Abstract
In cycling between the mammalian host and the tsetse fly vector, African trypanosomes undergo adaptive differentiation steps that are coupled to growth control. The signaling pathways underlying these cellular processes are largely unknown. Mitogen-activated protein kinases (MAPKs) are known mediators of growth and differentiation in other eukaryotic organisms. To establish the function of a MAPK homologue, TbMAPK2, in T. brucei, a null mutant was constructed. Bloodstream forms of a Δmapk2/Δmapk2 clone were able to grow normally and exhibited no detectable phenotype. When these cells were triggered to differentiate in vitro, however, they developed to the procyclic (fly midgut) form with delayed kinetics and subsequently underwent cell cycle arrest. Introduction of an ectopic copy of the TbMAPK2 gene into the null mutant restored its ability to differentiate and to divide. In contrast, a TbMAPK2 mutant, in which the T190 and Y192 residues of the activating phosphorylation site were replaced by A and F, was unable to restore the growth and differentiation phenotypes. Analysis of the DNA content and the nucleus/kinetoplast configuration of individual cells showed that the null mutant was arrested in all phases of the cell cycle and that 25–30% of the cells had failed to segregate their nucleus and kinetoplast correctly. This implies that cell cycle progression by the procyclic form depends on a constitutive stimulus exerted by the signaling cascade operating through TbMAPK2.
INTRODUCTION
The protozoan parasite Trypanosoma brucei, which causes human sleeping sickness and Nagana in domestic animals, depends on the tsetse fly for its dissemination. During cyclical transmission, trypanosomes undergo differentiation through an ordered series of distinct stages that are highly adapted to their respective environments (Vickerman, 1985). It is extremely important for the parasite to control growth and differentiation processes accurately as this is required for long-term survival in the mammalian host and successful transmission by the tsetse fly vector. Uncontrolled growth, or delayed or premature differentiation of the parasite, would either lead to rapid death of the host or the fly vector or to elimination of the parasite.
Throughout the life cycle of Trypanosoma brucei proliferating stages alternate with stages arrested in the G0 phase of the cell cycle (Mottram, 1994). At high parasite density in the blood, the proliferating long slender bloodstream form differentiates to the nondividing short stumpy form and thereby limits its growth in the mammalian host (reviewed by Matthews, 1999). When bloodstream forms are ingested by the tsetse, the short stumpy form, which is preadapted for survival in the fly, rapidly differentiates to the proliferating procyclic form in the fly midgut. The parasite continues its life cycle in the insect, finally giving rise to the nonproliferative metacyclic form in the salivary glands, which is capable of infecting a new host.
Differentiation of the bloodstream to the procyclic form can be induced in vitro by lowering the incubation temperature from 37 to 27°C and by the addition of cis-aconitate to the culture medium. Bloodstream form cells express a coat of variant surface glycoproteins (VSG) that is replaced by a different coat composed of two major classes of glycoproteins, the EP and GPEET procyclins, when cells differentiate to the procyclic form (Roditi et al., 1989; Ruepp et al., 1997). The short stumpy bloodstream form, which is arrested in the G0 phase of the cell cycle, differentiates rapidly and synchronously to the procyclic form (Ziegelbauer et al., 1990; Matthews and Gull, 1994; Vassella et al., 1997a). In contrast, differentiation of the proliferating long slender bloodstream form is asynchronous (Matthews and Gull, 1994) and proceeds via the short stumpy bloodstream form as an intermediate stage (Tasker et al., 2000). In the course of syringe passage between rodents bloodstream forms gradually lose their ability to differentiate to the stumpy form in vivo (and thus become monomorphic) but are still able to differentiate asynchronously to the procyclic form (Roditi et al., 1989; Matthews and Gull, 1994).
The different cellular events that occur during differentiation always appear in the same temporal order and with similar kinetics, making them suitable markers for mapping the different phases of this process (reviewed by Hendriks et al., 2000). Expression of procyclins and release of the VSG coat are considered to be early markers of differentiation. Repositioning of the kinetoplast (the genome of the single mitochondrion) to a nucleus proximal location and progression through S-phase are intermediate events. Expression of the procyclic-specific, cytoskeleton-associated protein CAP5.5 is a late marker of differentiation. Despite these useful markers, surprisingly little is known about the molecular mechanisms involved in these differentiation steps. Although cis-aconitate is an efficient trigger of differentiation in vitro, it is unlikely to be the signal for differentiation in the fly. Subjecting cells to acidic stress (Rolin et al., 1998) or treatment with proteases (Hunt et al., 1994; Sbicego et al., 1999) can also induce differentiation, but it is not known how these are translated into the differentiation signal.
Mitogen-activated protein kinases (MAPKs) play a central role in the regulation of cell growth and differentiation in eukaryotes (for review see Waskiewicz and Cooper, 1995). They are activated by stimuli such as extracellular factors or stress and form part of phosphorylation cascades that relay the external signal to the nucleus, thereby resulting in changes in gene expression. All MAPKs contain a conserved motif TXY in the regulatory loop that is phosphorylated by dual-specific threonine-tyrosine protein kinases (MEKs; Waskiewicz and Cooper, 1995). According to the central amino acid within this motif, they are further subdivided into extracellular-signal-regulated kinase (ERK), p38 and c-JUN NH2-terminal kinase. MAPKs phosphorylate numerous cellular proteins. These include cell surface proteins, cytoskeletal proteins, metabolic enzymes, components of signal transduction pathways and factors controlling transcription, mRNA stability, or translation (reviewed by Guan, 1994; and Whitmarsh and Davis, 2000). In contrast to higher eukaryotes, little is known about the role of MAP kinases in trypanosomatids and nothing about their specific substrates. In T. brucei, KFR1, an ERK homologue most closely related to the yeast kinases KSS1/FUS3, has been characterized biochemically (Hua and Wang, 1997). The kinase activity of the enzyme, which is higher in the bloodstream form than in the procyclic form, is decreased by serum starvation and induced by interferon-γ. LMPK, a MAP kinase homologue from a related parasite, Leishmania mexicana, is not required for the growth of promastigotes in the insect vector (Wiese, 1998). Null mutants are able to differentiate to amastigotes in infected macrophages but these cells are unable to grow (Wiese, 1998).
We have investigated the role of a new MAP kinase, TbMAPK2, in African trypanosomes. By generating a null mutant in bloodstream form trypanosomes and triggering these cells to differentiate to the procyclic form, we uncovered two TbMAPK2-specific phenotypes. Null mutants developed to the procyclic form with delayed kinetics and the newly differentiated cells were unable to divide. To our surprise these cells were arrested in all phases of the cell cycle. This indicates that procyclic form trypanosomes require sustained TbMAPK2 activity for progression through each phase of the cell cycle.
MATERIALS AND METHODS
Trypanosomes
Monomorphic bloodstream forms of T. brucei 427 (MITat 1.2; 221; Cross and Manning, 1973) and mutants derived from this clone were cultured according to Hesse et al. (1995) at 37°C/5% CO2. The GUSone cell line (Sbicego et al., 1999), in which the coding region of one copy of the EP1 procyclin gene was replaced by the Escherichia coli β-glucuronidase (GUS) gene, was used for generating TbMAPK2 deletion mutants. Proliferating bloodstream forms were harvested at ≤8 × 105 cells/ml, resuspended in modified DTM (Vassella and Boshart, 1996) at 1–2 × 106 cells/ml, and triggered to differentiate to the procyclic form at 27°C by the addition of 6 mM cis-aconitate to the culture medium (Brun and Schönenberger, 1981). Procyclic forms of the pleomorphic strain AnTat 1.1 (see Vassella and Boshart, 1996, for references) were cultured in SDM 79 supplemented with 10% fetal bovine serum and 10 mM glycerol (Brun and Schoenenberger, 1979; Vassella et al., 2000).
Isolation of the T. brucei MAPK2 cDNA and Sequence Analysis
A cDNA clone encoding T. brucei MAPK2 was serendipitously selected from a directional λgt22 cDNA expression library from procyclic forms of stock 427 (Liniger et al., 2001). The cDNA contained a long poly(A) stretch at the 3′ end but no miniexon sequence. The splice-acceptor site was therefore mapped by reverse transcription-PCR (Vassella et al., 1994) using a miniexon primer (5′-CGCTATTATTAGAACAGTTTCTGTAC-3′) and a TbMAPK2-specific primer (5′-AATCGTCTTTCCGTACTGGG-3′). Comparisons of TbMAPK2 with protein databases were performed using BLAST2.1 (Altschul et al., 1997) and FASTA3 (Pearson, 1990). Multiple alignments were generated by ClustalW 1.8 (Thompson et al., 1994). For profile database searches, the network services of SMART Version 3.1 (Schultz et al., 2000) and PROSITE (Hofmann et al., 1999) were used.
