Abstract
The human RNA-editing enzyme adenosine deaminase that acts on RNA (ADAR1) is expressed in two versions. A longer 150-kDa protein is interferon inducible and can be found both in the nucleus and cytoplasm. An amino-terminally truncated 110-kDa version, in contrast, is constitutively expressed and predominantly nuclear. In the absence of transcription, however, the shorter protein is also cytoplasmic and thus displays the hallmarks of a shuttling protein. The nuclear localization signal (NLS) of human hsADAR1 is atypical and overlaps with its third double-stranded RNA-binding domain (dsRBD). Herein, we identify regions in hsADAR1 that interfere with nuclear localization and mediate cytoplasmic accumulation. We show that interferon-inducible hsADAR1 contains a Crm1-dependent nuclear export signal in its amino terminus. Most importantly, we demonstrate that the first dsRBD of hsADAR1 interferes with nuclear localization of a reporter construct containing dsRBD3 as an active NLS. The same effect can be triggered by several other, but not all dsRBDs. Active RNA binding of either the inhibitory dsRBD1 or the NLS bearing dsRBD3 is required for cytoplasmic accumulation. Furthermore, hsADAR1's dsRBD1 has no effect on other NLSs, suggesting RNA-mediated cross talk between dsRBDs, possibly leading to masking of the NLS. A model, incorporating these findings is presented. Finally, we identify a third region located in the C terminus of hsADAR1 that also interferes with nuclear accumulation of this protein.
INTRODUCTION
Adenosine deaminases that act on RNA (ADARs) comprise a protein family that converts adenosines into inosines in structured or double-stranded RNA. If the edited RNA is an mRNA the observed base modification can lead to a codon exchange. Editing by ADARs can be specific, only affecting a single or a few adenosines in a given RNA, or it can lead to conversion of up to 50% of all adenosines present (reviewed by Bass, 1997). This latter type of hyperediting has been linked to the phenomenon of hypermutation occasionally found associated with some viral infections. The extent of RNA editing has been shown to be largely dependent on the form and structure of the substrate RNA (Lehmann and Bass, 2000).
ADAR-like enzyme activity has been detected in all metazoan tissues tested. Consistently, cDNAs encoding ADAR proteins have been cloned from several organisms, including Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, and several mammalian species (Bass et al., 1997). To date, three types of ADAR-like enzymes have been characterized. All show strong similarities to each other in their C-terminal ends where the catalytic deaminase domain is located, whereas considerable differences can be found in their central and amino-terminal regions (reviewed by Keegan et al., 2001). ADAR1 proteins contain three double-stranded RNA-binding domains (dsRBDs) in their central region and possess a relatively long amino-terminal end that harbors two Z-DNA binding domains (ZBDs). Although the dsRBDs are most likely involved in RNA binding and substrate recognition, the ZBDs have been suggested to assist in targeting the enzyme to transcriptionally active regions in chromatin (Liu et al., 1998). ADAR2 and ADAR3 proteins, in contrast, contain only two dsRBDs and have a short amino terminus that lacks any ZBDs (Gerber and Keller, 2001). Additionally, ADAR3 contains a single-stranded RNA-binding region at its amino-terminal end. Most interestingly, ADAR3 seems enzymatically inactive and has been implicated in regulating ADAR1 and ADAR2 enzyme function (Chen et al., 2000). Furthermore, alternatively spliced transcripts and differential promoter usage have been detected in several species for both ADAR1 and ADAR2 genes, most of which are giving rise to proteins with slightly altered enzyme activities (Liu et al., 1997).
Knockout mice indicate that both ADAR1 and ADAR2 enzymes are essential for normal development and life (Higuchi et al., 2000; Wang et al., 2000). However, despite the widespread expression of these proteins only few endogenous substrates are currently known. One of the best-studied examples is the mRNA-encoding subunit B of the glutamate-gated ion channel family, which is edited at a total of three sites. These three sites seem to be edited by different enzymes. One exonic site, termed the Q/R site, is predominantly edited by ADAR2, whereas editing at the R/G site can be performed by ADAR1 and ADAR2. An intronic site seems exclusively edited by ADAR1 (reviewed by Bass, 1997).
The fact that some editing sites are defined by base-paired regions formed between intronic and exonic sequences indicates that editing has to occur cotranscriptionally in the nucleus before intron removal takes place. Consistently, cell fractionation and cell-staining experiments confirmed that ADAR1 and ADAR2 enzymes are predominantly localized in the nucleus (O'Connell and Keller, 1994; O'Connell et al., 1995). Within the nucleus at least ADAR1 has been found associated with nascent transcripts (Eckmann and Jantsch, 1999). However, it has been demonstrated that two versions of hsADAR1 exist that are derived from alternative promotor use that give different distribution patterns in the cell. A full-length version of human (hs)ADAR1 is expressed from an interferon-inducible promoter, giving rise to a 150-kDa ADAR1-i version that is both nuclear and cytoplasmic. Use of a constitutive promoter, in contrast, produces a smaller, predominantly nuclear 110-kDa ADAR1-c protein (George and Samuel, 1999a,b).
Putative nuclear localization signals (NLSs) had been described in both mammalian and Xenopus ADAR1 proteins. However, in a recent study we could show that neither of these predicted NLSs is biologically active. Instead, we identified a short basic NLS in the amino-terminal end of Xenopus ADAR1 that is necessary and sufficient for nuclear import. Human ADAR1, in contrast, has an NLS that overlaps almost entirely with the third double-stranded RNA-binding domain (dsRBD). Mutations affecting RNA binding in this dsRBD have little or no influence on NLS activity, suggesting that RNA binding and NLS activity are separable functions (Eckmann et al., 2001). Furthermore, although Xenopus ADAR1 is constitutively nuclear, human ADAR1-i displays the characteristics of a shuttling protein: first, nuclear accumulation of hsADAR1-i is transcription dependent; and second, when expressed at low levels full-length hsADAR1-i is predominantly nuclear, whereas high levels of the protein lead to its cytoplasmic accumulation. Although this latter finding might also be explained by a saturable nuclear import mechanism, the fact that a reporter construct only containing the active NLS of hsADAR1 shows efficient nuclear accumulation even at high protein levels argues against this idea. Moreover, treatment of cells with leptomycin B (LMB), an inhibitor of Crm1-dependent nuclear export, leads to increased nuclear accumulation of overexpressed hsADAR1-i (Fukuda et al., 1997). It thus seems that hsADAR1-i can both be imported and exported from the nucleus. The finding that nuclear accumulation of endogenous hsADAR1 is transcription dependent also suggests that either one of those two processes can be controlled in a transcription- or RNA-dependent manner (Eckmann et al., 2001).
In this study, we have set out to determine the regions in full-length human ADAR1-i that regulate nuclear export and/or modulate nuclear import of this protein. We could identify three separable elements, all of which affect nuclear accumulation of hsADAR1-i via seemingly different mechanisms.
MATERIALS AND METHODS
Pyruvate Kinase (PK) Fusions for Transfection Assays
Regions to be tested for NLS activity were amplified using suitable primers containing HinDIII sites. The amplified fragments were cut with HinDIII and cloned in-frame upstream of the pyruvate kinase cDNA into one of the vectors described below. To amplify regions corresponding to regions of the human ADAR1 gene we used a previously cloned full-length hsADAR1 cDNA as a polymerase chain reaction (PCR) template (Eckmann et al., 2001). For Xenopus ADAR1, we used an already cloned cDNA encoding the entire xlADAR1 open reading frame (ORF) as a template (Eckmann and Jantsch, 1999).
