Abstract
Phospholipase D (PLD) has been suggested to mediate epidermal growth factor (EGF) signaling. However, the molecular mechanism of EGF-induced PLD activation has not yet been elucidated. We investigated the importance of the phosphorylation and compartmentalization of PLD1 in EGF signaling. EGF treatment of COS-7 cells transiently expressing PLD1 stimulated PLD1 activity and induced PLD1 phosphorylation. The EGF-induced phosphorylation of threonine147 was completely blocked and the activity of PLD1 attenuated by point mutations (S2A/T147A/S561A) of PLD1 phosphorylation sites. The expression of a dominant negative PKCα mutant by adenovirus-mediated gene transfer greatly inhibited the phosphorylation and activation of PLD1 induced by EGF in PLD1-transfected COS-7 cells. EGF-induced PLD1 phosphorylation occurred primarily in the caveolin-enriched membrane (CEM) fraction, and the kinetics of PLD1 phosphorylation in the CEM were strongly correlated with PLD1 phosphorylation in the total membrane. Interestingly, EGF-induced PLD1 phosphorylation and activation and the coimmunoprecipitation of PLD1 with caveolin-1 and the EGF receptor in the CEM were significantly attenuated in the palmitoylation-deficient C240S/C241S mutant, which did not localize to the CEM. Immunocytochemical analysis revealed that wild-type PLD1 colocalized with caveolin-1 and the EGF receptor and that phosphorylated PLD1 was localized exclusively in the plasma membrane, although some PLD1 was also detected in vesicular structures. Transfection of wild-type PLD1 but not of C240S/C241S mutant increased EGF-induced raf-1 translocation to the CEM and ERK phosphorylation. This study shows, for the first time, that EGF-induced PLD1 phosphorylation and activation occur in the CEM and that the correct localization of PLD1 to the CEM via palmitoylation is critical for EGF signaling.
INTRODUCTION
Epidermal growth factor (EGF) binds to a specific cell-surface receptor (EGF receptor, EGFR), stimulating cell growth and other important physiological functions (Carpenter and Cohen, 1990; Ullrich and Schlessinger, 1990). The EGF-induced activation of the EGFR leads to phosphatidylcholine hydrolysis, which increases phosphatidic acid (PA) levels (Cook and Wakelam, 1992; Kaszkin et al., 1992; Yeo and Exton, 1995). PA seems to be a potential second messenger (English et al., 1996) and has been implicated in a variety of cellular physiological processes, such as proliferation (Fukami and Takenawa, 1992; English et al., 1996), endocytosis (Shen et al., 2001), exocytosis (Cockcroft, 1992; Way et al., 2000), and cytoskeletal rearrangement (Ha and Exton, 1993; Honda et al., 1999). Therefore, phospholipase D (PLD), which is a phosphatidylcholine-hydrolyzing enzyme and generates PA, may be an important mediator of EGF signaling.
We reported previously that platelet-derived growth factor–induced activation of PLD might be an event downstream of the primary stimulation of PLC-γ1 and PKC (Lee et al., 1994). It has since been reported that in Swiss 3T3 cells, the stimulation of PLD by EGF requires PKC activation (Yeo and Exton, 1995). In contrast, it has also been reported that EGF-induced PLD activation occurs independently of PKC in Swiss 3T3cells and in A431 cells (Cook and Wakelam, 1992). Thus, the requirement for PKC in EGF-induced PLD activation remains controversial. Two isozymes of mammalian PLD, PLD1 and PLD2, have been cloned (Hammond et al., 1995; Colley et al., 1997; Park et al., 1997; Lopez et al., 1998). PLD1 is known to be regulated by PKC in vivo. Recently, it was reported that PLD1 PIM87 mutant, which is unresponsive to PKC, could not be activated in vivo by treatment with phorbol 12-myristate 13-acetate (PMA) and carbachol (Zhang et al., 1999). After cells were treated with PMA, PLD1 was found to associate with PKCα and to be multiply phosphorylated (Lee et al., 1997; Kim et al., 1999). Recently, both PLD1 and PLD2 were found to be activated by EGF stimulation (Slaaby et al., 1998), and this study also found that the EGFR is implicated in the phosphorylation of tyrosine 11 of PLD2. However, the molecular mechanism of PLD1 activation by EGF stimulation remains unknown.
