Abstract
Cohesion between sister chromatids is a prerequisite for accurate chromosome segregation during mitosis and meiosis. To allow chromosome condensation during prophase, the connections that hold sister chromatids together must be maintained but still permit extensive chromatin compaction. In Drosophila, null mutations in the orientation disruptor (ord) gene lead to meiotic nondisjunction in males and females because cohesion is absent by the time that sister kinetochores make stable microtubule attachments. We provide evidence that ORD is concentrated within the extrachromosomal domains of the nuclei of Drosophila primary spermatocytes during early G2, but accumulates on the meiotic chromosomes by mid to late G2. Moreover, using fluorescence in situ hybridization to monitor cohesion directly, we show that cohesion defects first become detectable in ordnull spermatocytes shortly after the time when wild-type ORD associates with the chromosomes. After condensation, ORD remains bound at the centromeres of wild-type spermatocytes and persists there until centromeric cohesion is released during anaphase II. Our results suggest that association of ORD with meiotic chromosomes during mid to late G2 is required to maintain sister-chromatid cohesion during prophase condensation and that retention of ORD at the centromeres after condensation ensures the maintenance of centromeric cohesion until anaphase II.
INTRODUCTION
Sister-chromatid cohesion is essential for accurate chromosome segregation during cell division (Lee and Orr-Weaver, 2001; Nasmyth, 2001). For proper kinetochore orientation, bipolar microtubule attachment and timing of the metaphase/anaphase transition to occur, sister chromatids must stay associated with each other from the time of their synthesis until anaphase (Amon, 1999; Cohen-Fix, 2001). Several gene products that control cohesion are conserved from yeast to humans and function during meiosis as well as mitosis (van Heemst and Heyting, 2000; Lee and Orr-Weaver, 2001; Uhlmann, 2001). A multiprotein complex, known as cohesin, appears to provide a structural link between sisters that must be severed to release cohesion during both mitosis and meiosis (Nasmyth et al., 2000; Cohen-Fix, 2001). However, cleavage of centromeric cohesin subunits by the endopeptidase separase must be inhibited during meiosis I (Klein et al., 1999; Watanabe and Nurse, 1999; Pasierbek et al., 2001). Although separase activity is required to release arm cohesion and allow the segregation of recombinant homologs during anaphase I (Buonomo et al., 2000; Siomos et al., 2001), maintenance of centromeric cohesion until anaphase II is essential for accurate segregation of the sister chromatids during the second meiotic division (Bickel and Orr-Weaver, 1996). At least a subset of meiotic cohesins at the centromeres are resistant to separase cleavage until anaphase II (Klein et al., 1999; Watanabe and Nurse, 1999; Pasierbek et al., 2001), although the mechanism by which their cleavage is prevented during meiosis I is not yet understood.
The Drosophila ORD protein is essential for normal sister-chromatid cohesion during meiosis. Several ord alleles have been isolated and characterized, and all result in aberrant meiotic chromosome segregation in males and females in genetic assays that monitor the fidelity of sex chromosome transmission (Mason, 1976; Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997). Moreover, the frequency and distribution of aneuploid gametes recovered from ordnull flies indicate that in the absence of ORD function, sister chromatids segregate randomly through both meiotic divisions (Bickel et al., 1997). These data support the conclusion that in meiotic cells lacking ORD activity, sister-chromatid cohesion is totally absent when kinetochores make microtubule attachments during prometaphase I. Consistent with this model, premature separation of sister chromatids before metaphase I has been documented cytologically in ord oocytes and spermatocytes (Goldstein, 1980; Lin and Church, 1982; Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997, 2002).
Here, we show that wild-type, as well as ORD tagged with green fluorescent protein (GFP), is concentrated within the extrachromosomal domains of the nucleus in primary spermatocytes during early G2 of the meiotic cell cycle. GFP-ORD protein redistributes within the nucleus and accumulates on the chromatin before the cells enter prophase I and the chromosomes condense. Using fluorescence in situ hybridization (FISH) to monitor the state of cohesion directly, we observe cohesion defects in ord spermatocytes shortly after the time when GFP-ORD accumulates on the chromosomes in wild-type cells. After chromosome condensation, GFP-ORD is detectable only at the centromeres and remains there until cohesion is lost at anaphase II. Our results suggest that association of ORD with spermatocyte chromosomes before condensation is required to maintain cohesion during meiosis I and that retention of ORD at the centromeres ensures the maintenance of centromeric cohesion until anaphase II.
MATERIALS AND METHODS
Fly Strains
Flies were raised at 25°C on standard cornmeal molasses media. Cytological analyses of wild-type spermatocytes were performed using testes from y/y+Y; cn bw sp flies. To generate ord5/Df larvae, y/Y; ord5 bw/CyO, y+ males were crossed to y/y; cn Df(2R) W1370/CyO, y+ virgins. Mutant ord larvae were selected by the presence of yellow mouth parts because they lack the y+ gene carried on the CyO balancer chromosome. In flies containing the P{w+mC ori Amp = gfp::ord} transposon, expression of GFP-ORD is controlled by the ord promoter and 5′-regulatory sequences. P{w+mC ori Amp = gfp::ord} is a CaSpeR 4 (Pirrotta, 1988) derivative that contains 6899 base pairs of genomic DNA encompassing the entire ord gene. Polymerase chain reaction (PCR) was used to engineer an XbaI site immediately after the initiator AUG of the ORD coding region and enhanced GFP (Clontech, Palo Alto, CA) was inserted at this site. This construct results in the expression of ORD protein tagged at its N terminus with GFP. Several independent insertion lines were established. For most lines, the presence of the GFP-ORD insertion rescued the meiotic segregation phenotype of ord1/ord3 males and females in our standard genetic assay (Kerrebrock et al., 1992). One rescuing insertion on the third chromosome (T076) was chosen to construct an ord10/ord10; P{gfp::ord}/P{gfp::ord} stock that was used for cytological analyses. Because the ord10 allele contains a nonsense mutation at codon 24 (Bickel et al., 1997), GFP-ORD is the only ORD protein in these cells.
Generation of ORD Antiserum
An EcoRI ord cDNA fragment corresponding to the C-terminal region of the ORD open reading frame (ORF) was cloned into pGEX1λt (Amersham Pharmacia, Piscataway, NJ). The resulting protein contained GST fused to the C-terminal 210 amino acids of ORD. After protein induction with isopropyl β-d-thiogalactoside, GST-ORD containing inclusion bodies were isolated and solubilized with 8 M urea and 2% SDS. After preparative SDS-PAGE, GST-ORD was electroeluted from the acrylamide slice and was concentrated. Immunogen was sent to Cocalico Biologicals (Reamstown, PA) to generate guinea-pig antiserum, GP43.
