Abstract
StmF mutants are chemotactic mutants that are defective in a cGMP phosphodiesterase (PDE) activity. We identified a novel gene, PdeD, that harbors two cyclic nucleotide–binding domains and a metallo-β-lactamase homology domain. Similar to stmF mutants, pdeD-null mutants displayed extensively streaming aggregates, prolonged elevation of cGMP levels after chemotactic stimulation, and reduced cGMP-PDE activity. PdeD transcripts were lacking in stmF mutant NP377, indicating that this mutant carries a PdeD lesion. Expression of a PdeD-YFP fusion protein in pdeD-null cells restored the normal cGMP response and showed that PdeD resides in the cytosol. When purified by immunoprecipitation, the PdeD-YFP fusion protein displayed cGMP-PDE activity, which was retained in a truncated construct that contained only the metallo-β-lactamase domain.
INTRODUCTION
Mutants in cGMP metabolism have implicated cGMP as intermediate for ligand-induced chemotaxis in Dictyostelium (Ross and Newell, 1979, 1981; Kuwayama et al., 1993). Mutants KI8 and KI10 show no ligand-induced cGMP response and cannot chemotax (Kuwayama et al., 1993). Streamer F (stmF) mutants are defective in cGMP-PDE activity (Van Haastert et al., 1982). These mutants show an elevated and prolonged cGMP response and prolonged association of myosin with the cytoskeleton (Ross and Newell, 1981; Liu and Newell, 1988, 1991, 1994). When grown as colonies on dense bacterial lawns, stmF mutants form aggregates with very pronounced aggregation streams, a phenotype that under these conditions is not displayed by wild-type cells.
Three PDE genes have been identified in Dictyostelium; two of those, RegA (Shaulsky et al., 1996; Thomason et al., 1998) and PDE3 (Kuwayama et al., 2001), belong to the large class of HD-domain PDEs that is commonly found in vertebrates (Mehats et al., 2002). The third enzyme, PdsA (Lacombe et al., 1986), is a class II PDE that is also found in yeast (Nikawa et al., 1987). None of these genes are associated with the stmF locus.
In our search for targets of cyclic nucleotides, we identified a gene, PdeD, with two putative cyclic nucleotide (cNMP)–binding domains and a binuclear Zn2+–binding domain. The latter domain forms the catalytic center of bacterial metallo-β-lactamases, which hydrolyze an amide bond in the β-lactam ring of carbapenem antibiotics (Carfi et al., 1995) and are a major cause for widespread bacterial antibiotic resistance (Payne, 1993). We show that pdeD-null mutants phenocopy stmF mutants. One of the cNMP-binding domains of PdeD most likely functions as an allosteric activator of the enzyme, whereas the metallo-β-lactamase homology domain catalyzes the hydrolysis of cGMP.
MATERIALS AND METHODS
Cell Growth and Development
D. discoideum strains NC4, XP55, and NP377 were grown in association with Klebsiella aerogenes on SM agar plates, and all other strains were grown in HL5 medium, supplemented with 5 μg/ml blasticidin or 20–200 μg/ml G418 for strains transformed with pBsrΔBam or neomycin selection markers, respectively. For developmental time courses, cells were harvested from bacterial plates or growth medium, washed with 10 mM NaH2PO4/K2HPO4 buffer, pH 6.5 (PB), plated at variable cell densities on PB agar (1.5% agar in PB), and incubated at 22°C.
Bioinformatics
The Dictyostelium genome and cDNA databases were screened with various sequences for cNMP-binding domains. In addition to the protein kinase (PK) A regulatory subunit (Mutzel et al., 1987), a 419–base pair (bp) fragment of another candidate gene was hit. Eight cycles of BLAST (Altschul et al., 1990) searches initiated with the 419-bp fragment and sequence assembly yielded contiguous DNA sequence of 3473 bp with at least fourfold coverage at any particular region. As of July 8, 2002, the sequence is available on the 80.3-kilobase (kb) contig JC3a201c05.r1, which was assembled by the Dictyostelium genome sequencing project Jena, Germany (http://genome.imb-jena.de/dictyostelium/) and is located on chromosome II (Gloeckner et al., 2002). The sequence shows an open reading frame (ORF) with two cNMPs when analyzed with SMART (Schultz et al., 1998) for PFAM domains (Bateman et al., 2000). The sequence upstream of the ORF was interrupted by two short AT-rich regions with stop codons in all reading frames. To determine whether these regions were introns, oligonucleotides were designed that flanked both regions simultaneously (CCCTGATATGATTAATTCAATCTCTACG and CCCGCTTCATGTAATACCACCG). These oligos were used to perform RT-PCR of mRNA of NC4 cells starved for 10 h by use of the Qiagen Onestep RT-PCR kit (Qiagen, Hilden, Germany). A band of 600 bp was obtained and sequenced. This showed the presence and position of a 96- and a 122-bp intron. The putative start codon could then be identified, which was preceded by a 1.56-kb AT-rich region and a 3-kb ORF with weak homology to a Brassica campestris pollen-coat protein. The start codon conformed well to the Dictyostelium Kozak sequence, and because introns >1 kb are very rare in Dictyostelium, the complete 2601-nucleotide coding sequence could be established with confidence. For reasons described below, we named the gene PdeD, and the sequence of its ORF is deposited in GenBank under accession number AY047363.
Gene Inactivation
Two DNA fragments of PdeD comprising nucleotides 2289–3109 and 1402–2195 were amplified by PCR using oligonucleotides that yielded a 5′-XbaI and 3′-BamHI site on the first fragment and 5′-BamHI and a 3′-EcoRI site on the second fragment. These fragments were cloned in tandem into the BamHI/EcoRI sites and XbaI/BamHI sites of pBsrΔBam (Sutoh, 1993). The construct was linearized with BamHI, which yielded the pBsrΔBam plasmid flanked by ∼800 bp of 5′ and 3′ PdeD DNA. Homologous recombination with this construct causes insertion of the entire plasmid at position 2195 and a deletion of 94 bp.
