Abstract
Centrioles and basal bodies are cylinders composed of nine triplet microtubule blades that play essential roles in the centrosome and in flagellar assembly. Chlamydomonas cells with the bld2-1 mutation fail to assemble doublet and triplet microtubules and have defects in cleavage furrow placement and meiosis. Using positional cloning, we have walked 720 kb and identified a 13.2-kb fragment that contains ε-tubulin and rescues the Bld2 defects. The bld2-1 allele has a premature stop codon and intragenic revertants replace the stop codon with glutamine, glutamate, or lysine. Polyclonal antibodies to ε-tubulin show peripheral labeling of full-length basal bodies and centrioles. Thus, ε-tubulin is encoded by the BLD2 allele and ε-tubulin plays a role in basal body/centriole morphogenesis.
INTRODUCTION
Centrioles are highly ordered structures composed of nine sets of triplet microtubules arranged in a turbine pattern. They possess various structural elaborations and are composed of >150 polypeptides (Preble et al., 2000). Centrioles are found in the microtubule-organizing center of most eukaryotic cells. They are required for the recruitment of the pericentriolar material to assemble a centrosome (Bobinnec et al. 1998) and are required for faithful completion of cytokinesis (Piel et al., 2001). Eukaryotic basal bodies are structurally similar to centrioles, but serve as templates for the assembly of cilia and flagella and as docking sites for molecular motors.
Centrosome duplication is closely tied to DNA replication. The activation of the cyclin E-Cdk2 complex at the G1-S phase transition of the cell cycle permits both DNA replication and centrosome duplication to proceed (reviewed in Hinchcliffe and Sluder, 2001). A subset of the targets of Cdk2 activity is involved in centrosome duplication but not DNA replication. These include the nucleolar protein nucleophosmin (B23) (Okuda et al., 2000) and the kinase Mps1p (Fisk and Winey, 2001; Stucke et al., 2002). The ZYG-1 kinase, which is necessary for centriole duplication in Caenorhabditis elegans, may also act in this pathway (O'Connell et al., 2001). Recently, Matsumoto and Maller (2002) have shown that a calcium spike may activate calmodulin-kinase II in Xenopus embryos to trigger centrosome duplication. However, little is known about the proteins needed for duplication. The replication of centrioles begins with the growth of new centrioles at right angles to the preexisting structures. Duplication requires γ-tubulin (Ruiz et al., 1999) and η-tubulin (Ruiz et al., 2000). Labeled α- and β-tubulin are incorporated into the growing centrioles (Kochanski and Borisy, 1990). Growth is believed to begin with a ring of singlet microtubules followed by doublet and triplet microtubules (Dippell, 1968).
The unicellular green alga Chlamydomonas reinhardtii provides an ideal system to address centriole assembly and function genetically. During interphase, basal bodies are found at the anterior end of the cell at the proximal ends of the two flagella. During mitosis, the flagella are resorbed and the basal bodies relocalize to the poles of the mitotic spindle as centrioles. Several mutations profoundly alter the assembly and function of basal bodies. A mutation in the UNI3 gene, which encodes δ-tubulin, results in the assembly of basal bodies that have doublet rather than triplet microtubules (Dutcher and Trabuco, 1998). These mutant cells have a defect in placement of the cleavage furrow (Dutcher and Trabuco, 1998). Similarly, depletion of δ-tubulin from Paramecium results in the assembly of basal bodies that have only doublet microtubules (Garreau De Loubresse et al., 2001).
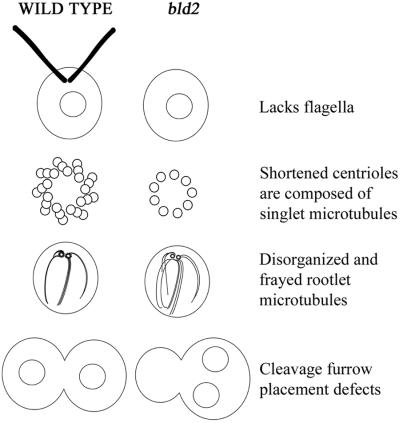
Mutations in the BLD2 gene also result in the assembly of abnormal basal bodies in Chlamydomonas (Goodenough and St. Clair, 1975). Most bld2-1 basal bodies have a ring of nine singlet microtubules and the cylinders are considerably shorter than in wild-type cells. A very few cells have longer cylinders that contain doublet and triplet microtubules, which seem to be fraying or disassembling (Goodenough and St. Clair, 1975). The abnormal basal bodies have other consequences for the cell (Figure 1). Cells with the bld2-1 allele are viable, but lack flagella. bld2-1 cells have disorganized cytoskeletal assemblies. Centrin, a calcium-binding protein, assembles into fibers that connect the two basal bodies to each other and to the nucleus in wild-type cells. These fibers fail to extend in bld2-1 cells (Ehler et al., 1995). The nucleus and the basal bodies lose their normal orientation. bld2-1 cells also have disorganized rootlet microtubules. Rootlet microtubules are a set of four microtubular bundles; two bundles contain two microtubules and two bundles contain four microtubules. These microtubule bundles are arranged in a cross-shaped pattern during interphase. During mitosis, the microtubule rootlets with four microtubules are found associated with the cleavage furrow (Holmes and Dutcher, 1989; Gaffal and el Gammel, 1990; Ehler and Dutcher, 1998). In contrast, the positioning of the cleavage furrow and the spindle is nearly random in bld2-1 cells, which is likely to be a consequence of the abnormal number and positioning of rootlet microtubules and centrin fibers (Ehler et al., 1995; Preble et al., 2001). These cells display a premature initiation of cytokinesis; bld2-1 cells assemble a cleavage furrow in metaphase rather than in telophase (Ehler et al., 1995). Homozygous bld2/bld2 cells show a profound meiotic defect. Although four cells are produced after meiosis, few viable meiotic progeny are produced (Ehler et al., 1995; Preble et al., 2001).
Figure 1.
Diagram of phenotypes of wild-type and bld2-1 cells. The primary defect in bld2-1 cells is likely to be shortened centrioles with singlet microtubules. The consequences of this defect are failure to assemble flagella, disorganized and frayed rootlet microtubules, and defects in cleavage furrow placement. Not shown are defects in centrin assembly and premature initiation of the cleavage furrow.
Database searches after the identification of δ-tubulin as the gene product of the UNI3 locus led to the identification of additional novel tubulins (reviewed in Dutcher, 2001). ε-Tubulin is present in multiple organisms (Chang and Stearns, 2000; Vaughan et al., 2000). Antibodies to ε-tubulin show that it is localized to centrioles in mammalian cells and suggest that ε-tubulin is associated with the mother centriole, but not the daughter centriole (Chang and Stearns, 2000). Other novel tubulins include ζ-tubulin from trypanosomatid flagellates (Vaughan et al., 2000) and η-tubulin (Ruiz et al., 2000).
