Abstract
The decisive events that direct a single polypeptide such as the prion protein (PrP) to be synthesized at the endoplasmic reticulum in both fully translocated and transmembrane forms are poorly understood. In this study, we demonstrate that the topological heterogeneity of PrP is determined cotranslationally, while at the translocation channel. By evaluating sequential intermediates during PrP topogenesis, we find that signal sequence-mediated initiation of translocation results in an interaction between nascent PrP and endoplasmic reticulum chaperones, committing the N terminus to the lumen. Synthesis of the transmembrane domain before completion of this step allows it to direct the generation of CtmPrP, a transmembrane form with its N terminus in the cytosol. Thus, segregation of nascent PrP into different topological configurations is critically dependent on the precise timing of signal-mediated initiation of N-terminus translocation. Consequently, this step could be experimentally tuned to modify PrP topogenesis, including complete reversal of the elevated CtmPrP caused by disease-associated mutations in the transmembrane domain. These results delineate the sequence of events involved in PrP biogenesis, explain the mechanism of action of CtmPrP-favoring mutations associated with neurodegenerative disease, and more generally, reveal that translocation substrates can be cotranslationally partitioned into multiple populations at the translocon.
INTRODUCTION
The pathogenesis of several neurodegenerative diseases such as bovine spongiform encephalopathy, Gerstmann-Straussler-Schienker disease, and Creutzfeldt-Jakob disease involves the prion protein (PrP) (Prusiner, 1998; Collinge, 2001). Although PrP is a common thread that links all of these disorders together, it is clear that prion diseases encompass a rather diverse set of clinical, pathological, and mechanistic manifestations. The most intensively studied aspect of these diseases has been the “protein-only” mode of transmission mediated by a misfolded form of PrP termed PrPSc. Although the molecular mechanisms remain to be elucidated, a central event in the transmission of prion diseases is the PrPSc-mediated conversion of normal cellular PrP into additional copies of PrPSc (Prusiner, 1997). Over time, the geometric rate of accumulation of PrPSc not only generates more transmissible agent but also is thought to lead to neurodegeneration by currently unknown mechanisms. Thus, a conceptual framework exists for studying the formation and properties of PrPSc, how it can propagate, and how its accumulation may lead to neurodegeneration in the transmissible forms of these diseases.
In contrast to the studies on the transmissible agent in prion diseases, relatively little is known about either the normal biogenesis and metabolism of PrP, or the pathogenic mechanisms that can lead to neurodegeneration. The observation that certain inherited mutations in PrP can lead to a neurodegenerative disease that neither seems to generate PrPSc nor is readily transmissible (Tateishi and Kitamoto, 1995; Tateishi et al., 1996; Hegde et al., 1999) has raised the possibility that the neurodegeneration seen in at least some forms of prion disease may be caused by mechanisms not involving PrPSc. These observations, coupled with the currently unknown normal function of PrP, have prompted investigations into aspects of PrP biology in addition to the mechanisms of PrPSc formation and propagation.
Studies examining the synthesis and translocation of PrP at the endoplasmic reticulum (ER) have revealed that it is capable of being made in three topological forms (Hegde et al., 1998a; Hölscher et al., 2001; Stewart and Harris, 2001). The majority of PrP is translocated completely across the ER membrane and is termed secPrP. The remaining PrP chains are made as single-spanning membrane proteins with either the N or C terminus translocated into the ER lumen, termed NtmPrP or CtmPrP, respectively. Remarkably, mutations that increase the generation of the CtmPrP form can cause neurodegenerative disease in either transgenic mice or in some naturally occurring heritable prion diseases (Hegde et al., 1998a, 1999). This CtmPrP-mediated neurodegeneration seems to act independently of PrPSc generation and is therefore not transmissible (Hegde et al., 1998a, 1999). In contrast, the ability of an organism to generate CtmPrP may influence its susceptibility to neurodegeneration upon accumulation of PrPSc (Hegde et al., 1999; Mishra et al., 2002), raising the possibility that the pathways of PrPSc- and CtmPrP-mediated neurodegeneration may converge on a common mechanism. Thus, deciphering the normal cellular mechanisms by which PrP topology is controlled may be of substantial importance in eventually understanding the pathogenesis of at least a subset of prion diseases.
Previous studies analyzing the topology of mutant PrPs have shown it to be sensitive to perturbations in either the signal sequence or the transmembrane domain (TMD) (Yost et al., 1990; Hegde et al., 1998a, 1999; Hölscher et al., 2001; Kim et al., 2001; Stewart et al., 2001; Stewart and Harris, 2001). Systematic analyses of which aspect(s) of PrP topology is influenced by signal and TMD mutants have indicated distinct roles for these two domains (Kim et al., 2001). Mutations in the signal sequence seem to primarily increase or decrease CtmPrP relative to both NtmPrP and secPrP. In contrast, mutations in the TMD region seem to generally increase or decrease both transmembrane forms relative to secPrP. Based on these observations, we have suggested that the signal sequence of PrP is the principal determinant of the localization of its N-terminus (either cytosolic or lumenal), whereas the TMD is the principal determinant of membrane integration (Kim et al., 2001). Thus, the characteristic ratio of PrP topological forms could potentially result from heterogeneity at the signal- and/or TMD-mediated steps during PrP translocation.
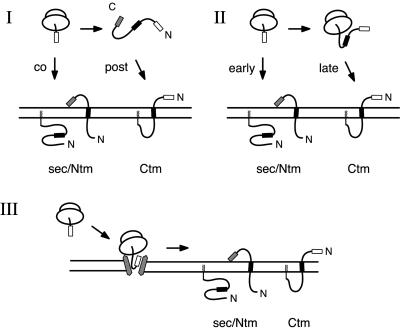
Several models of how this might occur have been suggested (Figure 1). Because mutations in the signal sequence, the element that mediates targeting of PrP to the ER, can influence its topology (Kim et al., 2001; Stewart et al., 2001), it is possible that at least a portion of the topological heterogeneity is mediated by modulation of the ER-targeting step. Indeed, it has been suggested (Hölscher et al., 2001) that the CtmPrP form can be generated, albeit inefficiently, by the posttranslational translocation of PrP chains that failed to target via its N-terminal signal sequence (Figure 1, model I). Alternatively, it may be possible for the TMD to act as an internal signal sequence that competes with the N-terminal signal in directing targeting of PrP (Hegde and Lingappa, 1999). In this scenario (Figure 1, model II), nascent chains that failed to target rapidly after synthesis of the N-terminal signal could target via the TMD, which would act as a signal anchor sequence to generate CtmPrP. Indeed, upon synthesis in vitro, the CtmPrP form seems to contain an uncleaved signal sequence, an observation that may be consistent with either of these two models (Stewart et al., 2001; Hegde, unpublished data).
Figure 1.
Possible models of PrP topogenesis. Shown are models depicting the generation of CtmPrP by posttranslational translocation (model I); by targeting at a late point in synthesis, perhaps via the transmembrane domain (model II); or by a cotranslational mechanism occurring entirely after the nascent chain has reached the translocon (model III). The N-terminal signal sequence is shown as an open box, the transmembrane domain as a black box, and the C-terminal signal for glycolipid anchor addition as a gray box (Stahl et al., 1987). The secPrP and CtmPrP forms can use the C-terminal signal to become modified by a glycolipid anchor (Stewart et al., 2001). This modification does not occur efficiently in the pancreatic rough microsomes used in this study (our unpublished observations; Stewart and Harris, 2001). However, the presence or absence of glycolipid anchor addition activity does not influence the translocation or topology of PrP (Stewart and Harris, 2001). Note that the CtmPrP form is shown containing an uncleaved signal sequence, as has been observed in vitro (Stewart et al., 2001). N-linked glycosylation sites, located at positions 181 and 197, are not shown for clarity. From the standpoint of topogenesis, glycosylation of PrP has no effect on the ratio of the three topologic forms generated at the ER (Hegde et al., 1998a).