The knockout (KO) construct was introduced into wild-type AX2 cells by electroporation, and transformed cells were selected by growth in the presence of 5 μg/ml blasticidin. Selected clones were screened for homologous recombination by two separate PCR reactions and analysis of Southern blots of genomic digests. Three of 200 clones tested from two separate transformations proved to carry a gene disruption. Three KO lines and three lines carrying different random vector integrations (RIs) were used for further analysis.
PdeD-YFP Fusion Constructs
PdeD gene fragments were created by PCR using oligonucleotides that generated 5′-BamHI and 3′-XhoI sites. These fragments corresponded to residues 2–867 for full-length PdeD, 524–867 for PdeDΔNΔlac, and 321–572 for PdeDΔNΔC. The fragments were cloned into the BamHI/XhoI-digested vector pB17SYFP, which is a derivative of pDXA-HC (Manstein et al., 1995) that contains the coding sequence for enhanced yellow fluorescent protein (YFP) (Miyawaki et al., 1997) downstream of the constitutive actin15 promoter. The constructs yield fusion proteins with YFP at the C-terminus of PdeD. The vectors were transformed into parent strain AX2 and the pdeD-null mutant clone KO96. Cells were selected in HL5 medium with 20 or 200 μg/ml G418.
Immunoprecipitation
Mouse monoclonal green fluorescent protein (GFP) antibody (αGFP) (25 μg) (Roche, Welwyn Garden City, U.K.) was incubated for 1 h at 4°C with a mixture of 250 μl each of slurries of protein G linked to Sepharose 4B (Sigma, St. Louis, MO) and protein A linked to Affiprep (Bio-Rad, Hercules, CA). The αGFP-linked matrix was washed with PBS (0.7% NaCl in PB) and resuspended to the original concentration. Dictyostelium cells, resuspended at 2 × 108 cells/ml in PBS, were lysed through Nucleopore filters. Lysates were cleared by centrifugation for 10 min at 14,000 × g, and 1 ml of cleared lysate was incubated for 4 h with 100 μl of αGFP-matrix suspension. The matrix was washed thoroughly with PDE assay buffer and resuspended in the same buffer.
Western Blot Analysis
Cleared lysates equivalent to 105 cells per lane were subjected to 8% SDS-PAA gel electrophoresis. Size-fractionated proteins were transferred to nitrocellulose and probed with a 1:1000 dilution of αGFP antibody. Detection was performed with the Supersignal chemoluminescence kit (Pierce, Rockford, IL) using a horseradish peroxidase conjugated goat-anti-mouse secondary antibody (Promega, Madison, WI) according to the manufacturer's instructions.
cGMP-PDE Assay and cGMP Response
To measure cGMP-PDE activity, cleared cell lysate or a suspension of matrix linked to αGFP immunoprecipitate was incubated for 30 min at 22°C with 10−8 M 3H-cGMP, 5 mM dithiothreitol, 0.2 mM IBMX, and 1 mM MgCl2 in 20 mM HEPES, pH 7.0 (Kuwayama et al., 2001). The reaction was terminated by boiling. 3H-5′-GMP was further hydrolyzed by incubation for 30 min at 37°C with 0.5 mg/ml Ophiophagus hannah snake venom to [3H]guanosine, which was separated from [3H]cGMP by adsorption of the latter to Dowex anion exchange resin.
To measure the cAMP-induced cGMP response, 108 cells/ml PB were stimulated with 10−7 M cAMP. Aliquots of cell suspension were rapidly mixed with an equal volume of 3.5% perchloric acid (vol/vol) at various intervals after stimulation. Lysates were neutralized with KHCO3, and cGMP levels were measured by radioimmunoassay.
RESULTS
Structure of PdeD
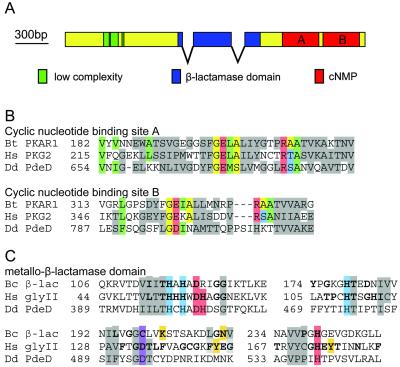
Screening of Dictyostelium cDNA and genome databases with consensus sequences for cNMP-binding domains yielded a novel gene, PdeD, with an ORF of 2601 bp. RT-PCR revealed that PdeD harbors two introns of 96 and 122 bp, respectively, at positions 1108 and 1565 (Figure 1A). Analysis of the domain architecture of the PdeD gene with SMART (Schultz et al., 1998) suggests subdivision of the gene into three regions: 1) an N-terminal region rich in low-complexity sequence and containing no known functional domains. Such N-terminal regions are common features of Dictyostelium genes (Mann and Firtel, 1991; Roelofs et al., 2001); 2) a middle region, which contains a PFAM metallo-β-lactamase domain (Bateman et al., 2000); and 3) a C-terminal region, which contains two PFAM cNMP-binding domains.
Figure 1.
PdeD gene structure and protein sequence. (A) Schematic of the ORF of PdeD indicating the position of the introns, the metallo-β-lactamase domain, and the cyclic nucleotide–binding motifs. (B) Alignment of the cNMP-binding motifs of PdeD, bovine PKA-RI, and human PKG2. Gray, conserved; green, hydrophobic pocket for purine binding; yellow, hydrogen bonding of backbone with exocyclic oxygens; red, side chain essential for hydrogen bond and ion-pair formation with ribose 2′-OH and exocyclic oxygens, respectively; blue, ser/thr that distinguishes guanine from adenine binding (Su et al., 1995). (C) Alignment of the β-lactamase domains of PdeD with those of the B. cereus β-lactamase 569/H/9 (Bc-βlac) and human glyoxylase II. Gray, conserved; bold letters, identical residues in either all β-lactamases or all glyoxylases; blue, binding to Zn2+(I); red, binding to Zn2+(II); purple, binding to water or to both Zn2+(I) and Zn2+(II); amber, binding to substrate (Carfi et al., 1995; Concha et al., 1996; Cameron et al., 1999).