MATERIALS AND METHODS
Physical Mapping of bld2
Several markers on the left arm of linkage group III were mapped in crosses with strain CC1952 and bld2 strains derived from 137c. The genetic markers in the strain in these crosses included bld2-3, pf15, maa2-1, and tua1-1. CC1952 contains many restriction fragment polymorphisms (Ranum et al., 1988) and single nucleotide polymorphisms (Vysotskaia et al., 2001) relative to the laboratory strain 137c (http://www.biology.duke.edu/chlamy). Two different crosses with CC1952 as one of the parents were performed. Full or partial tetrads (146) from a cross of bld2-3 nit2-1 tua1-1 × CC1952 yielded five progeny with recombination events between bld2-3 and nit2-1. Full or partial tetrads (109) from a cross of maa2 pf15-1 bld2-3 nit2-1 × CC1952 yielded 35 progeny with recombination between maa2 and bld2-3, and three progeny with recombination between bld2-3 and nit2-1. (These recombinants were lost and are not discussed further.)
Because the GP108 probe on linkage group III (a kind gift of Carolyn Silflow, University of Minnesota, St. Paul, MN) did not work consistently in Southern blots, a molecular marker close to GP108 was obtained. GP108 was used as a probe to isolate a clone from a lambda phage library. 211A, a l-kb PstI fragment, is located ∼14 kb from GP108 and gave an easily scored restriction fragment-length polymorphism (Preble, 1999). Genomic DNA was isolated from the 40 recombinant progeny as described by Dutcher and Trabuco (1998). Hybridization conditions were as described by Johnson and Dutcher (1991). Probes were labeled using [32P]dATP with the Multiprime DNA labeling system (Amersham Biosciences, Piscataway, NJ).
Screening the Bacterial Artificial Chromosome (BAC) Library and Fingerprinting
Filters containing a BAC library were purchased from Incyte Genomics (Palo Alto, CA). The BAC library is in a pBelloBAC11 vector that has been modified to include a 4.9-kb fragment containing the NIT8 gene that was cloned into the unique XhoI site. The vector contains T7 and Sp6 promoters on either side of the multiple cloning site. The library contains 15,360 clones spotted at high density in a grid pattern. The BAC filter was stripped and reprobed for all probes.
BAC ends were rescued using the vectorette polymerase chain reaction (PCR) method of Riley et al. (1990). RK532 (AAGGAGAGGACGCTCTCTGTCGAAGGTAAGGAACGGAC GAGAGAAGGGAGAG) and RK331 (CTCTCCCTTCTCCGCAAATCGATCT CGAGTCTAGAGTCGACGTCCTCTCCTT) were annealed at concentrations of 1 μm for each primer in 100 mM NaCl, 5 mM MgCl2, and 10 mM Tris-HCl, pH 8.0. The annealing mixture was boiled for 10 min, incubated at 65°C for 15 min, incubated at 37°C for 15 min, and placed at room temperature for 15 min. BAC DNA was digested with either PvuII, HaeIII, NaeI, BglI, SspI, or RsaI. Digests were placed at 65°C for 15 min to heat-kill enzymes. Then 20 ng of digested BAC DNA was placed in a 20-μl ligation with 1 μl of annealed vectorette mixture and 1.5 U of ligase (Promega, Madison, WI) under standard ligation conditions. PCR conditions were optimized using the Optiprime PCR Optimization kit (Stratagene, La Jolla, CA). PCR was performed with 1 μl of ligation mixture diluted 1:10, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.1 μg of vectorette primer, 0.1 μg of SP6 primer or 0.1 μg of T7 primer, and 2.5 U of Taq polymerase (Promega or Invitrogen) in 1× buffer at a final volume of 50 μl. To increase yields, 0.5% dimethyl sulfoxide was added to reactions with the SP6 primer and 5% formamide was added to reactions with the T7 primer. The primers are SP6 (CAAGCTATTTAGGTGACACTATAG), T7 (TAATACGACTCACTATAGGG), and vectorette (TCGATCTCGAGTCTA GAG). PCR parameters were as follows: one cycle at 94°C for 1 min; 35 cycles at 94°C for 1 min, 45°C for 2 min, and 72°C for 2 min; and 1 cycle at 94°C for 1 min, 45°C for 2 min, and 72°C for 10 min. Probes were digested with HindIII to remove BAC vector sequences before using as probes.
Cloning and Sequencing ε-Tubulin
Genomic DNA from bld2-1 cells was isolated as described previously (Johnson and Dutcher, 1991) and a 6026-base pair fragment was amplified using primers E37F (GGTG TACCATGACATTCAATTGTT) and E37R (CTACAAGGAT'CGACTTGTG AACT). The primers were derived from the sequence of Chlamydomonas BAC 40a20 (GenBank accession number AC087726). Additional 5′ DNA was amplified using primers 38F (AGTTTGTTGAGGCTGGAAAGAG) and 21R (ACCTGCTA TGCCTTGCTACG). Additional 3′ DNA was amplified using 47F (GCTGAAGTCGACAGCTCTGG) and 47R (CGGCACGTCCGTGCGGCTGC). Klentaq Long & Accurate polymerase (Barnes, 1994) was used with the following conditions: 35 cycles of 1 min at 94°C, 2 min at 45°C, and 10 min at 68°C. A final 30-min extension period at 68°C was included to favor the addition of an overhanging A base. The product was purified from a 1% agarose gel (gel purification kit; QIAGEN, Valencia, CA) and cloned into the PCR4-TOPO vector (TOPO TA Cloning kit; Invitrogen). Transformed plasmids were digested with EcoRI to verify the presence of the insert. Plasmid DNA was purified with a Plasmid Midi kit (QIAGEN) and then cycle sequenced in conjunction with the Protein and Nucleic Acid Sequencing Laboratory (Washington University, St. Louis, MO) by using primers listed in Table 1. Sequence data were aligned and analyzed with Sequencher (Gene Codes, Ann Arbor, MI). All sequences were verified by sequencing in both directions. The consensus sequence for polyA addition (TGTAA) was observed at nucleotide 6752.
Table 1.
Recombinants with physical markers in strains recombinant for the bld2-nit2 and bld2-maa intervals
| Region of recombination | GP205 | 211A | ef12e | 35d6T7 | 2e11T7 | 35d6SP6 | Rib43A |
|---|---|---|---|---|---|---|---|
| bld2-nit2 | NT | 0 /5 | 0 /5 | 0 /5 | 5 /5 | 5 /5 | 5 /5 |
| bld2-maa2 | 18/35 | 9 /35 | 0 /35 | NT | NT | NT | NT |
NT: Not tested.
