from Dr. Jon Monroe (James Madison University, Harrisonburg, VA). Binding of the primary antibodies was visualized using the Rabbit ExtrAvidin Alkaline Phosphatase kit and the fast 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium reagents from Sigma (St. Louis), according to the manufacturer's instructions.
Starch and Soluble Sugar Assays
Starch and soluble sugar (a combination of Glc, Suc, and Fru) levels were assayed using established procedures (Jones et al., 1977; Lin et al., 1988a) with modifications that have been described previously (Chia et al., 2000). One additional modification was used in this study. In the previous study (Chia et al., 2000), sugar content was assayed by drying down the ethanol fractions containing the soluble sugars, resuspending the sugars in deionized water, and then mixing aliquots of the resuspended sugars with Glc (HK) reagent (Sigma). In this study, sugar content was assayed by mixing aliquots of the ethanol fractions containing the soluble sugars directly with the Glc (HK) reagent. This procedural modification was made after control experiments revealed that the presence of low amounts of ethanol (e.g. 10%, v/v) in the Glc (HK) reagent does not interfere with the Glc assay. This modification simplified the assay by eliminating the time-consuming processes of drying down the ethanol fractions and resuspending the sugars. To generate a standard curve, known amounts of Suc and Glc were dissolved in ethanol and mixed with the Glc (HK) reagent.
Phloem Exudation Assays
The total amount of Suc, Glc, and Fru exuded from groups of three rosette leaves within a given time interval was assayed using an established procedure (Costello et al., 1982), with modifications that have been described previously (Chia et al., 2000).
Chlorophyll and Anthocyanin Assays
Chlorophyll (Wintermans and de Mots, 1965) and anthocyanin (Rabino and Mancinelli, 1986) levels were measured using established procedures, with modifications that have been described previously (Laby et al., 2000).
Plant Growth Curves
Wild-type and ram1 plants were grown in soil at 19°C to 20°C. Some of the plants were grown under continuous light at a light intensity of 70 to 80 μmol photons m−2 s−1 for the first 18 d and than at 100 to 110 μmol photons m−2 s−1 for the remainder of the experiment. Other plants were grown under an 8-h photoperiod at a light intensity of 60 to 90 μmol photons m−2 s−1 for the first 19 d and than at 140 to 180 μmol photons m−2 s−1 for the remainder of the experiment. Plants were grown under relatively low light intensities for the first 18 to 19 d because previous experience indicated that young seedlings appear healthier when grown under lower light intensities. At regular intervals, the shoot systems (corresponding to all above ground parts) of groups of five plants were harvested and weighed. Five to six groups of plants were weighed per time point.
ACKNOWLEDGMENTS
We thank Dr. Tim Caspar for providing us with the M2 seeds from which the ram mutants were isolated and Dr. Jon Monroe for giving us serum containing antibodies against the β-amylase protein. We thank Dr. Chris Somerville for helpful suggestions regarding early experiments. We also thank Stephen Benning and Seema Chandra for technical assistance and Donna Pattison and Lydia Sommerlad for suggestions regarding this manuscript.
Footnotes
This work was supported by the U.S. Department of Energy, Energy Biosciences Program (grant no. DE–FG03–98ER20300).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010723.
LITERATURE CITED
- Beck E, Ziegler P. Biosynthesis and degradation of starch in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:95–117. [Google Scholar]
- Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 1996;10:771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Smith P, Simpson CG. Arabidopsis consensus intron sequences. Plant Mol Biol. 1996;32:531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 1985;79:11–17. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin T-P, Monroe J, Bernhard W, Spilatro S, Preiss J, Somerville C. Altered regulation of β-amylase activity in mutants of Arabidopsis with lesions in starch metabolism. Proc Natl Acad Sci USA. 1989;86:5830–5833. doi: 10.1073/pnas.86.15.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DW, Yoder TJ, Reiter W-D, Gibson SI. Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta. 2000;211:743–751. doi: 10.1007/s004250000345. [DOI] [PubMed] [Google Scholar]
- Costello LR, Bassham JA, Calvin M. Enhancement of exudation from Fraxinus uhdei Wenz. (evergreen ash) using ethylenediaminetetraacetic acid. Plant Physiol. 1982;69:77–82. doi: 10.1104/pp.69.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussant J, Sadowski J, Rorat T, Mayer C, Laurière C. Independent regulatory aspects and posttranslational modifications of two β-amylases of rye. Plant Physiol. 1991;96:84–90. doi: 10.1104/pp.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussant J, Zbaszyniak B, Sadowski J, Wiatroszak I. Cereal β-amylase: immunochemical study on two enzyme-deficient inbred lines of rye. Planta. 1981;151:176–179. doi: 10.1007/BF00387820. [DOI] [PubMed] [Google Scholar]