Double cNMP-binding domains are typically found in the PKA regulatory subunit (PKA-R) and in PKG. In the latter protein, they are located upstream of the PK catalytic region (Francis and Corbin, 1999). cAMP-binding proteins, such as PKA-RI and PKA-RII, share a common folding pattern, as determined from cocrystal structures with cAMP (Diller et al., 2001; Su et al., 1995). Homologous cGMP-binding proteins are predicted to possess a similar structure (Francis and Corbin, 1999). Critical residues for cNMP binding in both cNMP domains of PdeD were aligned with equivalent residues in bovine PKA-RI and human PKG2 (Figure 1B). The major determinant for nucleotide specificity is Ala210 (site A) and Ala334 (site B) in bovine PKA-RI. The equivalent position in cGMP-binding proteins is a Ser or Thr (blue), which would allow hydrogen bond formation with the C2-NH2 group of the guanine base (Shabb et al., 1991). Site A in PdeD harbors a serine at the position equivalent to Ala210, suggesting that this site preferentially binds cGMP (Figure 1B). Site B carries a Lys811 at the position equivalent to Ala334, which is not likely to contribute to either cAMP or cGMP binding. Site B shows further deviations from the consensus binding motif. The essential arginine (in red) that forms an ion-pair with the equatorial exocyclic phosphate oxygen of cNMP (equivalent to Arg209 and Arg333 in PKA-RI) (Su et al., 1995) is replaced by His. In addition, an insertion of three residues directly precedes this key residue in site B. Additional residues at this position are absent in all characterized cNMP-binding proteins.
The metallo-β-lactamase domain contains a binuclear Zn2+-binding motif that was first found in the bacterial class B β-lactamases. These enzymes catalyze the hydrolysis of an amide bond in the β-lactam ring of penicillin-type antibiotics (Carfi et al., 1995; Concha et al., 1996). A similar domain is found in glyoxylase II. Here, it hydrolyses the thiolester bond between gluthathione and 2-hydroxycarboxylic acid during inactivation of toxic methylglyoxal, a side product of glycolysis (Cameron et al., 1999). The crystal structure for both enzymes has been solved. Figure 1C shows the alignment of the PdeD metallo-β-lactamase homology region with that of Bacillus cereus β-lactamase II (Bc-βlac) and human glyoxylase II. Apart from the conserved His and Asp residues that are involved in metal binding, the PdeD sequence does not conform to any of the other enzymes with respect to residues that are specifically conserved for that class of enzyme (in bold), particularly those that are involved in substrate binding (in amber). The yeast and Dictyostelium class II phosphodiesterases, PDE1 and PdsA, also harbor the highly conserved HxHxDHxxG motif, which is the most characteristic feature of metallo-β-lactamase domain. However, unlike PdeD, these proteins share little homology with this domain elsewhere in the protein or with PdeD itself. It is therefore not possible to deduce the function of the PdeD from its sequence with any confidence.
Developmental Regulation and Disruption of the PdeD Gene
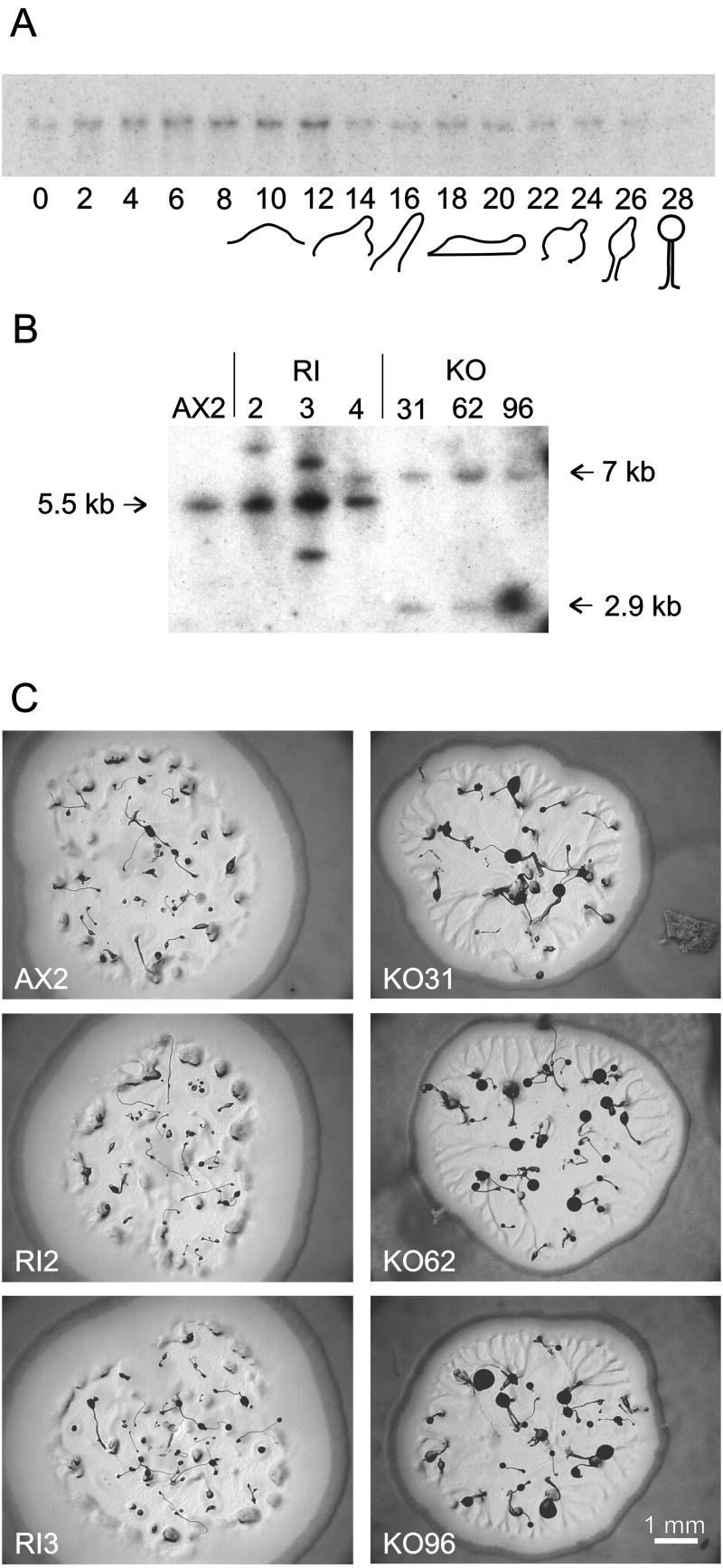
To gain more insight into the function of PdeD, we studied its developmental regulation and disrupted the gene. A Northern blot of PdeD probed to RNA isolated during the 28-h developmental life cycle shows that PdeD is transcribed into an mRNA of ∼2.9 kb during growth and development, with the highest level of expression during aggregation (Figure 2A).