Genomic DNA from three intragenic revertants (bld2-1R1, bld2-1R9, and bld2-1R11) was isolated, and a PCR product was amplified with primers E39F (ACGAAACACCAGCCTACAGC) and E39R (TCGCTCACCTTGAGTGTACG) and directly sequenced in conjunction with the Protein and Nucleic Acid Sequencing Laboratory (Washington University).
Transformation of bld2 Cells
Cells were grown in Sager and Granick II medium with stirring at 25°C to 8 × 106 cells/ml. Cells were harvested by centrifugation, treated with freshly made gametic lysin (Dutcher, 1995), and transformed using agitation with glass beads with 1 μg BAC DNA, which was purified using Plasmid Maxi kit (QIAGEN) with the protocol of Maatman et al. (1996). Digested BAC DNA was treated with 1.5 U of enzyme for 3 h and the enzyme was inactivated according to manufacturer's protocols. Transformed cells (3 × 107) were placed in16 × 150-mm tubes with 20 ml of Sager and Granick II medium. Cells were grown for 4 d and the top 5 ml of medium was transferred to a fresh tube of medium. On the observation of swimming cells, cells were diluted and plated for single colonies. Single colonies that produced swimming cells were analyzed.
Genetic Protocols
Cells were grown as described previously (Holmes and Dutcher, 1989). Matings were performed as described in Dutcher (1995). Revertants were isolated as described in Preble et al. (2001).
Antibodies to ε-Tubulin
Three peptides (KMDPGGFTSAME, (KLTRLRPRGL, and (KQYRALEGATQ) from the carboxy terminus of ε-tubulin were synthesized by Research Genetics (Huntsville, AL). The lysine residue (underlined) was added for conjugation to a carrier protein. The peptides were conjugated to hen egg albumen by using glutaraldehyde. After dialysis against phosphate-buffered saline, the conjugated peptides were used to raise antibodies in rabbits (Cocalico Biologicals, Reamstown, PA).
Three types of cells were used for staining. Ten-minute-old dikaryons formed between wild-type and bld2-1 rgn1-1 cells were used as the cell walls were removed and provided excellent resolution. Wild-type and bld2-1 vegetative cells were examined after treatment with gametic lysin (Dutcher, 1995). Immunofluorescence staining was performed as described previously (Marshall and Rosenbaum, 2001) by using the antibodies epsilon 15, epsilon 16, and epsilon 17 and the mouse monoclonal 6-11B-1, which recognizes acetylated α-tubulin (LeDizet and Piperno, 1986). Spindle microtubules were visualized with a mouse monoclonal antibody (mAb), DM1A, that recognizes the carboxy terminus of α-tubulin (Neomarkers, Fremont, CA). Alexa 594 and Alexa 488 mouse and rabbit secondary antibodies were obtained from Molecular Probes (Eugene, OR). Samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Images were collected using an Axioscope microscope and Axiovision camera (Carl Zeiss, Thornwood, NY). Images were exported to Photoshop 5.5 (Adobe Systems, Mountain View, CA).
To test for specificity of the antibodies, antibodies were incubated overnight at 4°C with 0.7 mg/ml peptide. Antibody epsilon 15 was incubated with peptide 15, 16, and 17. Antibodies epsilon 16 and 17 were incubated with their cognate peptides. Dikaryons were fixed for immunofluorescence and stained with rabbit and mouse secondary antibodies. No labeling with the rabbit secondary was observed when the antibody was incubated with its cognate peptide, but staining with both secondary antibodies was observed when the irrelevant peptide was used (our unpublished data).
RESULTS
BLD2 Gene Encodes ε-Tubulin
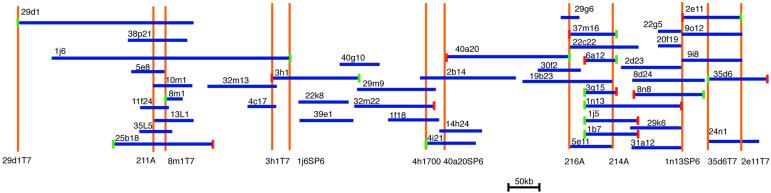
The BLD2 gene maps to linkage group III between NIT2 and MAA2 (Preble et al., 2001). We crossed bld2-3 nit2 maa2 cells by the highly polymorphic strain CC1952 (BLD2 NIT2 MAA2) (Gross et al., 1988), to generate 235 tetrads. We recovered eight tetrads with recombination events between bld2 and nit2 and 35 tetrads with recombination events between bld2 and maa2-1. Three physical markers that map to this region, 211A, GP205, and Rib43A, recombine with bld2, whereas the ef12e marker fails to recombine (Table 2). This generates the order MAA2 GP205-211A-[ef12e, BLD2]-Rib43A NIT2. BLD2 is 2.2 map units from NIT2 and 2.3 map units from 211A. The probes 211A and ef12e were used to begin a walk using a BAC library (see MATERIALS AND METHODS; Incyte Genomics). The completed walk covers 720 kb and spans 4.5 map units (Figure 2). To determine where in the walk the BLD2 gene is located, we screened for rescue of the aflagellate phenotype of bld2-1 cells by using transformation with DNA from eight BAC clones that represented the minimum tiling path of the walk. Two of the BAC clones (19b23 and 40a20) generated cells that were able to swim because the cells were able to assembly two flagella (Table 3; n = 15). These BAC clones are large (118 and 180 kb) and contain multiple genes.
Table 2.
Rescue of the aflagellate phenotype of bld2-1 cells with DNA from chromosome walk (number of transformants per 108 cells)
| 1j6 | 3h1 | 32m22 | 4i21 | 40a20 | 19b23 | 1n13 | 2e11 | |
|---|---|---|---|---|---|---|---|---|
| No. of independent rescue events | 0 | 0 | 0 | 0 | 4 | 11 | 0 | 0 |
Figure 2.
Diagram of BAC clones mapped to linkage group III surrounding the BLD2 locus. The total chromosome walk covers 720 kb and 4.5 map units. Dark blue lines represent the BAC clones and their length as determined by fingerprinting gels (Marra et al., 1997). Orange lines represent the position of the probes used in the walk and their names are given below the line. For example, 40a20SP6 represents a probe generated from the sequence on the SP6 side of the BAC 40a20. Small red boxes are the SP6 side of the BAC and small green boxes are on the T7 side of the walk. BACs 11 m13, 1h1, 18k12, 22a5, 7n18, 27c21, 8122, 8f21, 8d23, and 8g22 were found in this region, but were not placed on the map.