- Hagenimana V, Vézina L-P, Simard RE. Distribution of amylases within sweet potato (Ipomoea batatas L.) root tissue. J Agric Food Chem. 1992;40:1777–1783. [Google Scholar]
- Hildebrand DF, Hymowitz T. Role of β-amylase in starch metabolism during soybean seed development and germination. Physiol Plant. 1981;53:429–434. [Google Scholar]
- Jones MGK, Outlaw WH, Lowry OH. Enzymic assays of 10−7 to 10−14 moles of sucrose in plant tissues. Plant Physiol. 1977;60:379–383. doi: 10.1104/pp.60.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz AR, Kirchheim B. Genetic resources in Arabidopsis. Arabidopsis Inform Serv. 1987;24:1–111. [Google Scholar]
- Kreis M, Williamson M, Buxton B, Pywell J, Hejgaard J, Svendsen I. Primary structure and differential expression of β-amylase in normal and mutant barleys. Eur J Biochem. 1987;169:517–525. doi: 10.1111/j.1432-1033.1987.tb13640.x. [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Lao NT, Schoneveld O, Mold RM, Hibberd JM, Gray JC, Kavanagh TA. An Arabidopsis gene encoding a chloroplast-targeted β-amylase. Plant J. 1999;20:519–527. doi: 10.1046/j.1365-313x.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- Lever M. Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal Biochem. 1977;81:21–27. doi: 10.1016/0003-2697(77)90594-2. [DOI] [PubMed] [Google Scholar]
- Lin T-P, Caspar T, Somerville C, Preiss J. Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol. 1988a;86:1131–1135. doi: 10.1104/pp.86.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-P, Spilatro SR, Preiss J. Subcellular localization and characterization of amylases in Arabidopsis leaf. Plant Physiol. 1988b;86:251–259. doi: 10.1104/pp.86.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami B, Degano M, Hehre EJ, Sacchettini JC. Crystal structures of soybean β-amylase reacted with β-maltose and maltal: active site components and their apparent roles in catalysis. Biochemistry. 1994;33:7779–7787. [PubMed] [Google Scholar]
- Mita S, Hirano H, Nakamura K. Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana: a recessive mutation causing enhanced expression of a gene for β-amylase. Plant Physiol. 1997a;114:575–582. doi: 10.1104/pp.114.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K. Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for β-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J. 1997b;11:841–851. doi: 10.1046/j.1365-313x.1997.11040841.x. [DOI] [PubMed] [Google Scholar]
- Mita S, Suzuki-Fujii K, Nakamura K. Sugar-inducible expression of a gene for β-amylase in Arabidopsis thaliana. Plant Physiol. 1995;107:895–904. doi: 10.1104/pp.107.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Salminen MD, Preiss J. Nucleotide sequence of a cDNA clone encoding a β-amylase from Arabidopsis thaliana. Plant Physiol. 1991;97:1599–1601. doi: 10.1104/pp.97.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Ohto M, Yoshida N, Nakamura K. Sucrose-induced accumulation of β-amylase occurs concomitant with the accumulation of starch and sporamin in leaf-petiole cuttings of sweet potato. Plant Physiol. 1991;96:902–909. doi: 10.1104/pp.96.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H-G, Giraudat J, den Boer B, Moonan F, Loos WDB, Hauge BM, Goodman HM. Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell. 1989;1:699–705. doi: 10.1105/tpc.1.7.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Akazawa T. Enzymic mechanism of starch breakdown in germinating rice seeds. 8. Immunohistochemical localization of β-amylase. Plant Physiol. 1979;64:337–340. doi: 10.1104/pp.64.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S-M, Chang T-C, Juang R-H, Su J-C. Starch phosphorylase inhibitor is β-amylase. Plant Physiol. 1988;88:1154–1156. doi: 10.1104/pp.88.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986;81:922–924. doi: 10.1104/pp.81.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsuka A, Nong VH, Kadokawa H, Kim C-S, Itoh Y, Fukazawa C. Residues essential for catalytic activity of soybean β-amylase. Eur J Biochem. 1994;221:649–654. doi: 10.1111/j.1432-1033.1994.tb18777.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjölund RD. Identification and characterization of a phloem-specific β-amylase. Plant Physiol. 1995;109:743–750. doi: 10.1104/pp.109.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans JFGM, de Mots A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim Biophys Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Nakamura K. Molecular cloning and expression in Escherichia coli of cDNA encoding the subunit of sweet potato β-amylase. J Biochem. 1991;110:196–201. doi: 10.1093/oxfordjournals.jbchem.a123556. [DOI] [PubMed] [Google Scholar]
- Ziegler P. Cereal beta-amylases. J Cereal Sci. 1999;29:195–204. [Google Scholar]
- Ziegler P, Beck E. Exoamylase activity in vacuoles isolated from pea and wheat leaf protoplasts. Plant Physiol. 1986;82:1119–1121. doi: 10.1104/pp.82.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]