To test ADAR1 fragments for NLS activity in tissue culture cells the fragments were cloned in frame with a pyruvate kinase-encoding ORF in a pcDNA3 derivative described previously (Eckmann et al., 2001). The vector provided its own AUG for accurate expression upstream of a unique HindIII site used for the insertion of the fragments of interest. The pyruvate kinase reporter protein was fused to six tandemly arranged myc-epitopes at its C terminus. For green fluorescent protein (GFP) tagging, the GFP open reading frame was inserted upstream of the myc tags. Insertion of the GFP ORF allowed in-frame expression of a PK-GFP fusion protein. However, stop codons at the 3′ end of the GFP ORF prevented expression of the myc-tags.
Production of Chimeric Fusion Proteins and Deletions
Chimeric constructs or deletion constructs were made by fusing two independent PCR fragments via a unique restriction site. To do this, each of the two fragments used for chimera production was amplified with a HindIII primer on one side and a XhoI- or XbaI-containing primer on the other side. The PCR fragments were subsequently cut with the enzymes found on either end of the fragments and inserted in a single ligation reaction into the HindIII site of the above-described PK-myc or PK-GFP vectors. After transformation of Escherichia coli, clones containing the expected fragments in the correct orientation were identified by restriction mapping. Putative positive clones were verified by sequencing.
Tissue Culture and Transfection Assays
HeLa or mouse 3T3 cells grown on coverslips were transfected using Tfx-20 transfection reagent, following the manufacturer's instructions (Promega, Madison, WI). Subsequently, cells were fixed, permeabilized, and stained using monoclonal antibody 9E10 as described previously (Jantsch and Gall, 1992). GFP-tagged proteins were either visualized directly after fixation using an appropriate filter set. Alternatively, cells were stained using a polyclonal anti-GFP rabbit serum as a primary antibody. Secondary antibodies were either labeled with fluorescein isothiocyanate (FITC) or tetramethyl rhodamin isothiocyanate. Microscopic images were taken on a fluorescence microscope (Carl Zeiss, Jena, Germany) equipped with filters for FITC, GFP, rhodamine, and 4,6-diamidino-2-phenylindole (DAPI) by using an ORCA cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ). Images were imported into Photoshop 5 (Adobe Systems, Mountain View, CA) with the help of a QED plugin module (QED-imaging, Pittsburgh, PA).
Drug Treatment of Tissue Culture Cells
To inhibit transcription, actinomycin D (AMD) (Sigma-Aldrich, St. Louis, MO) was added to the tissue culture medium at a final concentration of 40 μg/ml 5–10 h before harvesting of the cells. To inhibit nuclear export, leptomycin B (a kind gift of Minoru Yoshida, University of Tokyo, Tokyo, Japan) was added at a final concentration of 10 ng/ml to the tissue culture cells 5–8 h before fixation. Interferon-α or interferon-γ was added to a final concentration of 1000 U/ml and 5 ng/ml, respectively, for 5 h before fixation. Ocadaic acid was added to a final concentration of 10 μM for 20 min. After removal of ocadaic acid cells were washed and incubated for an additional 6 h before fixation and staining.
RESULTS
A Leucine-rich Export Signal That Overlaps with the First Z-DNA Binding Domain in hsADAR-i
We previously showed that the third dsRBD of hsADAR1 is the active NLS of this protein. Fusion of this dsRBD to either myc- or GFP-tagged chicken pyruvate kinase (PK-myc or PK-GFP) was sufficient to confer nuclear localization to the resulting protein. In contrast, full-length hsADAR1 protein was predominantly cytoplasmic but did enrich in the nucleus upon LMB treatment (Eckmann et al., 2001). We therefore set out to identify potential leucine-rich export signals in hsADAR1 that could suppress nuclear localization of the full-length protein. To do this, various regions of the hsADAR1 cDNA were added in frame to a construct encoding the third dsRBD of hsADAR1 that was N-terminally fused to PK-myc or PK-GFP (Figure 1, A and B). These constructs were transfected into HeLa cells and the intracellular distribution of the translation product was determined by immunofluorescence staining using either an anti-myc or anti-GFP antibody or by autofluorescence of the GFP protein. Using this assay, we found that the amino-terminal part of hsADAR-i containing aa 8–205 (construct hsNLS55 in Figure 1B) could confer cytoplasmic localization to the reporter construct (Figure 2). Further deletions within this fragment allowed us to identify a leucine-rich fragment located between aa 122 and 153 that was sufficient for cytoplasmic accumulation of the resulting protein (construct hsNLS80 in Figures 1B and 2). This fragment overlapped with the first Z-DNA binding domain (Z-alpha) located within hsADAR1-i (Herbert et al., 1998). To verify that this region was a leucine-rich NES that could interact with Crm1, several further experiments were performed. First, we tested whether cytoplasmic accumulation of construct hsNLS 80 was LMB sensitive. As expected, LMB treatment of cells transiently expressing this construct did result in its rapid accumulation in the nucleus (Figure 2). Second, introduction of leucine-to-alanine mutations within this region also led to nuclear accumulation of this construct. Constructs hsNLS82 mutating leucines 147 and 150 into alanines show strong nuclear accumulation, whereas hsNLS103 carrying an additional leu133ala mutation is exclusively nuclear (Figures 1B and 2; our unpublished data). As mentioned, the fragment encoded by construct hsNLS 80 partially overlaps with Z-alpha, the structure of which has been determined. Interestingly, only leucine 133 seems to be exposed, whereas both leucines 147 and 150 are predicted to be buried on the inside of this domain (Schwartz et al., 1999). We therefore tested the importance of these three leucines in the context of the entire Z-alpha domain. Thus, the entire Z-alpha domain carrying either no mutation (construct hsNLS131), a single leu133ala mutation (construct hsNLS130), or the triple mutation leu133/147/150ala (construct hsNLS132) were fused to the active NLS dsRBD3. As expected, the wild-type Z-alpha domain found in hsNLS131 conferred cytoplasmic localization that was sensitive to LMB treatment (our unpublished data). The single leu133ala mutation, however, was equally distributed between the nucleus and cytoplasm, whereas the triple mutation found in hsNLS132 showed an almost exclusive nuclear localization (Figures 1B and 2). These data suggest that leucine residues 147 and 150 are important for nuclear export and might thus be partially exposed under conditions that trigger export of hsADAR1.
Figure 1.