Figure 2.
Developmental regulation and gene disruption of PdeD. (A) Developmental regulation. D. discoideum NC4 cells were washed free of bacteria and incubated on PB agar at 2 × 106 cells/cm2 until mature fruiting bodies had formed. RNA was isolated at 2-h intervals, and Northern transfers were probed with a [32P]dATP-labeled PdeD probe. RNA markers of 0.28–6.6 kb (Promega) were run on the same gel and stained with ethidium bromide to estimate the size of the pdeD mRNA, which appeared to be ∼2.9 kb. (B) Genomic digests of putative gene disruptants. AX2 cells were transformed with a linear construct of vector pBsrΔBam flanked by ∼800 bp of 5′ and 3′ DNA of the PdeD gene. Clones 31, 62, and 96 were selected by two PCR reactions as putative PdeD gene knockouts (KO) and clones 2, 3, and 4 as random integrants (RI). Genomic DNA of the KO and RI clones was double-digested with XhoI and NsiI, and Southern transfers were probed with PdeD DNA as described above. In AX2 and RI cells, a 5.5-kb band should be present, with additional bands of unknown size marking the random vector integrations. Insertion of pBsrΔBam in the PdeD gene by homologous recombination should yield bands of 2.9 and 7 kb. (C) The morphology of colonies of parent strain AX2, two RI and three pdeD KO clones grown on Klebsiella aerogenes lawns. Note the streaming phenotype and significantly larger fruiting bodies of the KO clones.
The PdeD gene was inactivated by homologous recombination to generate cell line pdeD. Construct integration was verified by two separate PCR reactions and by Southern analysis of genomic digests (Figure 2B). Three KO constructs and three clones with randomly integrated vectors (RI) were selected for further analysis. All KO and RI clones formed normal-looking aggregates, slugs, and fruiting bodies when cells were plated on nonnutrient agar. However, the KO cells showed an abnormal morphology when plated out clonally on bacterial lawns (Figure 2C). Colonies of the parent strain AX2 and the control RI lines showed mound-shaped aggregates at some distance from the growth edge and fruiting bodies toward the center of the colony. Formation of aggregation streams is not evident at this high cell density. The KO clones showed marked formation of large aggregation streams. In addition, aggregate and fruiting body size was larger than in the control cell lines.
This phenotype resembles the phenotype of a class of chemotactic mutants called streamers, which fall into different complementation groups (Ross and Newell, 1979). None of the mutated genes have been identified, but the most thoroughly characterized stmF mutants are defective in a cGMP-stimulated cGMP-PDE activity (Van Haastert et al., 1982) that was first identified in a mutant defective in cAMP phosphodiesterase (Dicou and Brachet, 1980). Absence of the cGMP phosphodiesterase results in an elevated and prolonged cGMP response on stimulation with chemoattractant, which is assumed to cause the streamer phenotype (Ross and Newell, 1981).
The cGMP Response and cGMP-PDE Activity in the pdeD-null Mutant
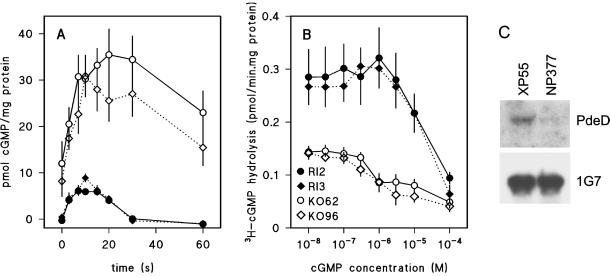
We first measured whether the pdeD mutant showed the characteristic elevated and prolonged cGMP response of the stmF mutants. Figure 3A shows that the cGMP response in the pdeD KO lines was 3 times higher than in the RI lines. Moreover, cGMP levels were still elevated in the KO lines at 30 s after stimulation, when in the control lines, cGMP was already back to the basal level. This almost exactly mimics the kinetics of the cGMP response in the stmF mutants (Ross and Newell, 1981) and suggests that pdeD encodes the cGMP-PDE that is defective in these mutants.
Figure 3.
Absence of cGMP-PDE in pdeD-null mutants and PdeD transcripts in stmF. (A) cGMP response. Two pdeD KO cell lines and two control RI lines were developed on PB agar until aggregation territories had formed. Cells were stimulated with 10−7 M cAMP, and accumulated cGMP levels were determined by radioimmunoassay at the indicated time points. Data were standardized on the protein content of the cell suspension. Means and SEs of two experiments, each consisting of six time courses, are presented. (B) cGMP-PDE activity. KO and RI cells were harvested and lysed while forming territories. Hydrolysis of 3H-cGMP was determined in the presence of increasing concentrations of cGMP in lysates that were diluted to degrade not more than 30% of the 3H-cGMP. Data were standardized on the protein content of the cleared lysate. Means and SEs of three experiments performed in triplicate are presented. (C) PdeD mRNA. Total RNA was isolated from stmF mutant NP377 and parent strain XP55 after 8 h of starvation. A Northern transfer was first probed with 32P-labeled PdeD DNA, then stripped and reprobed with the 1G7 rRNA gene, which is constitutively expressed throughout development (Williams et al., 1987).