Immunolocalization of ORD
Testes were dissected from third instar larvae or young adults in saline testes buffer containing 183 mM KCl, 47 mM NaCl, 10 mM Tris-HCl, pH 6.8, and 1 mM EDTA (Gatti and Baker, 1989). Each set of testes was transferred to saline testes buffer containing 2 mM Pefabloc (Sigma, St. Louis, MO) on a precleaned Superfrost Plus slide (VWR, West Chester, PA). Adult testes were cut with tungsten needles before squashing. A siliconized 18-mm coverslip was gently lowered onto the testes to squash them and the preparation was quick frozen in liquid nitrogen. On removal from liquid nitrogen, the coverslip was quickly removed and the slide was immediately placed in 90% MeOH/20 mM EGTA (at −30°C) for ∼5–15 min. Squashes were then fixed for 5 min at room temperature in 1× PHEM (Starr et al., 1998; 60 mM Pipes, 25 mM HEPES, pH 7.0, 10 mM EGTA, and 4 mM MgSO4) containing 4% formaldehyde (Ted Pella, Redding, CA). Slides were rinsed three times in phosphate-buffered saline (PBS; 130 mM NaCl, 7 mM NaH2PO4, and 3 mM NaH2PO4) and stored (up to 1 h) in PBS. Before the addition of antibody, slides were incubated three times for 5 min each in PBS/0.1% Triton-X 100 (PBT), and then rinsed three times with PBS. The tissue was blocked in 5% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA), 2% bovine serum albumin (BSA), 0.2× Superblock/PBS (Pierce, Rockford, IL), and 0.01% NaAzide for 1.0 h at room temperature. All subsequent antibody incubations were performed at room temperature in a humidified chamber unless noted otherwise. After each antibody incubation, slides were rinsed three times and washed for three 8-min periods in PBS.
To stain for ORD, squashes were incubated for 1.0 h in guinea-pig ORD antiserum (GP43) diluted 1:1000 in 0.2× Superblock. For ORD/EAST double-labeling experiments, polyclonal mouse EAST antisera ED3 and ED4 (Wasser and Chia, 2000) were diluted 1:1000 in 0.2× Superblock containing GP43 antiserum (also diluted 1:1000). To detect GFP-ORD, affinity-purified rabbit anti-GFP antibodies (Molecular Probes, Eugene, OR) were diluted 1:1000 in 0.2× Superblock (except for Figure 3C, 1:250 dilution and Figures 1I, 3E and 6K, 1:500 dilution) and used in a 1-h antibody incubation. Cy3 affinity-purified anti-guinea-pig and anti-rabbit antibodies (Jackson ImmunoResearch Laboratories) were used to detect ORD and GFP-ORD, respectively, except for ORD/GFP-ORD double-labeling experiments where Cy5 affinity-purified anti-rabbit antibodies (Jackson ImmunoResearch Laboratories) were used to detect GFP-ORD. Alexa 488 anti-mouse antibodies (Molecular Probes) were used to visualize EAST protein. MEI-S332 immunofluorescence was performed as described by Tang et al. (1998), and MEI-S332 protein was visualized using Cy5 affinity-purified anti-guinea-pig antibodies (Jackson ImmunoResearch Laboratories). All secondary antibodies were diluted in PBS/0.5% BSA, and incubations were performed for 45 min in the dark. Tubulin or nuclear lamin staining was performed after ORD, EAST, GFP, and/or MEI-S332 primary and secondary antibody incubations were completed. Anti-tubulin rat monoclonal antibodies YL1/2 and YOL1/34 (Sera-Lab, Loughborough, UK) were used together, each at a dilution of 1:5. Mouse monoclonal nuclear lamin antibodies (T40; a gift from H. Saumweber) were used at a dilution of 1:50. Squashes were incubated for 30–45 min in PBS/0.5% BSA containing the appropriate antibodies. Alexa 488 anti-rat antibodies (Molecular Probes) were used to detect tubulin, and Cy5 anti-mouse antibodies (Jackson ImmunoResearch Laboratories) were used to detect lamin. To visualize DNA, slides were stained for 10 min with 1 μg/ml 4′6-diamidino-2-phenylindole (DAPI; Molecular Probes) in PBS followed by three rinses in PBS. An 18-mm coverslip containing 5 μl of Prolong Antifade reagent (Molecular Probes) was lowered onto the tissue and was allowed to dry overnight.
Figure 3.
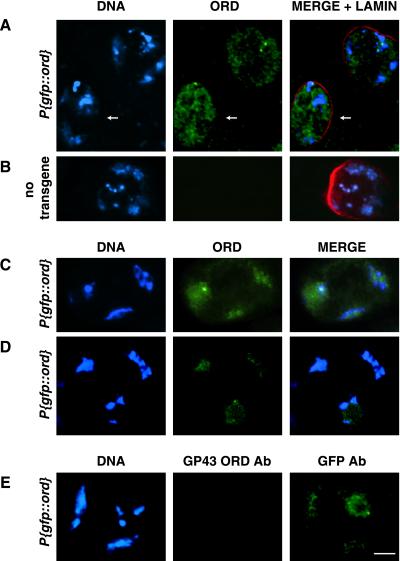
ORD accumulates on chromatin in late G2 spermatocytes. Localization of GFP-ORD in spermatocytes from P{gfp::ord} (A, C, D, and E) or nontransgenic (B) males. ORD (green), DNA (blue), nuclear lamin (red). (A) Immunofluorescence detection of GFP-ORD in S4 spermatocytes using GFP antibodies. Nuclear distribution of ORD appears more homogeneous (arrow) than in earlier stages (compare with Figure 1G), and colocalization with chromatin is visible in a slightly older cell (unlabeled, top right). Image is a single section from a deconvolved z-series. (B) GFP immunofluorescence signal is absent in a nontransgenic S4 primary spermatocyte. Image represents a full projection of a nondeconvolved z-series. (C) GFP-ORD signal detected with GFP antibody coincides with DNA in an S5 spermatocyte. (D) GFP fluorescence in a live S6 spermatocyte. (E) Late G2 P{gfp::ord} primary spermatocyte costained with GP43 ORD (red) and GFP (green) antibodies. All images are same scale. Bar, 10 μm.
Figure 1.