Construction of Cassettes for Deletion or Ectopic Expression of the TbMAPK2 Gene
The TbMAPK2 cDNA clone was used to screen a λEMBL3 library, constructed from genomic DNA of T. brucei stock 227 partially digested with Sau 3A (Carrington et al., 1987). A HindIII/BamHI fragment of 1.2-kb containing sequences upstream of the TbMAPK2 gene, including the first 108 base pairs of the open reading frame (ORF), was isolated from the genomic clone 111 and subcloned into pBluescript SK+ (Stratagene, La Jolla, CA) to generate pBS-111a. A 2.8-kb BamHI/KpnI fragment encompassing the last 476 base pairs of the ORF and downstream sequences was isolated and subcloned into pBluescript SK+ to give rise to pBS-111b.
Two promoterless constructs (pMAPK2koHYGR and pMAPK2koBLER) were designed to delete sequentially both alleles of TbMAPK2 by homologous recombination. Each construct contains sequences flanking TbMAPK2 including the complete 5′ untranslated region (UTR) and the last 32 base pairs of the 3′ UTR, respectively. The 3′ flanking sequence was amplified from pBS-111b using primer MK3′ (5′-TAGGATCCACTCAACGTTAGT), which binds to the 3′ UTR of TbMAPK2, and a Bluescript-specific primer. Underlined sequences indicate a BamHI site introduced to facilitate cloning. A 2.4-kb fragment was cloned between the BamHI and KpnI sites of pBluescript SK+ to generate pBS-3′ flank. The 5′ flanking sequence was amplified from clone 111a using the primer pair MAPK-XbaI (5′-CGTCTAGATGATGAGATCAATGG-3′), which binds to the 5′ end of the insert, and MAPK-HindIII (5′-CCGAAGCTTATTTCCTTAAACTC-3′), which binds to the 5′ UTR of TbMAPK2. Relevant restriction sites used for cloning are underlined. The PCR product was digested with XbaI and HindIII to release a DNA fragment of 1.2 kb. The hygromycin- and phleomycin-resistance genes were released from pKOH or pKOP (Ruepp et al., 1997), respectively, by cleavage with HindIII and BamHI. The 5′ flanking sequence and the relevant resistance gene were cloned between the XbaI and BamHI sites of pBS-3′ flank by a three-component ligation to give pMAPK2koHYGR and pMAPK2koBLER, respectively.
For ectopic reexpression of TbMAPK2, the plasmid pGAPRONE-MAPK2 was constructed. The complete ORF of the gene was amplified by PCR using the primer pair MAP-ATG (5′-CCGCTCGAGTATGGACATACCA-3′) and MAP-TAG (5′-CGGAATTCCTAGTCACCCTTTG-3′) and clone 111 as template. The PCR product was cloned between the SalI and EcoRI sites of a derivative of the original pGAPRONE construct (Furger et al., 1997), in which the HindIII and BamHI sites flanking the GARP gene had been replaced by SalI and EcoRI sites, respectively. The neomycin-resistance gene (NEO) of pGAPRONE-MAPK2 was replaced by the puromycin-resistance gene (PAC) as follows: the plasmid was linearized with NheI, and the ends were regenerated to blunt ends by Klenow treatment and cleaved with NotI to release the neomycin-resistance gene. The PAC gene was released from pGAPRONEΔ164EP1Pur (Ruepp et al., 1997) by cleavage with PinAI and NotI and cloned into the filled in site and the NotI site of pGAPRONE-MAPK2. For construction of pGAPRONE-MAPK2 (T190A, Y192F), the oligonucleotides 5′-GATCAATGTACGCAGACC-TCTGCGCTCGCTGAATTCGTTGTAACTAGGTGGTATCGACCAC-CTGAAGTGTTAGGCATGGGATCCCAT-3′ and 5′-CGATGGGATCCCATGCCTAACACTTCAGGTGGTCGATACCACCTAGTTACAAC-GAATTCAGCGAGCGCAGAGGTCTGCGTACAT-3′ containing complementary sequences were annealed and cloned between the BclI and ClaI sites of pGAPRONE-MAPK2. Mutations are underlined.
Stable Transformation
Stable transformation of bloodstream or procyclic form cells (Li and Gottesdiener, 1996) and selection of independent clones in microtiter plates (Vassella et al., 2000) were performed as described, except that transformed procyclic form cells were supplemented with 5 × 105 nontransformed cells/ml during selection in microtiter plates. Bloodstream forms were selected with 0.1 μg/ml puromycin, 1.5 μg/ml phleomycin, or 1.0 μg/ml hygromycin and procyclic forms with 1 μg/ml phleomycin or 20 μg/ml hygromycin. For stable transformation, pMAPK2koHYGR and pMAPK2koBLER were linearized with XbaI and XhoI and pGAPRONE-MAPK2 was linearized with KpnI and NotI.
Nucleic Acid and Protein Analyses
Northern blot and Southern blot analyses were performed using standard procedures (Sambrook et al., 1989). Multiprime labeled probes used for hybridization were generated from the coding regions of TbMAPK2, or from a PstI fragment of plasmid 9B1 derived from the β-tubulin gene (Schneider et al., 1988). Hybridization signals were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
For immunoblot analysis, total cell protein extracts were separated on a 12% polyacrylamide gel and transferred to Immobilon P (Millipore Corp., Bedford, MA). A polyclonal antibody directed against KFR1 (provided by C.C. Wang) was used at a dilution of 1:2000 as described (Hua and Wang, 1994).
Enzyme Assay, Cytological Assays, and Immunofluorescence
For the GUS activity assay, logarithmically growing bloodstream form trypanosomes were harvested, washed twice with colorless medium lacking phenol red and haemin according to Sbicego et al. (1999), and resuspended at 106 cells/ml in colorless medium. At different time points after triggering differentiation at 27°C, 100 μl aliquots were withdrawn and mixed with 100 μl reaction buffer in microtiter plates, containing 1 mM 4-methylumbelliferyl β-d-glucuronide (MUG) substrate (Molecular Probes Europe BV, Leiden, The Netherlands), 0.82 M Tris-HCl, pH 8.0, 0.6% SDS, and 0.3 mg/ml BSA, and incubated for 60 min at 37°C. The fluorescent product was quantified using a Spectra MAX 340 (Molecular Devices, Menlo Park, CA) set at 355-nm excitation and 460-nm emission wavelengths. Each measurement was performed in duplicate.
5-Bromo-2′deoxyuridine (BrdU) incorporation into the kinetoplast and nucleus of dividing trypanosomes was performed as described (Woodward and Gull, 1990; Vassella et al., 1997a). Cell smears were air-dried and fixed with acetone at −20°C for 10 min. Incorporation of BrdU was analyzed by immunofluorescence using an anti-BrdU mAb (hybridoma supernatant, obtained from the Developmental Studies Hybridoma Bank of the University of Illinois, Urbana, IL) used at a dilution of 1:2 and a TRITC-conjugated anti-mouse secondary antibody (Sigma, St. Louis, MO) used at a dilution of 1:400. Cells were counterstained with the DNA binding dye 4,6-diamino-2-phenylindole (DAPI). Analysis of the nuclear DNA content by flow cytometry was performed as described (Vassella et al., 1997a).
Expression of EP and GPEET procyclins and VSG on the surface of acetone-fixed cells was determined by immunofluorescence using the anti-EP mAb TRBP1/247 (Richardson et al., 1988) at 1:500, anti-GPEET K1 antiserum (Ruepp et al., 1997) at 1:500 and polyclonal anti-VSG 221 antiserum (obtained from George Cross, Rockefeller University, New York) at 1:1000. TRITC-conjugated anti-mouse antibody (Sigma) was used at 1:400 and FITC-conjugated anti-rabbit antibody (Sigma) at 1:2000. Expression of CAP5.5 was determined on formaldehyde-fixed cells permeabilized with Triton X-100 (Vassella et al., 1997b) using anti-CAP5.5 antiserum (Matthews and Gull, 1994; provided by K. Gull, University of Manchester, Manchester, United Kingdom) diluted 1:2 and a FITC-conjugated anti-rat secondary antibody (DAKO, Carpinteria, CA) diluted 1:500.