To test this directly, we measured cGMP-PDE activity in lysates of KO and RI cell lines in the presence of increasing concentrations of cGMP. Figure 3B shows that in the control RI cell lines, unlabeled cGMP induced a modest stimulation of 3H-cGMP hydrolysis between 3 × 10−8 and 10−6 M before it showed significant competition with 3H-cGMP at 10−5 M. In the KO cell lines, total 3H-cGMP hydrolysis was strongly reduced and no stimulation by cGMP was evident. This suggests that the two KO lines lack a cGMP-PDE activity. The remaining activity is most likely the previously characterized cGMP phosphodiesterase PDE3, which is not stimulated by cGMP (Kuwayama et al., 2001).
To obtain further evidence that stmF mutants are defective in PdeD, we probed mRNA extracted from stmF mutant NP377 and its parent strain XP55 (Ross and Newell, 1981) with 32P-labeled PdeD DNA. Figure 3C shows that PdeD mRNA is greatly reduced in stmF mutant NP377.
Expression of PdeD-YFP Fusion Constructs
The loss of cGMP-PDE activity from the pdeD-null cell lines can in theory be caused by an (indirect) inhibitory effect of PdeD on the expression of the gene that actually encodes a cGMP-PDE. We have tried to express His-tagged PdeD and GST-PdeD fusion proteins in Escherichia coli to measure the enzyme activity of purified PdeD directly. Although the tagged proteins were expressed in E. coli, we have not yet been able to isolate them in native form.
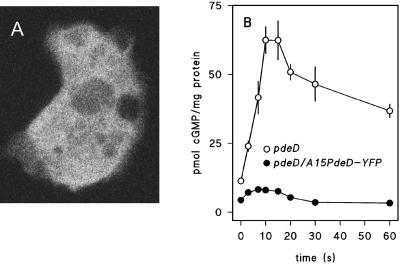
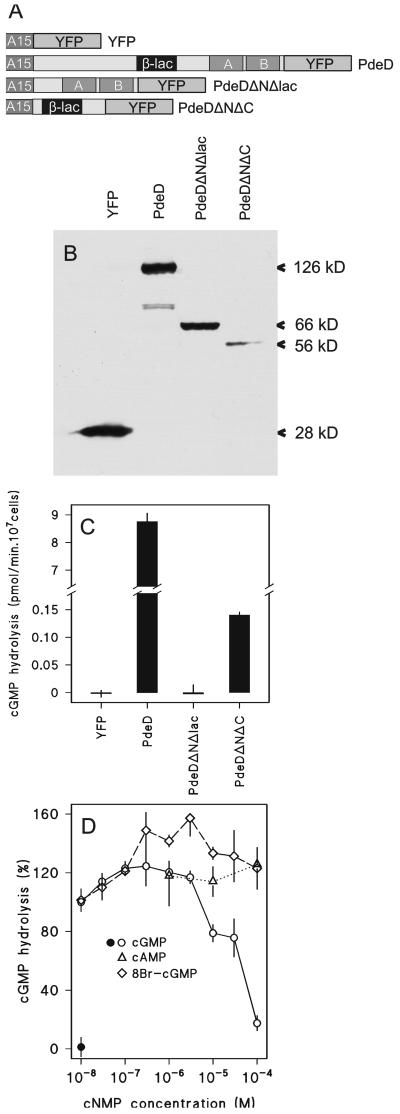
As an alternative approach, we fused the PdeD gene to the gene for enhanced YFP (Miyawaki et al., 1997) and expressed the fusion construct under control of the constitutive actin 15 promoter (A15) in the pdeD-null mutant. Confocal microscopy of pdeD/A15PdeD-YFP cells shows that the protein is present in the cytosol (Figure 4A). Expression of PdeD-YFP brought the cAMP-induced cGMP response in the pdeD-null mutant back to the level that is normal for wild-type cells, indicating that the phenotype of the null mutant is caused by loss of PdeD (Figure 4B). Two truncated forms of PdeD, ΔNΔlac with only the cNMP-binding domains and ΔNΔC with only the β-lactamase domain, were also expressed as YFP fusion proteins under the A15 promoter. A15YFP-transformed cells were included as control (Figure 5A). Figure 5B shows Western blots of the expressed proteins detected with GFP antibody. The YFP construct yielded the highest level of protein expression, followed by the full-length PdeD-YFP construct. The ΔNΔlac-YFP construct was also expressed to reasonable levels, but the ΔNΔC-YFP construct was expressed rather poorly. Lysates from cells transformed with full-length PdeD-YFP showed by far the highest cGMP-PDE activity (Figure 5C). No activity was found in either ΔNΔlac-YFP or A15-YFP lysates, but the ΔNΔC-YFP lysates showed significant cGMP-PDE activity despite the low levels of protein expression. The activity was lost within a few hours from the ΔNΔC-YFP lysates, whereas it was stable for at least 1 day for the full-length constructs. This led us to suppose that protein expression from the ΔNΔC-YFP construct may seem so low because the protein is unstable. Nevertheless, the ΔNΔC-YFP protein, with only the β-lactamase domain present, still displayed cGMP-PDE activity, whereas the ΔNΔlac-YFP protein, which was expressed to much higher levels, showed none. This strongly suggested that the PDE activity resides in the β-lactamase domain.
Figure 4.
Expression of PdeD-YFP in pdeD-null cells. (A) PdeD-YFP expression. A fusion construct of YFP with full-length PDE was expressed under the actin 15 promoter in pdeD KO clone 96. An optical section of YFP fluorescence in a living pdeD/A15PdeD-YFP cell was obtained with a Leica DMRBE confocal laser scanning microscope. (B) cGMP response. pdeD and pdeD/A15PdeD-YFP cells were harvested when forming aggregation territories, stimulated with 10−7 M cGMP, and assayed for accumulated cGMP levels. Means and SEs of two experiments, each consisting of six time courses, are presented.