In marked contrast to either of these mechanisms that involve differential targeting, it is also plausible that topological heterogeneity is entirely generated at a posttargeting step, once nascent PrP is at the translocon (Figure 1, model III). Such a dynamic process of determining membrane protein orientation within the translocon has been demonstrated to be possible in the case of artificially constructed membrane proteins (Goder et al., 1999; Goder and Spiess, 2001). In the present study, we discriminate between these models and identify the key steps during PrP translocation that contribute to its topogenesis.
MATERIALS AND METHODS
Plasmid Constructions
All constructs for cell-free transcription and translation were made using the SP64 vector (Invitrogen, Carlsbad, CA). Plasmids containing the coding regions for PrP, PrP(AV3), PrP(G123P), N7-PrP, and N9-PrP have been described previously (Hegde et al., 1998a; Kim et al., 2001). PrP, PrP(AV3), and PrP(G123P) lacking the C-terminal GPI anchor addition signal (Figure 2B) were generated by deletion of codons 222 through 254 by digestion with StuI and EcoRI, treatment with mung bean nuclease, and recircularizing the plasmid. PrP(Δ62–85) was made by digesting the PrP plasmid with BstXI and recircularizing the plasmid. PrP(Δ53–95) was made by digesting PrP with Bsu36I and KpnI, treating with Klenow fragment of DNA polymerase, and recircularizing the plasmid. The PrP(+120) construct in which an insertion was introduced into the N terminus of PrP (Figure 5) has been described previously (Yost et al., 1990).
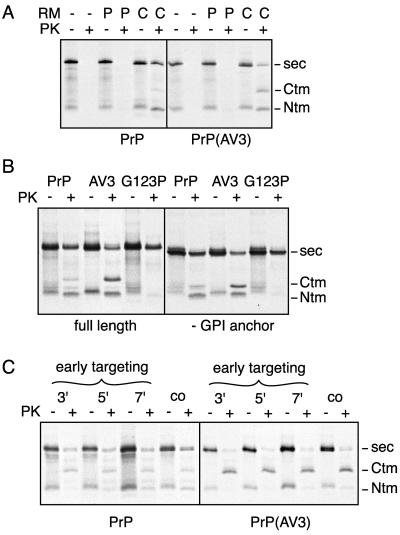
Figure 2.
Generation of PrP topological heterogeneity at a posttargeting step. (A) PrP and PrP(AV3) were synthesized in a rabbit reticulocyte lysate and translocation was allowed to proceed either cotranslationally (C) or posttranslationally (P) by including ER-derived RMs during or after the translation, respectively, as described in MATERIALS AND METHODS. A control reaction lacking RMs altogether (−) was also prepared in parallel. The samples were subsequently divided for analysis of PrP topology by PK protection assays. After treatment with PK, secPrP remains undigested, whereas CtmPrP and NtmPrP yield fragments of ∼18 and 14 kDa, respectively, that remain protected from digestion (the positions of each form are indicated to the right of the autoradiograph). (B) Cotranslational translocation of PrP, PrP(AV3), and PrP(G123P) were compared with these same constructs lacking the C-terminal GPI anchor addition signal. Topology was assessed by the PK protection assay as in A, and the positions of the three topological forms indicated to the right of the autoradiograph. Note that the size of the full-length and CtmPrP fragment, but not the NtmPrP fragment, is decreased by deletion of the GPI anchor. (C) Translocation reactions of PrP or PrP(AV3) were allowed to proceed for either 3, 5, or 7 min, after which the RMs, along with any targeted nascent chains, were isolated by sedimentation and resuspended in fresh translation extract lacking additional transcript or RMs, and containing an inhibitor of translational initiation. Translation of these pretargeted nascent chains was allowed to continue for an additional 30 min, and the topology achieved by PrP was assessed by PK protection assays. For comparison, a standard cotranslational translocation reaction (co) of each construct was analyzed in parallel.
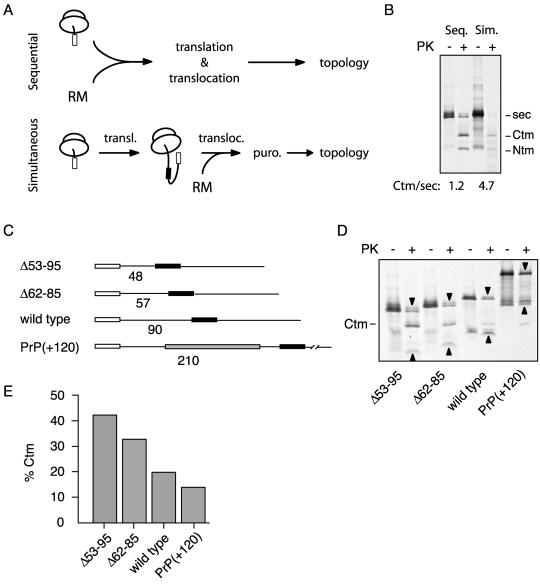
Figure 5.
PrP topology is influenced by the temporal relationship between the signal and TMD. (A) Schematic diagram of the experimental protocol for presenting the signal and TMD to the translocon sequentially versus simultaneously (see MATERIALS AND METHODS for details). In the sequential mode, translocation occurs cotranslationally, allowing the signal to be presented to the translocon before the TMD. In the simultaneous mode, ribosome-associated nascent chains containing both the signal and TMD are prepared before translocation is initiated by the addition of RMs. In both cases, topology is assessed after translocation is completed and the ribosome is released by the addition of puromycin. (B) Topology of PrP(AV3) after having been translocated by the sequential versus simultaneous mode of presentation. The ratio of the CtmPrP to secPrP forms for each mode is indicated below the autoradiograph. (C) Line diagrams of the deletion and insertion constructs that change the length (indicated below each diagram) of the domain separating the signal sequence (white box) from the TMD (black box). The insertion (gray box) consists of 120 amino acids from the protein Globin. (D) Translocation and topology of the constructs in C were assessed. The positions of the secPrP (downward arrowhead) and NtmPrP (upward arrowhead) forms, whose migrations are altered due to the deletion or insertion, are indicated. The position of CtmPrP is indicated to the left of the autoradiograph. The percentage of synthesized PrP made as CtmPrP was quantitated and shown in E.