Shotgun sequencing of the walk showed that these two BAC clones shared 57 kb of sequence. Gene prediction programs suggest the presence of nine to 11 open reading frames that are shared between BAC 19b23 and BAC 40a20 (GenBank accession number AC090433 and AC087726; Wu, Lin, Jia, Li, Stormo, Dutcher, and Roe, unpublished data). Expressed sequence tags for six genes are also present; they encode ε-tubulin, arginine methytransferase, and Nopp140/CBF5/dyskerin as well as three novel proteins. ε-Tubulin was an excellent candidate for the BLD2 gene. HindIII cuts ε-tubulin, whereas XhoI and EcoRI cut only outside of the open reading frame. Transformants were recovered with 19b23 BAC DNA that was digested with either XhoI or EcoRI (two transformants from 107 cells for each), whereas transformations with DNA cut with HindIII failed (zero from 2 × 108 cells). We transformed bld2-1 cells with a 13.2-kb plasmid (pSTL1) that contains the coding region for ε-tubulin as well as 5795 base pairs upstream of the start codon and 3768 base pairs downstream of the stop codon. We obtained two transformants using 107 cells. Transformants with the BAC clones or the plasmid clone showed nearly complete rescue of the flagellar phenotype (see below).
ε-Tubulin from the bld2-1 strain was sequenced and compared with the wild-type ε-tubulin sequence (Figures 3A and 4D). The bld2-1 allele has changes from C to T at positions 35 and 36 of the predicted open reading frame. These changes result in a third position synonymous change in isoleucine (I8) and the introduction of a premature stop codon (TAG) after amino acid eight (Figure 3B).
Figure 3.
Nucleotide and amino acids changes in bld2 alleles and alignment of ε-tubulin from multiple organisms. (A) Nucleotide and protein sequence for the first 30 bp of the coding region of ε-tubulin from wild-type cells. (B) From bld2-1 cells, 5743 bp of sequence were obtained. The nucleotide and protein sequences for the first 30 bp of the coding region are shown. There were two C-to-T transition mutations, which are indicated by underlines. (C) Nucleotide and protein sequence for the first 30 bp of the coding region is shown for three intragenic revertant alleles. In each revertant the TAG stop codon is changed to an amino acid. (D) Protein coding sequence of ε-tubulin predicted using Genie trained on 50 Chlamydomonas genes (Kulp et al., 1996) and aligned with ε-tubulin sequences from human, mouse, Trypanosoma, Plasmodium, and Paramecium (see accession numbers in Figure 4). The peptides used for making antibodies are underlined (MDPGGFTSAME, QYRALEGSTQ, and LTRLRPRG). Light gray indicates conservation of the amino acid in all ε-tubulins, medium gray indicates conservation in >50% of the ε-tubulins, dark gray indicates similarity, and white indicates the lack of conservation.
Additional evidence was provided by the selection of revertants of the aflagellate phenotype associated with the bld2-1 allele at 32°C. We isolated 19 strains that show complete reversion of the flagellar defect. These strains have wild-type numbers of flagella based on 300 cells. In 40–65 tetrads for each strain, we failed to recover Bld2 meiotic products, which suggests the event was intragenic. We sequenced ε-tubulin from three of the 19 alleles and find three different substitutions (Figure 3C). bld2-1R1 changes the TAG stop codon to CAG to restore glutamine, the amino present in the wild-type allele. bld2-1R9 changes the TAG stop codon to GAG to substitute glutamate (Q9E). bld2-1R11 changes the TAG stop codon to AAG to substitute lysine (Q9K).
The coding region of ε-tubulin was predicted using the Genie algorithm (Kulp et al., 1996) that was trained on 50 Chlamydomonas genes. The prediction was refined using similarity to other tubulin genes. One additional intron was introduced to the Genie prediction based on similarity to other tubulins. When this predicted coding region is compared with ε-tubulin from mouse, there is 51% identity and 71% similarity at the amino acid level (Figure 3D; GenBank accession number AAM23012; AF502577). Multiple alignment of ε-tubulin from human, mouse, Chlamydomonas, Paramecium, Plasmodium, and Leishmania suggests that most of the protein is conserved among these organisms. However, there are several regions that are not conserved. These are amino acids 53–62, which are not conserved in most tubulins. Amino acids 220–250, which would form helices H7 and H8 in α- and β-tubulin, are not conserved. The carboxy tail, which is variable in all tubulins, is also variable among the ε-tubulins.
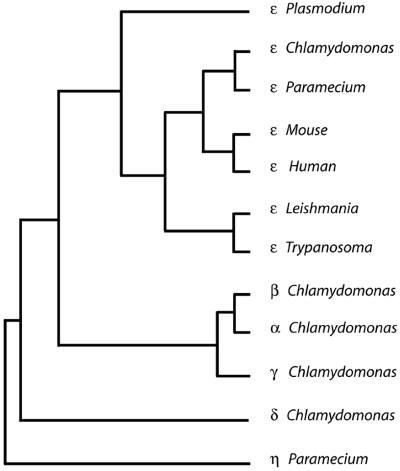
A phenogram was generated following a multiple alignment using ClustalW and the Phylip neighbor joining algorithm; the tree is shown in Figure 4 (Thompson et al., 1994; Felsenstein, 1996). ε-Tubulin from Chlamydomonas is clearly more similar to human and mouse ε-tubulin than to α, β, γ, or δ-tubulin from Chlamydomonas. ε-Tubulins from Trypanosoma, Leishmania, and Plasmodium are more distantly related to human and mouse than are Chlamydomonas and Paramecium ε-tubulin. ε-Tubulin from Danio (BM101764), Xenopus (AW147718; BI444418), Ciona (AV864303; AV863339), Rattus (AW143775), and Oryctolagus (C83345) are available in expressed sequence tag databases. Thus, ε-tubulin is conserved in organisms with triplet microtubules in their centrioles and basal bodies with the exception of Drosophila melanogaster.
Figure 4.
Phenogram of ε-tubulins. ClustalW was used to align five ε-tubulin sequences, the α, β, γ, and δ-tubulin genes from Chlamydomonas, and η-tubulin from Paramecium, which served as an outgroup for the construction of the tree (Thompson et al., 1994). This alignment was analyzed using the Phylip package (Felsenstein, 1996) to obtain a distance matrix using a PAM matrix and the tree was calculated using neighbor joining. A consensus tree was determined with α-tubulin from Chlamydomonas (A53298), β-tubulin from Chlamydomonas (UBKM), γ-tubulin from Chlamydomonas (AAB71841), δ-tubulin from Chlamydomonas (T07903), ε-tubulin from Chlamydomonas (AF502577), ε-tubulin from human (Q9UJT0), ε-tubulin from mouse (BAB26636), ε-tubulin from Paramecium (CAD20554), ε-tubulin from Plasmodium (AF216743), ε-tubulin from Trypanosoma (AAF32302), and η-tubulin from Paramecium (CAB99490).