Figure 5.
Expression and activity of full-length and truncated YFP-PdeD constructs. (A) Schematic of four fusion constructs of the actin 15 promoter, full-length or partial PdeD sequence, and YFP. Transformants of either of the four constructs were grown under selection with 200 μg/ml G418 to obtain YFP expression for the PdeDΔNΔC construct, which showed no YFP fluorescence under normal selection conditions. (B) Western blots of expressed YFP fusion proteins. Cleared lysates of vegetative cells transformed with the four YFP constructs were size-fractionated by SDS-PAGE and immunoblotted with mouse monoclonal αGFP antibodies. The size of the bands as deduced from their position relative to protein markers is indicated. (C) cGMP-PDE activity in transformants. Cleared lysates were assayed for hydrolysis of 10−8 M 3H-cGMP at 10-, 30-, 100-, 300-, and 1000-fold dilutions. Activities were calculated from dilutions that did not hydrolyze >30% of the substrate. Means of two experiments performed in duplicate are presented. (D) Immunoprecipitation of YFP fusion proteins. Cleared lysates of YFP (closed symbol) and PdeD-YFP (open symbols) -transformed cells were incubated with αGFP antibody linked to a mixed protein A/protein G coupled matrix. The matrix was thoroughly washed, then incubated with 10−8 M 3H-cGMP and the indicated concentrations of cGMP, 8-Br-cGMP, and cAMP for 30 min and assayed for 3H-5′-GMP. Data are expressed as percentage of 3H-cGMP hydrolysis obtained from the PdeD-YFP immunoprecipitate in the absence of added cyclic nucleotides. Means of two experiments performed in duplicate are presented.
The full-length PdeD-YFP and YFP proteins were immunoprecipitated with anti-GFP antibody. There was no activity associated with the YFP immunoprecipitate. The cGMP-PDE activity in the PdeD-YFP immunoprecipitate was ∼5% of the lysate from which it was derived. The immunoprecipitated activity was slightly activated by submicromolar cGMP concentrations and more strongly by 8-Br-cGMP, which is a good activator but a poor substrate for the stmF cGMP-PDE (Kesbeke et al., 1985). cAMP was a very poor competitor for 3H-cGMP hydrolyzing activity. This yields convincing evidence that PdeD encodes a cGMP-specific phosphodiesterase activity.
DISCUSSION
A novel gene, PdeD, was identified from the sequencing project of Dictyostelium discoideum chromosome II (Gloeckner et al., 2002). PdeD encodes a protein with two cNMP-binding domains and a binuclear Zn2+-binding motif, which is common to the metallo-β-lactamases. The same gene was recently identified, but not functionally characterized, from the genome sequencing project by Goldberg et al. (2002) and named GbpA. These authors also detected three other proteins with cNMP-binding motifs (GbpB–D). We also found GbpB; knockout and overexpression studies show that this gene, which we have named PdeE, encodes a cAMP phosphodiesterase (Meima, Weening, and Schaap, P., unpublished observations).
The cNMP-binding site A of PdeD harbors a characteristic Ser for cGMP binding and has all the essential residues for binding to the purine, ribose, and cyclic phosphate moieties of cGMP (Figure 1B). Binding site B shows several deviations in critical residues as well as an insertion of three amino acids in a region that in PKA-R has critical interactions with both the ribose and cyclic phosphate ring of cAMP (Su et al., 1995). Site B may therefore not be functional.
The function of the histidine-rich binuclear Zn2+-binding motif was not immediately obvious. The domain forms the catalytic center of three unrelated enzymes: metallo-β-lactamase, glyoxylase II, and a less-well-characterized bacterial arylsulfatase, which hydrolyze amide, thiolester, and sulfate ester bonds, respectively (Barbeyron et al., 1995; Carfi et al., 1995; Cameron et al., 1999). A histidine-rich metal-binding domain is also essential for catalysis in the cNMP phosphodiesterases, but the configuration of the histidines in this domain is entirely different (Xu et al., 2000). However, two low-affinity PDEs, PdsA from Dictyostelium (Lacombe et al., 1986) and PDE1 from S. cerevisiae (Nikawa et al., 1987), harbor a histidine-rich motif that is similar to that of the metallo-β-lactamases, indicating that this motif can also mediate the hydrolysis of cNMPs. The observation that pdeD-null mutants phenocopied stmF mutants, which are defective in cGMP-PDE, suggested that the hydrolytic activity encoded by its β-lactamase domain degrades cGMP.
The PdeD Gene Is Most Likely Defective in stmF Mutants
The stmF mutants provided the first evidence that cGMP might mediate induction of chemotaxis by the Dictyostelium chemoattractants cAMP and folic acid (Ross and Newell, 1979, 1981). Selected for their propensity to form long aggregation streams when grown clonally, they were later shown to be defective in a cGMP-PDE activity (Van Haastert et al., 1982; Coukell and Cameron, 1986). As a consequence, they show a prolonged cGMP response on stimulation with chemoattractant, which was correlated with a prolonged period but reduced rate of cell movement (Ross and Newell, 1981; Newell and Liu, 1992; Segall, 1992).
The PdeD mutants also showed extensive streaming when plated clonally with bacteria (Figure 2C), and further biochemical analysis (Figure 3) showed that their cGMP response was similarly prolonged and their cGMP-PDE activity similarly reduced, as is the case for the stmF mutants. The stmF genetic locus maps to chromosome II (Coukell and Cameron, 1985), and so does the PdeD sequence. In combination with the observation that the stmF mutant NP377 expressed almost no PdeD mRNA (Figure 3C), this suggests that the defective gene in the stmF mutants is PdeD. However, we have not been able to find mutations in the promoter of the stmF PdeD gene, which could account for reduced transcription. Final evidence has to await complementation of the stmF mutants by PdeD and identification of the genetic lesion.