Cell-free Translation and Translocation Assays
In vitro transcription with SP6 polymerase, translation in rabbit reticulocyte lysate containing canine pancreatic rough microsomal membranes (RMs), and assessment of PrP topology by digestion with proteinase K (PK) (0.5 mg/ml on ice for 60 min) were as described previously (Hegde et al., 1998a, and references therein). Cotranslational glycosylation of PrP was inhibited by inclusion in the translation reaction of a tripeptide inhibitor of glycosylation (Ac-Asn-Tyr-Thr-COOH, 100 μM final concentration), which has no discernible effect on either the translation or the topogenesis of PrP (Hegde et al., 1998a). Posttranslational translocation reactions (Figure 2A) were performed by first translating in the absence of RMs for 10 min, addition of 100 μM aurin tricarboxylic acid to inhibit initiation, incubation for an additional 20 min, and the addition of 100 μM emetine to inhibit further translation. RMs were then added at 1 eq/10-μl translation reaction and incubated at 32°C for 30 min. Subsequent analysis for translocation by protection from PK digestion was performed as for cotranslational translocation reactions. For Figure 2C, translation/translocation reactions were treated with 100 μM aurin tricarboxylic acid to inhibit further translational initiation after 3, 5, or 7 min of translation and rapidly chilled on ice. The RMs from the reaction were isolated by centrifugation (4 min at 50,000 rpm in a TLA100.1 rotor, Beckman Instruments, Palo Alto, CA) through a 100-μl cushion containing 0.5 M sucrose, 100 mM KAc, 50 mM HEPES, pH 7.4, 5 mM MgCl2, and resuspended in the original volume of fresh translation mix lacking RMs or transcript, and containing 100 μM aurin tricarboxylic acid. Translation was continued for 30 min at 32°C before analysis of topology by the PK protection assay. Translocation intermediates of 61, 93, 112, 137, and 180 residues in length were generated by translation of transcripts prepared from PrP (or mutant) plasmids digested with BstXI, KpnI, NgoMIV, Nsi1, or HincII, respectively. After translation for 30 min at 32°C, the RMs were isolated by sedimentation (4 min at 50,000 rpm in a TLA100.1 rotor through a 100-μl cushion containing 0.5 M sucrose, 100 mM KAc, 50 mM HEPES, pH 7.4, 5 mM MgCl2), and resuspended in 250 mM sucrose, 100 mM KAc, 50 mM HEPES, pH 7.4, 5 mM MgCl2. In preliminary experiments, these conditions were identified as being sufficient to sediment RMs (and their associated ribosome-nascent chain complexes), but not ribosomes or polysomes (our unpublished data; Matlack and Walter, 1995). Thus, ribosome-nascent chains that are not associated with RMs (e.g., those synthesized in the absence of RMs) were observed not to sediment under these conditions (our unpublished data). Protease protection assays of these translocation intermediates were with 0.5 mg/ml PK for 60 min on ice. Translocation reactions in which the two topogenic elements of PrP were presented to the translocon “simultaneously” (Figure 5) were performed using a ribosome-associated 254-mer of PrP. This was generated by transcription and translation of an NheI-digested plasmid encoding PrP(AV3) in which a silent NheI site was engineered at the stop codon. This truncated transcript is predicted to contain the entire coding region of PrP, but lacking the stop codon. After translation for 30 min at 32°C in the absence of RMs, emetine was added to inhibit further translation before the addition of RMs at 1 eq/10 μl. After incubation for 15 min at 32°C to allow translocation, the nascent chains were released with 1 mM puromycin for 15 min at 32°C before assessment of topology by PK digestion as described above. The “sequential” reaction was performed in parallel and involved the cotranslational inclusion of RMs from the beginning of the translation reaction. Subsequent treatments were the same as for the “simultaneous” reaction.
Chemical Cross-linking Studies
Translocation intermediates prepared and isolated as described above were treated with 0.5 mM disuccinimidyl suberate (added from a freshly prepared 20× stock solution in dimethyl sulfoxide) for 30 min at 25°C. The cross-linker was quenched with 0.1 M Tris, 0.1 M glycine, pH 8.0, transferred to ice, and subsequently adjusted to 10 mM EDTA and 1% saponin to disassemble the ribosomes, release the nascent chains, and extract the lumenal proteins. In preliminary experiments (our unpublished data), these extraction conditions were determined to result in the extraction of >90% of lumenal proteins (as assessed by both Coomassie staining and immunoblots for GRP94, BiP, and protein disulfide isomerase), while extracting <2% of membrane proteins (as assessed with immunoblots for TRAM and Sec61β). The microsomes in the sample were isolated by sedimentation through a 100-μl cushion containing 0.5 M sucrose, 100 mM KAc, 50 mM HEPES, pH 7.4, 5 mM MgCl2, as described above. The supernatant containing the lumenal cross-links was removed and precipitated with 15% trichloroacetic acid, the precipitate washed once with acetone, and dissolved in 1% SDS, 0.1 M Tris, pH 8. The membrane pellet was dissolved directly in 1% SDS, 0.1 M Tris, pH 8. Where indicated in the figure legends, proteins in the supernatant or pellet fractions were also subsequently analyzed by immunoprecipitation before analysis by SDS-PAGE.
Miscellaneous
Samples were analyzed by SDS-PAGE on 12% Tris-Tricine minigels, except those in Figure 3G, which were on 15% Tris-Glycine gels. Immunoprecipitations of PrP were performed using the 3F4 monoclonal antibody (mAb) as described previously (Hegde et al., 1998a). Polyclonal antiserum against pancreas-specific protein disulfide isomerase (PDIp) was the generous gift of M. Lan (Louisiana State University, Baton Rouge, LA) and has been characterized previously (DeSilva et al., 1997). Polyclonal antiserum (SPA-890) against a ubiquitously expressed isoform of PDI was from Stressgen (Victoria, British Columbia, Canada). Polyclonal antiserum against the C terminus of Sec61α was the generous gift of Kennan Kellaris and Reid Gilmore (University of Massachusetts Medical School, Worcester, MA). Polyclonal antiserum against the C terminus of TRAM was the generous gift of Kent Matlack and Peter Walter (University of California, San Francisco, San Francisco, CA). Figures were prepared by digitizing autoradiographs by using a UMAX Powerlook III flatbed scanner (UMAX Technologies, Dallas, TX) and by using Adobe Photoshop and Illustrator software (Adobe Systems, Mountain View, CA).
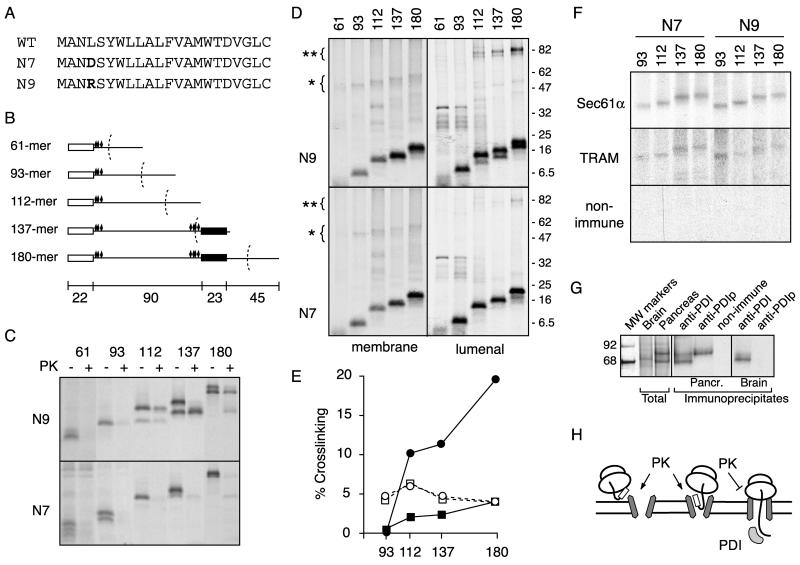
Figure 3.