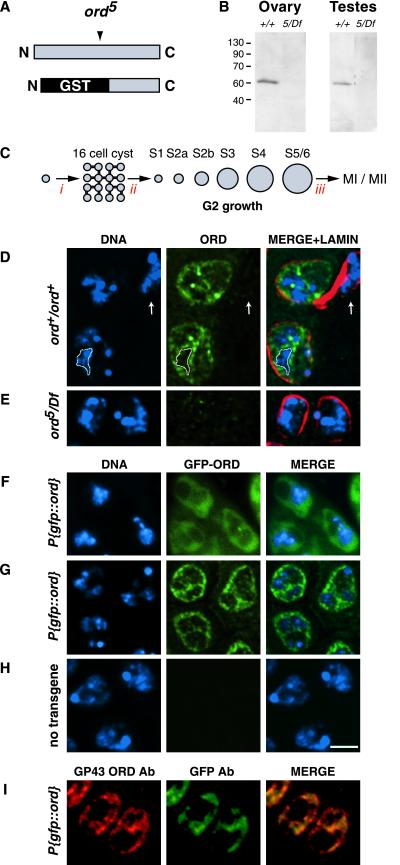
ORD localization in early G2 primary spermatocytes. (A) Schematic of full-length ORD protein showing relative position of the ord5 nonsense mutation and the GST-ORD immunogen (bottom) against which GP43 antibodies were raised. (B) Western blot of Drosophila ovary (15 μg/lane) and testis (4 sets/lane) extracts probed with GP43 ORD antiserum. (C) Spermatocyte development in D. melanogaster (not to scale). A spermatagonial cell undergoes four incomplete mitotic divisions (i) to generate a cyst of 16 primary spermatocytes. After S phase (ii), spermatocytes proceed through an extensive G2 phase that can be divided into seven stages (Cenci et al., 1994). Subsequently, chromosome condensation occurs (iii) and cells undergo meiosis I and II. (D and E) Fixed testes squashes from wild-type and ord5/Df third instar larvae were immunostained using GP43 ORD antiserum. DNA (blue), ORD (green), nuclear lamin (red). Images represent either a single section (D) or full projection (E) of a deconvolved z-series. The extrachromosomal distribution of ORD is emphasized by the outlined region in D, which surrounds a single chromosomal territory. ORD staining is absent in the somatically derived cyst cell (arrow). (F-H) GFP-ORD localization in S2a primary spermatocytes. DNA (blue), GFP-ORD (green). (F) GFP fluorescence in live spermatocytes from a P{gfp::ord} third instar larva. GFP-ORD is primarily nuclear, as determined by phase contrast microscopy (not shown). (G and H) Immunofluorescence detection of GFP-ORD in fixed testes squashes using anti-GFP antibody. (G) P{gfp::ord} transgenic spermatocytes, single section from a deconvolved z-series. (H) Nontransgenic control spermatocytes, full projection of a deconvolved z-series. (I) P{gfp::ord} transgenic spermatocytes costained with GP43 ORD (red) and GFP (green) antibodies. Both antibodies detect the same distribution of ORD protein at this stage. All images are same scale. Bar, 5 μm.
Analysis of GFP-ORD in Living Spermatocytes
Wild-type and ord10/ord10; P{gfp::ord}/P{gfp::ord} third instar larvae and adults were dissected in Shields and Sang M3 insect medium (Sigma) containing 10% fetal calf serum (Life Technologies/BRL, Grand Island, NY). Testes were transferred to Shields and Sang M3 insect medium and 10% fetal calf serum supplemented with 1.0 μg/ml Hoechst 33342 (Molecular Probes) on a precleaned Superfrost slide (VWR), opened with tungsten needles, and gently squashed using an 18-mm coverslip. Coverslips were immediately sealed to the slides with vasoline:lanolin:paraffin (1:1:1), and cells were visualized by epifluorescence to monitor GFP localization.
Generation of FISH Probes
An X chromosome probe (X het) directed against the 359-base pair 1.686 g/cm3 satellite repeat located near the centromere of the X chromosome was generated as described previously (Bickel et al., 2002). To generate an X-chromosome arm probe (X arm), bacteria artificial chromosome DNA spanning 3C1–6 (RPCI-98 34.O.3; BACPAC Resources, Oakland, CA) was fragmented as described by Dernburg (Dernburg, 2000). Approximately 10 μg of digested DNA was labeled with 60 U of terminal deoxynucleotidyl transferase (NE Biolabs, Beverly, MA) in a 100-μl reaction containing 135 μM dTTP and 67.5 μM Cy3-dUTP (Amersham Pharmacia). The probe was precipitated in 2 M ammonium acetate to remove unincorporated nucleotides, resupended in Tris-EDTA, and stored at −20°C in the dark.
FISH Analysis
For the analysis of cohesion on interphase chromatin, testes were dissected from either yw/Y; ord+/ord+ or y/Y; ord5 bw/Df third instar larvae and were squashed and fixed as described in the immunofluorescence section above. A modified fixation method was used to analyze centromeric cohesion on condensed meiotic chromosomes. Testes from either yw/Y; ord+/ord+, y/Y; ord5 bw/Df, or y/Y; ord10 bw/Df adults were dissected in 0.7% NaCl and were placed in 0.5% Na citrate for 10 min. Testes then were transferred to 5.0 μl of 45% acetic acid/2% formaldehyde (Ted Pella) on a siliconized 18-mm coverslip where they were opened with tungsten needles and fixed for 3 min. Squashing was performed by lowering a precleaned Superfrost slide (VWR) onto the coverslip and pressing firmly downward for ∼5 s. The slides were then placed and stored (up to 2 h) in liquid nitrogen. On removal from liquid nitrogen, coverslips were removed and slides were placed in PBS.
Before hybridization, fixed slides (both procedures) were rinsed in PBS, incubated twice for 10 min in 70% EtOH, once for 10 min in 100% EtOH, and were permitted to air dry at room temperature. To rehydrate, the squashes were incubated in 2× SSC/0.1% Tween 20 (SSCT) for 30 min with two changes of buffer. Slides were then incubated in 25% formamide/2× SSCT for 10 min, followed by another 10 min wash in 50% formamide/2× SSCT. The tissue was covered in 500 μl of 50% formamide/2× SSCT and was allowed to prehybridize for at least 3 h at 37°C in a humidified chamber. Ten microliters of probe solution containing Fluorogreen-labeled 359-base pair X het probe diluted to 0.5 ng/μl and Cy3-labeled X arm probe diluted to ∼10 ng/μl in hybridization buffer (3× SSC, 50% formamide, 10% Dextran sulfate) were added to each slide. Siliconized coverslips were placed over the tissue and sealed to the slides with rubber cement. Probe and chromosomal DNA were denatured at 94°C for 2 min (Boekl slide moat). After denaturation, slides were placed in a humidified chamber and were hybridized overnight at 37°C. After hybridization, coverslips were removed in 37°C prewarmed 50% formamide/2× SSCT and were incubated in 50% formamide/2× SSCT at 37°C for 2 h with one change of buffer. Slides were placed in 25% formamide/2× SSCT and incubated at room temperature for 10 min, followed by three 10-min washes in 2× SSCT without formamide. The tissue was counterstained and mounted as described in the immunofluorescence section above. Scoring of FISH signals was performed on full projections of z-series. Single FISH signals or two closely associated signals (within 0.3 μm) were scored as together. Two signals separated by a distance greater than 0.3 μm were scored as separated.
Microscopy and Image Analysis
Confocal microscopy was performed on a confocal microscope equipped with UV, Ar, Kr/Ar, and He/Ne lasers (TCS SP2; Leica, Deerfield, IL). All images were collected using a 63X Plan-APO objective and sequential scanning mode. Single channel TIFF images were combined and cropped using Openlab 3.0 software (Improvision, Lexington, MA). Epifluorescence microscopy was performed on a Zeiss Axioplan2 microscope (Jena, Germany) using a 63X Plan-APOCHROMAT objective. Single-channel images were collected with a Hamamatsu ORCA-ER camera (Hamamatsu, Japan) controlled by Openlab 3.0 software. Registration differences between channels were eliminated using the registration module of Openlab 3.0 software and Tetraspeck fluorescently labeled beads (Molecular Probes). Deconvolution was performed using Volocity 1.3 software (Improvision).