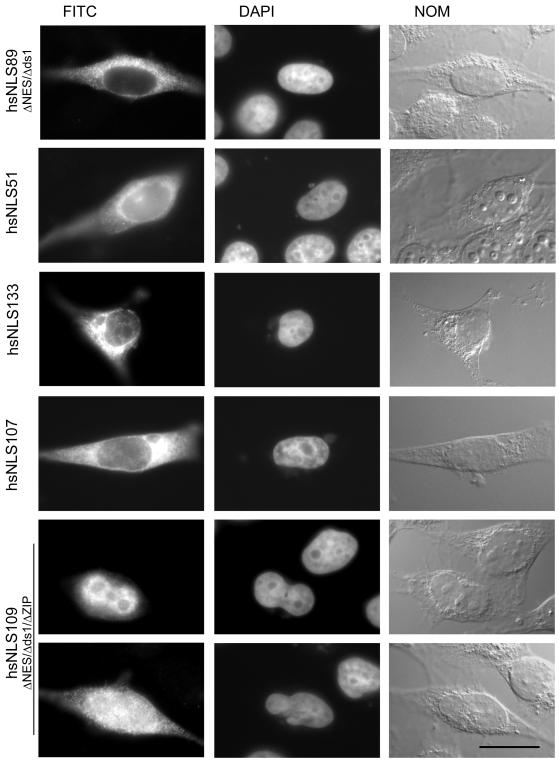
Regions in hsADAR1 that regulate nuclear import and export. (A) Fragments tested for their influence on nuclear accumulation were inserted in a unique HindIII site downstream of an AUG start codon and upstream of a pyruvate kinase-encoding ORF. The pyruvate kinase ORF was fused to either seven myc tags or GFP. (B) Schematic representation of the human ADAR1-i protein. Dotted boxes represent the Z-DNA binding domains and gray boxes the three dsRBDs. The hatched box at the end of the protein depicts the deaminase domain. The minimal NLS of hsADAR1 is indicated by the black triangle. The leucine-rich nuclear export signal is marked as “Expo.” The MOI, overlapping the first dsRBD, is labeled MOI and a putative leucine-zip region that leads to cytoplasmic accumulation of hsADAR1 is labeled ZIP. Regions tested as PK fusion proteins are drawn to scale underneath. Thick green lines represent fragments that accumulate in the nucleus thin red lines those that show cytoplasmic accumulation. Construct numbers are indicated in front of each construct. hsNLS80, labeled with a red star, is the minimal construct showing cytoplasmic accumulation. The amino acid sequence of this NES is shown in the enlargement. Construct hsNLS82 mutates leucines 147 and 150 into alanines, whereas construct hsNLS103 carries an additional L133A mutation. Both constructs are nuclear. Constructs hsNLS130 and hsNLS132 carry the single L133A or the triple L133, 147, 150A mutations, respectively, in the context of the full-length Z-DNA binding domain. The minimal NES fused to an SV40 NLS is also cytoplasmic (hsNLS106). (C) The first dsRBD in hsADAR1 interferes with nuclear import. hsNLS65 marked by a red star is predominantly cytoplasmic. Constructs labeled in gray are found in both the nuclear and cytoplasmic compartments. A mutation in dsRBD1 (hsNLS83) or dsRBD3 (hsNLS87) leads to reduced cytoplasmic accumulation, whereas a mutation in both dsRBDs (hsNLS94) leads to complete nuclear accumulation. Mutations are indicated by small arrows. Construct hsNLS95 has a fragment of the lacZ protein inserted between dsRBDs 1 and 3. Fusion of the MOI region to either an SV40 NLS (hsNLS98) or the basic bipartite NLS of Xenopus ADAR1 (hsNLS92) does not interfere with the activity of these NLSs. Similarly, fusion of both dsRBD1 and dsRBD3 to an SV40 NLS did not interfere with the SV40 NLS activity. Other dsRBDs can interfere with nuclear accumulation to a variable degree. dsRBDs 1 or 2 of the Xlrbpa protein lead to equal distribution between the nucleus and cytoplasm (hsNLS105 and hsNLS96), whereas the first dsRBD of Xenopus ADAR1 leads to predominantly cytoplasmic accumulation (hsNLS84). dsRBD2 of hsADAR1 does not interfere with nuclear accumulation (hsNLS66). (D) A putative b-zip dimerization domain interferes with nuclear accumulation. Construct 108 is cytoplasmic. A C-terminal deletion (hsNLS107) leads to a slight increase in nuclear accumulation (depicted in orange). The entire deaminase domain can confer cytoplasmic accumulation when fused to an SV40NLS (hsNLS133). The candidate dimerization region is shown enlarged and hydrophobic residues in a heptad periodicity are marked bold. However, construct 110 carrying the double mutationV906P and I913A is still predominantly cytoplasmic. The triple deletion construct hsNLS109 is both nuclear and cytoplasmic and is hence depicted in a faded green.
Figure 2.
A leucine-rich export signal in hsADAR1-i. hsNLS35 is exclusively nuclear, whereas hsNLS80 containing the minimal NES fused to the NLS of hsADAR1 is almost exclusively cytoplasmic. Cyoplasmic accumulation of hsNLS80 is LMB sensitive (hsNLS80+LMB). Mutation of two (our unpublished data) or three (hsNLS103) leucines into alanines inhibits NES activity. Mutation of a single leucine 133 in the context of the entire Z-DNA binding domain only has a moderate effect on NES activity (hsNLS130), whereas NES activity is almost completely abolished in the triple mutation hsNLS132. The NES is transferable to an SV40 NLS (hsNLS106) where it is also sensitive to leptomycin B (hsNLS106+LMB). FITC channel shows localization of the PK fusion protein reporter after detection with an appropriate antibody. DAPI channel shows the localization of the nucleus, whereas the whole cell outline can be seen in the Nomarski optics channel (NOM). Bar, 20 μm.
As mentioned above, hsADAR1 contains an atypical NLS that overlaps entirely with its third dsRBD. It therefore seemed conceivable that the newly identified NES would only act on a construct bearing the NLS found in hsADAR1. Thus, to test whether NES activity was transferable, the fragment of interest was fused upstream of a basic, monopartite simian virus 40 (SV40) NLS, which by itself would lead to nuclear accumulation of the PK-GFP reporter construct (our unpublished data) (Hodel et al., 2001). As predicted, addition of the leucine-rich NES of hsADAR to the SV40 NLS-containing construct resulted in its cytoplasmic accumulation (construct hsNLS106 in Figure 2). Cytoplasmic accumulation was sensitive to LMB treatment, indicating that the fragment in question did show NES activity independent of the type of NLS present in the reporter construct (Figure 2).
Xenopus ADAR1.1 Seemingly Lacks a Leucine-rich NES
X. laevis ADAR1.1 is highly homologous to human ADAR1-i. Yet Xenopus but not human ADAR1 is constitutively nuclear, even when overexpressed in HeLa cells or after actinomycin-D treatment (Eckmann et al., 2001). We therefore compared the region containing the leucine-rich export signal of mammalian ADAR1 proteins with that of Xenopus, Fugu, and zebrafish proteins. Interestingly, although this region is highly conserved between rat and human ADAR1 it is less conserved in Xenopus or fish ADAR1. Specifically, two leucine residues at position 147 and 150 in human ADAR1 are replaced by phenylalanine and isoleucine, respectively, in the Xenopus protein. Moreover, all basic residues found in this region in mammalian ADARs are missing in the corresponding Xenopus sequence, suggesting that the region in the Xenopus protein does not give rise to a functional NES (Figure 3). In Fugu and zebrafish ADAR1 the NES seems even less conserved.
Figure 3.