Analysis of serial PrP translocation intermediates. (A) Sequences of the N7 and N9 signal sequence mutants of PrP that increase and decrease the relative proportion of CtmPrP, respectively. (B) Schematic diagram of translation intermediates of the indicated lengths (in amino acid residues). The approximate relative positions of the signal sequence (white box), transmembrane domain (black box), ribosome (dotted arc), and lysine residues (diamonds) are indicated. Lengths of each domain in amino acid residues are indicated on the scale below the diagrams. (C) Translocation intermediates of the indicated lengths were prepared for the N7- and N9-PrP constructs as described in MATERIALS AND METHODS, and analyzed for protection from cytosolically added PK. (D) Same intermediates as in C were analyzed by cross-linking with a homobifunctional lysine reactive cross-linker as described in MATERIALS AND METHODS. After cross-linking, samples were fractionated into lumenal and membrane cross-links (as described in MATERIALS AND METHODS) and analyzed separately. The position of prominent cross-links to membrane protein(s) of ∼35–40 kDa is indicated with an asterisk, and the position of prominent cross-links to lumenal proteins of ∼60 kDa is indicated with a double asterisk. (E) Amounts of the 35–40-kDa membrane protein cross-links (open symbols) and 60-kDa lumenal protein cross-links (closed symbols) were quantified (as a percentage of total membrane targeted nascent chains) and plotted as a function of chain length. Circles represent data from the N9 translocation intermediates, and squares from the N7 intermediates. (F) Membrane protein cross-links of the indicated sizes from the N7 and N9 intermediates were analyzed by immunoprecipitation with antibodies against Sec61α, TRAM, or an irrelevant antibody as indicated. (G) PrP 180-mer intermediates were assembled using RMs from either canine pancreas or mouse brain, as indicated, and subjected to chemical cross-linking as in D. Shown on the left are the total lumenal protein cross-links observed in brain or pancreas RMs. These samples were subjected to immunoprecipitation by using antibodies against PDIp, bovine liver PDI, or nonimmune serum as indicated. Molecular weight markers are indicated to the left. (H) Schematic diagram summarizing the protease protection and cross-linking results is shown: early translocation intermediates are accessible to PK and do not cross-link to lumenal proteins, whereas later intermediates that cross-link to lumenal proteins (predominantly PDI and PDIp) are found to be inaccessible to protease digestion. All translocation intermediates are depicted as being adjacent to the translocon, as suggested by the cross-linking data.
RESULTS
Topological Heterogeneity of PrP Is Generated at a Posttargeting Step
PrP topogenesis can be reconstituted in a rabbit reticulocyte lysate-based cell-free translation system containing ER-derived microsomes from canine pancreas (Hegde et al., 1998a). Although PrP is a glycoprotein, inhibition of its cotranslational glycosylation does not influence either its translation or topogenesis. Thus, both the wild type and various mutants of PrP achieve the exact same ratio of topological forms in the presence or absence of glycosylation (Hegde et al., 1998a). However, interpretation of the protease protection assays for PrP topology is more complicated in the presence of glycosylation due to heterogeneity of glycosylation site usage. For this reason, a tripeptide inhibitor of glycosylation (see MATERIALS AND METHODS) was included in the translation reactions presented in this study to prevent cotranslational glycosylation of PrP, thereby simplifying the banding patterns on SDS-PAGE.
To begin discriminating among the general models of PrP topogenesis shown in Figure 1, we first determined whether PrP, and in particular CtmPrP, could be generated by posttranslational translocation. We found that wild-type PrP was unable to generate any of the topological forms when the ER-derived rough microsomes were added posttranslationally to the translation reaction (Figure 2A). Because the amount of CtmPrP is rather low for wild-type PrP, even during cotranslational translocation, we also tested the transmembrane domain mutant PrP(AV3), which generates substantially more CtmPrP (Hegde et al., 1998a). Even for this mutant, we could not detect significant amounts of posttranslationally translocated PrP in any of the topological forms (Figure 2A).
Because the C-terminal hydrophobic segment involved in glycolipid anchor addition has been suggested to potentially function as an alternate targeting signal for posttranslational translocation (Hölscher et al., 2001), we also tested the effects of deleting this domain on the translocation and topology of wild-type PrP, PrP(AV3), and PrP(G123P), a TMD mutant incapable of being made in the transmembrane forms of PrP (Hegde et al., 1998a). As shown in Figure 2B, neither the topology nor overall translocation efficiency for any of these substrates was influenced by deletion of the C-terminal hydrophobic domain. Consistent with these observations, replacement of the entire C-terminal domain of PrP, including the glycolipid anchor addition signal, with sequences coding for the green fluorescent protein still resulted in the protein being made in all three topological forms in very similar ratios as wild-type PrP (our unpublished data). Thus, together with Figure 2A and the observation that disruption of the N-terminal signal sequence abrogates generation of all three topological forms (Kim et al., 2002), our results argue against the involvement of posttranslational translocation mechanisms in determining PrP topology.
Because cotranslational targeting could, in principle, be mediated by either the N-terminal signal sequence or the TMD, we next examined whether differential targeting by these two domains might play a role in PrP topogenesis (e.g., as in model II of Figure 1). We therefore wished to ascertain whether chains that had targeted to the membrane by a means other than the TMD would nonetheless be able to generate all three topological forms, particularly CtmPrP. To do this, we first determined in preliminary experiments the length of time required to synthesize up to residues 109–112 (recognized by the 3F4 mAb; Rogers et al., 1991) and residues 138–141 (recognized by the 13A5 mAb; Rogers et al., 1991), epitopes that flank the TMD (residues 113–135). We found that at least 5 min of translation is required before these two epitopes (and therefore the TMD) even begin to be synthesized (our unpublished data). This identified a time point before which any targeting of nascent chains would necessarily have occurred via a mechanism not involving the TMD.
We next conducted translations for varying periods of time (both shorter than and longer than 5 min), and isolated the membrane-targeted nascent chains. Chains targeted at time points earlier than or equal to 5 min have presumably targeted via either the N-terminal signal sequence, or alternatively, due to the natural affinity of the ribosome for the translocation channel at physiological salt conditions (Kalies et al., 1994; Potter and Nicchitta, 2000). Although the nascent chains probably represent a heterogeneous mixture of lengths, they share in common the feature that they could not have targeted via the TMD, which has not yet been synthesized. These membrane-targeted nascent chains, were then allowed to complete their synthesis and translocation in fresh translation extract in the presence of an inhibitor of translational initiation. We found that PrP or PrP(AV3) synthesized in this manner, in which it had been forced to target to the microsomes before synthesis of the TMD, nonetheless was able to generate CtmPrP at levels comparable with a standard translocation reaction (Figure 2C). These results indicate that each of the topological forms of PrP can arise from a translocation intermediate generated by targeting to the ER membrane at a point before synthesis of the TMD. This is consistent with previous observations that disruption of the targeting function of the PrP signal sequence abrogates the generation of all of the topological forms (Rutkowski et al., 2001; Kim et al., 2002). Additionally, the TMD in PrP seems to be incapable of serving as either an internal signal sequence or signal anchor, because its placement in a heterologous context is insufficient to mediate targeting or translocation of the reporter (DeFea et al., 1994). Taken together, these previous observations and the data in Figure 2 argue that the key events in generating topological heterogeneity of PrP occur at steps in translocation after signal-mediated targeting to the translocon, in a manner consistent with model III (Figure 1).