RESULTS
A T. brucei Protein Kinase Containing the Signature of Extracellular-Signal–Regulated Kinases
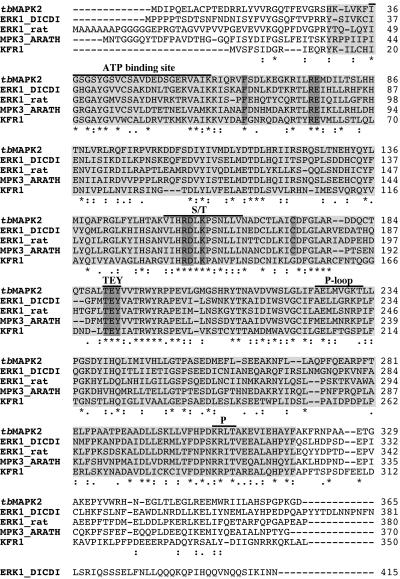
A cDNA clone serendipitously selected from an expression library from procyclic forms of T. brucei matched a genomic sequence in the database encoding a MAP kinase-like protein (accession no. Z54341; Wilson, K. and Boothroyd, J.C, unpublished results). The cDNA contained a short 3′ untranslated region (UTR) of 47 nucleotides and a poly(A) tail. All trypanosomal mRNAs contain a spliced leader at their 5′ end that is joined to the protein-coding exon by trans-splicing (Agabian, 1990). The splice-acceptor site of the MAP kinase mRNA was mapped by reverse transcription-PCR to a position 60 nucleotides upstream of the start of the major open reading frame (ORF). The ORF potentially encodes a protein with a length of 365 amino acids and a predicted molecular mass of 42 kDa. By using profile database searches, all the conserved amino acid residues that define the catalytic domain of protein kinases were identified in a region that encompasses the sequences from amino acid positions 30–318 (Figure 1, shaded light gray). A sequence motif that is diagnostic for serine-threonine protein kinases was also found in this domain (Figure 1, S/T). The signature motif of MAP kinases (F-X10-R-E-X77-R-D-X-K-X14-C), which is absent from all other classes of protein kinases (Dorin et al., 1999), is also present in this sequence and maps to amino acid positions 65–173. In addition, the TEY activation site of the regulatory loop of ERKs was found 17 amino acids downstream of the signature motif (Figure 1, shaded dark gray). In agreement with these predictions, Blast searches of databases revealed the highest similarities to ERKs from various eukaryotes. The protein kinase shares 44% amino acid sequence identity with ERK1 from Dictyostelium discoideum (Gaskins et al., 1994), 43% identity with ERK3 from Arabidopsis thaliana (Mizoguchi et al., 1993), 40–41% identity with ERK1/2 from mammals and 40–42% identity with the different yeast homologues. The other MAPK from T. brucei, KFR1 (Hua and Wang, 1994), is more distantly related (39% identity) as is the case for LMPK from Leishmania major (35% identity; Wiese, 1998) or for the MAPK homologues from Plasmodium falciparum (30–36% identity). However, the protein kinase shares the highest amino acid sequence identity with an unusual MAPK-like gene product from the database of Leishmania major (CAB94009; 69% identity) which has a TQY sequence in the regulatory loop instead of the conserved TEY motif. An alignment of the T. brucei homologue with ERKs from A. thaliana, D. discoideum, and Rattus norvegicus and KFR1 is shown in Figure 1, demonstrating that the conserved residues are present at the same positions in all these sequences. Based on these findings, the protein kinase gene was classified as belonging to the ERK group of MAP kinases. Since this is the second MAP kinase described in trypanosomes, it was named T. brucei MAPK2 (TbMAPK2). Although TbMAPK2 is most similar in amino acid sequence to ERK1 and ERK2 from various organisms, it lacks the characteristic docking/cytosolic retention motif common to this subgroup of ERKs (Tanoue et al., 2001).
Figure 1.
The T. brucei MAPK homologue contains the signature of ERK kinases. The amino acid sequence is displayed by Clustal W alignment with the corresponding sequences of ERKs from Arabidopsis thaliana (MPK3_ARATH, EMBL accession no. Q39023; Mizoguchi et al., 1993), Dictyostelium discoideum (ERK1_DICDI, EMBL accession no. P42525; Gaskins et al., 1994), Rattus norvegicus (ERK1_rat, EMBL accession no. P21708; Marquardt and Stabel, 1992), and KFR1 from T. brucei (EMBL accession no. L10997; Hua and Wang, 1994). Asterisks, colons, and dots underneath the alignment indicate identical, conserved, and semiconserved amino acids, respectively. The catalytic domain is shaded light gray and sequences highly diagnostic for ERK kinases (Dorin et al., 1999) are shaded dark gray. Domains with known functions are overlined: these include the ATP binding site, a domain diagnostic for serine/threonine kinases (S/T), the P-loop domain, and the phosphorylation site of protein kinase A and protein kinase C (P).
Generation of a TbMAPK2 Null Mutant in Bloodstream Form Trypanosomes
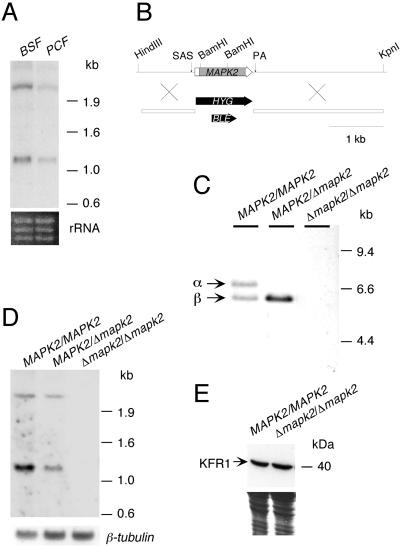
We wanted to investigate whether TbMAPK2 was involved in growth or differentiation processes of trypanosomes. To generate a null mutant, it was important to determine the genomic organization of TbMAPK2 and to investigate if the expression of the gene was developmentally regulated. Southern blot analysis revealed that TbMAPK2 is a single copy gene (unpublished data). Northern blot analysis revealed the presence of two TbMAPK2-specific transcripts of 2.5 and 1.3 kb, the latter corresponding to the length of the isolated cDNA clone (Figure 2A). The two transcripts might be alternatively spliced or alternatively polyadenylated products or might be derived from different alleles. The relative amounts of the two mRNA species were similar in the bloodstream and the procyclic form, but the steady state level of both mRNA species together was approximately threefold higher in the bloodstream form than in the procyclic form.
Figure 2.
Nucleic acid analyses of TbMAPK2 mutants. (A) Steady state level of TbMAPK2 mRNA. Total RNA (10 μg) extracted from bloodstream form (BSF) or procyclic form (PCF) trypanosomes was compared by Northern blot analysis using a TbMAPK2-specific probe (top panel). The rRNA bands were used as internal control for sample loading (bottom panel). (B) Deletion constructs. The TbMAPK2 locus is shown in the top part of the figure and the targeting constructs for deletion of both alleles of TbMAPK2 are shown below (drawn to scale). The ORFs of the kinase and the resistance genes are indicated by horizontal arrows, the splice-acceptor site (SAS) and polyadenylation site (PA) by vertical arrows, the catalytic region by a gray box, and sequences flanking the TbMAPK2 gene in the knock-out constructs by open boxes. Relevant restriction sites used for cloning are shown. (C) Southern blot analysis of TbMAPK2 mutants. Genomic DNA was digested with XhoI and XbaI, which separate the two TbMAPK2 alleles α and β, and hybridized with a radiolabeled probe from the ORF of TbMAPK2. (D) Northern blot analysis of TbMAPK2 mutants. Total RNA extracted from bloodstream form trypanosomes of the wild-type and TbMAPK2 mutants were hybridized with sequences from TbMAPK2 (top panel) or, as a control for sample loading, with a β-tubulin–specific probe (bottom panel). (E) Top panel: immunoblot analysis of bloodstream forms of the wild-type and the null mutant using a rabbit antiserum against KFR1 (Hua and Wang, 1997). Equal cell numbers were loaded per lane. Bottom panel: after electroblotting, the residual proteins in the polyacrylamide gel were stained with Coomassie blue, confirming that similar amounts of proteins were loaded per lane.
Trypanosomes are diploid organisms. To generate a null mutant by homologous recombination, two targeting constructs, pMAPK2koHYGR and pMAPK2koBLER, were made. These contained antibiotic-resistance genes cloned between sequences flanking the ORF of TbMAPK2 (Figure 2B). The constructs were used to stably transform bloodstream forms of a transgenic trypanosome clone, GUSone, in which the coding region of one EP procyclin gene had been replaced by E. coli β-glucuronidase (GUS; Sbicego et al., 1999). In these cells the expression of GUS occurs in parallel to that of EP procyclin when cells are triggered to differentiate to the procyclic form. The advantage of using GUSone cells for gene disruption is that the kinetics of differentiation of the mutant cell lines can be easily monitored in a simple one-step enzyme reaction in microtiter plates (Sbicego et al., 1999).