Phenotypes of Transformed bld2-1 Cells
bld2-1 mutant cells have a variety of phenotypes that include the inability to assemble full-length basal bodies or to build flagella. The abnormal number and placement of microtubules and the abnormal placement of centrin fibers result in the misplacement of the cleavage furrow and generation of anucleate and binucleate cells. There is a failure to produce viable meiotic progeny. We have examined several of these phenotypes in four independent meiotic progeny with the bld2-1 gene and the wild-type BLD2 transgene. The distribution of flagellar number of these transformed cells resembles the distribution seen in wild-type cells. The number of cells with two flagella ranged from 87 to 95%, which is similar to the distribution for wild-type cells (89–95%) (n = 300 for each) at 14, 21, 25, or 32°C.
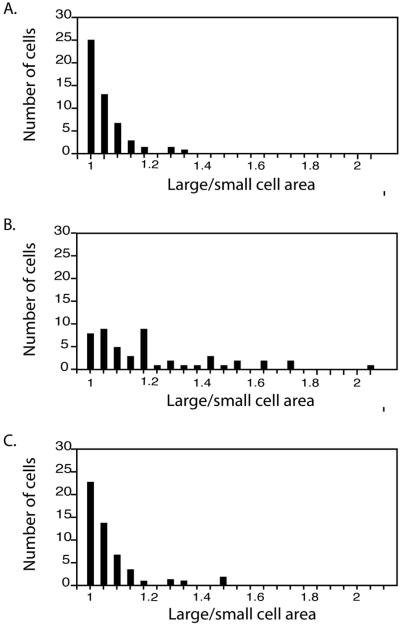
We measured the area of 50 recently divided cells, which is an indicator of the placement of the cleavage furrow. Wild-type cells produced two daughters of equal size, whereas bld2-1 cells produce daughters of different sizes (Preble et al., 2001). The distributions of the four transformants resemble those from wild-type cultures (Figure 5). There are few anucleate and binucleate cells (0 and 0.5%, respectively) after staining with the DNA dye 4,6-diamidino-2-phenylindole (n = 200) compared with bld2-1 cultures that have 3 and 7%, respectively. There is no meiotic defect in crosses of bld2-1 TG × bld2-1TG or in crosses of bld2-1 × bld2-2 TG and bld2-1 × bld2-3 TG. Of the meiotic progeny 94–97% were viable in 37–42 tetrads of the four tested strains. In summary, all of the tested phenotypes were rescued. Rescue of the flagellar assembly, the multiple nuclei, and the meiotic defects was observed for bld2-2 and bld2-3 alleles (our unpublished data) (Preble et al., 2001).
Figure 5.
Reversion of cleavage furrow placement defect in bld2-1 transformants. Recently divided pairs of daughter cells were photographed and the areas measured. The ratio of the larger to the smaller cell is plotted for 50 cells. (A) Wild-type cells. (B) bld2-1 cells. (C) bld2-1 cells with a BLD2 transgene. Wild-type and bld2-1 cells with a BLD2 transgene produced similar distributions, whereas both were significantly different from the bld2-1 cells.
Rescue of bld2-4 Meiotic and Lethal Phenotype
The bld2-4 allele was generated by insertional mutagenesis in bld2-2/BLD2 diploid cells (Preble et al., 2001). Unlike the bld2-1 cells, this allele has a lethal phenotype when examined in haploid cells. This allele also shows a dominant meiotic defect rather than the recessive phenotype observed for the bld2-1 allele. Greater than 99% of meiotic products from zygotes heterozygous for the wild-type BLD2 and the mutant bld2-4 alleles die (Preble et al., 2001). We introduced the 13.2-kb plasmid that rescues the phenotypes of the bld2-1 allele and find that it rescues the mitotic lethality associated with the bld2-4 allele. Two copies of the BLD2 transgene rescue the dominant meiotic defect. Tetrads from crosses of bld2-4 TG1 × BLD2 TG2, where TG1 and TG2 represent two BLD2 transgenes that are unlinked to each other or to the BLD2 locus, have two (n = 16), three (n = 14), or four (n = 12) viable progeny. Microscopic examination of the progeny 24 h after meiosis was completed revealed that most cells (122) had completed three to four cell divisions. After 72 h, these cells continued to divide. Thirty-nine cells failed to divide and seven cells completed one cell cycle, but did not divide further. These 46 cells are likely to carry the bld2-4 allele without a BLD2 transgene. In tetrads with two viable progeny, both progeny have two copies of the transgene and wild-type levels of viability in subsequent meiotic crosses to a bld2-2 parent (n = 4). In tetrads with four viable progeny, two of the cells show lethality in subsequent crosses and each of the progeny that shows lethality in crosses has at least a one transgene. Thus, the lethal phenotype is the failure to complete mitosis. In addition, the presence of two copies of the BLD2 transgene has no obvious effects on the cells.
Localization of ε-Tubulin to Basal Bodies
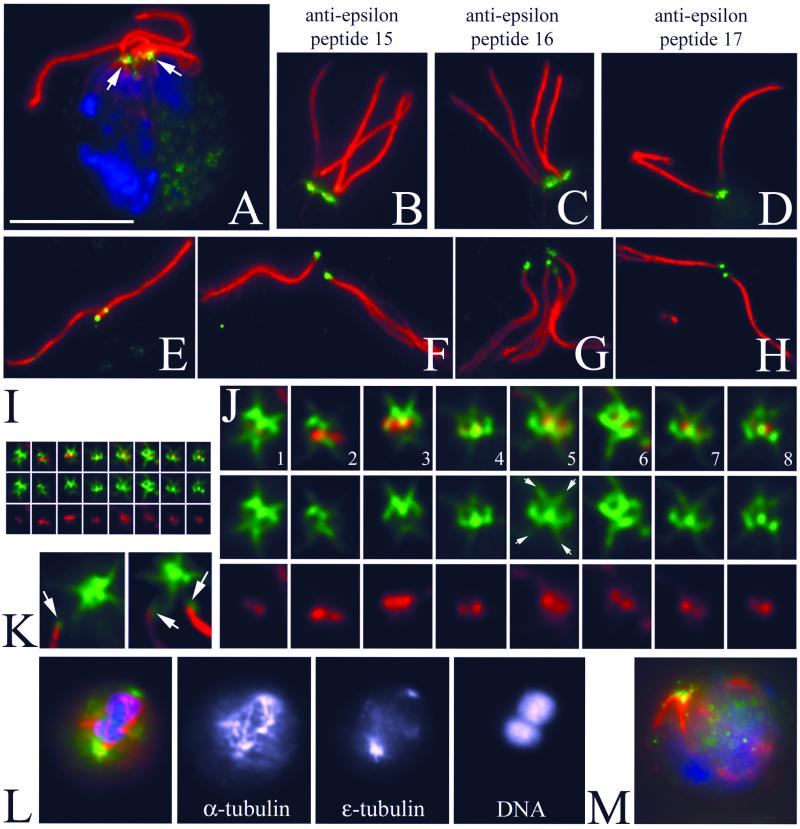
Polyclonal antibodies were raised to three different peptides in the carboxy terminus of Chlamydomonas ε-tubulin. We observe no staining of the flagella or cell body with any of the three preimmune sera to ε-tubulin beyond the low background observed with the secondary antibody by itself (our unpublished data). Each of the three antibodies showed similar patterns of staining, which provides further evidence that the staining is specific. Images A–K and M are labeled with a mAb to acetylated α-tubulin to visualize the basal bodies, rootlet microtubules, and flagella in red. The individual rabbit antibodies to peptides in ε-tubulin are detected with a green secondary antibody. Many of the images come from 10-min-old dikaryons, which are newly mated cells. Dikaryons were used for two reasons. First, the cell wall is removed during mating and sharper images could be obtained. Second, it allows us to compare levels of staining of basal bodies from BLD2 and bld2-1 rgn1-1 cells in the same cytoplasm. We have seen similar patterns of staining in vegetatively growing cells (our unpublished data).