A Posttargeting Role for PrP Signal Sequence in Initiating N Terminus Translocation
After targeting to the ER translocon, PrP nascent chains must subsequently be segregated into a population that has the N terminus in the lumen (eventually giving rise to secPrP and NtmPrP), versus a population with the N terminus in the cytoplasm (as is seen with CtmPrP). On the basis of the analysis of various signal sequence mutations, we have speculated previously that this domain may play a role in determining localization of the N terminus (Kim et al., 2001). To examine this putative step in PrP topogenesis, we prepared and examined a series of translocation intermediates of two signal sequence mutants of PrP, termed N7 and N9, which differ at a single amino acid (Figure 3A). Although both signals seem to be equally functional in their targeting role (Kim et al., 2002), the N7 mutant generates substantially more CtmPrP than the N9 mutant (∼35 vs. ∼5%; Kim et al., 2001). We therefore reasoned that by comparing the sequence of events occurring at the translocon for these two mutants, we could gain insight into the specific step(s) that may lead to determination of the final topology of PrP.
Translocation intermediates of between 61 and 180 residues (Figure 3B) were assembled, the microsomes containing these intermediates were isolated (see MATERIALS AND METHODS), and the state of the nascent chain was probed by either protease protection or cross-linking assays. Protease protection assays (Figure 3C) revealed that early translocation intermediates (of 61 and 93 residues) of both N7 and N9 were accessible to cytosolically added PK. However, at later points in synthesis (beginning at 112 residues), the N9 but not N7 construct achieved a state where a substantial proportion of nascent chains was not accessible to cytosolic PK. In addition, we observed that the 137- and 180-mer intermediates of the N9 construct have a greater proportion of chains with their signal sequences cleaved than the corresponding N7 intermediates. Given that signal cleavage occurs on the lumenal side of the ER membrane, this observation, together with the protease protection assays, suggest that the N9 intermediates have initiated translocation into the lumen with greater efficiency than the matched N7 intermediates.
These same translocation intermediates were analyzed in parallel by chemical cross-linking with a lysine-reactive homo-bifunctional reagent. After cross-linking, the nascent chains were released from the ribosome with EDTA, and the products were fractionated by saponin extraction to separate cross-links to integral membrane proteins, which are not extracted by saponin, from cross-links to lumenal proteins, which are efficiently extracted by saponin. Analysis of the cross-links to integral membrane proteins revealed that for each of the translocation intermediates, both constructs cross-linked equally efficiently to protein(s) of ∼35–40 kDa (Figure 3D, indicated by the single asterisks). Quantitation of these cross-links (Figure 3E) indicated that at each intermediate for both constructs, ∼5% of total chains were cross-linked to these membrane proteins of ∼35–40 kDa. Immunoprecipitation studies confirmed that these bands predominantly represent cross-links to the core translocon component Sec61α, and to a lesser degree TRAM (Figure 3F), suggesting that both substrates are docked at the translocation channel.
In contrast, substantial differences between N7 and N9 could be observed for cross-links to ER lumenal proteins. Beginning at the 112-mer intermediate, N9 was seen to cross-link much more strongly than N7 to protein(s) of ∼60 kDa (Figure 3D, indicated by the double asterisks; quantitated in Figure 3E). Purification of the predominant lumenal cross-linking partner of ∼60 kDa (Hegde, unpublished data) revealed it to be a previously characterized pancreatic homolog of the protein disulfide isomerase, termed PDIp (DeSilva et al., 1997; Volkmer et al., 1997). Antibodies against this protein were able to specifically immunoprecipitate the primary lumenal cross-linked adduct with PrP (Figure 3G). In addition, the slightly lower molecular weight protein also seems to be a member of the protein disulfide isomerase family and could be immunoprecipitated by a commercially available polyclonal antibody raised against purified bovine liver PDI (Figure 3G). Cross-links to PDI were also observed using microsomes isolated from other tissues such as mouse brain (Figure 3G), indicating that the interaction between PrP and PDIp is not unique to the pancreatic homolog of PDI, but to members of the PDI family in general. These data establish that the principal lumenal cross-links observed in the analysis of various translocation intermediates of PrP (Figure 4) are members of the PDI family of ER lumenal molecular chaperones.
Figure 4.
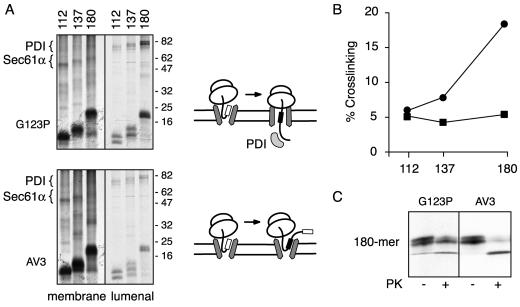
Analysis of TMD mutants by cross-linking. Translocation intermediates of the indicated lengths of the PrP(AV3) and PrP(G123P) mutants were prepared and analyzed by cross-linking as in Figure 3D. The positions of cross-links to Sec61α and PDI are indicated. Schematic diagrams of the results are indicated to the right of the autoradiographs: upon increased synthesis, PrP(G123P) but not PrP(AV3) displays increased cross-linking to PDI. The relative efficiencies of cross-linking to PDI for the PrP(G123P) (circles) and PrP(AV3) (squares) translocation intermediates was quantitated and plotted in B. (C) Translocation intermediates of 180 residues for PrP(G123P) and PrP(AV3) were analyzed by protease protection as in Figure 3C.
Taken together, the proteolysis and cross-linking experiments suggest that until the synthesis of between ∼93 and 112 residues, the nascent N7 and N9 signal mutants of PrP are similar in their biogenesis: both have targeted to the membrane and docked at the translocon (as indicated by cross-links to translocon proteins), but have not yet initiated translocation into the ER lumen (as suggested by cytosolic accessibility of the nascent chain and lack of cross-linking to ER lumenal chaperones). At 112 residues, however, the N9 nascent chain seems to begin initiation of translocation, resulting in its shielding from the cytosol and concomitant access to the lumen (Figure 3H, diagram). As nascent chain length increases, stronger cross-links to lumenal chaperones are seen, perhaps indicating that a higher proportion of nascent chains have initiated translocation. In contrast, the CtmPrP-favoring N7 mutant seems to either be inefficient or protracted in its initiation of translocation. The majority of these nascent chains therefore remain accessible to the cytosol, cross-linking to lumenal chaperones is poor (although at longer nascent chain lengths, such cross-links are observed), and signal sequence cleavage is poor. The data in Figure 3 suggest that determinants in the PrP signal sequence influence the initiation of translocation, the efficiency of which correlates inversely with the eventual generation of CtmPrP.
TMD-mediated Interference of N Terminus Translocation
In addition to the signal sequence, the TMD also plays a key role in PrP topogenesis. Mutations within this domain can diminish or enhance generation of the transmembrane forms of PrP (Yost et al., 1990; Hegde et al., 1998a, 1999; Kim et al., 2001). To understand what role the TMD might play during PrP translocation, we used cross-linking to analyze translocation intermediates of topology-altering mutants within the TMD. The two mutants chosen for analysis were PrP(G123P) (Figure 2B), which abolishes formation of the transmembrane forms, and PrP(AV3) (Figure 2B), which generates increased amounts of the transmembrane forms. We analyzed three lengths of translocation intermediates: 112 amino acids, at which point the TMD (and hence the mutation) has not yet been synthesized; 137 amino acids, when the TMD has been fully synthesized but remains within the ribosomal tunnel; and 180 amino acids, a point when the TMD has emerged from the ribosome and can potentially interact with the translocation apparatus.