PLD1 has been identified in diverse subcellular organelles, including the endoplasmic reticulum (ER), Golgi, endosomes, lysosomes, and plasma membrane (Brown et al., 1998; Jones et al., 1999; Kim et al., 1999; Roth et al., 1999; Toda et al., 1999). ADP-ribosylation factor (ARF), which is a well-known activator of PLD1, is required for many vesicular processes between the Golgi, ER, and plasma membranes (Jones et al., 1999). Conversely, PKC-regulated PLD1 was found to be restricted to the caveolin-enriched microdomains within the plasma membrane (Kim Y et al., 2000). Thus, PLD1 may be differentially regulated at different locations by different activators. However, the intracellular location of EGF-stimulated PLD1 remains unknown.
Caveolae are plasma membrane invaginations rich in glycosphingolipids and cholesterol (Anderson, 1998). Many intracellular signaling molecules, including Gα subunits, Ha-Ras, endothelial nitric oxide synthase, and the Src family of tyrosine kinases, are localized to these microdomains (Okamoto et al., 1998). Many acylated proteins are known to be localized to caveolae; accordingly, it has been suggested that posttranslational modifications of lipids, such as myristoylation and palmitoylation, are required and play an important part in the molecular mechanism of localization to the caveolae (Okamoto et al., 1998). The growth factor–induced Ras/Raf-1 and MAP kinase pathways are two well-understood processes that occur in caveolae (Mineo et al., 1996; Liu et al., 1997). Moreover, the entire pathway from platelet-derived growth factor stimulation to MAP kinase activation is known to function in isolated caveolae (Liu et al., 1997). Although PLD1 is known to be localized to caveolae (Kim et al., 1999), it remains to be determined how PLD1 can be localized to caveolae and whether the regulatory machinery of PLD1 activation by EGF stimulation is also localized in caveolae.
In this study, we investigated these issues concerning the molecular mechanism and subcellular compartmentalization of EGF-induced PLD1 activation. For the first time, we show that PKCα-dependent phosphorylation is required for EGF-induced PLD1 activation and that the correct localization of PLD1 to the caveolae via palmitoylation is critical for EGF signaling.
MATERIALS AND METHODS
Materials
Phenylmethylsulfonylfluoride, leupeptin, and aprotinin were obtained from Roche Molecular Biochemicals; paraformaldehyde and anti-actin antibody from Sigma (St. Louis, MO); [3H]palmitic acid and [32P]orthophosphate from Dupont NEN (Boston, MA); [3H]myristic acid and the chemiluminescence kit (ECL system) from Amersham International (Buckinghamshire, U.K.); Silica Gel 60 TLC plates from Merck (Darmstadt, Germany); immobilized protein A and rhodamine-conjugated anti-mouse antibody from Pierce (Rockford, IL); DMEM and LipofectAmine from Gibco-BRL (Grand Island, NY); fetal bovine serum from PAA Laboratories, Inc. (Parker Ford, PA); and horseradish peroxidase–conjugated goat anti-rabbit IgG or anti-mouse IgG, IgM, and IgA from Kirkegaard and Perry Laboratories, Inc. (Gaithersburg, MD). The antibody against the C-terminal region of PLD1 was made and purified as described previously (Lee et al., 1997). Anti-PKCα monoclonal antibody (mAb), anti-PKCδ mAb, anti-BiP/GRP78 mAb, and anti-EGFR polyclonal antibody were purchased from Transduction Laboratories (Lexington, KY). Anti-phospho-Erk mAb was from New England Biolabs (Beverly, MA), anti-Raf-1 polyclonal antibody from Upstate Biotechnology (Lake Placid, NY), and anti-caveolin-1 polyclonal antibody from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture
COS-7 cells were cultured at 37°C in a humidified 5% CO2 atmosphere in high-glucose DMEM supplemented with 10% heat-inactivated fetal bovine serum.
In Vivo Assay of PLD
Transfections and assays of PLD were performed as previously described (Kim et al., 1999). Briefly, PLD1 was transiently overexpressed at 5 × 105 cells per 60-mm dish. Transfection was performed with LipofectAmine according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were starved for 24 h and then incubated with 5 μCi [3H]myristic acid for 3 h. The cells were then treated with EGF in the presence of 1.5% ethanol, as described in the figure legends. Lipids were extracted and separated by Silica Gel 60 TLC (chloroform:methanol:acetic acid, 90:10:10 by volume). A Fuji BAS-2000 image analyzer (Fuji Film, Tokyo, Japan) was used to determine the quantities of labeled phosphatidylethanol and total lipids.
Infection of COS-7 Cells with Dominant Negative PKC Adenoviruses
The adenovirus expression vector for dominant negative (DN)-PKCα (DN-PKCα AdV) or DN-PKCδ (DN-PKCδ AdV) has been described previously (Ohba et al., 1998; Kuroki et al. 1999). Three hours after the transfection, COS-7 cells were infected with DN-PKCα AdV of DN-PKCδ AdV for 6 h in DMEM supplemented with 10% fetal bovine serum. After the virus was removed, the cells were incubated for an additional 24 h in DMEM supplemented with 10% fetal bovine serum and reincubated for 24 h in serum-free DMEM.