In intact dikaryons (Figure 6A), ε-tubulin is localized at the base of the two pairs of flagella and displays a fibrous appearance. All three peptide antibodies label the same pattern (Figure 6, B–D). In detached flagella, we observed a dot of staining associated with the proximal end. This end can be identified because the flagella have begun to fray from the distal end (Figure 6, E–H). Cell bodies with detached flagella also retain ε-tubulin and its distinctive pattern can be observed more clearly (Figure 6I). In these samples, the basal bodies appear as a pair of dots labeled with the antibody to acetylated α-tubulin. The ε-tubulin antibodies label a region that is peripheral to the paired dots in a figure-eight pattern (Figure 6, I–K). Additional localization occurs as four spurs, which radiate in the region of the striated fibers (Figure 6J, especially 5, 6, and 7). Collectively, the staining on the proximal end of the flagella and the basal body pattern creates the more complicated pattern seen in intact cells (Figure 6A). During mitosis, antibodies to ε-tubulin label spots at the poles of the spindle; spindle microtubules are labeled with the DMA1mAb (Figure 6L).
Figure 6.
Immunofluorescence of Chlamydomonas with anti-ε-tubulin peptide antibodies labels an unusual pattern associated with basal bodies and striated fibers. (A) Triple fluorescence of a Chlamydomonas dikaryon: DNA is labeled with 4,6-diamidino-2-phenylindole (blue), the mouse monoclonal 6-11B-1 identifies acetylated α-tubulin (red), and ε-tubulin is labeled with the rabbit polyclonal p15 peptide antibody (green). Bar, 10 μm. Magnification is conserved in the subsequent panels, unless noted. One of the basal body apparatuses is from the wild-type parent and the other is from the rgn1 bld2-1 parent. (B–D) Three independent antipeptide antibodies label the basal bodies in an identical manner. Red is acetylated α-tubulin and green is anti-ε-tubulin p15 (B), p16 (C), or p17 (D). (E–H) ε-Tubulin is associated with the proximal end of flagella detached from Chlamydomonas cell bodies. The staining appears as a discrete spot that is opposite the end of the flagella that has begun to fray (especially note F and G). E and F are labeled with anti-p15 ε-tubulin antibody. G and H are stained with anti-p17 antibody. (I and J) Staining of a basal body associated structure that is left when flagella become detached from the Chlamydomonas cell body. The basal bodies are labeled with acetylated α-tubulin (red) and appear as a pair of dots. The ε-tubulin (green) seems to surround the acetylated α-tubulin dots, creating a figure-eight–shaped cage. Additional staining extends in a “cross-shaped” pattern, reflecting association of ε-tubulin with the striated fibers (arrows in 5). (I) Montage of basal body images shown at the same magnification as the previous panels. (J) Threefold enlargement of this montage. (K) Enlarged image of detached flagella adjacent to the cell body showing the regions of staining associated with the proximal tip of the flagellum (arrows) and retained in the cell body. (L) Merged image of a telophase cell showing ε-tubulin staining (green) at the poles of the spindle (red). Single-channel images with staining of α-tubulin, ε-tubulin, and DNA. (M) bld2-1 cell showing staining with ε-tubulin in the basal body region (green), disorganized rootlet microtubules (red), and no flagella.
rgn1-1 is an extragenic suppressor that partially rescues the flagellar and basal body assembly defects of bld2 cells (Preble et al., 2001). In the basal body apparatus from the double mutant bld2-1 rgn1-1, we observed staining that is similar in its pattern to that of wild-type cells and the staining is similar in intensity to the staining seen in wild-type basal body apparatus (Figure 6, B and C). This suggests that at least the carboxy terminus of ε-tubulin is present in the double mutant and it is assembled into structures. This is confirmed when we examined bld2-1 cells and observed staining in the mutant alone, which indicates that the bld2-1 mutant strain has product (Figure 6M).
DISCUSSION
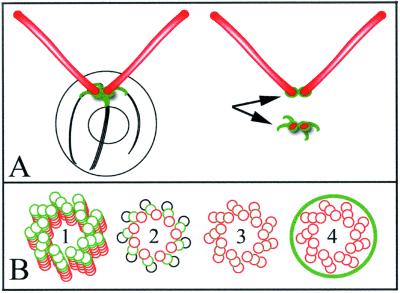
ε-Tubulin was first described after database searches for new tubulin family members (Chang and Stearns, 2000; Vaughan et al., 2000). Antibodies raised to human ε-tubulin suggest that ε-tubulin is localized to the centrioles of mammalian cells and that ε-tubulin is involved in centriole maturation because it does not localize to the daughter centriole in the G1 and S phases of the cell cycle (Chang and Stearns, 2000). In Chlamydomonas, ε-tubulin localizes to a cage surrounding the mature basal bodies and to fibers that run along the rootlet microtubules (Figure 7A). We do not see staining of the probasal bodies in whole cells as has been observed for the Vfl1 protein (Silflow et al., 2001). The staining of the fibers may obscure distinct probasal body staining. However, in isolated flagella–basal body apparatuses (Figure 6K), there are two regions of staining. One region is found in association with the flagella and a second is found with the fibers and cage of the deflagellated cell; this staining is diagrammed in Figure 7A. The staining in the deflagellated cells is likely to include the probasal bodies. Double staining with the epitope-tagged Vfl1 protein may help to resolve this issue in the future.
Figure 7.