Comparison of the cross-linking patterns for PrP(AV3) and PrP(G123P) revealed one principal difference (Figure 4, A and B). Although the cross-links to the lumenal protein PDI increased with increasing nascent chain length for PrP(G123P), they remained relatively constant for PrP(AV3). Thus, at the 112-mer intermediate, a certain proportion of the nascent chains have initiated N-terminal translocation, resulting in cross-linking to PDI (at ∼5% efficiency; Figure 4, A and B). Synthesis of another 25 or 68 residues (i.e., up to the 137-mer and 180-mer intermediates, respectively) results in increasing proportions of nascent PrP(G123P) chains cross-linking to PDI (with up to ∼20% efficiency). In contrast, continued translation of PrP(AV3) (from the 112-mer to 180-mer intermediate) did not result in a higher proportion of cross-links to PDI. Cross-links to membrane proteins of the translocon were comparable for both AV3 and G123P, indicating that despite differences in PDI cross-linking, both substrates are at the translocation channel to a similar extent.
One interpretation of these cross-linking data is that upon synthesis of a functional TMD, it has the potential to interfere with the initiation of N terminus translocation for those nascent chains that have not already done so. Such a model would explain the lack of additional cross-linking to PDI upon synthesis of the PrP(AV3) TMD (by residue 137), whereas a nonfunctional TMD such as PrP(G123P) shows the increase in cross-linking that would accompany N-terminus translocation of additional nascent chains. This suggests that during the period of chain growth from 112 to 180 residues, the PrP(G123P) substrate is better able to initiate translocation of the N terminus of PrP than the PrP(AV3) substrate. Analysis of the 180-mer intermediates of PrP(G123P) and PrP(AV3) by proteolysis corroborated this conclusion. Although the PrP(G123P) intermediate was largely protected from cytosolic protease, the PrP(AV3) was substantially more accessible (Figure 4C). Thus, synthesis of a nonfunctional TMD [in the case of PrP(G123P)] results in increased access of nascent chains to the ER lumen and decreased exposure to the cytosol, whereas synthesis of a functional TMD seems to interfere with this process, resulting in decreased lumenal access and increased cytosolic exposure. A schematic diagram representing this idea is shown to the right of the autoradiograph in Figure 4A.
We further examined the relationship between the TMD and N-terminus translocation in two ways. In the first experiment, we asked whether the final topology of PrP would be altered if the signal and TMD were presented to the translocon simultaneously, as opposed to the sequential manner in which they are ordinarily presented during cotranslational translocation (Figure 5A). If the time between the synthesis of the signal and emergence of the TMD is important for the signal to initiate N-terminal translocation without interference, we reasoned that taking away this temporal advantage might result in increased CtmPrP relative to secPrP. To present the signal and TMD to the translocon simultaneously, we prepared ribosome-associated nascent chains containing full-length PrP. These were then presented to RMs, after which the nascent chains were released from the ribosome with puromycin before analysis of the topology that was achieved. A cotranslational translocation reaction of the same construct was performed in parallel for the “sequential” mode of presentation of the signal and TMD. When this analysis was performed on PrP(AV3), we observed that the simultaneous mode of presentation resulted in lower overall translocation efficiency (Figure 5B), as is commonly observed with longer nascent chains (Perara et al., 1986; Roitsch and Lehle, 1988). However, of the nascent chains that were translocated, a higher proportion of them was made in the CtmPrP topology with the simultaneous mode of presentation than with the sequential mode. Indeed, the CtmPrP-to-secPrP ratio changed from 1.2 with the sequential to 4.7 with the simultaneous mode of translocation (Figure 5B).
In another type of experiment designed to explore the temporal relationship between the signal and TMD interactions with the translocon, we examined the behavior of constructs in which the number of residues between the signal sequence and TMD was varied (Figure 5C). Targeting of these constructs should be mediated by the N-terminal signal sequence, because PrP is unable to be translocated efficiently in the absence of a functional signal Rutkowski et al., 2001; Kim et al., 2002). After targeting, however, the number of residues separating the signal from the TMD determines the length of time that the signal has to initiate N-terminal translocation before emergence of the TMD. Given that the initiation of N-terminal translocation is an important determinant of PrP topology (Figure 3) and that the TMD can interfere with this step (Figure 4), we reasoned that changing the number of residues separating these two domains should have a predictable effect on topology. As seen in Figure 5D, the percentage of PrP made in the CtmPrP form varies inversely with the number of amino acid residues separating the signal and TMD. This indicates that as the time period between interaction of the signal with the translocon and emergence of the TMD is increased, the ability to make CtmPrP decreases. It seems that by giving the signal sequence more time before synthesis of the TMD, it is better able to initiate translocation of the N terminus, which precludes the generation of CtmPrP. Taken together, the observations presented in Figure 5 are consistent with the notion that the timing of the initiation of N-terminus translocation, mediated by the signal sequence, in relation to the emergence of the TMD is an important aspect of PrP topogenesis.
Reversal of Disease-associated TMD Mutants
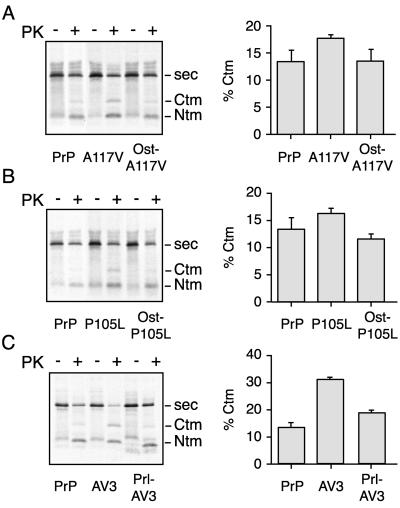
A variety of naturally occurring mutations in PrP are associated with genetic forms of prion disease (Prusiner, 1997; Prusiner, 1998; Collinge, 2001). Two of these mutations (an alanine-to-valine substitution at position 117 and a proline-to-leucine substitution at position 105) increase the hydrophobicity of a residue adjacent to or within the TMD and result in a slight increase in the generation of CtmPrP when analyzed in the in vitro translocation system (Hegde et al., 1998a; Figure 6, A and B). The data presented thus far provide a plausible hypothesis for how these mutants might act. On its emergence from the ribosome, the mutant TMD, being slightly more hydrophobic than wild type, is able to more effectively interfere with the signal-mediated N-terminal translocation of chains that have not yet completed this step. This slight competitive advantage, in a manner similar to but less extreme than PrP(AV3), would result in the slightly higher percentage of CtmPrP observed.
Figure 6.
Reversal of disease-associated TMD mutants of PrP. The translocation and topology of wild-type PrP was compared with TMD mutants of PrP containing either the PrP signal sequence or the signal sequence from another protein (either osteopontin or prolactin, as indicated). (A–C) Analyses for the PrP(A117V), PrP(P105L), and PrP(AV3) TMD mutants. Shown to the right of each autoradiograph is a quantitation of the amount of CtmPrP from an experiment in which the analysis was performed in triplicate.