Quantification of the level of GFP-ORD protein within individual nuclei from single fields of cells was performed on deconvolved volumes. The GFP-ORD signal intensity (0–216 units) within a volumetric region of interest (VROI) was calculated using the measurements function of Volocity 1.4.4. Total signal = (mean intensity/voxel) × total number of voxels within VROI. The average value (mean intensity/voxel) for several regions with no visible signal in each field was used to determine the background signal due to mechanical noise for that field. This value was subtracted from mean intensity/voxel values for each nuclear VROI before multiplying by the total volume. Because the amount of DNA remains constant throughout G2, we used the quantified DAPI signal within each nucleus to normalize the GFP-ORD values. The average normalized GFP-ORD value for each stage is shown in Figure 4C for three separate fields of cells. Figure 4, A and B, shows a subset of cells within field 1.
Figure 4.
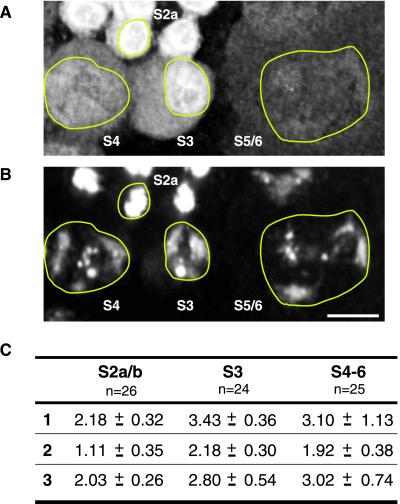
Quantification of ORD protein in spermatocyte nuclei during G2. (A) Immunofluorescence detection of GFP-ORD and (B) DAPI staining of DNA in P{gfp::ord} spermatocytes. Nuclei are outlined in yellow. Bar, 10 μm. ORD staining appears to diminish during spermatocyte growth. (C) Three-dimensional reconstructions of deconvolved z-series from three different fields of cells were used to calculate the total amount of ORD protein within the nuclei of spermatocytes at different stages of G2 growth. GFP-ORD values were normalized against the average DNA signal for the corresponding stage. The average of the normalized GFP-ORD values is shown for each stage. The number of cells used to calculate the average is indicated below each stage designation. A subset of cells from field 1 is shown above, with specific stages labeled. Units are arbitrary.
RESULTS
Extrachromosomal Localization of ORD during Early G2
To study the expression and localization of ORD in meiotic cells, we generated guinea-pig antiserum (GP43) against a GST fusion protein containing the C-terminal 210 amino acids of the ORD protein (Figure 1A). Immunoblot analysis of wild-type ovary and testis extracts indicates that GP43 antibodies recognize a single band that migrates at ∼60 kDa (Figure 1B), slightly larger than the predicted molecular mass of ORD (55 kDa) calculated from its primary sequence. This band is absent in gonadal extracts prepared from ord5/Df flies (Figure 1B). The ord5 nonsense allele (Bickel et al., 1996) encodes a truncated ORD protein that is missing the C-terminal fragment used as the immunogen (Figure 1A). Therefore, lack of signal in ord5/Df extracts indicates that the GP43 antiserum is ORD specific.
Within the Drosophila testis, germ-line stem cell division gives rise to a spermatagonial cell that subsequently undergoes four mitotic divisions in which cytokinesis is incomplete, thereby producing a cyst of 16 interconnected primary spermatocytes (see Figure 1C). After synchronous DNA replication, the primary spermatocytes within each cyst enter an extensive G2 growth phase (80–90 h; Lindsley and Tokuyasu, 1980; Fuller, 1993) that precedes chromosome condensation and the meiotic divisions. Based on a number of morphological criteria, Cenci et al. (1994) have divided G2 progression into seven intervals (S1, S2a, S2b, S3, S4, S5, and S6, see Figure 1C) that can be distinguished cytologically. Unlike meiotic progression in Drosophila oocytes, synaptonemal complex formation does not occur during male meiosis and homologous chromosomes do not undergo recombination. Instead, spermatocytes use an alternative mechanism to ensure pairing of homologs during meiosis I (McKee, 1996).
Figure 1D shows the subcellular distribution of ORD protein in stage S2b primary spermatocytes when fixed squashes are immunostained with GP43 ORD antiserum. Because premeiotic S phase occurs soon after completion of the fourth spermatogonial mitotic division (Cenci et al., 1994), most small 16-cell cysts have entered G2. Double staining with nuclear lamin antibodies indicates that the most intense ORD signal is present within the nucleus (Figure 1D). ORD is enriched near the nuclear periphery, with the majority of ORD signal lying interior to nuclear lamin staining. In addition, ORD is concentrated within projections that extend into the nuclear interior (Figure 1D). Although some ORD appears to colocalize with chromatin in young G2 spermatocytes, a majority of the ORD signal within the nucleus corresponds to regions adjacent to but not overlapping with the DAPI signal. We refer to such areas as extrachromosomal domains. Concentration of ORD within extrachromosomal domains was also observed when testes were detergent extracted during squash preparation or sample fixation (our unpublished data).
Little or no immunostaining is detectable in ord5/Df spermatocytes (Figure 1E), confirming that the signal observed in wild type is specific. In addition, ORD staining is absent in the two somatically derived cyst cells that surround each cyst of germ cells (Figure 1D, arrow). Absence of ORD signal in cyst cells indicates that ORD expression is germ cell specific within the testis.
Because ORD is essential for meiotic sister-chromatid cohesion, we were surprised that it did not extensively colocalize with the spermatocyte chromosomes, but instead was found predominantly within extrachromosomal spaces of the nucleus. However, epifluorescence analysis of GFP localization in spermatocytes expressing a P{gfp::ord} transgene confirmed our immunofluorescence observations (Figure 1F). In P{gfp::ord} flies, expression of the transgene is controlled by wild-type ord regulatory sequences. The transgene complements strong ord mutations (our unpublished data) and indicates that GFP-ORD protein is functional. Moreover, by immunoblot analysis with GP43 antiserum, the relative level of GFP-ORD in transgenic testes extracts is similar to that of endogenous ORD in wild-type testes extracts (our unpublished data).
We examined live squash preparations of ord mutant spermatocytes in which the P{gfp::ord} transgene provided the only source of ORD protein. In young G2 spermatocytes, GFP-ORD localizes predominantly to the nucleus (Figure 1F) where it exhibits an extrachromosomal distribution pattern similar to that observed with GP43 ORD immunostaining. However, GFP fluorescence is significantly weaker than the signal we observe using GP43 antibodies, most likely because the ORD signal is amplified during the indirect immunodetection procedure. We also visualized GFP-ORD localization in fixed squashes using anti-GFP antibodies and again observed nuclear staining that was largely extrachromosomal (Figure 1G). Immunolocalization of GFP-ORD protein in P{gfp::ord} spermatocytes using both GP43 ORD and GFP antibodies resulted in nearly identical staining patterns (Figure 1I), suggesting that both antibodies detect the same population of ORD molecules during early G2. No GFP signal was detected in live (not shown) or fixed preparations (Figure 1H) from flies lacking the P{gfp::ord} transgene. In addition, GFP-ORD was not visible in cyst cells (our unpublished data). Thus, using three different methods, we observe that ORD resides primarily in the spaces surrounding meiotic chromosomes during early G2. We also detected the same localization pattern in spermatogonial mitotic cysts (our unpublished data). Therefore, although ORD is primarily nuclear when germ cells undergo mitotic divisions, as well as during premeiotic S phase and early G2, ORD protein is not extensively associated with chromatin during these stages.