Xenopus ADAR1 lacks a leucine-rich NES. Displayed is the region containing the NES of hsADAR1-i after a multiple sequence alignment of the entire protein with zebrafish (dr), Fugu (fu), rat (rn), and Xenopus (xl) ADAR1 proteins. Amino acid positions in the corresponding proteins are indicated. Conserved leucines are printed bold, whereas other conserved hydrophobic residues are boxed. The three leucines mutated in hsNLS103 that have been proven to be essential for NES activity are printed in underlined italics at the positions marked by an arrow. L147 and L150 are not conserved in Xenopus ADAR1. Similarly the positions corresponding to L144 and L126 in the human protein are not conserved in any of the fish sequences. Additionally, fish and Xenopus sequences differ by several insertions and gaps from the mammalian ADAR1 sequences.
As mentioned, the leucine-rich NES of human ADAR1-i is capable to transfer an SV40 NLS-containing fragment to the cytoplasm (hsNLS106 in Figure 2). This indicates that, if present, a leucine-rich NES would be strong enough to compete with the basic NLS found in Xenopus ADAR1.1. Thus, both sequence comparison and biological data indicate that Xenopus ADAR1.1 lacks a functional NES.
A dsRBD Can Interfere with NLS Activity in Human ADAR1-i
To test whether the identified NES was indeed responsible for the observed cytoplasmic localization of ectopically expressed hsADAR1-i we deleted the corresponding region from full-length hsADAR1-i. The resulting construct, termed hsADAR1-i/ΔNES, was fused to PK-myc and tested for its intracellular distribution. Much to our surprise this construct still showed prominent cytoplasmic localization (hsNLS81 in Figure 4). Removal of the PK part of the construct did not alter its intracellular localization, indicating that the observed cytoplasmic distribution was not caused by the PK part of the protein (our unpublished data). Furthermore, LMB treatment did not alter the dominant cytoplasmic localization of the ΔNES construct, indicating that cytoplasmic accumulation was not caused by a second Crm1-dependent export signal. We therefore screened additional regions of the hsADAR1-i protein for their ability to inhibit nuclear accumulation of the PK-GFP reporter construct containing the third dsRBD of hsADAR1 as an active NLS.
Figure 4.
A dsRBD can modulate nuclear accumulation of hsADAR in an RNA-binding–dependent manner. Deletion of the NES from hsADAR1 does not restore nuclear accumulation (hsNLS81). Similarly, a region containing dsRBDs 1–3 (hsNLS38) or dsRBDs 1 and 3 alone (hsNLS65) also show cytoplasmic accumulation. A single point mutation that interferes with RNA binding of either dsRBD1 (hsNLS83) or dsRBD3 (hsNLS87) leads to increased nuclear localization. A double mutation interfering with RNA binding in both dsRBDs (hsNLS94) leads to strong nuclear accumulation. Mutated dsRBDs are indicated by bold italics. hsNLS95 has dsRBDs 1 and 3 separated by 49 unstructured amino acids of the lacZ protein. The cytoplasmic accumulation of this construct indicates that dsRBD1 does not interfere with nuclear accumulation by inhibiting the folding of dsRBD3. Bar, 20 μm.
Initially, the minimal NLS was gradually extended N terminally (Figure 1C). Using this assay we could show that a fragment covering dsRBDs 1 through 3 (hsNLS38) did give rise to predominantly cytoplasmic localization (Figure 4). In some cells nuclear signals were also discernible that never exceeded an estimated 10% of the observed cytoplasmic signal. Further mapping experiments showed that construct hsNLS65, only containing dsRBD1 and 3, was the minimal construct giving a predominantly cytoplasmic distribution (Figure 4). Deletion into dsRBD1 from either end immediately restored the nuclear localization caused by the dsRBD3-resident NLS. This indicated that the entire dsRBD1 was necessary to cause cytoplasmic accumulation of the reporter construct. Cytoplasmic accumulation of hsNLS65 was insensitive to LMB treatment, indicating that this dsRBD was not a substrate for Crm1-mediated nuclear export (our unpublished data).
dsRBD Cross Talk Requires RNA Binding
Considering that the active NLS of hsADAR1-i was found to be contained within dsRBD3, and given the fact that a fragment within dsRBD1 could prevent nuclear accumulation, raised the obvious question of whether RNA binding was required for the activity of either domain. We had shown previously that RNA binding of the third dsRBD was not required for NLS activity (Eckmann et al., 2001). Therefore, we tested whether the first dsRBD required its RNA-binding capability to allow its interference with nuclear accumulation caused by the NLS within dsRBD3. To do this histidine 531 located in dsRBD1 was mutated into an alanine. This histidine residue is highly conserved in dsRBDs and several studies have shown that its mutation impairs RNA binding dramatically (Bycroft et al., 1995; Krovat and Jantsch, 1996). Interestingly, the resulting construct hsNLS83 containing a mutated dsRBD1 and a wild-type dsRBD3 showed a relatively strong nuclear staining compared with hsNLS65, containing both wild-type domains (Figure 4). Even more surprisingly, construct hsNLS87, carrying a wild-type dsRBD1 together with the above-described histidine-to-alanine mutation at position 754 in dsRBD3, also showed an increased nuclear accumulation comparable with that of hsNLS65. Additionally, within nuclei hsNLS87 showed an enriched nucleolar staining. Finally, a double mutation inhibiting RNA binding of both the first and third dsRBD showed an almost exclusively nuclear localization (hsNLS94). These data clearly demonstrated that although RNA binding was not required for NLS activity of dsRBD3, it was essential for the cytoplasmic accumulation caused by dsRBD1. Moreover, the fact that mutations affecting RNA binding in either dsRBD1 or dsRBD3 had comparable effects and that the double mutation was entirely nuclear suggests that cytoplasmic localization caused by dsRBD1 might depend on a common RNA bound by both dsRBDs. These data also suggest that dsRBD1 could lead to cytoplasmic accumulation by masking the NLS present in dsRBD3 through binding to an RNA. In principle, however, RNA binding might also be required to unravel an otherwise hidden nuclear export signal in dsRBD1. Because the data obtained so far seemed compatible with both a cytoplasmic-anchoring function of dsRBD1 and a nuclear export function of the same domain we decided to call the corresponding region in dsRBD1 modulator of import (MOI).
However, while making our constructs, dsRBDs that would normally be separated from each other by several dozens of amino acids were artificially brought into proximity. It was therefore conceivable that dsRBD1 might interfere with the folding of dsRBD3 by bringing it into an artificial context. To eliminate this possibility, a flexible spacer from the E. coli β-galactosidase protein was introduced between dsRBDs 1 and 3. The resulting construct still did accumulate in the cytoplasm, indicating that the inhibitory function of dsRBD1 on dsRBD3 was not caused by misfolding of the latter (Figure 4).
dsRBD Identity Is Important for Cytoplasmic Accumulation
As shown, cytoplasmic accumulation of construct hsNLS65 containing both dsRBD1 and 3 of hsADAR1 is RNA-binding dependent. We therefore wanted to know whether other dsRBDs could also act as MOIs, and if so, whether this activity would correlate with the RNA-binding ability of the dsRBD.