The first transformation was performed with the deletion construct pMAPK2koHYGR and the hygromycin-resistant clone MAPK2/Δmapk2::HYG GUS NEO was selected for Southern blot analysis. Digestion of genomic DNA with XbaI and XhoI separates both allelic loci of TbMAPK2 owing to a restriction site polymorphism. Southern blot analysis of genomic DNA from GUSone wild-type cells, digested with these enzymes, revealed two DNA fragments hybridizing to a TbMAPK2-specific probe, whereas only one fragment was detected in the mutant, demonstrating that one allele had been deleted in this clone (Figure 2C). Both forms of TbMAPK2 mRNAs could still be detected in Northern blots, however, ruling out the possibility that the two transcripts were derived from different alleles (Figure 2D). This clone was subjected to a second round of transformation using the deletion construct pMAPK2koBLER. One clone, Δmapk2::HYG/Δmapk2::BLE GUS NEO, in which the second copy of TbMAPK2 had been deleted (Figure 2C), was selected for further analysis in vitro. TbMAPK2-specific mRNA was not detected in this clone (Figure 2D), confirming that both copies of the kinase gene were deleted. Bloodstream forms of the null mutant had a population doubling time indistinguishable from that of the wild type (Figure 3A) and no detectable alterations in cell morphology. To investigate whether (over)expression of other ERK homologues might compensate for the deletion of TbMAPK2, immunoblot analysis was performed using an antiserum against KFR1. However, the levels of KFR1 were similar in wild-type and null mutant cells (Figure 2E).
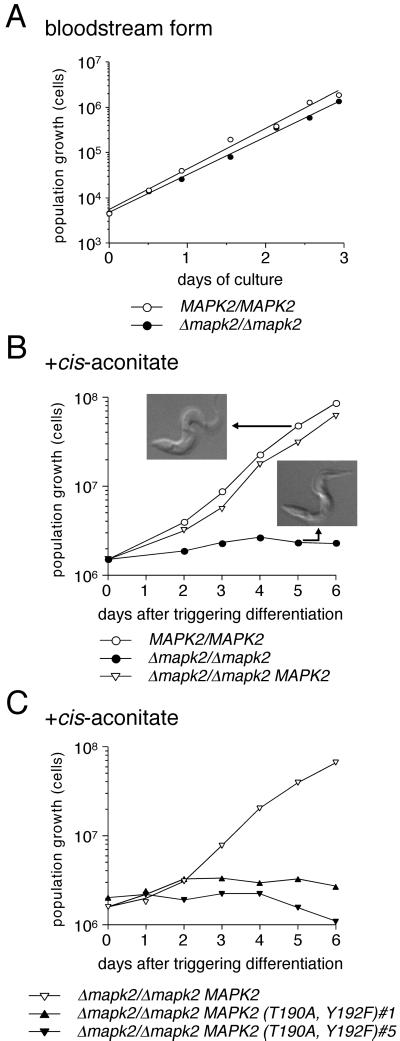
Figure 3.
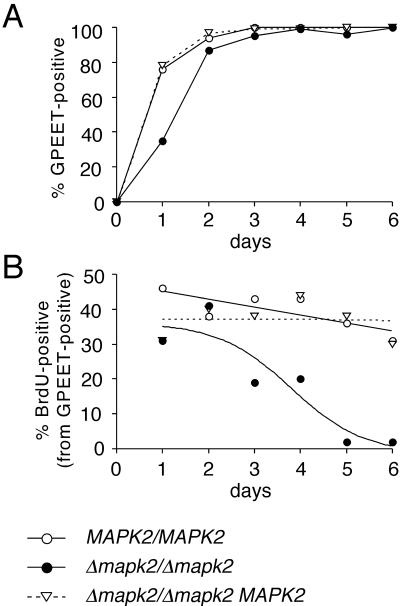
Stage-specific growth inhibition of the TbMAPK2 null mutant. (A) Population growth of bloodstream form trypanosomes of the wild-type (MAPK2/MAPK2 GUS NEO) and the null mutant (Δmapk2:: HYG/Δmapk2:: BLE GUS NEO). (B and C) Population growth upon triggering differentiation (+cis-aconitate) of (B) the same clones as above and the add-back mutant (Δmapk2:: HYG/Δmapk2:: BLE GUS NEO MAPK2 PAC) and (C) the add-back mutant together with two clones in which the TEY activation site of TbMAPK2 was mutated (Δmapk2:: HYG/Δmapk2:: BLE GUS NEO MAPK2 (T190A, Y192F) PAC 1 and 5). Cells triggered to differentiate to the procyclic form were diluted at daily intervals to ensure logarithmic growth. The population growth was calculated as cell density multiplied by the cumulative dilution factors. A representative experiment from six independent experiments is shown, together with Normarski images of formaldehyde-fixed wild-type or null mutant cells, harvested 5 d after exposure to the differentiation signal.
Deletion of TbMAPK2 Results in Delayed Differentiation and Growth Inhibition
Bloodstream form trypanosomes can be triggered to differentiate to the procyclic form in vitro by subjecting the cells to a drop in temperature and the addition of cis-aconitate to the culture medium (Brun and Schönenberger, 1981). When Δmapk2/Δmapk2 bloodstream forms were exposed to the differentiation signal, the majority of cells showed morphological changes characteristic for the procyclic form (Figure 3B), including repositioning of their kinetoplast, the mitochondrial genome, to a nucleus proximal location (unpublished data). In marked contrast to the wild type, however, the null mutant underwent subsequent growth arrest (Figure 3B), and an increased proportion of cells showed aberrant morphologies (unpublished data). It was theoretically possible that growth inhibition of this mutant was due to secondary mutations unrelated to TbMAPK2 deletion. Before we embarked on a detailed phenotypic analysis of the null mutant, we reintroduced one copy of TbMAPK2 into an ectopic locus of the null mutant in order to verify whether growth could be restored. A puromycin-resistant clone (Δmapk2::HYG/Δmapk2::BLE GUS NEO MAPK2 PAC) was obtained in which the first gene of the pair of procyclin genes in the EP/PAG1 locus (Roditi and Clayton, 1999) had been replaced by the TbMAPK2 gene and the second gene by the puromycin-resistance gene. Correct integration of the TbMAPK2 add-back construct was confirmed by Southern blot analysis (unpublished data). On Northern blots, a single species of mRNA was detected that corresponded to the predicted length of the transcript of ∼1400 nucleotides (unpublished data). In this clone, the steady state level of TbMAPK2 mRNA was 5–10 fold higher than in the wild-type, presumably due to transcription from the strong procyclin promoter upstream of the ectopic TbMAPK2 gene. In marked contrast to the null mutant, the TbMAPK2 add-back mutant was able to grow after triggering differentiation to the procyclic form (Figure 3B). The population doubling time of this clone (21.6 h, r = 1.00), calculated from the slope of the linear regression from days 2–6 in Figure 3B, was almost identical to that obtained for the wild type (20.9 h, r = 0.99). Thus, growth inhibition of the null mutant is due to deletion of the TbMAPK2 gene.
Activation of MAP kinases requires phosphorylation at both threonine and tyrosine residues of the TEY site in the regulatory loop by dual-specific MEK (Waskiewicz and Cooper, 1995). To investigate whether activation of TbMAPK2 is essential for growth of the parasite, a TbMAPK2 mutant was constructed in which the T190 and Y192 residues in the activation domain were substituted by alanine and phenylalanine, respectively, and integrated into the procyclin locus of the TbMAPK2 null mutant. Two independent bloodstream form clones (Δmapk2/Δmapk2 MAPK2 (T190A, Y192F) 1 and 5) in which the mutated TbMAPK2 gene had integrated correctly (as shown by PCR analysis of genomic DNA) were selected for further analysis. Northern blots confirmed that the mutated gene was expressed in these cells (unpublished data). When the Δmapk2/Δmapk2 MAPK2 (T190A, Y192F) clones 1 and 5 were triggered to differentiate to the procyclic form, they ceased to proliferate (Figure 3C), as was the case for the null mutant. This suggests that phosphorylation of TbMAPK2 by MEK is essential for the parasite to establish procyclic form cultures.