Model of ε-tubulin localization and function within the basal body and centriole. (A, left) When intact Chlamydomonas are labeled with the ε-tubulin antibody, small fibrous structures at the base of the flagella are labeled, which reflects association with the basal bodies. (A, top right) A small spot of staining is associated with the proximal end of detached flagella. (A, bottom right) Chlamydomonas cell bodies with flagella detached show labeling of a small distinctive structure consisting of a figure-eight–shaped cage surrounding the two basal body and/or probasal body spots, with four spikes of staining extending along the striated fibers. (B) bld2-1 cells have disrupted triplet microtubules and basal bodies/centrioles have singlet microtubules. This observation, coupled with the cage-like pattern of ε-tubulin staining suggest several models by which ε-tubulin stabilizes triplet microtubules. These include 1) ε-tubulin serving as a plus end cap to the basal body microtubules, 2) ε-tubulin contributing to the B (or B and C) subfiber microtubule lattice as a subunit, 3) ε-tubulin creating the unusual junction of the B (or B and C) subfiber microtubule structure with the A microtubule, or 4) ε-tubulin forming a cage around the entire centriole that confers stability or assembly properties to this structure.
The ε-tubulin antibody fiber staining pattern is similar in appearance to the labeling observed with SF-assemblin-green fluorescent protein (Lechtreck et al., 2002). This coiled coil protein is found in striated fibers that emanate off the basal bodies in Chlamydomonas. RNAi experiments suggest a role of striated-fiber-assemblin in flagellar assembly. If ε-tubulin localization were dependent on SF-assemblin, a flagellar assembly defect might be expected upon depletion of SF-assemblin. A cage of staining around centrioles has been observed in mammalian somatic cells with antibodies to ninein (Ou and Rattner, 2000). Similarly, kinesin II, which is encoded by the FLA10 gene in Chlamydomonas, forms a horseshoe-shaped ring around the basal bodies (Cole et al., 1998). Both these patterns are similar to the “cage-like” localization of ε-tubulin around the basal bodies/centrioles of Chlamydomonas.
The finding that the BLD2 gene encodes ε-tubulin suggests it is needed for the assembly and/or maintenance of the doublet microtubules in the basal body or centriole. Many of the electron microscopic images of basal bodies in the bld2-1 mutant strain from Goodenough and St. Clair (1975) showed that the basal bodies had only singlet microtubules and the basal bodies were significantly reduced in length from the wild-type length of 250 nm. Transition zones (∼200 nm in length) were absent. In rare images, there were structures that had doublet and triplet microtubules that seemed to be fraying or disassembling. Thus, the Bld2 gene product may play a role in stabilizing doublet and triplet microtubules.
Inclan and Nogales (2001) used molecular threading programs to place the sequence of human ε-tubulin onto the crystal structures of α- and β-tubulin (Nogales et al., 1999). They found that ε-tubulin shared the greatest similarity with minus ends of α- and β-tubulin. ε-Tubulin is most dissimilar in the M-loop, which is needed for longitudinal contacts between the adjacent protofilaments. These observations led them to propose that ε-tubulin may not form longitudinal contacts with other tubulin molecules and may reside as a cap at the plus end of microtubules, which might include the distal end of the basal body or centriole (Figure 7B1). This is an attractive model given the phenotype of the bld2-1 basal bodies. ε-Tubulin could serve to stabilize the doublet and triplet microtubules. Other alternative models remain possible. ε-Tubulin may contribute to the B-subfiber microtubule lattice as a subunit (Figure 7B2), or may be involved the formation of the junction between the A and the B subfibers and between the B and C fibers (Figure 7B3). The B and C subfibers of the triplet microtubules form a junction with the adjacent microtubule and the inner and outer junctions differ from one another. The inner junction is staggered by one-half of a dimmer, whereas the outer junction is in register (Song and Mandelkow, 1995). Thus, one might imagine that the presence of ε-tubulin might be responsible for the staggering. In addition, the inner junction seems to be more stable than the outer junction (Song and Mandelkow, 1995). A fourth possibility is raised by the pattern of staining with the antibodies to ε-tubulin peptides. This pattern suggests that ε-tubulin may encircle the basal bodies (Figure 7B4). Given the similarity to the localization of ninein and CEP110 (Ou et al., 2002), these proteins may assemble into a cage surrounding the basal body and centriole to promote assembly. Ultimately, immunoelectron microscopy will be needed to resolve the localization within the basal bodies.
Centrioles seem to be necessary to recruit pericentrosomal material and for the fidelity of cytokinesis. Centrosomes, and perhaps centrioles, are needed for progression into the S phase of the cell cycle. When antibodies to glutamylated tubulin are injected into cells, the centrioles disassemble and the pericentriolar material disperses (Bobinnec et al., 1998). Centrosomes removed by microsurgery before the formation of the spindle result in the loss of centrosomes, but cells remain able to assemble spindles and segregate chromosomes (Hinchcliffe et al., 2001). Laser ablation of one centrosome in mitotic cells results in the loss of the microtubule aster from the pole lacking a centrosome. Regardless of the timing of removal ∼40% of the cells have defects in completing cytokinesis (Khodjakov et al., 2000; Hinchcliffe et al., 2001; Piel et al., 2001). However, all of the treated cells are unable to enter into the next S phase. Ablation of only one centrosome shows that the G1 arrest does not simply result from the presence of errors in the preceding division (Khodjakov and Rieder, 2001).
We have identified three additional alleles at the BLD2 locus. bld2-2 and bld2-3 are weak partial revertants of the bld2-1. Cells with either of these two alleles assemble a single flagellum on one of 500 cells. The bld2-4 allele was created by insertional mutagenesis in diploid cells (Preble et al., 2001) and is phenotypically most different. This allele has a lethal phenotype in haploid cells. We have shown that the lethality is rescued by the introduction of the gene for ε-tubulin. It remains a formal possibility that the bld2-4 allele has a synthetic lethal phenotype that results from the deletion of ε-tubulin and a nearby gene and that the synthetic lethality is abolished by the addition of ε-tubulin.
The lethality associated with the bld2-4 allele seems in conflict with the finding that the bld2-1 allele has a premature stop codon, which may suggest that it is a null allele. We suggest that there is an internal initiation of protein synthesis downstream of the premature stop codon or readthrough of the nonsense codon. This has been documented previously in Chlamydomonas. The PF14 gene in Chlamydomonas encodes RSP3, a component of the radial spoke in the flagellar axoneme. Analysis of the pf14-1 mutation shows it contains a premature stop codon. Two-dimensional gel electrophoresis of isolated flagellar protein showed that a truncated RSP3 protein was incorporated into the flagellar axoneme in the pf14-1 allele, but it was not fully functional (Williams et al., 1989). The staining that we observe of basal body complexes from bld2-1 rgn1-1 cells suggest that this allele can make some ε-tubulin. Thus, the phenotypes observed in bld2-1 cells are unlikely to result from a null allele, but rather from a reduction in the amount of protein or a truncation of the protein. We think it is likely that ε-tubulin is an essential gene that is needed for the assembly of basal bodies and centrioles. In the future, its essential nature will be useful for probing the role of the centriole in cell division.