Thus, chimeric constructs containing individual dsRBDs of various proteins fused to the third dsRBD of human ADAR1 were made. Specifically, we used the first dsRBD of xlADAR1, the first or the second dsRBD of the X. laevis RNA-binding protein A (Xlrbpa), and the second dsRBD of hsADAR1. As a control, the first dsRBD of hsADAR was included. The domains all show different RNA-binding capacities in vitro. The second dsRBD of Xlrbpa is by far the strongest RNA-binder available in our laboratory (Krovat and Jantsch, 1996). In contrast dsRBD1 of xlADAR1 and dsRBD1 of Xlrbpa show almost no RNA-binding ability (Brooks et al., 1998; Jantsch, unpublished data). Similarly, dsRBD2 of hsADAR1 has been reported to be a weak RNA binder (Liu and Samuel, 1996). Most interestingly, all domains except dsRBD2 of hsADAR1 could mediate cytoplasmic accumulation of the chimeric construct. The extent of cytoplasmic accumulation varied among constructs but did not correlate with the RNA-binding capacity of the dsRBD (Figure 5). Construct hsNLS65 containing the original dsRBDs 1 and 3 of hsADAR1 showed the strongest cytoplasmic accumulation with only minor amounts of protein present in the nucleus. Cytoplasmic staining of similar intensity could be observed when dsRBD1 of xlADAR1 was fused to dsRBD3 of hsADAR1 (construct hsNLS84). This Xenopus domain is most closely related to dsRBD1 in human ADAR1 but is a relatively weak RNA binder. In contrast, constructs hsNLS105 and hsNLS96 containing the first and second dsRBDs of Xlrbpa, respectively, both showed moderate cytoplasmic accumulation, giving rise to similar signal intensities in the cytoplasmic and nuclear compartments (Figure 5). Because dsRBD2 of Xlrbpa is a very strong RNA binder and dsRBD1 of this protein shows almost no RNA-binding ability, this clearly indicates that RNA-binding does not correlate with the amount of cytoplasmic accumulation observed. Moreover, construct hsNLS66 containing dsRBDs 2 and 3 of hsADAR1 showed an exclusively nuclear accumulation. Taken together, our data indicate that although RNA binding seems required for MOI activity, the type of dsRBD present in a protein seems more important than its RNA-binding strength.
Figure 5.
Some but not all dsRBDs can modulate nuclear accumulation. hsNLS65 containing both dsRBD1 and dsRBD3 of hsADAR is predominantly cytoplasmic. Also dsRBD1 of Xenopus ADAR1 can interfere with nuclear accumulation, leading to a predominantly cytoplasmic accumulation of the reporter construct (hsNLS84). dsRBDs 1 and 2 of the Xlrbpa protein, in contrast, show only moderate interference with nuclear accumulation, leading to an equal distribution of the protein in both the nucleus and cytoplasm (hsNLS105 and hsNLS96). dsRBD2 of hsADAR1 does not interfere with nuclear accumulation (hsNLS66). Bar, 20 μm.
MOI Activity Is Transcription Independent
Endogenous hsADAR1 is located to the nucleus in a transcription-dependent manner. The protein is normally localized to the nucleus, whereas treatment of cells with the transcriptional inhibitor AMD results in translocation of the protein to the cytoplasm (Eckmann et al., 2001). This type of transcription-dependent nuclear localization is the hallmark of many shuttling proteins (Pinol-Roma and Dreyfuss, 1992; Michael et al., 1995). The mechanism underlying this phenomenon, however, is poorly understood. We therefore wondered whether constructs containing dsRBD1 and dsRBD3 would also show transcription-dependent nuclear accumulation and move to the cytoplasm in the absence of transcription. Because hsNLS65, containing both a wild-type dsRBD1 and dsRBD3 was already predominantly cytoplasmic even during ongoing transcription we primarily focused on constructs hsNLS83, hsNLS87, and hsNLS94 containing mutations in dsRBD1, dsRBD3, or both domains, respectively. As previously stated, any of these mutations leads to increased nuclear accumulation with maximum nuclear localization being reached by the double mutation hsNLS94. However, AMD treatment did not alter the nucleocytoplasmic distribution of any of these constructs, suggesting that the transcription-dependent shuttling behavior of hsADAR1 does not solely depend on its dsRBDs (our unpublished data).
MOI Activity Is Not Transferable to Other NLSs
As mentioned, dsRBD1 of hsADAR1 might mediate cytoplasmic accumulation by either masking the dsRBD3-resident NLS or, alternatively, by acting as a nuclear export signal. To get a hint which of these two possibilities would be more likely we tested whether dsRBD1 would also have MOI activity when fused to different NLSs. We expected that masking of an NLS would be very specific for the type of NLS present, whereas NES activity should also be transferable to other NLSs. We therefore fused dsRBD1 of hsADAR1 to either the bipartite type NLS of xlADAR1 or to the SV40 basic NLS (Eckmann et al., 2001; Hodel et al., 2001). However, the resulting constructs showed exclusively nuclear localization, indicating that MOI activity of dsRBD1 was not transferable to other NLSs (Figure 6).
Figure 6.
MOI activity is specific for a dsRBD-NLS. To test whether the first dsRBD would also interfere with the nuclear accumulation of constructs containing other NLSs the MOI-containing region was either fused to a fragment containing the bipartite NLS of Xenopus ADAR1 (hsNLS92) or to an SV40 NLS (hsNLS98). In either case, no MOI activity could be detected and all construct showed strong accumulation in the nucleus. Similarly, fusion of both dsRBD1 and dsRBD3 to an SV40 NLS (hsNLS134) also showed strong nuclear accumulation, indicating that even a combination of two dsRBDs was unable to anchor an SV40 NLS in the cytoplasm or to export it efficiently from the nucleus. Note that the presence of the first dsRBD does lead to an accumulation of the protein in the nucleolus. The FITC channel shows the localization of the fusion protein. DAPI shows position of nuclei, whereas the whole cell images can be seen in the Nomarski (NOM) images. Bar, 20 μm.
Our finding that cytoplasmic accumulation was dependent on the presence of the two actively RNA-binding dsRBDs 1 and 2 also opened the possibility that a putative nuclear export activity might only be exhibited by constructs containing a combination of two dsRBDs. To test for this possibility we fused dsRBDs 1 and 3 to an SV40 NLS and analyzed the cellular distribution of the resulting construct (hsNLS134). However, hsNLS134 was predominantly nuclear (Figure 6). These results support the idea that MOI activity acts by masking an NLS rather than by mediating nuclear export.
A Putative Leucine-Zipper Region Prevents Nuclear Accumulation of hsADAR1
To test whether the leucine-rich NES and the dsRBD1-resident MOI were the only regions within hsADAR1 that would lead to cytoplasmic accumulation of the protein we deleted the corresponding regions giving a Δ-NES/Δ-MOI construct. Surprisingly, this construct still showed considerable cytoplasmic accumulation (hsNLS89 in Figure 7). We therefore extended our search for regions interfering with nuclear accumulation of hsADAR1 toward the C terminus of the protein.
Figure 7.