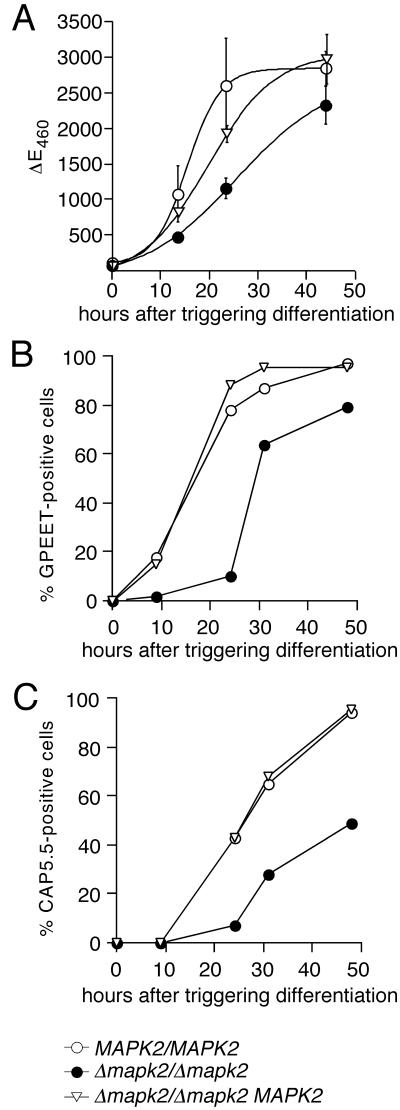
The null mutant might either be blocked in differentiation or, alternatively, might be able to undergo differentiation but be unable to grow as procyclic form trypanosomes. We therefore assessed the expression profile of markers of the early and late phases of differentiation. We first determined activation of GUS expression as an early marker of differentiation. As shown in Figure 4A, Δmapk2/Δmapk2 trypanosomes were able to express high levels of GUS enzyme activity, but the kinetics of appearance of this marker was delayed by 10–12 h relative to the GUSone wild-type or the add-back mutant (Δmapk2/Δmapk2 MAPK2). This result was confirmed in two further experiments (unpublished data). In individual cells, GUS expression mirrors that of EP procyclin during differentiation (Sbicego et al., 1999). GPEET procyclin appears on the surface a few hours later than EP procyclin and is regulated independently (Vassella et al., 2000). We therefore investigated the appearance of GPEET-positive cells by immunofluorescence (Figure 4B). GPEET-positive cells from the wild-type and the add-back appeared with similar kinetics, whereas those from the null mutant were delayed by ∼13 h (calculated from the time points at which 50% of the cells were positive for GPEET in the different cultures), consistent with the results obtained with the GUS activity. The same also held true for the expression of CAP5.5, a marker of late phase differentiation, which appears with a lag of ∼8 h relative to EP procyclin (Matthews and Gull, 1994; Figure 4C). The expression levels of GPEET and CAP5.5 were not affected by the deletion of TbMAPK2, because differentiating cells of the wild-type, knock-out, or add-back mutant, stained with the different antibodies, exhibited similar fluorescence intensities (unpublished data). In conclusion, in the null mutant the appearance of each of these markers was delayed by approximately 12 h relative to the wild-type or the add-back clone. This was confirmed in a total of seven differentiation experiments using the different markers. By days 3–4, differentiation of the null mutant was complete, as indicated by maximal expression of the entire set of differentiation markers in all cells. In addition, all cells had completely shed their variant surface glycoprotein (VSG) coat at that time point (unpublished data). Thus, the null mutant seems to be able to undergo complete differentiation, but it develops to the procyclic form with markedly delayed kinetics. The same also holds true for the Δmapk2/Δmapk2 MAPK2 (T190A, Y192F) clones 1 and 5, which differentiated with kinetics similar to those of null mutant cells (unpublished data), demonstrating that phosphorylation of TbMAPK2 is also required for this developmental process.
Figure 4.
The null mutant expresses early and late differentiation markers with slow kinetics. (A) Kinetics of GUS expression upon triggering differentiation with cis-aconitate at 27°C. Substrate conversion was assayed in duplicate in a SpectraMax 340 (Molecular Devices) at 460 nm. The mean (±SD) from three independent experiments is presented. (B and C) Kinetics of appearance of GPEET procyclin (B) and CAP5.5 (C) in cells triggered to differentiate. The percentage of positive cells was determined by immunofluorescence using specific antibodies. Differentiating cells of the wild-type, knock-out, or add-back mutant, stained with the different antibodies, exhibited similar fluorescence intensities (unpublished data). At least 100 cells were counted per sample.
To uncouple differentiation from growth, we attempted to produce null mutants in established procyclic forms of the pleomorphic strain AnTat1.1. We were able to obtain single knock-out clones (MAPK2/Δmapk2::HYG), but it was not possible to delete the second copy of TbMAPK2. MAPK2/Δmapk2 cells that were stably transformed with the second construct (pMAPK2koBLER) and selected with phleomycin in the absence of hygromycin, always integrated the phleomycin-resistance gene into the locus containing the hygromycin-resistance gene, thereby deleting the latter. This was confirmed by Southern blot analysis of 10 independent clones (unpublished data). In contrast, 10 independent transformants that were selected with both antibiotics integrated the phleomycin-resistance gene incorrectly and retained the second copy of TbMAPK2 (unpublished data). This supports the conclusion that TbMAPK2 is also required by fully differentiated procyclic forms.
The Null Mutant Undergoes Cell Cycle Arrest
When long slender forms are triggered to differentiate to the procyclic form, they transiently express markers of the short stumpy bloodstream form and presumably also undergo transient cell cycle arrest in the G0 phase in which the short stumpy form is held (Tasker et al., 2000). Release of the cell from a quiescent state is one of the major functions of MAP kinases in different systems (Lavoie et al., 1996). We therefore investigated if the reason for the failure of the null mutant to grow as procyclic forms was due to irreversible arrest in G0. To test this hypothesis, trypanosomes were triggered to differentiate and, at daily intervals, aliquots were pulse labeled for 6 h with the thymidine analogue BrdU, which is incorporated into the genome of cells proceeding through S-phase. Acetone-fixed trypanosomes were double-labeled with antibodies directed against BrdU and GPEET procyclin (Figure 5, A and B). To exclude cells that were still replicating as bloodstream forms from the analysis, the percentage of BrdU-positive cells was determined only from those expressing GPEET. Twenty-four hours after exposure to the differentiation signal, 30–40% of GPEET-positive cells of the wild-type and the add-back mutant had incorporated BrdU during the short labeling period (Figure 5B), which corresponds to one quarter of the generation time of procyclic forms. More than half of this population remained BrdU-negative, because these cells had not proceeded through S-phase during the labeling period. Extending the incubation period to 24 h resulted in the labeling of >90% of these cells (unpublished data). A similar percentage of double-positive cells was also obtained for the Δmapk2/Δmapk2 clone (Figure 5B). This clearly demonstrates that most null mutant cells are able to progress efficiently through the first cell division as procyclic form trypanosomes.
Figure 5.
Growth inhibition of the null mutant is due to cell cycle arrest. Bloodstream form trypanosomes were triggered to differentiate by addition of cis-aconitate at 27°C. At daily intervals, aliquots were pulse-labeled with BrdU for 6 h. Acetone-fixed cells were double-stained with antibodies against GPEET procyclin and BrdU. (A) Kinetics of appearance of GPEET-positive cells. (B) Percentage of BrdU-positive cells of the subpopulation expressing GPEET during the time course of the experiment. At least 100 cells were counted per sample. After 4–5 d ≥90% of the null mutant cells had undergone cell-cycle arrest (4 independent experiments).
At later time points after exposure to the differentiation signal, the percentage of BrdU-positive cells from the null mutant dropped progressively. By day 3, ∼20% of the cells were BrdU positive, varying slightly between experiments, and by days 4–5 virtually no BrdU-positive cells were detected (Figure 5B). In contrast, the proportion of BrdU-positive cells from the wild-type and the add-back clone remained at a constant level between 30 and 40% during the course of the experiment.
Cell Cycle Position of Arrested Cells
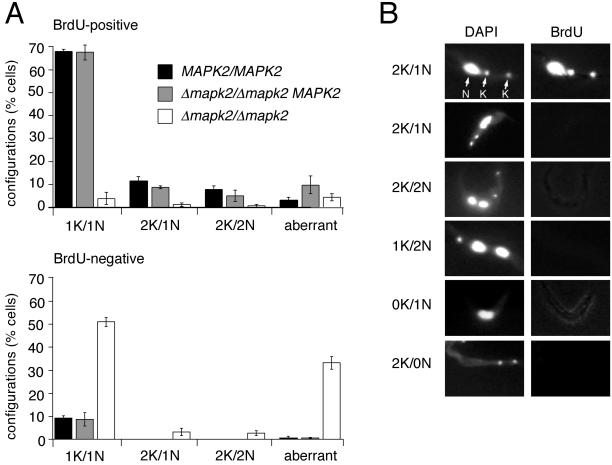
In trypanosomes, replication and division of the nucleus and the kinetoplast occurs in a temporally ordered manner (Woodward and Gull, 1990). Cells in the interphase of the cell cycle contain one kinetoplast (K) and one nucleus (N). Dividing trypanosomes first segregate their daughter kinetoplasts before they segregate their daughter nuclei. As a consequence, cells with the 2K/1N configuration are still in the G2 phase of their nuclear cell cycle (Woodward and Gull, 1990). After nuclear division has occurred, cells show the 2K/2N configuration and, after cytokinesis, two daughter cells emerge, both showing the 1K/1N configuration. Thus, by determining the proportion of cells with the different kinetoplast/nucleus configurations it is possible to map the position in the cell cycle in which the null mutant was arrested.