Colocalization of ORD and EAST
Enrichment of ORD in the spaces between meiotic chromosomes was reminiscent of the localization pattern reported for EAST, a Drosophila protein implicated in the assembly of an expandable nuclear endoskeleton (Wasser and Chia, 2000). Overexpression of EAST in the polyploid nuclei of larval salivary glands and in diploid male germline cells results in the expansion of an EAST-containing extrachromosomal domain, accompanied by changes in the spacing of chromosomes (Wasser and Chia, 2000). Therefore, we used confocal microscopy to determine whether the subnuclear distribution of ORD in primary spermatocytes coincided with that of the endogenous EAST protein.
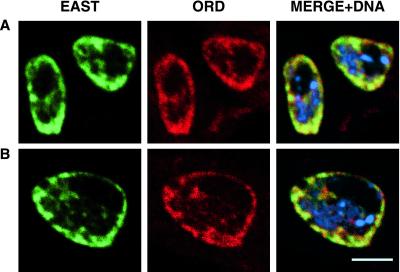
During stages S1-S3, ORD and EAST exhibit extensive nuclear colocalization, especially near the nuclear periphery (Figure 2, A and B). The staining patterns of the two proteins also coincide within projections that extend into the nuclear interior. Although ORD and EAST display remarkably similar localization patterns, there are sites at which only one of the two proteins is detected. Moreover, ORD localization appears normal in spermatocytes lacking EAST protein and EAST staining is unaffected by ord mutations that eliminate ORD activity (our unpublished data). Therefore, ORD and EAST do not depend upon one another for correct subnuclear targeting in primary spermatocytes. However, colocalization of EAST and ORD confirms our assessment that ORD is concentrated within extrachromosomal domains of the spermatocyte nucleus during early G2 and raises the possibility that localization of ORD at this stage might be supported by attachment to a nuclear endoskeleton.
Figure 2.
ORD and EAST colocalize in early G2 primary spermatocytes. Fixed testes squashes from wild-type third instar larvae were immunostained with GP43 ORD and EAST polyclonal antisera and subjected to confocal analysis. EAST (green), ORD (red), DNA (blue). (A) S2a spermatocytes. (B) A single S2b spermatocyte. Bar, 5 μm.
ORD Accumulates on the Chromosomes by Late G2
Although a large proportion of GFP-ORD protein does not colocalize with DNA during early G2 (S1-S3), GFP-ORD staining on the meiotic chromosomes becomes apparent as spermatocytes progress through G2. The GFP-ORD localization pattern appears more uniform within the nuclei of early S4 cells than during preceding stages (Figure 3A, arrow). As spermatocytes mature, GFP-ORD staining becomes concentrated on the chromosomes, which occupy distinct territories near the nuclear periphery (Figure 3, C and D). The chromatin-associated GFP-ORD signal is fairly homogeneous, suggesting that ORD protein localizes along the entire length of the chromatids. However, bright foci of staining are also visible (Figure 3, A and C) and may correspond to enrichment at centromeric regions (see below).
Our localization results suggest that accumulation of ORD protein on the chromosomes during spermatogenesis is an active process that begins during mid to late G2 (stage S4). However, the intensity of nuclear GFP-ORD staining appears to decrease as spermatocytes grow. Therefore, an alternative possibility is that ORD protein occupying extrachromosomal domains is selectively degraded, thereby revealing low levels of GFP-ORD already associated with the chromatin. To test this model, we quantified the GFP-ORD immunofluorescence signal within individual nuclei at different developmental stages (Figure 4). Using three-dimensional reconstructions of deconvolved z-series that included multiple stages of spermatocytes, we compared the total amount of GFP-ORD protein within different nuclei by summing the intensity values for voxels within a selected region of interest. As an internal control, we calculated the total DAPI signal/nucleus and confirmed that the level of DNA remained relatively constant throughout G2. We then normalized the GFP-ORD intensity values/nucleus against the average DNA signal/nucleus for each stage. Calculated values for three fields of cells are shown in Figure 4C. A subset of cells from field 1 is shown in Figure 4, A and B. Our results indicate that the total amount of nuclear GFP-ORD protein is greater in the mid to late G2 stages (S3–6) than in early G2 (S2a/b). These results are not consistent with selective degradation of an extrachromosomal pool of ORD exposing low levels of chromatin-associated ORD. Instead, our data support the conclusion that changes in the GFP-ORD staining pattern during spermatogenesis reflect the redistribution of GFP-ORD protein as it moves from extrachromosomal spaces onto the chromosomes.
Interestingly, although GP43 antiserum recognized ORD and GFP-ORD located within extrachromosomal domains of S1-S3 spermatocyte nuclei (Figure 1, D and I), GP43 nuclear signal was minimal at later stages. Figure 3E shows a P{gfp::ord} spermatocyte costained with GP43 and GFP antibodies. Although chromosomal GFP-ORD is detectable with GFP antibodies, GP43 does not appear to recognize GFP-ORD and results in a signal comparable with that observed in ord5/Df spermatocytes (our unpublished data). Furthermore, foci of GFP-ORD detected with GFP antibodies on condensed meiotic chromosomes were not observed by GP43 immunofluorescence (Figure 6K). These observations suggest that after ORD associates with chromatin, the C terminus of ORD is less accessible to GP43 antibodies. Masking of the C terminus when ORD associates with chromatin is consistent with our previous genetic analysis of mutant ord alleles, which suggested that this region of ORD mediates interactions that are required for ORD activity (Bickel et al., 1996; Bickel and Orr-Weaver, 1998). One possibility is that C-terminal interactions are required for ORD to associate with chromatin. Although ORD could be interacting directly with DNA, no obvious DNA-binding motifs are found within the ORD coding region (Bickel et al., 1996). More likely, protein-protein interactions drive the association of ORD with chromatin during mid to late G2 and reduce the ability of GP43 antibodies to bind ORD.
Figure 6.