The C-terminal deaminase domain of hsADAR1 interferes with nuclear accumulation. hsNLS89 deleting both the NES and the first dsRBD of hsADAR1 still shows almost exclusive cytoplasmic localization. A fragment extending from the NLS of hsADAR1 to the very C terminus (hsNLS51) is also cytoplasmic. Fusion of the entire deaminase domain to an SV40 NLS (hsNLS133) also resulted in cytoplasmic accumulation, indicating that the deaminase domain can confer cytoplasmic accumulation to various NLSs. hsNLS107 contains a short fragment from the amino-terminal end of the deaminase domain fused to the NLS of hsADAR1. The resulting fragment also shows strong cytoplasmic accumulation. A construct deleting this region in addition to the NES and dsRBD1 accumulates in the nucleus to a variable degree. In some cells the protein is almost exclusively in the nucleus, whereas in other cells the protein is both in the nucleus and cytoplasm (hsNLS109, top and bottom). Bar, 20 μm.
Doing so, we could identify a region located in the N-terminal half of the deaminase domain that did interfere with nuclear accumulation of a dsRBD3-NLS–containing reporter construct. This fragment is ∼100 amino acids in length. Closer investigation of this region allowed us to identify several hydrophobic amino acids in heptad periodicity with the potential to form a leucine-zipper-like dimerization domain (Figure 1D). However, to this point, mutation of two hydrophobic residues into alanines has no effect on nuclear accumulation of the resulting constructs. Thus, the presence of a leucine-zipper element in this region remains to be proven. Nonetheless, a possible way a leucine-zipper or any other protein–protein interaction domain could lead to cytoplasmic accumulation would be by forming homodimers with cytoplasmic hsADAR1-i. We had shown previously that LMB treatment leads to increased nuclear accumulation of both endogenous and ectopically expressed hsADAR1. We therefore reasoned that if dimer formation with endogenous hsADAR1 might prevent nuclear accumulation of hsNLS51, a construct containing the C-terminal inhibitory region, LMB treatment should cause at least partial nuclear enrichment. However, LMB treatment had no effect on the intracellular distribution of this construct (Figure 7).
We also considered the possibility that cytoplasmic localization might be caused via interaction with a cytoplasmic anchor protein, similar to the one described for the interaction between nuclear factor-κB and inhibitory κB (reviewed by Mattaj and Englmeier, 1998). Interaction with such a cytoplasmic anchor might be sensitive to several factors, allowing a dynamic regulation of nuclear entry. Interferon stimulation had already been shown to induce hsADAR1 expression and cause cytoplasmic accumulation of the protein. We thus tested the influence of human interferon-α, interferon-γ, or the phosphatase inhibitor ocadaic acid on this construct. However, none of these treatments had an effect on the primarily cytoplasmic localization of hsNLS51 or any other construct containing the C-terminal fragment. Nonetheless, additional deletion of the amino-terminal part of the deaminase domain, giving rise to a ΔNES/ΔMOI/Δdeminase construct did give rise to a construct showing increased nuclear accumulation. Nuclear accumulation was not complete and varied from cell to cell. In ∼20% of transfected cells the protein could be found predominantly in the nucleus (Figure 7). In the remaining 80% the protein was distributed about equally in the nucleus and cytoplasm, suggesting that yet another region in hsADAR1 might interfere with the nuclear accumulation of the protein (Figure 7). Alternatively, it is also possible that the deletion of the putative zip-region was not large enough thus still leaving some crucial regulatory elements in the remaining construct.
Formally, we also had to consider the possibility that our deletion constructs would expose parts of the deaminase domain that would normally not be exposed, thereby creating artificial domains that would interfere with the atypical NLS of hsADAR1. Thus, to determine whether the entire deaminase domain could regulate nuclear accumulation independent of the type of NLS present we fused the entire domain to an SV40 NLS (hsNLS133). The resulting fusion construct, hsNLS133, showed an almost exclusive cytoplasmic accumulation, suggesting that the deaminase domain was able to confer cytoplasmic accumulation independent of the NLS present (Figure 7).
Thus, taken together our data show that hsADAR1-i contains three regions that interfere with nuclear accumulation of this protein via seemingly different mechanisms. It also shows that the constitutively expressed hsADAR1-c shares at least two of these regulatory regions with hsADAR1-i. Similar regulatory elements might even be present in ADAR2 proteins.
DISCUSSION
Evolution of an Amino-terminal Nuclear Export Signal in Mammalian ADAR1-i
In this study we could show that nuclear entry of hsADAR1 is regulated at several levels. At its amino terminus hsADAR1-i contains a leucine-rich nuclear export signal. This export signal is Crm1 dependent and can be inhibited by LMB treatment. Recently, another group has identified the same region as a Crm1-dependent nuclear export signal in human ADAR1 (Poulsen et al., 2001). Poulsen et al. (2001) could also show that a fragment of hsADAR1 containing this sequence can physically interact with Crm1. However, the NES identified by Poulsen et al. (2001) is located between amino acids 125 and 137 and is thus smaller than the minimal NES identified by us (Poulsen et al., 2001). Because mutagenesis of leucines 147 and 150 leads to increased nuclear localization we would suggest that the NES is located between amino acids 125 and 147 in hsADAR1. In fact, construct hsNLS80 containing amino acids 122–153 contains the minimal fragment identified by us that can mediate nuclear export of a reporter construct. Interestingly, the NES activity is transferable and works also in combination with a basic SV40 NLS.
We had shown previously that Xenopus ADAR1 is exclusively nuclear and that the NLSs found in human and Xenopus ADAR1 proteins are different. The fact that the human NES can translocate a construct containing a basic NLS to the cytoplasm, together with the finding that Xenopus ADAR1 is constitutively nuclear, suggests that Xenopus ADAR1 lacks an active NES. In fact, alignment of the corresponding sequences from mouse, human, and Xenopus reveals that these are only partially conserved. Although a high degree of conservation can be found in the amino-terminal part, the C-terminal part of the NES is less conserved in Xenopus. Remarkably, both the spacing and identity of hydrophobic residues is different in the Xenopus sequence. The sequence is even less conserved in more distantly related species such as Fugu or zebrafish. Taken together, this suggests that a functional NES has only evolved in the vertebrate line somewhere between amphibia and mammals. A functional NES is apparently not essential for enzyme function. The NES is also absent from ADAR2 proteins and from the constitutively expressed shorter version of hsADAR1. Thus, a nuclear export signal seems to have coevolved with a strongly inducible interferon-responsive promoter. This in turn raises the question whether hsADAR1-i might also have a cytoplasmic function possibly after viral infection.
It should also be mentioned that the NES overlaps with one of the two Z-DNA binding domains found in ADAR1 proteins. It had been proposed that these domains facilitate association of the protein with transcriptionally active regions (Herbert et al., 1998). The fact that both domains overlap raises the possibility that the Z-DNA binding domains help the protein to associate with viral DNAs in the cytoplasm. Interestingly, our mutagenesis data suggest that leucine residues 147 and 150 that are predicted to be buried within the structure of the Z-DNA binding domain are important for NES activity. This seemingly contradictory finding can be interpreted in two ways. On the one hand it might be possible that the Z-DNA binding domain undergoes a conformational change when bound to the export receptor Crm1, thereby leading to exposure of amino acids that are hidden when the domain is bound to Z-DNA. Alternatively, the mutations introduced by us might interfere with the overall structure of the Z-DNA binding domain, thereby also influencing the function of the NES.
Masking of a Nuclear Localization Signal by RNA Binding?