ACKNOWLEDGMENTS
We thank Dr. David Kulp, Dr. Gary Stormo, and Robin Matlib for the training of Genie on Chlamydomonas genes, and Dr. Wallace Marshall for advice on immunofluorescence staining protocols. We are grateful to Dr. Ursula Goodenough for continued interest in the bld2-1 mutation. We thank members of the Dutcher laboratory in Colorado and in St. Louis for useful comments through the course of this work. This work was supported by a grant from the National Institutes of Health (GM-32843 to S.K.D.) and a training grant position to A.M.P. (5T32-07135).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0205. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0205.
REFERENCES
- Barnes WM. PCR amplification of up to 35 kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Eddé B, Bornens M. Centriole disassembly in vivo and its effects on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Stearns T. Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2:30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis eleganssensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippell RV. The development of basal bodies in Paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 1995;47:531–539. doi: 10.1016/s0091-679x(08)60857-2. [DOI] [PubMed] [Google Scholar]
- Dutcher SK. The tubulin fraternity: alpha to eta. Curr Opin Cell Biol. 2001;13:49–54. doi: 10.1016/s0955-0674(00)00173-3. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonasand encodes δ-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;6:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler LL, Dutcher SK. Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 1998;40:193–207. doi: 10.1002/(SICI)1097-0169(1998)40:2<193::AID-CM8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ehler LL, Holmes JA, Dutcher SK. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtiimutant lacking basal bodies. Genetics. 1995;141:945–960. doi: 10.1093/genetics/141.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Gaffal KP, el-Gammal S. Elucidation of the enigma of the metaphase band of Chlamydomonas reinhardtii. Protoplasma. 1990;156:139–148. [Google Scholar]
- Garreau De Loubresse N, Ruiz F, Beisson J, Klotz C. Role of delta-tubulin and the C-tubule in assembly of Parameciumbasal bodies. BMC Cell Biol. 2001;2:4. doi: 10.1186/1471-2121-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, St. Clair HS. BALD-2: a mutation affecting the formation of doublet and triplet sets of microtubules in Chlamydomonas reinhardtii. J Cell Biol. 1975;66:480–491. doi: 10.1083/jcb.66.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CH, Ranum LP, Lefebvre PA. Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr Genet. 1988;13:503–508. doi: 10.1007/BF02427756. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement for a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Holmes JA, Dutcher SK. Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci. 1989;94:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Inclan YF, Nogales E. Structural models for the self-assembly and microtubule interactions of gamma-, delta- and epsilon-tubulin. J Cell Sci. 2001;114:413–422. doi: 10.1242/jcs.114.2.413. [DOI] [PubMed] [Google Scholar]
- Johnson DE, Dutcher SK. Molecular studies of linkage group XIX of Chlamydomonas reinhardtii: evidence against a basal body location. J Cell Biol. 1991;113:339–346. doi: 10.1083/jcb.113.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–224. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. A. generalized hidden Markov model for the recognition of human genes in DNA. Proc Int Conf Intell Syst Mol Biol. 1996;4:134–142. [PubMed] [Google Scholar]
- Lechtreck KF, Rostmann J, Grunow A. Analysis of ChlamydomonasSF-assemblin by GFP-tagging and expression of antisense constructs. J Cell Sci. 2002;115:1511–1522. doi: 10.1242/jcs.115.7.1511. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Cytoplasmaic microtubules containing acetylated alpha tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J Cell Biol. 1986;103:13–22. doi: 10.1083/jcb.103.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatman R, Whitaker E, Mcminimy D, Gossler A. Purification of bacterial artifical chromosome (BAC) DNA using Qiagen plasmid kits. Qiagen News. 1996;4:10–11. [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;11:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Maller JL. Calcium, calmodulin, and CaMKII requirement for the initiation of centrosome duplication in Xenopusegg extracts. Science. 2002;295:499–502. doi: 10.1126/science.1065693. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. eleganszyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Okuda M, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Ou YY, Mack GJ, Zhang M, Rattner JB. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J Cell Sci. 2002;115:1825–1835. doi: 10.1242/jcs.115.9.1825. [DOI] [PubMed] [Google Scholar]
- Ou Y, Rattner JB. A subset of centrosomal proteins are arranged in a tubular configuration that is reproduced during centrosome duplication. Cell Motil Cytoskeleton. 2000;47:13–24. doi: 10.1002/1097-0169(200009)47:1<13::AID-CM2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Preble AM. A Genetic and Molecular Dissection of Basal Body Assembly and Function. Ph.D. Thesis. Boulder, CO: University of Colorado; 1999. [Google Scholar]
- Preble AM, Giddings TH, Jr, Dutcher SK. Basal bodies and centrioles: their function and structure. Curr Top Dev Biol. 2000;49:207–233. doi: 10.1016/s0070-2153(99)49010-6. [DOI] [PubMed] [Google Scholar]
- Preble AM, Giddings TH, Dutcher SK. Extragenic bypass suppressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics. 2001;157:163–181. doi: 10.1093/genetics/157.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Thompson MD, Schloss JA, Lefebvre PA, Silflow CD. Mapping flagellar genes in Chlamydomonasusing restriction fragment length polymorphisms. Genetics. 1988;120:109–122. doi: 10.1093/genetics/120.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosomes (Y.A.C) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F, Beisson J, Rossier D, Dupuis-Williams P. Basal body duplication in Parameciumrequires gamma tubulin. Curr Biol. 1999;9:43–46. doi: 10.1016/s0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Krzywicka A, Klotz C, Keller A, Cohen J, Koll F, Balavoine G, Beisson J. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, eta-tubulin. Curr Biol. 2000;10:1451–1454. doi: 10.1016/s0960-9822(00)00804-6. [DOI] [PubMed] [Google Scholar]
- Silflow CD, LaVoie M, Tam L-W, Tousey S, Sander M, Wu W-C, Borodovsky M, Lefebvre PA. The Vfl1 protein in Chlamydomonaslocalizes in a rotationally asymmetric pattern at the distal end of the basal bodies. J Cell Biol. 2001;153:63–74. doi: 10.1083/jcb.153.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Mandelkow E. The anatomy of flagellar microtubules: polarity, seam, junctions, and lattice. J Cell Biol. 1995;128:81–94. doi: 10.1083/jcb.128.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S, Attwood T, Navarro M, Scott V, McKean P, Gull K. New tubulins in protozoal parasites. Curr Biol. 2000;10:R258–R259. doi: 10.1016/s0960-9822(00)00414-0. [DOI] [PubMed] [Google Scholar]
- Vysotskaia VS, Curtis DE, Voinov AV, Kathir P, Silflow CD, Lefebvre PA. Development and characterization of genome-wide single nucleotide polymorphism markers in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2001;127:386–389. [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14is an ochre allele. J Cell Biol. 1989;109:235–245. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]