To distinguish between proliferating and arrested cells, trypanosomes were labeled with BrdU for 24 h. During this labeling period, which corresponds to one generation time, we would expect that almost all replicating cells would become BrdU positive, while cells which had arrested before the labeling period would remain BrdU negative. Cells were triggered to differentiate and subsequently cultured for 3 d in the absence and 1 d in the presence of BrdU. Acetone-fixed cells were labeled with anti-BrdU mAb and counterstained with DAPI. BrdU-positive or -negative cells were scored individually for their nucleus/kinetoplast configurations. As expected, ≥90% of the wild type and the add-back mutant incorporated BrdU, and these showed the normal distribution of cells with 1K/1N (70%), 2K/1N (10%), and 2K/2N (8%) configurations (Woodward and Gull, 1990; Figure 6, top panel). The only discernible difference between the two cultures was a higher percentage of aberrant forms (scored as 0K/1N, 0K/2N, 1K/2N, or xK/yN, y ≥ 3) in the add-back (10%) than in the wild type (3%). Virtually all the BrdU-negative wild-type or add-back cells had the 1K/1N configuration, and cells with the 2K/1N or 2K/2N configurations were never found in these cultures (Figure 6, bottom panel). This is to be expected, because cells showing these configurations have just emerged from S-phase and would therefore be BrdU positive.
Figure 6.
Null mutant cells are arrested at multiple phases of the cell cycle. (A) Percentage of dividing (BrdU-positive) and nondividing (BrdU-negative) cells showing different kinetoplast/nucleus configurations as indicated in the figure. Cells with 0K/1N, 0K/2N, 1K/2N and xK/yN, y ≥ 3 configurations were scored as aberrant cells. Trypanosomes were triggered to differentiate, cultured for 3 d and then incubated for an additional day in the presence of BrdU. Acetone-fixed cells were labeled with a monoclonal anti-BrdU antibody and counterstained with DAPI. Bloodstream forms of all three clones showed only a low percentage (≤5%) of aberrant cells. At least 600 cells were counted per sample. (B) Examples of nondividing null mutant cells with aberrant configurations. Cells were stained with DAPI (left panel) and labeled with the monoclonal anti-BrdU antibody (right panel).
In contrast to the wild type and the add-back mutant, 90% of the null mutant cells had undergone arrest and, among these, 25–30% showed aberrant configurations (Figure 6, bottom panel). From the arrested cells with normal configurations, 89% showed the 1K/1N, 6% the 2K/1N, and 5% the 2K/2N configurations (Figure 6A, bottom panel, and Figure 6B), reminiscent of the distribution in proliferating wild-type and the TbMAPK2 add-back cultures (Figure 6A, top panel). This suggests that the null mutant had undergone arrest at multiple phases of the cell cycle. Among the arrested cells with aberrant configurations, 47% showed the 1K/2N configuration, 21% had lost their nucleus (xK/0N configuration, termed zoid according to Matthews et al., 1994), 15% had lost their kinetoplast (0K/xN, termed akinetoplast), and 17% were multinucleated (xK/yN, y ≥ 3; see Figure 6B). These findings suggest that cytokinesis is also severely impaired in the null mutant.
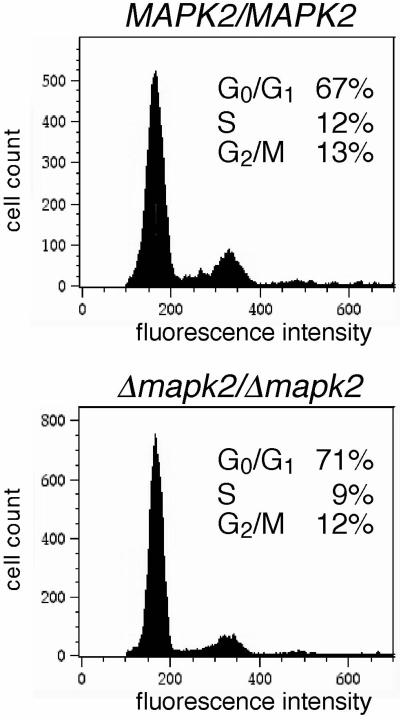
To confirm that null mutant cells had undergone nonphase-specific arrest, the DNA content of propidium iodide–stained cells was measured by flow cytometry. Cells were gated on fluorescence pulse width vs. area measurement to exclude cell doublets (and cells that had segregated their daughter nuclei) from the analysis. Measurements by flow cytometry revealed a similar distribution of cells in G1 (2n), S (2n-4n), and G2-M (4n) in proliferating wild-type cultures and arrested Δmapk2/Δmapk2 cultures (Figure 7). This confirms that the null mutant had undergone arrest in multiple phases of the cell cycle. Among the cells with aberrant configurations, only the akinetoplasts (0K/1N), which comprise 4–5% of the total population, were gated, whereas the 1K/2N cells, the zoids and the multinucleated cells were excluded from the analysis (see above). Thus, in the gated population, aberrant cells are unlikely to be overrepresented in a specific phase of the cell cycle.
Figure 7.
Nuclear DNA content of wild-type (MAPK2/MAPK2) and null mutant (Δmapk2/Δmapk2) cells. Trypanosomes were triggered to differentiate and subsequently cultured in DTM medium for 4 d. Cells were fixed with 70% ethanol at −20°C, stained with propidium iodide, and analyzed by flow cytometry. Cells with the DNA content 2n-4n were gated on fluorescence pulse width vs. area measurement to exclude cell doublets from the analysis. The percentage of cells in the different cell-cycle phases was calculated using the Watson Pragmatic Model of FlowJo (v3.4) software (Tree Star, Inc., San Carlos, CA).
DISCUSSION
A TbMAPK2 deletion mutant, lacking both alleles of the single copy gene, was constructed in bloodstream form trypanosomes. The null mutant exhibited no detectable phenotype in this stage, but was unable to grow after differentiation to the procyclic form. Compared with the wild type, the null mutant differentiated to the procyclic form with markedly delayed kinetics. The appearance of both early and late markers of differentiation was delayed by ∼12 h. This phenotype was due to TbMAPK2 deletion, because the kinetics of differentiation of the add-back mutant was similar to that of the wild type. If the differentiation “clock” runs more slowly in the null mutant, we would expect later events to be delayed more than early events. Because appearance of procyclin and CAP5.5 were delayed to the same extent, this rather suggests that cells are retarded in passing a particular checkpoint in differentiation but are then free to proceed through the subsequent steps at a normal pace. This checkpoint would be before the onset of procyclin expression. Generating TbMAPK2 null mutant cells in procyclic form trypanosomes was not possible, indicating that the inability of Δmapk2/Δmapk2 cells to grow as procyclic forms was not due to a block in differentiation. Thus, the growth and differentiation phenotypes of the null mutant are not coupled. Both these processes seem to be controlled by activated TbMAPK2, because introduction of a kinase mutant, in which the TEY activation domain was replaced by the amino acids AEF, into the null mutant did not restore its ability to differentiate with fast kinetics and to proliferate thereafter. This indicates that growth and differentiation are controlled by one or several signaling cascades operating through TbMAPK2. Searches of database libraries revealed four putative MEKs in T. brucei, but it remains to be shown if one of these is able to phosphorylate TbMAPK2.
Null mutant cells that had undergone differentiation were able to progress efficiently through S-phase of the first cell cycle as procyclic forms, as indicated by a high proportion of BrdU/GPEET double-positive cells 1–2 d after triggering differentiation. Although null mutant cells incorporated BrdU, the cell density did not increase during this incubation period (see Figure 3). The apparent discrepancy between these results may be explained by the finding that 30–40% of bloodstream forms normally fail to differentiate to the procyclic form and die subsequently (Matthews et al., 1994). In contrast to the wild type, differentiated null mutant cells underwent cell cycle arrest. Again, this phenotype was completely restored in the add-back mutant. Analysis of the DNA content by flow cytometry and the nucleus/kinetoplast configuration of procyclic forms both revealed that the null mutant arrested in all phases of the cell cycle. Moreover, 25–30% of the null mutant cells either showed the 1K/2N configuration, had lost their kinetoplast or nucleus, or were multinucleated, indicating that cytokinesis was also impaired to some extent. Proliferating bloodstream forms are only receptive for the differentiation signal when they reach a particular window in G1, in which the stumpy form is held (Matthews et al., 1994). Thus, it can be formally excluded that null mutant cells were arrested at different phases of the cell cycle as procyclic forms because they were already in these phases before differentiation.