We could also show that dsRBDs can lead to cytoplasmic accumulation of hsADAR1. Because we could not clearly decide whether cytoplasmic accumulation was caused by increased nuclear export or reduced nuclear import, we chose to call the region covering the first dsRBD that was causing increased cytoplasmic accumulation of our reporter construct MOI. Several facts, however, lead us to believe that the MOI acts by interfering with import rather than by stimulating export. First, MOI activity is specific for the dsRBD-type NLS found in the third dsRBD of hsADAR1 and cannot be transferred to either the basic NLS of Xenopus ADAR1 or an SV40 NLS. Most importantly, however, RNA-binding activity of the NLS is required for cytoplasmic accumulation. The fact that mutation of the import signal leads to a decrease in cytoplasmic accumulation strongly argues for a function of the MOI in inhibiting import. Because RNA binding of either dsRBD1 or dsRBD3 is required we propose that dsRBD1 masks the import activity of dsRBD3 by binding to an RNA. To prevent import, RNA binding would have to take place in the cytoplasm. Such a mechanism would be most easily conceivable if both dsRBDs would bind to the same RNA. One dsRBD could then cover the NLS-constituting surface on dsRBD3. Besides RNA binding this would require a protein–protein interaction between the NLS and MOI-containing dsRBDs. Alternatively, RNA binding might simply lead to a conformational change of the NLS, thereby preventing binding of a suitable transportin molecule. In this scenario the role of the MOI-containing dsRBD would be to enhance overall RNA-binding affinity via cooperative binding. This would imply that only strong RNA binders can act as inhibitors, whereas dsRBDs with weak RNA-binding activity should show little or no MOI activity. In fact, our experiments showed that other dsRBDs could also exhibit MOI activity and cause cytoplasmic accumulation of a dsRBD3-containing reporter construct. However, the amount of protein localized to the cytoplasm did not correlate with the RNA-binding strength of the second dsRBD present. dsRBDs1 and 2 of the Xlrbpa protein, for instance, show very different RNA-binding behaviors but have comparable MOI activities. Moreover, dsRBD2 of hsADAR1 lacks MOI activity completely. Taken together, these data are compatible with a model in which RNA-binding activity is required to initiate or stabilize dsRBD–dsRBD interaction. In addition, however, protein–protein contacts would be required to mask the NLS in dsRBD3. If only some, but not all, dsRBDs are capable of such a protein–protein interaction this would explain why some dsRBDs fail to show MOI activity (Figure 8).
Figure 8.
Cytoplasmic anchoring of hsADAR1 via RNA binding. Nuclear entry of hsADAR1 is mediated via its third dsRBD. When unbound this dsRBD is freely accessible for import factors and can act as an NLS (gray sphere). However, RNA binding of the first and third dsRBD can mask the NLS or lead to a conformational change in the third dsRBD (gray box), thus preventing binding of import factors and subsequent nuclear entry. Nuclear export of hsADAR1-i is mediated via an NES (small box at the N terminus of hsADAR1-i) that is absent from hsADAR1-c. Additionally, homo- or heterodimer formation of the deaminase domain could possibly anchor the protein to the cytoplasm. Similar regulatory mechanisms for nuclear entry might also exist in ADAR2 proteins.
It should be mentioned that while this article was being written an interaction between dsRBDs and exportin-5, a novel export factor, was reported (Brownawell and Macara, 2002). In this study it could also be shown that the interaction between dsRBDs and exportin-5 can be inhibited by double-stranded RNAs. Although we formally cannot exclude that dsRBD1 of hsADAR-i exhibits export activity we believe that several facts argue against this notion: First, the fact that mutations in the dsRBD3 resident NLS lead to reduced cytoplasmic accumulation of a fragment containing both dsRBD1 and 3 suggests that the interaction between both dsRBDs, rather than the activity of a single dsRBD, is important for cytoplasmic accumulation. Second, if dsRBD1 of hsADAR1 was an export signal one would expect this signal to be transferable to other NLSs. Our experiments, however, suggest that this is not the case. Along these lines we also considered the possibility that export activity of dsRBD1 might depend on active RNA binding and could therefore be stimulated by the presence of the actively RNA-binding dsRBD3. However, our data show that even fusion of both dsRBD1 and dsRBD3 to an SV40 NLS does not lead to cytoplasmic accumulation of the resulting construct (hsNLS 134). This finding is compatible with the idea that dsRBD1 acts by masking a dsRBD3-resident NLS rather than by exhibiting export activity. However, we cannot exclude the possibility that such a construct would shuttle very rapidly between the nucleus and cytoplasm with the majority of protein being localized to the nucleus at all times, thus giving the impression that the protein was strictly nuclear.
Deaminase Domain as a Third Regulator for Nuclear Accumulation?
As indicated, we also obtained evidence for the involvement of the deaminase domain in the regulation of nuclear concentration of hsADAR1. The minimal fragment causing cytoplasmic accumulation could be mapped to the amino-terminal part of the deaminase domain and contained a putative leucine zipper-like dimerization motif. However, mutagenesis of two hydrophobic residues within the putative heptad periodicity did not restore nuclear accumulation. Interestingly, the entire deaminase domain can confer cytoplasmic accumulation to an unrelated SV40 NLS, indicating that the observed activity was intrinsic to the deaminase domain and not caused by artificial misfolding of this domain. Furthermore, this result suggests that the deaminase domain does not act by masking the atypical NLS of hsADAR but can function independently of the type of NLS present. However, it is unclear, whether the C-terminal region that leads to cytoplasmic accumulation is acting by inhibiting nuclear import, by anchoring the protein to a cytoplasmic component, or by stimulating or even mediating nuclear export. It should be noted, however, that adenosine deaminases that act on t-RNAs (ADATs) can form heterodimers (Gerber and Keller, 1999). Molecularly, ADATs resemble truncated ADAR molecules that only contain deaminase domains. This suggests that deaminase domains might be capable to interact with each other and that cytoplasmic accumulation of a deaminase domain-containing reporter construct might be caused by interaction of the latter with another ADAR or ADAR-like molecule. LMB treatment, however, had no effect on the localization of our reporter constructs. Because we could show previously, that LMB treatment leads to increased nuclear accumulation of endogenous hsADAR1 this suggests that the inhibitory effect of the deaminase domain on nuclear entry is not caused via its physical interaction with endogenous hsADAR1. Similarly, treatment of cells with interferon-α, interferon-γ, or ocadaic acid also had no effect on the localization of our reporter construct. It thus remains to be determined how the deaminase domain of hsADAR1 or the N-terminal part of it can interfere with nuclear accumulation.
Why Is Nuclear Accumulation So Highly Regulated?
An obvious question being raised by the complex results obtained herein would be why nuclear accumulation of hsADAR1 is regulated through so many signals. A possible explanation might be found by considering the ortho- and paralogues of ADARs. ADAR2 molecules, for instance, lack the amino-terminal part that harbors the leucine-rich export signal found in hsADAR1 (Gerber and Keller, 2001). Similarly, this region also seems to be absent from hsADAR1-c, the constitutively expressed, shorter version of hsADAR1 (Patterson and Samuel, 1995). It is thus conceivable that ADAR2 or hsADAR1-c molecules regulate their nuclear concentration exclusively via the interaction of a dsRBD-resident NLS and an MOI identical to the regulatory mechanism shown by us to occur in our reporter constructs (Figure 8). Although at this stage it is not clear how ADAR2 molecules regulate nuclear entry it is worth considering that these proteins only contain two dsRBDs. Interestingly, multiple sequence alignments indicate that the first dsRBD of ADAR2 molecules is most homologous to the first, MOI containing, dsRBD found in ADAR1. Additionally, the second dsRBD of ADAR2s is most homologous to the NLS-bearing dsRBD3 of ADAR1 molecules. Taken together, this supports the idea that ADAR2 also regulates its nuclear concentration via interaction of its two dsRBDs.