We therefore reasoned that the manifestation of these TMD mutants might be preemptively avoided if the function of the signal sequence in initiating N-terminal translocation could be made more efficient or to occur earlier in translocation. Recently, a comparison of signal sequences from a variety of different substrates suggested that they differ substantially in their timing and efficiency of initiating N-terminal translocation (Kim et al., 2002). Of the various signals analyzed, some, such as the signal sequences from the proteins osteopontin or prolactin, were more efficient than the PrP signal at carrying out this step. We asked whether, by simply replacing the PrP signal sequence with that of osteopontin or prolactin, the increased CtmPrP conferred by the disease-associated TMD mutants could be reversed.
Remarkably, in the case of both PrP(A117V) and PrP(P105L), the levels of CtmPrP could be normalized to wild-type levels if the osteopontin signal sequence was used (Figure 6, A and B). Even the substantial increase in CtmPrP conferred by the PrP(AV3) mutant, which causes severe and rapid onset of neurodegenerative disease in transgenic mice (Hegde et al., 1998a), could be completely reversed to wild-type levels by using the signal sequence from prolactin (Figure 6C). In this case, the prolactin signal has been well documented in several studies (Crowley et al., 1993; Jungnickel and Rapoport, 1995; Rutkowski et al., 2001) to initiate efficient N-terminal translocation at a very early point after targeting (after synthesis of only ∼70 residues). Thus, the increased generation of CtmPrP seen with disease-associated mutations in the TMD of PrP can be masked by modulation of an earlier, signal-mediated step in translocation. This finding further substantiates the idea that the action of the TMD in determining PrP topology is intimately dependent on the efficiency and timing of the posttargeting function of the signal sequence in initiating N-terminus translocation.
DISCUSSION
The data presented in this study allow one to construct a plausible working model of the mechanisms involved in directing PrP topology. The first step, cotranslational targeting of nascent PrP via the N-terminal signal sequence to their sites of translocation at the membrane, seems to be shared among each of the topological forms. Not only is each of the topological forms capable of arising from a nascent chain targeted before the synthesis of the TMD (Figure 2C) but also disruption of the targeting function of the signal abrogates the generation of all of the topological forms (Kim et al., 2002). This is consistent with the observation that in a heterologous context, the TMD cannot function as either an internal signal sequence or a signal anchor to mediate translocation (DeFea et al., 1994). Furthermore, translocation by an alternative posttranslational pathway seems to be very inefficient (Hölscher et al., 2001; our unpublished observations). Although forced usage of such an alternative targeting strategy may, under some experimental conditions, inefficiently generate CtmPrP, it need not be invoked to explain the generation of CtmPrP.
Once at the translocon, an initial population of topologically homogeneous nascent chains eventually gives rise to the different topological forms observed. The data in this study suggest that this is achieved through two sequential partitioning events that each involve interactions between the nascent chain and the translocon. The first event, controlled by the signal sequence, segregates nascent chains into two populations: 1) a subset that initiates N-terminus translocation into the ER lumen at a point preceding the emergence of the TMD, and 2) the remainder of nascent chains for which the TMD fully emerges from the ribosome (by ∼165 residues of synthesis, assuming that ∼30 residues are within the ribosomal tunnel) before the N terminus has initiated translocation and contacted lumenal chaperones. Each of these sets of nascent chains is subsequently subjected to a second partitioning event, controlled in part by the TMD.
For population 1 mentioned above, the N terminus has already been committed to the ER lumen by the time the TMD is synthesized and enters the translocation channel. Thus, the efficiency of integration of the TMD into the lipid bilayer presumably determines what fraction of these nascent chains eventually become secPrP (if the TMD fails to integrate into the lipid bilayer) versus NtmPrP. Integration of a transmembrane domain is thought to be controlled by both the Sec61 complex and features of the TMD (Heinrich et al., 2000).
For population 2 mentioned above, two topological elements (the signal and TMD) are simultaneously present in the vicinity of the translocon. Because the net charge differential of residues flanking the TMD is heavily positive (+5) on the N-terminal side, the orientation predicted (Hartmann et al., 1989; Sipos and von Heijne, 1993) to be preferred by the TMD (type II or Ncyt/Cexo) is contradictory to that preferred by the signal (whose action is generally to initiate translocation of the N terminus). The outcome of “competition” between these two elements determines the fraction of these nascent chains partitioned to become CtmPrP (due to dominance of the TMD). It therefore seems that disease-associated mutations in the TMD that result in increased generation of CtmPrP act by providing the TMD with a slight competitive advantage during this second partitioning step.
Thus, the proportion of PrP made as CtmPrP is controlled by a combination of two successive cotranslational partitioning events. Because mutations in the TMD act at the second of these two events, they can be preemptively neutralized by modulation of the earlier, signal-mediated step (Figure 6). Taken together, these data indicate that the timing and/or efficiency of the signal sequence in initiating N-terminal translocation sets an upper limit on the proportion of chains that have the potential to become CtmPrP, whereas features of the TMD determine the extent to which this potential is realized.
The ability to modulate the generation of CtmPrP by changing the signal sequence allows the altered topology of otherwise pathogenic mutants to be reversed to wild-type levels. This insight should allow the dissociation of the effects of altered topology on neurodegeneration from other potential effects of the mutation itself. It will be of substantial interest to determine whether in transgenic mice, the severe neurodegenerative phenotype of mutants such as PrP(AV3) (Hegde et al., 1998a) can be completely or partially alleviated simply by changing the signal sequence to one that reduces the generation of CtmPrP. If this proves true then the posttargeting, signal-mediated step in PrP translocation may represent a point for intervention of at least a subset of prion diseases.
At present, the mechanisms by which features of a signal sequence are recognized to regulate the initiation of N-terminal translocation are not known. Although both Sec61 and the TRAM protein have been implicated in signal sequence recognition (High et al., 1993; Jungnickel and Rapoport, 1995; Voigt et al., 1996; Mothes et al., 1998), it is unclear how these and/or other components of the translocon differentially interact with various signal sequences to achieve the observed diversity of function. Whether there are other components of the translocon that, although not essential for translocation per se, influence the timing or efficiency of events such as signal-mediated initiation of translocation remains to be determined. In support of such a notion, it seems that proper PrP topogenesis requires membrane proteins in addition to Sec61, TRAM, and the signal recognition particle-receptor (Hegde et al., 1998b). The identification and characterization of such putative trans-acting factors implicated in regulating PrP topogenesis may therefore shed light on more general aspects of translocation such as control of the initiation of translocation.
On the basis of the data in the present study, the functional relevance of the interaction between the N terminus of PrP with PDI orthologues in the ER lumen remains unclear. It is possible that by binding the N terminus upon its exposure to the ER lumen, PDI (and/or other lumenal proteins) either actively pulls the nascent chain in, or prevents its slippage out of the lumen, thereby providing directionality to the transport process. Consistent with this idea, PrP translocation into proteoliposomes containing total ER membrane proteins (but lacking lumenal proteins) generates substantially less of the secPrP and NtmPrP forms (but normal levels of CtmPrP) compared with unfractionated microsomes (Hegde et al., 1998b). Although the basis of this deficit is not presently clear, one possibility is that a lack of lumenal proteins results in inefficient N-terminal translocation. If this is the case, it is tempting to speculate that generation of the potentially toxic CtmPrP form could be modulated by conditions of ER stress due to the titration of lumenal chaperones by increased levels of unfolded proteins.