We were surprised by the finding that most procyclic form cells showed normal morphology, were motile, and survived for relatively long periods in culture, although they were arrested in all phases of the cell cycle. Nonphase-specific arrest is not unique for trypanosomes, however, because incubation of proliferating T cells (Eichhorn et al., 1993) or B cells (Vaickus et al., 1989; Higaki et al., 1994) with antibodies directed against major histocompatibility complex class II antigens also resulted in growth arrest in all phases of the cell cycle. Moreover, arrest was reversible in these cells (Eichhorn et al., 1993), indicating that they had not lost their vital cell functions. Inhibitors of tyrosine protein kinases abolished antibody-induced growth arrest, suggesting that this process is mediated by tyrosine kinases or phosphatases (Eichhorn et al., 1993), but it is not known whether the MAP kinase pathway is involved.
To our knowledge, this is the first example of a MAP kinase knock-out that resulted in nonphase-specific arrest. In other cell systems, overexpression of dominant negative mutants of MAP kinase pathways or addition of specific inhibitors of MEK to cultured cells always resulted either in specific arrest in G1 (Weber et al., 1997; Talarmin et al., 2000) at the entry into S (Rescan et al., 2001) or in G2 (Wright et al., 1999). Mammalian MAP kinases control cell cycle progression by activating cyclin D1-cdk4 or cyclin B-CDC2 complexes (Cheng et al., 1998; Wright et al., 1999). These are unlikely to be the sole targets of TbMAPK2, however, which rather seems to be required at multiple phases of the cell cycle. In higher eukaryotes, MAPKs have also been shown to phosphorylate a variety of other proteins involved in cellular growth (reviewed by Whitmarsh and Davis, 2000). These include proteins regulating gene expression, e.g., transcription factors, factors controlling mRNA stability, eukaryotic translation factor-4E, or proteins implicated in modulating chromatin structure. MAPKs also control growth and differentiation by inhibition of phosphodiesterase 4, thus modulating intracellular cAMP levels (Hoffmann et al., 1999). Recently, rat ERK1 has been shown to phosphorylate carbamoyl phosphate synthetase II (Graves et al., 2000), a key enzyme in the de novo synthesis of pyrimidines. To investigate whether growth arrest in the TbMAPK2 null mutant was caused by low intracellular concentrations of pyrimidines, Δmapk2/Δmapk2 cells were cultured in the presence of orotate, a membrane-permeable product of dihydroorotate dehydrogenase operating downstream of carbamoyl phosphate synthetase II (Seymour et al., 1997). It was not possible, however, to restore growth of the null mutant under these conditions. Likewise, addition of lipophilic 8-(4-chlorophenylthio)-cAMP to the culture medium had no effect (unpublished results).
In higher eukaryotes, MAPK has been shown to contribute to the regulation of cellular functions and growth in different stages of differentiation. In contrast, TbMAPK2 seems to be restricted to one life cycle stage. A deletion mutant of the MAP kinase LMPK in L. mexicana revealed a phenotype very reminiscent of that of TbMAPK2 in T. brucei. The null mutant grew normally as the promastigote form and was also able to differentiate to the amastigote form (via the nondividing metacyclic form), but the latter stage was unable to grow (Wiese, 1998). Triggering differentiation of the Leishmania null mutant resulted in a fourfold increase in cell density before cells stopped proliferating. The authors speculated that promastigotes could undergo several cell divisions before entering the differentiation program to amastigotes, and this would be responsible for the increase in cell density (differentiation in this system is also asynchronous). Because their results were only based on cell counts—no cell cycle and differentiation markers were used—it is also possible that, by analogy to the TbMAPK2 null mutant, freshly differentiated amastigotes could divide several times before undergoing arrest. It was not investigated if the LMPK null mutant differentiated with delayed kinetics or if growth arrest of amastigotes was nonphase specific. It would be interesting to know if growth and differentiation processes in different trypanosomatids might operate by the same mechanism(s).
Despite the finding that TbMAPK2 is not required by the bloodstream form, the steady state level of TbMAPK2 mRNA in this stage was threefold higher than in the procyclic form of the parasite. Northern blot analysis of the pleomorphic strain AnTat1.1 revealed that the expression level of TbMAPK2 mRNA in the long slender form was similar to that in the short stumpy bloodstream form, whereas synchronously differentiating cells, exposed to cis-aconitate for 2 h, showed a twofold reduction in the expression level. Finally, 6 h after triggering differentiation, cells had reached the expression level of procyclic forms (E.V., unpublished results). If the gene had no function in the bloodstream form, it would be difficult to understand why the mRNA levels were specifically upregulated in this stage. A plausible explanation for these apparently contradictory results is that TbMAPK2 is functionally redundant in the bloodstream form (but not in the procyclic form) and can be compensated by (over)expression of other kinases. A potential candidate for such a functional homologue might be KFR1, which exhibits ∼10-fold more kinase activity in extracts from the bloodstream form than from the procyclic form (Hua and Wang, 1997). However, expression of KFR1 was not upregulated in the null mutant. Searches of database libraries revealed six additional candidate MAP kinases in T. brucei that might also be able to functionally replace TbMAPK2 in the bloodstream form.
Many protozoan parasites have to control growth and differentiation processes adequately to be able to survive in the different environments they encounter during their life cycle. One way to control growth of different life-cycle stages individually in response to extracellular signals might be by expressing different sets of MAP kinases for each proliferative stage. TbMAPK2 may control growth of procyclic form trypanosomes in the tsetse midgut, LMPK growth of Leishmania amastigotes in macrophages, and it is possible that other MAP kinases might be required for growth of other life-cycle stages in these and other related parasites.
ACKNOWLEDGMENTS
We thank Keith Gull (University of Manchester), C.C. Wang (University of California), and George Cross (Rockefeller University) for antibodies and Dirk Dobbelaere for critical reading of the manuscript. This research was supported by grants from the Swiss National Science Foundation (31-63987.00), the Stanley Thomas Johnson Foundation and the Novartis and Roche Research Foundations to I.R. and by a grant from the Swiss National Science Foundation (31-64900.01) to E.V.
Abbreviations used:
- BrdU
5-bromo-2′deoxyuridine
- DAPI
4,6-diamino-2-phenylindole
- ERK
extracellular-signal-regulated kinase
- GUS
β-glucuronidase
- MAPK
mitogen-activated protein kinase
- ORF
open reading frame
- UTR
untranslated region
- VSG
variant surface glycoprotein
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–02–0093. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–02–0093.