ORD remains at the centromeres of condensed meiotic chromosomes until cohesion is released at anaphase II. Immunolocalization of GFP-ORD on fixed testes squashes from P{gfp::ord} (A-H, and K) or nontransgenic (I and J) adult flies. (A, B, C, E, G, and I) show GFP-ORD (green) and DNA (blue) during meiotic progression. Distinct GFP-ORD foci are visible on condensed chromosomes from prometaphase I until sister chromatids segregate at anaphase II (compare A, B, C, and E, with G). GFP-ORD colocalizes with MEI-S332 (red) at the centromeres (C and E). All images are either partial (A, B, C, and E) or full (G and I) projections of deconvolved z-series and are same scale. Bar, 2 μm. (D, F, H, and J) Corresponding whole cell images showing DNA (blue) and tubulin (red). Arrow in H designates the chromatin mass shown in G. (K) Nucleus of a telophase I P{gfp::ord} transgenic spermatocyte stained with both GP43 ORD (red) and GFP (green) antibodies. Four chromosomal foci are detected by GFP antibodies, but are not recognized by GP43 ORD antibodies. Bars, 5 μm.
Cohesion Deteriorates during Late G2 in ord Spermatocytes
In genetic assays, the frequency and classes of aneuploid gametes that arise in ordnull flies indicate that sister chromatids segregate randomly during both meiotic divisions (Bickel et al., 1997). Therefore, in flies lacking ORD activity, meiotic cohesion is absent before the chromosomes make stable microtubule attachments. In orcein-stained squash preparations, gross cohesion defects have been observed in ord primary spermatocytes during prometaphase I (Goldstein, 1980; Lin and Church, 1982; Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997). However, the state of cohesion in ord mutants before the chromosomes condense has not been investigated.
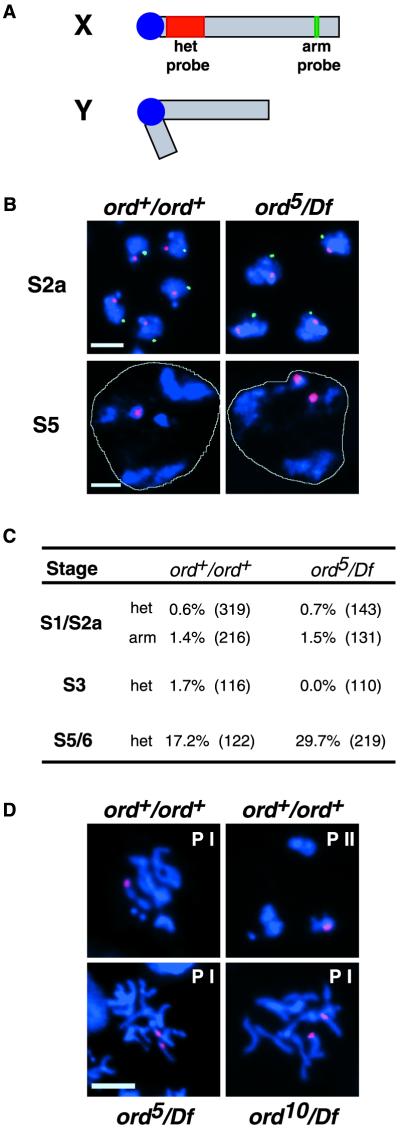
To determine when meiotic cohesion defects first become evident in ord males, we performed FISH experiments using two X chromosome probes (Figure 5A). Hybridization with a probe that recognizes the 359-base pair satellite repeat on the X chromosome (designated X het) allowed us to monitor sister cohesion within the heterochromatin near the centromere (red) and a second nonrepetitive probe (X arm; cytological location 3C1–6) provided an assay for arm cohesion (green). After DNA replication, a primary spermatocyte contains two X chromatids and thus a single arm and centromere proximal FISH signal within each nucleus would indicate that sister X chromatids are held together along their entire length. However, if arm or centromere proximal cohesion is not established or is lost prematurely, two separated hybridization signals will be visible for a single probe.
Figure 5.
Cohesion defects in ordnull spermatocytes become manifest in late G2. (A) Schematic representation of sex chromosomes indicating the X chromosome probes used for FISH analysis. Centromere-proximal probe directed against 359-base pair satellite repeat is shown in red, and the arm probe spanning region 3C1–6 (BACR34O03) is green. (B) Full projection z-series of wild-type and ord5/Df spermatocytes hybridized with het (red) and arm (green) probes. DNA is shown in blue and a white line designates the perimeter of a single nucleus in the S5 spermatocyte panels. In both genotypes, single FISH signals for each probe are apparent during the S2a stage. Two het signals in a single ord5/Df spermatocyte nucleus at stage S5 indicate that sister chromatids have prematurely separated. Arm signal is not visible in older spermatocytes. Bars, 5 μm. (C) Percentage of cells displaying separated sister FISH signals at various stages of G2 growth in wild-type and ord5/Df spermatocytes. Three to four slides were scored for each genotype, and the total number of cells scored is shown in parentheses. (D) The centromere-proximal FISH probe (red) was used to assay cohesion after DNA (blue) condensation. Prophase figures from wild-type (top) as well as ord5/Df and ord10/Df spermatocytes (bottom) are shown. Prophase I and II are designated by PI and PII, respectively. The nucleus of a single spermatocyte is shown in each panel. Bar, 5 μm.
During early G2, the hybridization of X chromosome probes to spermatocyte chromatin resulted in single arm and het signals in the nuclei of both wild-type and ord5/Df cells (Figure 5B), indicating that sister chromatids are held together at their arms and near their centromeres at this time in both genotypes. Our results are consistent with the GFP reporter analysis of live wild-type spermatocytes by Vazquez et al. (2002), which indicates that sister chromatids are associated along their entire length during early G2. Using the nonrepetitive X arm probe, we were unable to obtain a reliable FISH signal in mature, late G2 spermatocytes, most likely because of the expanded volume occupied by the chromosomes within the nucleus at these stages. However, Vazquez et al. (2002) have observed that sister chromatid arms, but not centromeres, separate shortly after the formation of chromosome territories during G2 (S3) in wild-type spermatocytes. Therefore, maintenance of arm cohesion does not appear to be required for normal meiotic segregation during male meiosis and suggests that disruption of centromeric cohesion is the primary defect in male ord mutants.
Interestingly, separated X het signals were rare in ord mutant spermatocytes until S5/6 (Figure 5B), when 30% of ord5/Df nuclei exhibited cohesion defects near the centromere (Figure 5C). Although the incidence of separated sisters in wild-type S5/6 spermatocytes was higher than expected, the number of ord5/Df cells with cohesion defects was significantly greater (2 × 2 χ2 contingency analysis, 0.01<P<0.02). Because ord+ males exhibit minimal levels of meiotic nondisjunction (<1%) in genetic tests (Bickel et al., 1997; Miyazaki and Orr-Weaver, 1992), separated X het FISH signals in mature (S5/6) wild-type spermatocytes are unlikely to reflect true defects in centromeric cohesion. Instead, we believe that our squash procedure may have resulted in artificially elevated levels of separated FISH signals (17%) in late G2 cells. Alternatively, because our probe recognizes heterochromatin that lies near the centromere but not within the centromere, the incidence of cohesion defects that we detect in wild-type spermatocytes may reflect an extension of the separation of chromatid arms observed by Vazquez et al. (2002). In either case, our results indicate that ord5/Df spermatocytes exhibit centromere proximal cohesion defects ∼13% more frequently than wild-type cells.