Is cotranslational partitioning into multiple nascent populations a property unique to PrP biogenesis? Evidence that other naturally occurring membrane proteins may use similar mechanisms during their biogenesis has been provided by studies of the MDR1 protein. In this protein, the orientation favored by the eighth transmembrane (TM) segment is highly dependent on the manner in which it is presented to the translocon (Moss et al., 1998). This step is in turn dependent on both the action of the previous transmembrane segment (TM7b), as well as determinants in the intervening sequence. Because TM7b seems to be heterogeneous in its ability to partition into the lipid bilayer upon its entry into the translocon, the orientation taken by TM8 as well as the final protein is heterogeneous (Skach et al., 1993; Moss et al., 1998). Thus, for both TM8 in MDR1 and the TMD in PrP, information regarding the orientation it should take at the membrane is partially encoded by sequences in or around the transmembrane domain, and partially by the action of the previous topogenic element. It seems that PrP topogenesis may recapitulate, in a simplified form, certain events that occur during the biogenesis of substantially more complex membrane proteins. Thus, the present study provides insight into not only the mechanism of PrP topology determination and how this can be influenced in certain neurodegenerative disease, but also into the general question of how successive events during translocation can functionally interact and are coordinated to determine a protein's topology at the ER membrane.
ACKNOWLEDGMENTS
We thank Jeff Salerno and Devarati Mitra for help with some of the constructs, Michael Lan for antisera to PDIp, Kent Matlack and Peter Walter for antisera to TRAM, Kennan Kellaris and Reid Gilmore for antisera to Sec61α, and various members of the Hegde laboratory for stimulating discussions. This work was supported by the Intramural Research Program of the National Institutes of Health.
Abbreviations used:
- ER
endoplasmic reticulum
- PDI
protein disulfide isomerase
- PK
proteinase K
- PrP
prion protein
- RM
rough microsome
- TMD
transmembrane domain
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0293. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0293.
REFERENCES
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- DeFea KA, Nakahara DH, Calayag MC, Yost CS, Mirels LF, Prusiner SB, Lingappa VR. Determinants of carboxyl-terminal domain translocation during prion protein biogenesis. J Biol Chem. 1994;269:16810–16820. [PubMed] [Google Scholar]
- DeSilva MG, Notkins AL, Lan MS. Molecular characterization of a pancreas-specific protein disulfide isomerase, PDIp. DNA Cell Biol. 1997;16:269–274. doi: 10.1089/dna.1997.16.269. [DOI] [PubMed] [Google Scholar]
- Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol. 1999;147:257–265. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Lingappa VR. Regulation of protein biogenesis at the endoplasmic reticulum membrane. Trends Cell Biol. 1999;9:132–137. doi: 10.1016/s0962-8924(99)01504-4. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998a;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prusiner SB, Lingappa VR. Transmembrane and genetic prion diseases share a common pathway of neurodegeneration involving transmembrane prion protein. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Voigt S, Lingappa VR. Regulation of protein topology by trans-acting factors at the endoplasmic reticulum. Mol Cell. 1998b;2:85–91. doi: 10.1016/s1097-2765(00)80116-1. [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- High S, Martoglio B, Gorlich D, Andersen SS, Ashford AJ, Giner A, Hartmann E, Prehn S, Rapoport TA, Dobberstein B, Brunner J. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- Hölscher C, Bach UC, Dobberstein B. Prion protein contains a second endoplasmic reticulum targeting signal sequence located at its C terminus. J Biol Chem. 2001;276:13388–13394. doi: 10.1074/jbc.M007331200. [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A post-targeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;71:489–503. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kalies KU, Gorlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Rahbar R, Hegde RS. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J Biol Chem. 2001;276:26132–26140. doi: 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Walter P. The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. J Biol Chem. 1995;270:6170–6180. doi: 10.1074/jbc.270.11.6170. [DOI] [PubMed] [Google Scholar]
- Mishra RS, Gu Y, Bose S, Verghese S, Kalepu S, Singh N. Cell surface accumulation of a truncated transmembrane prion protein in GSS P102L. J Biol Chem. 2002;277:24554–24561. doi: 10.1074/jbc.M200213200. [DOI] [PubMed] [Google Scholar]
- Moss K, Helm A, Lu Y, Bragin A, Skach WR. Coupled translocation events generate topological heterogeneity at the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2681–2697. doi: 10.1091/mbc.9.9.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W, Jungnickel B, Brunner J, Rapoport TA. Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J Cell Biol. 1998;142:355–364. doi: 10.1083/jcb.142.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perara E, Rothman RE, Lingappa VR. Uncoupling translocation from translation: implications for transport of proteins across membranes. Science. 1986;232:348–352. doi: 10.1126/science.3961485. [DOI] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J Biol Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M, Serban D, Gyuris T, Scott M, Torchia T, Prusiner SB. Epitope mapping of the Syrian hamster prion protein utilizing chimeric and mutant genes in a vaccinia virus expression system. J Immunol. 1991;147:3568–3574. [PubMed] [Google Scholar]
- Roitsch T, Lehle L. Post-translational translocation of polypeptides across the mammalian endoplasmic reticulum membrane is size and ribosome dependent. Eur J Biochem. 1988;174:699–705. doi: 10.1111/j.1432-1033.1988.tb14154.x. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Lingappa VR, Hegde RS. Substrate-specific regulation of the ribosome-translocon junction by N-terminal signal sequences. Proc Natl Acad Sci USA. 2001;98:7823–7828. doi: 10.1073/pnas.141125098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- Skach WR, Calayag MC, Lingappa VR. Evidence for an alternate model of human P-glycoprotein structure and biogenesis. J Biol Chem. 1993;268:6903–6908. [PubMed] [Google Scholar]
- Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- Stewart RS, Drisaldi B, Harris DA. A transmembrane form of the prion protein contains an uncleaved signal peptide and is retained in the endoplasmic reticulum. Mol Biol Cell. 2001;12:881–889. doi: 10.1091/mbc.12.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RS, Harris DA. Most pathogenic mutations do not alter the membrane topology of the prion protein. J Biol Chem. 2001;276:2212–2220. doi: 10.1074/jbc.M006763200. [DOI] [PubMed] [Google Scholar]
- Tateishi J, Kitamoto T. Inherited prion diseases and transmission to rodents. Brain Pathol. 1995;5:53–59. doi: 10.1111/j.1750-3639.1995.tb00577.x. [DOI] [PubMed] [Google Scholar]
- Tateishi J, Kitamoto T, Kretzsxhmar H, Mehraein P. Immunhistological evaluation of Creutzfeldt-Jakob disease with reference to the type PrPres deposition. Clin Neuropathol. 1996;15:358–360. [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer J, Guth S, Nastainczyk W, Knippel P, Klappa P, Gnau V, Zimmerman R. Pancreas specific protein disulfide isomerase, PDIp, is in transient contact with secretory proteins during late stages of translocation. FEBS Lett. 1997;406:291–295. doi: 10.1016/s0014-5793(97)00288-3. [DOI] [PubMed] [Google Scholar]
- Yost CS, Lopez CD, Prusiner SB, Myers RM, Lingappa VR. Non-hydrophobic extracytoplasmic determinant of stop transfer in the prion protein. Nature. 1990;343:669–672. doi: 10.1038/343669a0. [DOI] [PubMed] [Google Scholar]