REFERENCES
- Agabian N. Trans-splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Schoenenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Brun R, Schönenberger M. Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z Parasitenkd. 1981;66:17–24. doi: 10.1007/BF00941941. [DOI] [PubMed] [Google Scholar]
- Carrington M, Roditi I, Williams RO. The structure and transcription of an element interspersed between tandem arrays of mini-exon donor RNA genes in Trypanosoma brucei. Nucleic Acids Res. 1987;15:10179–10198. doi: 10.1093/nar/15.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Sexl V, Sherr CJ, Roussel MF. Assembly of cyclin D-dependent kinase and titration of p27kip1 regulated by mitogen-activated protein kinase kinase (MEK1) Proc Natl Acad Sci USA. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GAM, Manning JC. Cultivation of Trypanosoma brucei ssp. in semi-defined media. Parasitology. 1973;67:315–331. doi: 10.1017/s0031182000046540. [DOI] [PubMed] [Google Scholar]
- Dorin D, Alano P, Boccaccio I, Cicéron L, Dörig C, Sulpice R, Parzy D, Dörig C. An atypical mitogen-activated protein kinase (MAPK) homologue expressed in gametocytes of the human malaria parasite Plasmodium falciparum. J Biol Chem. 1999;274:29912–29920. doi: 10.1074/jbc.274.42.29912. [DOI] [PubMed] [Google Scholar]
- Eichhorn M, Prospero TD, Heussler VT, Dobbelaere DAE. Antibodies against major histocompatibility complex class II antigens directly inhibit the growth of T cells infected with Theileria parva without affecting their state of activation. J Exp Med. 1993;178:769–776. doi: 10.1084/jem.178.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, Schürch N, Kurath U, Roditi I. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol Cell Biol. 1997;17:4372–4380. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins C, Maeda M, Firtel RA. Identification and functional analysis of a developmentally regulated extracellular signal-regulated kinase gene in Dictyostelium discoideum. Mol Cell Biol. 1994;14:6997–7012. doi: 10.1128/mcb.14.10.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LM, Guy HI, Kozlowski P, Huang M, Lazarowski E, Pope RM, Collins MA, Dahlstrand GN, Earp HS, 3rd, Evans DR. Regulation of carbomoyl phosphate synthetase by MAP kinase. Nature. 2000;403:328–332. doi: 10.1038/35002111. [DOI] [PubMed] [Google Scholar]
- Guan K-L. The mitogen activated protein kinase signal transduction pathway: from the cell surface to the nucleus. Cell Signal. 1994;6:581–589. doi: 10.1016/0898-6568(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Hendriks E, van Deursen FJ, Wilson J, Sarkar M, Timms M, Matthews KR. Life-cycle differentiation in Trypanosoma brucei: molecules and mutants. Biochem Soc. 2000;28:531–536. doi: 10.1042/bst0280531. [DOI] [PubMed] [Google Scholar]
- Hesse F, Selzer PM, Mühlstädt K, Duszenko M. A novel cultivation technique for long-term maintenance of bloodstream form trypanosomes in vitro. Mol Biochem Parasitol. 1995;70:157–166. doi: 10.1016/0166-6851(95)00027-x. [DOI] [PubMed] [Google Scholar]
- Higaki Y, Hata D, Kanazashi S, Horiguchi Y, Yamaoka K, Ohshima Y, Kim K-M, Hike T, Mayumi M. Mechanisms involved in the inhibition of growth of a human B lymphoma cell line, B104, by anti-MHC class II antibodies. Immunol Cell Biol. 1994;72:205–214. doi: 10.1038/icb.1994.31. [DOI] [PubMed] [Google Scholar]
- Hoffmann R, Baillie GS, MacKenzie SJ, Yarwood SJ, Houslay MD. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 1999;18:893–903. doi: 10.1093/emboj/18.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua SB, Wang CC. Differential accumulation of a protein kinase homolog in Trypanosoma brucei. J Cell Biochem. 1994;54:20–31. doi: 10.1002/jcb.240540104. [DOI] [PubMed] [Google Scholar]
- Hua SB, Wang CC. Interferon-gamma activation of a mitogen-activated protein kinase, KFR1, in the bloodstream form of Trypanosoma brucei. J Biol Chem. 1997;272:10797–10803. doi: 10.1074/jbc.272.16.10797. [DOI] [PubMed] [Google Scholar]
- Hunt M, Brun R, Köhler P. Studies on compounds promoting the in vitro transformation of Trypanosoma brucei from bloodstream to procyclic forms. Parasitol Res. 1994;80:600–606. doi: 10.1007/BF00933009. [DOI] [PubMed] [Google Scholar]
- Lavoie JN, Rivard N, L'Allemain G, Pouysségur J. A temporal and biochemical link between growth factor-activated MAP kinases, cyclin E1 induction and cell cycle entry. Prog Cell Cycle Res. 1996;2:49–58. doi: 10.1007/978-1-4615-5873-6_5. [DOI] [PubMed] [Google Scholar]
- Li FS, Gottesdiener KM. An efficient method for stable transfection of bloodstream form Trypanosoma brucei. Nucleic Acids Res. 1996;24:534–535. doi: 10.1093/nar/24.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liniger M, Bodenmüller K, Pays E, Gallati S, Roditi I. Overlapping sense and antisense transcription units in Trypanosoma brucei. Mol Microbiol. 2001;40:869–878. doi: 10.1046/j.1365-2958.2001.02426.x. [DOI] [PubMed] [Google Scholar]
- Marquardt B, Stabel S. Sequence of a rat cDNA encoding the ERK1-MAP kinase. Gene. 1992;120:297–299. doi: 10.1016/0378-1119(92)90109-3. [DOI] [PubMed] [Google Scholar]
- Matthews K. Developments in the differentiation of Trypanosoma brucei. Parasitol Today. 1999;15:76–80. doi: 10.1016/s0169-4758(98)01381-7. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Gull K. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J Cell Biol. 1994;125:1147–1156. doi: 10.1083/jcb.125.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K. ATMPKs: a gene family of plant MAP kinases in Arabidopsis thaliana. FEBS Lett. 1993;336:440–444. doi: 10.1016/0014-5793(93)80852-l. [DOI] [PubMed] [Google Scholar]
- Mottram JC. Cdc2-related protein-kinases and cell-cycle control in trypanosomatids. Parasitol Today. 1994;10:253–257. doi: 10.1016/0169-4758(94)90136-8. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Rescan C, Coutant A, Talarmin H, Theret N, Glaise D, Guguen-Guillouzo C, Baffet G. Mechanism in the sequential control of cell morphology and S phase entry by epidermal growth factor involves distinct MEK/ERK activations. Mol Biol Cell. 2001;12:725–738. doi: 10.1091/mbc.12.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol. 1988;31:203–216. doi: 10.1016/0166-6851(88)90150-8. [DOI] [PubMed] [Google Scholar]
- Roditi I, Clayton C. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol Biochem Parasitol. 1999;103:99–100. doi: 10.1016/s0166-6851(99)00124-3. [DOI] [PubMed] [Google Scholar]
- Roditi I, et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolin S, Hancocq Quertier J, Paturiaux Hanocq F, Nolan DP, Pays E. Mild acid stress as a differentiation trigger in Trypanosoma brucei. Mol Biochem Parasitol. 1998;93:251–262. doi: 10.1016/s0166-6851(98)00046-2. [DOI] [PubMed] [Google Scholar]
- Ruepp S, Furger A, Kurath U, Kunz Renggli C, Hemphill A, Brun R, Roditi I. Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J Cell Biol. 1997;137:1369–1379. doi: 10.1083/jcb.137.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sbicego S, Vassella E, Kurath U, Blum B, Roditi I. The use of transgenic Trypanosoma brucei to identify compounds inducing the differentiation of bloodstream forms to procyclic forms. Mol Biochem Parasitol. 1999;104:311–322. doi: 10.1016/s0166-6851(99)00157-7. [DOI] [PubMed] [Google Scholar]
- Schneider A, Hemphill A, Wyler T, Seebeck T. Large microtubule-associated protein in T. brucei has tandemly repeated, near-identical sequences. Science. 1988;241:459–462. doi: 10.1126/science.3393912. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour KK, Yeo AET, Rieckmann KH, Christopherson RI. dCTP levels are maintained in Plasmodium falciparum subjected to pyrimidine deficiency or excess. Ann Trop Med Parasitol. 1997;91:603–609. doi: 10.1080/00034989760699. [DOI] [PubMed] [Google Scholar]
- Talarmin H, Rescan C, Cariou S, Glaise D, Zanninelli G, Bilodeau M, Loyer P, Guguen-Ghillouzo C, Baffet G. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signaling pathway in the regulation of G1 phase progression in proliferating hepatocytes. Mol Cell Biol. 2000;19:6003–6011. doi: 10.1128/mcb.19.9.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker M, Wilson J, Sarkar M, Hendriks E, Matthews K. A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol Biol Cell. 2000;11:1905–1917. doi: 10.1091/mbc.11.5.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaickus L, Jones VE, Morton CL, Whitford K, Bacon RN. Antiproliferative mechanism of anti-class II monoclonal antibodies. Cell Immunol. 1989;119:445–458. doi: 10.1016/0008-8749(89)90257-8. [DOI] [PubMed] [Google Scholar]
- Vassella E, Boshart M. High molecular mass agarose matrix supports growth of bloodstream forms of pleomorphic Trypanosoma brucei strains in axenic culture. Mol Biochem Parasitol. 1996;82:91–105. doi: 10.1016/0166-6851(96)02727-2. [DOI] [PubMed] [Google Scholar]
- Vassella E, Braun R, Roditi I. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 1994;22:1359–1364. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 1997a;110:2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- Vassella E, Strässer K, Boshart M. A mitochondrion-specific dye for multicolour fluorescent imaging of Trypanosoma brucei. Mol Biochem Parasitol. 1997b;90:381–385. doi: 10.1016/s0166-6851(97)00171-0. [DOI] [PubMed] [Google Scholar]
- Vassella E, van Den Abbeele J, Bütikofer P, Kunz Renggli C, Furger A, Brun R, Roditi I. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. Developmental cycles and biology of pathogenetic trypanosomes. Br Med Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Cooper JA. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- Weber JD, Raben DM, Phillips PJ, Baldassare JJ. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D1 in G1 phase. Biochem J. 1997;326:61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. A central control for cell growth. Nature. 2000;403:255–256. doi: 10.1038/35002220. [DOI] [PubMed] [Google Scholar]
- Wiese M. A mitogen-activated protein (MAP) kinase homologue of Leishmania mexicana is essential for parasite survival in the infected host. EMBO J. 1998;17:2619–2628. doi: 10.1093/emboj/17.9.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- Wright JH, Munar E, Jameson DR, Andreassen PR, Margolis RL, Seger R, Krebs EG. Mitogen-activated protein kinase kinase activity is required for the G2/M transition of the cell cycle in mammalian fibroblast. Proc Natl Acad Sci USA. 1999;96:11335–11340. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer K, Quinten M, Schwarz H, Pearson TW, Overath P. Synchronous differentiation of Trypanosoma brucei from bloodstream to procyclic forms in vitro. Eur J Biochem. 1990;192:373–378. doi: 10.1111/j.1432-1033.1990.tb19237.x. [DOI] [PubMed] [Google Scholar]