Our FISH data suggest that after DNA replication and during early G2, sister chromatids remain associated along their entire length in ord primary spermatocytes. However, without ORD activity, cohesion near the centromere is lost prematurely. Interestingly, defects in cohesion within heterochromatin become visible in mutant cells shortly after the time that we first observe GFP-ORD protein accumulating on the chromosomes in wild-type spermatocytes. Although cohesion defects in ordnull spermatocytes are low during late G2, FISH analysis (Figure 5D) confirms our previous genetic analysis (Bickel et al., 1997) that cohesion is absent after chromosomes undergo condensation. These data are consistent with the model that ORD must load onto the chromosomes during G2 to stabilize cohesion between sisters during the process of condensation. In the absence of functional ORD protein, sister chromatids separate prematurely as the chromosomes compact. We propose that the redistribution of ORD protein during mid to late G2 is required to maintain the association of sister chromatids during chromosome condensation in prophase I.
ORD Remains at the Centromere until Cohesion Is Lost at Anaphase II
As chromosomes begin to condense during S6, the intensity of the GFP-ORD signal decreases. However, distinct foci of GFP-ORD staining are visible on condensed chromosomes undergoing prometaphase I congression (Figure 6, A and C). We never observed more than eight signals per nucleus, suggesting that each spot might correspond to the centromeric constriction of each pair of sisters. Association of ORD with meiotic centromeres was confirmed by colocalization of GFP-ORD with MEI-S332, a centromeric cohesion protein (Figure 6, C and E). MEI-S332 loads onto centromeres during prometaphase I and is required to maintain centric cohesion from anaphase I until anaphase II (Kerrebrock et al., 1995; Moore et al., 1998; Tang et al., 1998). Although the ORD and MEI-S332 signals aligned closely, they did not overlap completely.
We do not attribute failure to detect GFP-ORD on condensed chromatid arms (Figure 6A) to limited antibody accessibility, because we observed the same pattern when monitoring GFP fluorescence in unfixed spermatocytes (our unpublished data). These observations suggest that the majority of ORD molecules dissociate from the chromosomes during condensation, whereas a subset remains at the centromeres.
During the first meiotic division, GFP-ORD persists at sister centromeres and GFP-ORD foci remain visible on telophase I chromosomes (Figure 6B). Although we can detect GFP-ORD staining on metaphase II centromeres (Figure 6E), no GFP-ORD signal is detectable on meiotic chromosomes during anaphase II or later (Figure 6G). Therefore, ORD persists at sister centromeres until centric cohesion is released.
DISCUSSION
Our FISH analysis of sister-chromatid cohesion in wild-type spermatocytes agrees well with the results of Vazquez et al. (2002). During early G2, sister chromatids are connected along their entire length. In addition, cohesion at the centromere is maintained throughout G2. Using FISH, we are unable to monitor arm cohesion during late G2, but the experiments of Vazquez et al. (2002) clearly demonstrate that arm cohesion is released in Drosophila spermatocytes by late G2. One explanation for this unexpected finding is that, unlike Drosophila females, males do not undergo meiotic recombination and, therefore, arm cohesion is not essential to maintain chiasmata and ensure correct meiosis I segregation. However, cohesion at the centromere is absolutely essential in both sexes to ensure proper segregation during both meiotic divisions.
We have shown that GFP-ORD accumulates on meiotic chromosomes shortly before cohesion defects near the centromere become detectable in ord mutant spermatocytes. By FISH analysis, moderate defects in cohesion are apparent in ordnull spermatocytes before chromosome condensation. However, genetic assays indicate that centromeric cohesion is completely lost before metaphase I in flies lacking ORD activity (Bickel et al., 1997). Our cytological analysis of cohesion defects in prophase I ordnull spermatocytes supports this conclusion. These data argue that association of ORD with spermatocyte chromosomes during G2 is required to prevent premature separation of sister centromeres before and during chromosome condensation in meiosis I. In addition, continued centromeric localization of ORD during the first meiotic division suggests that ORD is required to stabilize sister-chromatid cohesion at the centromere until anaphase II. This hypothesis is supported by genetic evidence that weak ord alleles disrupt the maintenance of meiotic cohesion between anaphase I and II (Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997).
Disappearance of ORD signal from chromatid arms during condensation is similar to that described for metazoan cohesins during mitosis (Losada et al., 1998; Sumara et al., 2000; Waizenegger et al., 2000; Warren et al., 2000). In addition, retention of ORD at meiotic centromeres until anaphase II mimics the behavior of the meiosis-specific cohesin subunit, Rec8 (Klein et al., 1999; Watanabe and Nurse, 1999; Pasierbek et al., 2000). Because no Rec8 ortholog has been identified in the Drosophila genome, one possibility is that ORD functions as a meiotic cohesin subunit. However, in sharp contrast to the behavior of cohesins, ORD does not appear to accumulate on spermatocyte chromosomes until well after S phase. Although defects in cohesion are not evident in ord spermatocytes until late G2, we cannot rule out the possibility that ORD function is required when cohesion is established or during early G2. Catenation of the sister chromatids could be masking cohesion defects at this time. However, the extensive redistribution of ORD in mature primary spermatocytes during mid to late G2 argues that ORD stabilizes meiotic cohesion by a novel mechanism not previously described. Although ORD appears to associate with chromatid arms and centromeres during late G2, our results combined with those of Vazquez et al. (2002) indicate that ORD activity does not maintain arm cohesion in mature wild-type spermatocytes. However, ORD function is essential to maintain centromeric cohesion until anaphase II. One possibility is that accumulation of ORD on chromosome arms before condensation somehow contributes to the stabilization of centromeric cohesion during prophase I.
ORD is essential for meiotic cohesion in both males and females (Mason, 1976; Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997). Our data are consistent with the model that ORD is required to maintain cohesion during the compaction of meiotic chromosomes and to prevent the release of centromeric cohesion until anaphase II. The observation that orcein-stained bivalents in ord spermatocytes appear less condensed than wild-type (Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997) supports the hypothesis that ORD also facilitates normal condensation. Spermatocyte chromosomes condense in meiosis I just before their segregation. In contrast, oocyte chromosomes compact during assembly of the synaptonemal complex (Carpenter, 1975), well before meiotic chromosome segregation occurs. Decreased levels of meiotic recombination in ord females (Mason, 1976; Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997) suggest that ORD performs an essential role during prophase compaction of meiotic chromosomes in females as well as males. In addition, ORD activity is required to maintain arm cohesion and stabilize chiasmata in Drosophila oocytes until anaphase I (Bickel et al., 2002). After metaphase I, both ORD and MEI-S332 activity are required in both sexes to prevent the release of centromeric cohesion until anaphase II. Although MEI-S332 can localize to meiotic centromeres in the absence of ORD protein, MEI-S332 is unable to maintain cohesion in ordnull flies (Bickel et al., 1998). Moreover, additional genetic experiments suggest that a balance of ORD and MEI-S332 activity is required for proper regulation of meiotic cohesion in Drosophila (Bickel et al., 1998). Together, ORD and MEI-S332 may prevent cleavage of centromeric cohesins until anaphase II.