The PdeD Catalytic Activity Resides in Its Metallo-β-Lactamase Domain
The full-length PdeD gene and PdeD truncations that either lacked the β-lactamase domain or contained only this domain were expressed in Dictyostelium cells as YFP fusion proteins (Figure 5). Only the full-length protein and the protein that contained the β-lactamase domain showed activity, indicating that the PDE-catalytic activity is provided by the β-lactamase domain. The full-length protein was purified by immunoprecipitation with GFP antibodies. The purified PdeD showed cGMP-PDE activity that was stimulated by cGMP and by 8-Br-cGMP, as was previously described for the enzyme lacking in stmF mutants (Kesbeke et al., 1985). In our hands, the stimulation by cGMP was less pronounced than described by Kesbeke et al., which is perhaps a result of the use of different parental strains. In the original biochemical characterization of the enzyme, stimulation by cGMP was also not noted (Dicou and Brachet, 1980). 8-Br-cGMP induced a more significant stimulation, because it is a poor substrate (Kesbeke et al., 1985). cAMP was no competitor for 3H-cGMP hydrolysis. This indicates that the PdeD gene product is a cGMP-specific phosphodiesterase.
The cNMP-Binding Domain A of PdeD May Mediate Allosteric Activation
The cGMP-PDE that is lost in stmF mutants is allosterically activated by cGMP. Studies with cGMP analogues showed that cyclic nucleotide specificity of the activator and catalytic sites of the enzyme are not identical (Kesbeke et al., 1985). Both sites are specific for cGMP, do not bind cAMP, and do not tolerate modification of the exocyclic oxygens. In addition, however, the catalytic site does not tolerate modification of cGMP at N1H/C6O, C8, and O3′, whereas the activator site does not tolerate modification of C2-NH2 and the 2′-OH. The latter two positions would characteristically interact with Ser681 and Glu671 in cNMP-binding domain A of PdeD (Figure 1B). Binding site B does not contain the conserved Ser/Thr at the equivalent position and is, as discussed above, probably not functional as a cGMP-binding site.
8-Br-cGMP is a very poor PdeD substrate but a good agonist for the activator site. This also suggests that the activator site is homologous to eukaryotic cNMP-binding domains, which bind cyclic nucleotides in the syn conformation that is enforced by the bulky bromine at the 8-position of the purine ring (de Wit et al., 1982). The nucleotide specificity of the catalytic domain does not resemble that of any known cGMP-binding protein. Elucidation of the crystal structure of PdeD will be necessary to understand its interaction with the substrate and the manner by which the cNMP-binding domain activates catalytic activity.
ACKNOWLEDGMENTS
We thank Tomo Abe for confocal microscopy, Duke Näthke for advice on immunoprecipitation, and Kees Weijer for the gift of vector pB17SYFP. Sequence data for D. discoideum was obtained from the Genome Sequencing Center Jena website at http://genome.imb-jena.de/dictyostelium/. The German part of the D. discoideum Genome Project is carried out by the Institute of Biochemistry I, Cologne, and the Department of Genome Analysis, IMB Jena, with support by the Deutsche Forschungsgemeinschaft (No. 113/10–1 and 10–2). This research was funded by Wellcome Trust University Award Grant 057137.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0285. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0285.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Focal Alignment Search Tool. J Mol Biol. 1990;25:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barbeyron T, Potin P, Richard C, Collin O, Kloareg B. Arylsulphatase from Alteromonas carrageenovora. Microbiology. 1995;141:2897–2904. doi: 10.1099/13500872-141-11-2897. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AD, Ridderstrom M, Olin B, Mannervik B. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure Fold Des. 1999;7:1067–1078. doi: 10.1016/s0969-2126(99)80174-9. [DOI] [PubMed] [Google Scholar]
- Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha NO, Rasmussen BA, Bush K, Herzberg O. Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis. Structure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- Coukell MB, Cameron AM. Genetic locus (stmF) associated with cyclic GMP phosphodiesterase activity in Dictyostelium discoideum maps in linkage group II. J Bacteriol. 1985;162:427–429. doi: 10.1128/jb.162.1.427-429.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coukell MB, Cameron AM. Characterization of revertants of stmF mutants of Dictyostelium discoideum: evidence that stmF is the structural gene of the cGMP-specific phosphodiesterase. Dev Gen. 1986;6:163–177. doi: 10.1002/dvg.1020060303. [DOI] [PubMed] [Google Scholar]
- de Wit RJ, Hoppe J, Stec WJ, Baraniak J, Jastorff B. Interaction of cAMP derivatives with the “stable” cAMP-binding site in the cAMP-dependent protein kinase type I. Eur J Biochem. 1982;122:95–99. doi: 10.1111/j.1432-1033.1982.tb05852.x. [DOI] [PubMed] [Google Scholar]
- Dicou EL, Brachet P. A separate phosphodiesterase for the hydrolysis of cyclic guanosine 3′,5′-monophosphate in growing Dictyostelium discoideum amoebae. Eur J Biochem. 1980;109:507–514. doi: 10.1111/j.1432-1033.1980.tb04822.x. [DOI] [PubMed] [Google Scholar]
- Diller TC, Madhusudan, Xuong N, Taylor SS. Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: crystal structure of the type II beta regulatory subunit. Structure. 2001;9:73–82. doi: 10.1016/s0969-2126(00)00556-6. [DOI] [PubMed] [Google Scholar]
- Francis SH, Corbin JD. Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit Rev Clin Lab Sci. 1999;36:275–328. doi: 10.1080/10408369991239213. [DOI] [PubMed] [Google Scholar]