We could also show that at least frogs seem to lack both the leucine-rich NES and the atypical dsRBD-resident NLS. dsRBD-resident nuclear import and MOI signals in ADAR1 might thus have evolved somewhere between the amphibian and mammalian lineages. The presence of additional export signals might thus represent evolutionary leftovers. Finally, the C-terminal deaminase domain can also regulate nuclear accumulation of hsADAR1. Although we do not know the mechanism by which this domain is acting it is tempting to speculate that this domain might also regulate the nuclear accumulation of ADATs.
Our data also raise the question of why nucleocytoplasmic distribution of hsADAR1 needs to be so tightly regulated. Two principle views can be envisaged. On the one hand, nuclear ADAR concentration might have to be kept low, to prevent inadvertent editing of structured endogenous RNAs. On the other hand, ADARs might also have a cytoplasmic function either to edit endogenous substrates or viral RNAs. The latter idea is supported by the fact that the interferon-induced hsADAR1-i version is predominantly found in the cytoplasm (Patterson and Samuel, 1995).
A Single NLS in hsADAR1
As mentioned, another study has also identified the leucine-rich eport signal at the amino-terminal end of hsADAR1-i (Poulsen et al., 2001). Additionally, the study by Poulsen et al. (2001) provided evidence for the existence of both an N-terminal and a C-terminal NLS in hsADAR1-i. Moreover, in their study Poulsen et al. (2001) show that an hsADAR1 molecule lacking the NES was exclusively nuclear, whereas the same fragment was predominantly cytoplasmic in our hands. Although the cytoplsmic accumulation can be easily explained by the regulatory elements identified herein, it also has to be considered that the study by Poulsen et al. (2001) was performed in mouse cells. We therefore repeated several of our experiments in mouse 3T3 cells. However, no differences to the results obtained in HeLa cells could be observed. We also extended our search for the presence of an additional amino-terminal NLS. Despite an extensive search we could not obtain any data supporting the existence of an additional amino-terminal NLS in hsADAR1-i. We also failed to reproduce strong nuclear accumulation of an N-terminally deleted hsADAR1 molecule lacking the NES. The different results obtained by our study and the one performed by Poulsen et al. (2001) might thus be attributable to the different reporter systems used. In our system, we relied on a pyruvate kinase GFP/myc reporter construct that had been used in many different studies investigating nuclear transfer. Moreover, in all of our experiments we used overlapping fragments that always gave comparable results. We are thus very confident that our system mimics the true regulation of nuclear entry of hsADAR1-i.
Implications for Shuttling
As had been shown previously, hsADAR1 is a nuclear enzyme when expressed constitutively and at low levels. Overexpression of hsADAR1-i upon interferon induction, however, leads to cytoplasmic accumulation of the protein (Patterson and Samuel, 1995). Additionally, we could show that overexpression of either full-length or amino-terminally truncated (ΔNES) hsADAR1-c also leads to cytoplasmic accumulation of these proteins. Moreover, transcriptional inhibition, also leads to cytoplasmic accumulation of the protein (Eckmann et al., 2001). This latter finding strongly suggests that hsADAR1 is a shuttling protein. However, it is not clear how transcriptional inhibition causes increased cytoplasmic export of any shuttling protein. The fact, however, that two different dsRBDs are involved in mediating and regulating nuclear import of hsADAR1 strongly suggests that these domains regulate the nucleocytoplasmic distribution of hsADAR1 by directly interacting with RNAs and by responding to different RNA levels.
As we could show, mutations that reduce the RNA-binding ability of either dsRBD1 or dsRBD3 in hsADAR1 both lead to increased nuclear accumulation of hsADAR1. This suggests that binding of both dsRBDs to cytoplasmic RNAs prevents nuclear import. Although this does not explain why, in the absence of transcription, the protein accumulates in the cytoplasm it is still tempting to speculate that binding to a nuclear RNA in turn might also tether the protein to the nucleus. If, for instance, a nuclear RNA would be present at low levels and could thus be saturated very easily, overexpressed hsADAR1 would always tend to accumulate in the cytoplasm. Similarly, inhibition of transcription might reduce the nuclear concentration of such an anchoring RNA and therefore lead to cytoplasmic accumulation of endogenous hsADAR1. However, further experiments will have to be performed to address this question in more detail.
ACKNOWLEDGMENTS
We thank Dr. Pamela Silver (Dana Faber Cancer Institute, Boston, MA) and Dr. Walter Keller (Biocenter Basel) for generous gifts of anti-GFP and anti-mammalian ADAR1 antibodies, respectively. Minoru Yoshida (University of Tokyo, Tokyo, Japan) kindly provided leptomycin. A.S. was supported by the Austrian University foundation grant H42-2000. This work was supported by the Austrian Science Foundation grant SFB 1706 (to M.J.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0161. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0161.
REFERENCES
- Bass BL. RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- Brooks R, Eckmann CR, Jantsch MF. The double-stranded RNA-binding domains of Xenopus laevis ADAR1 exhibit different RNA-binding behaviors. FEBS Lett. 1998;434:121–126. doi: 10.1016/s0014-5793(98)00963-6. [DOI] [PubMed] [Google Scholar]
- Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft M, Grunert S, Murzin AG, Proctor M, St. Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Jantsch MF. The RNA-editing enzyme ADAR1 is localized to the nascent ribonucleoprotein matrix on Xenopus lampbrush chromosomes but specifically associates with an atypical loop. J Cell Biol. 1999;144:603–615. doi: 10.1083/jcb.144.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell. 2001;12:1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene. 1999a;229:203–213. doi: 10.1016/s0378-1119(99)00017-7. [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci USA. 1999b;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- Herbert A, Schade M, Lowenhaupt K, Alfken J, Schwartz T, Shlyakhtenko LS, Lyubchenko YL, Rich A. The Z-alpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res. 1998;26:3486–3493. doi: 10.1093/nar/26.15.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- Jantsch MF, Gall JG. Assembly and localization of the U1-specific snRNP C protein in the amphibian oocyte. J Cell Biol. 1992;119:1037–1046. doi: 10.1083/jcb.119.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- Krovat BC, Jantsch MF. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J Biol Chem. 1996;271:28112–28119. doi: 10.1074/jbc.271.45.28112. [DOI] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1, and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- Liu Y, George CX, Patterson JB, Samuel CE. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- Liu Y, Herbert A, Rich A, Samuel CE. Double-stranded RNA-specific adenosine deaminase: nucleic acid binding properties. Methods. 1998;15:199–205. doi: 10.1006/meth.1998.0624. [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel CE. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- O'Connell MA, Keller W. Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc Natl Acad Sci USA. 1994;91:10596–10600. doi: 10.1073/pnas.91.22.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol Cell Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]