Although ORD sequence homologs have not been identified, ORD-like activity may be essential in other organisms. Consistent with this hypothesis, recent findings indicate that in vertebrates, Drosophila, and yeast, securin proteins are unrelated at the sequence level but exhibit functional similarities that are essential for proper regulation of cohesion (Funabiki et al., 1996; Ciosk et al., 1998; Zou et al., 1999; Leismann et al., 2000). Because of the small number of chromosomes in Drosophila, viable gametes are recovered even if meiotic cohesion is completely abolished and sister segregation is randomized. Therefore, the study of cohesion in Drosophila meiosis offers an opportunity to unravel aspects of regulation that may not be accessible in other model systems, and continued molecular analysis of ORD function will provide critical information about the regulation of meiotic cohesion in metazoans.
We propose that our analysis of ORD function during Drosophila spermatogenesis has uncovered a novel aspect of how the maintenance of cohesion must be coordinated with the extensive compaction of chromosomes during prophase. We show that accumulation of ORD on meiotic chromosomes during mid to late G2 is required to maintain sister-chromatid cohesion before and during prophase condensation. In addition, our results support the model that retention of ORD at the centromeres after condensation ensures the maintenance of centromeric cohesion until anaphase II. We believe that these findings provide the first description of an activity that is required to maintain centromeric cohesion from late G2 until anaphase II during meiosis in a metazoan.
ACKNOWLEDGMENTS
We thank H. Saumweber for providing T40 nuclear lamin monoclonal antibodies, M. Wasser and W. Chia for providing EAST ED3/ED4 antisera and easthop-7 flies, and T. Tang and T. L. Orr-Weaver for providing MEI-S332 antibodies. We thank H. Webber for generating the ord10 stock containing the P{gfp::ord} insertion used for cytology and S. Randall of Improvision for advice and support with imaging. We also thank C. R. McClung, R. Sloboda, T.L. Orr-Weaver, J. Vazquez, and members of the Bickel laboratory for providing helpful comments on the manuscript. The GST-ORD construct used for antibody production and the transgenic P{gfp::ord} lines were generated in the laboratory of T.L. Orr-Weaver in work funded by the March of Dimes. E.M.B. is supported by a National Institutes of Health training grant (GM-08704). This work was funded by the National Institutes of Health (grant GM59354 to S.E.B.) and by the March of Dimes (grant 5-FY98–738 to S.E.B).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0332. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0332.
REFERENCES
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Bickel SE, Moore DP, Lai C, Orr-Weaver TL. Genetic interactions between mei-S332 and ord in the control of sister-chromatid cohesion. Genetics. 1998;150:1467–1476. doi: 10.1093/genetics/150.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel SE, Orr-Weaver TL, Balicky EM. The sister-chromatid cohesion protein ORD is required for chiasma maintenance in Drosophila oocytes. Curr Biol. 2002;12:925–929. doi: 10.1016/s0960-9822(02)00846-1. [DOI] [PubMed] [Google Scholar]
- Bickel SE, Orr-Weaver TL. Holding chromatids together to ensure they go their separate ways. BioEssays. 1996;18:293–300. doi: 10.1002/bies.950180407. [DOI] [PubMed] [Google Scholar]
- Bickel SE, Orr-Weaver TL. Regulation of sister-chromatid cohesion during Drosophila meiosis. In: Zirkin BR, editor. Germ Cell Development, Division, Disruption, and Death. New York: Springer-Verlag; 1998. pp. 37–48. [Google Scholar]
- Bickel SE, Wyman DW, Miyazaki WY, Moore DP, Orr-Weaver TL. Identification of ORD, a Drosophila protein essential for sister-chromatid cohesion. EMBO J. 1996;15:1451–1459. [PMC free article] [PubMed] [Google Scholar]
- Bickel SE, Wyman DW, Orr-Weaver TL. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 1997;146:1319–1331. doi: 10.1093/genetics/146.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Carpenter ATC. Electron microscopy of meiosis in Drosophila melanogaster females. Chromosoma. 1975;51:157–182. doi: 10.1007/BF00319833. [DOI] [PubMed] [Google Scholar]
- Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic, and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J Cell Sci. 1994;107:3521–3534. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O. The making and breaking of sister chromatid cohesion. Cell. 2001;106:137–140. doi: 10.1016/s0092-8674(01)00439-1. [DOI] [PubMed] [Google Scholar]
- Dernburg AF. In situ hybridization to somatic chromosomes. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 24–55. [DOI] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 71–147. [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis is required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gatti M, Baker B. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Goldstein LSB. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. Chromosoma. 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. MEI-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Lee JY, Orr-Weaver TL. The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol. 2001;17:753–777. doi: 10.1146/annurev.cellbio.17.1.753. [DOI] [PubMed] [Google Scholar]
- Leismann O, Herzig A, Heidmann S, Lehner CF. Degradation of Drosophila PIM regulates sister chromatid separation during mitosis. Genes Dev. 2000;14:2192–2205. doi: 10.1101/gad.176700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HP, Church K. Meiosis in Drosophila melanogaster III. The effect of orientation disruptor (ord) on gonial mitotic and the meiotic divisions in males. Genetics. 1982;102:751–770. doi: 10.1093/genetics/102.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D, Tokuyasu TL. Spermatogenesis. In: Ashburner M, Wright T, editors. The Genetics and Biology of Drosophila. New York: Academic Press; 1980. pp. 225–294. [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM. Orientation disruptor (ord): a recombination-defective and disjunction-defective meiotic mutant in Drosophila melanogaster. Genetics. 1976;84:545–572. doi: 10.1093/genetics/84.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee BD. The license to pair: identification of meiotic pairing sites in Drosophila. Chromosoma. 1996;105:135–141. doi: 10.1007/BF02509494. [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid misbehavior in Drosophila ord mutants. Genetics. 1992;132:1047–1061. doi: 10.1093/genetics/132.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP, Page AW, Tang TT-L, Kerrebrock AW, Orr-Weaver TL. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J Cell Biol. 1998;140:1003–1012. doi: 10.1083/jcb.140.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing, and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Siomos MF, Badrinath A, Pasierbek P, Livingstone D, White J, Glotzer M, Nasmyth K. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr Biol. 2001;11:1825–1835. doi: 10.1016/s0960-9822(01)00588-7. [DOI] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TT, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. Chromosome cohesion and segregation in mitosis and meiosis. Curr Opin Cell Biol. 2001;13:754–761. doi: 10.1016/s0955-0674(00)00279-9. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Heyting C. Sister chromatid cohesion and recombination in meiosis. Chromosoma. 2000;109:10–26. doi: 10.1007/s004120050408. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Belmont AS, Sedat JW. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol. 2002;12:1473–1483. doi: 10.1016/s0960-9822(02)01090-4. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Wasser M, Chia W. The EAST protein of Drosophila controls an expandable nuclear endoskeleton. Nat Cell Biol. 2000;2:268–275. doi: 10.1038/35010535. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]