- Gloeckner G, et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature. 2002;418:79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- Goldberg JM, Bosgraaf L, Van Haastert PJM, Smith JL. Identification of four candidate cGMP targets in Dictyostelium. Proc Natl Acad Soc USA. 2002;99:6749–6754. doi: 10.1073/pnas.102167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesbeke F, Baraniak J, Bulgakov R, Jarstorff B, Morr M, Petridis G, Stec WJ, Seela F, Van Haastert PJM. Cyclic nucleotide specificity of the activator and catalytic sites of a cGMP-stimulated cGMP phosphodiesterase from Dictyostelium discoideum. Eur J Biochem. 1985;51:179–186. doi: 10.1111/j.1432-1033.1985.tb09083.x. [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Ishida S, Van Haastert PJM. Non-chemotactic Dictyostelium discoideum mutants with altered cGMP signal transduction. J Cell Biol. 1993;123:1453–1462. doi: 10.1083/jcb.123.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama H, Snippe H, Derks M, Roelofs J, Van Haastert PJ. Identification and characterization of DdPDE3, a cGMP-selective phosphodiesterase from Dictyostelium. Biochem J. 2001;353:635–644. doi: 10.1042/0264-6021:3530635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe M-L, Podgorski GJ, Franke J, Kessin RH. Molecular cloning and developmental expression of the cyclic nucleotide phosphodiesterase gene of Dictyostelium discoideum. J Biol Chem. 1986;261:16811–16817. [PubMed] [Google Scholar]
- Liu G, Newell PC. Evidence that cyclic GMP regulates myosin interaction with the cytoskeleton during chemotaxis of Dictyostelium. J Cell Sci. 1988;90:123–129. doi: 10.1242/jcs.90.1.123. [DOI] [PubMed] [Google Scholar]
- Liu G, Newell PC. Evidence that cyclic GMP may regulate the association of myosin II heavy chain with the cytoskeleton by inhibiting its phosphorylation. J Cell Sci. 1991;98:483–490. doi: 10.1242/jcs.98.4.483. [DOI] [PubMed] [Google Scholar]
- Liu G, Newell PC. Regulation of myosin regulatory light chain phosphorylation via cyclic GMP during chemotaxis of Dictyostelium. J Cell Sci. 1994;107:1737–1743. doi: 10.1242/jcs.107.7.1737. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. A developmentally regulated, putative serine/threonine protein kinase is essential for development in Dictyostelium. Mech Dev. 1991;35:89–101. doi: 10.1016/0925-4773(91)90060-j. [DOI] [PubMed] [Google Scholar]
- Manstein DJ, Schuster H-P, Morandini P, Hunt DM. Cloning vectors for the production of proteins in Dictyostelium discoideum. J Cell Biol. 1995;162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- Mehats C, Andersen CB, Filopanti M, Jin SLC, Conti M. Cyclic nucleotide phosphodiesterases and their role in endocrine cell signaling. Trends Endocrinol Metab. 2002;13:29–35. doi: 10.1016/s1043-2760(01)00523-9. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Mutzel R, Lacombe M-L, Simon M-N, De Gunzburg J, Veron M. Cloning and cDNA sequence of the regulatory subunit of cAMP-dependent protein kinase from Dictyostelium discoideum. Proc Natl Acad Sci USA. 1987;84:6–10. doi: 10.1073/pnas.84.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PC, Liu G. Streamer F mutants and chemotaxis of Dictyostelium. Bioessays. 1992;14:473–478. doi: 10.1002/bies.950140708. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Sass P, Wigler M. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3629–3636. doi: 10.1128/mcb.7.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DJ. Metallo-beta-lactamases: a new therapeutic challenge. J Med Microbiol. 1993;39:93–99. doi: 10.1099/00222615-39-2-93. [DOI] [PubMed] [Google Scholar]
- Roelofs J, Meima M, Schaap P, Van Haastert PJM. The Dictyostelium homologue of mammalian soluble adenylyl cyclase encodes a guanylyl cyclase. EMBO J. 2001;20:4341–4348. doi: 10.1093/emboj/20.16.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FM, Newell PC. Genetics of aggregation pattern mutations in the cellular slime mold Dictyostelium discoideum. J Gen Microbiol. 1979;115:289–300. doi: 10.1099/00221287-115-2-289. [DOI] [PubMed] [Google Scholar]
- Ross FM, Newell PC. Streamers: chemotactic mutants of Dictyostelium discoideum with altered cyclic GMP metabolism. J Gen Microbiol. 1981;127:339–350. doi: 10.1099/00221287-127-2-339. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JE. Behavioral responses of streamer F mutants of Dictyostelium discoideum: effects of cyclic GMP on cell motility. J Cell Sci. 1992;101:589–597. doi: 10.1242/jcs.101.3.589. [DOI] [PubMed] [Google Scholar]
- Shabb JB, Buzzeo BD, Ng L, Corbin JD. Mutating protein kinase cAMP-binding sites into cGMP-binding sites: mechanism of cGMP selectivity. J Biol Chem. 1991;266:24320–24326. [PubMed] [Google Scholar]
- Shaulsky G, Escalante R, Loomis WF. Developmental signal transduction pathways uncovered by genetic suppressors. Proc Natl Acad Sci USA. 1996;93:15260–15265. doi: 10.1073/pnas.93.26.15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Dostmann WR, Herberg FW, Durick K, Xuong NH, Ten Eyck L, Taylor SS, Varughese KI. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science. 1995;269:807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Thomason PA, Traynor D, Cavet G, Chang WT, Harwood AJ, Kay RR. An intersection of the camp/pka and two-component signal transduction systems in Dictyostelium. EMBO J. 1998;17:2838–2845. doi: 10.1093/emboj/17.10.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJM, Van Lookeren Campagne MM, Ross FM. Altered cGMP phosphodiesterase activity in chemotactic mutants of Dictyostelium discoideum. FEBS Lett. 1982;147:149–152. doi: 10.1016/0014-5793(82)81029-6. [DOI] [PubMed] [Google Scholar]
- Williams JG, Ceccarelli A, McRobbie S, Mahbubani H, Kay RR, Early A, Berks M, Jermyn KA. Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell. 1987;49:185–192. doi: 10.1016/0092-8674(87)90559-9. [DOI] [PubMed] [Google Scholar]
- Xu RX, et al. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]