Construction of Expression Plasmids
The splice-overlap extension method (Ho et al., 1989) was used to generate the C240S/C241S mutant using the following oligonucleotides: forward primer, 5′-CCGGAATTCACATGGCAAGTTAAGAG-3′/reverse primer, 5′-CCATGGCCACTGCTATTCACAC-3′ and forward primer, 5′-GTGTGAATAGCAGTGGCCATGG-3′/reverse primer, 5′-CCGGAATTCTTTGTCTACAAGAAGGACG-3′. Next, the C240S/C241S mutant was amplified using forward primer 5′-CCGGAATTCACATGGCAAGTTAAGAG-3′ and reverse primer 5′-CCGGAATTCTTTGTCTACAAGAAGGACG-3′. The EcoRI fragment of the PCR product was then exchanged with the EcoRI fragment of the wild-type (WT) PLD1 cDNA, and the whole PLD1 C240S/C240S cDNA was cloned into the pCDNA3.1 vector. The mutations were verified by sequence analysis.
Immunoprecipitation and Immunoblot Analysis of PLD1
Immunoprecipitation and immunoblot analysis were performed as described previously (Kim Y, et al., 2000). Briefly, the cells were lysed in 1 ml of lysis buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 10 mM NaCl, 1% Triton X-100, and 1% sodium cholate) containing protease inhibitors (0.5 mM PMSF, 1 μg/ml leupeptin, and 5 μg/ml aprotinin) and phosphatase inhibitors (30 mM NaF, 1 mM Na3VO4, 30 mM Na4O7P2). After centrifugation (150,000 × g for 30 min), equal amounts of soluble extract were incubated with 2 μg of anti-PLD antibody precoupled to immobilized protein A agarose resin. The immunoprecipitated proteins were then separated in 6–16% gradient SDS-polyacrylamide gels. Anti-PLD antibody, anti-EGFR mAb, anti-caveolin-1 polyclonal antibody, or the culture supernatant of a hybridoma cell line secreting anti-phospho-PLD1 was used as primary antibody.
Labeling of COS-7 Cells with [3H]palmitate
Forty-three hours after transfection, COS-7 cells were washed with serum-free DMEM and then incubated with 1 mCi/ml [9,10-3H]palmitic acid for 5 h at 37°C. Cells were then washed in ice-cold phosphate-buffered saline (PBS), harvested by scraping, and lysed in lysis buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 10 mM NaCl, 1% Triton X-100, and 1% sodium cholate) containing protease inhibitors (0.5 mM PMSF, 1 μg/ml leupeptin, and 5 μg/ml aprotinin). The cell lysates were then clarified by centrifugation and immunoprecipitated with 2 μg of anti-PLD antibody precoupled to protein A agarose. Immunoprecipitates were washed 5 times with lysis buffer, eluted with 5 × nonreducing Laemmli sample buffer by boiling for 6 min, subjected to SDS-PAGE, and then transferred to a nitrocellulose membrane. The PLD1 band was excised, and radioactivity was monitored by Cerenkov counting.
Isolation of Caveolin-enriched Membranes
Caveolin-enriched membranes (CEMs) were prepared as previously described (Kim et al., 1999), with some modification. In brief, COS-7 cells overexpressing PLD1 in a 100-mm dish were starved for 24 h and then treated with 100 nM of EGF for 0.5 min. The cells were then washed with PBS and scraped into 2 ml of 500 mM sodium carbonate, pH 11.0. The cell suspension was then homogenized with a Dounce homogenizer and a Polytron tissue grinder and lysed by sonication. The lysed homogenate was adjusted to 40% sucrose by adding 80% sucrose in MBS buffer (25 mM MES-NaOH, pH 6.5, and 150 mM NaCl) and placed in the bottom of a centrifuge tube. Four milliliters of 30% and then 4 ml of 5% sucrose in MBS buffer were layered on top. The sample was centrifuged at 100,000 × g for 6 h, and twelve 1-ml fractions were collected from the top of the tube.
Immunocytochemical Analysis
COS-7 cells were grown on coverslips and were cotransfected with pGFP-PLD1 WT or pGFP-PLD1 C240S/C241S mutant and with pDsRed1-c1-caveolin-1 (RFP-caveolin-1) or pDsRed1-C1-EGFR (RFP-EGFR). The cells were fixed in 4% (wt/vol) paraformaldehyde for 30 min at 37°C, washed with PBS, and mounted on slides. GFP- or RFP-tagged proteins were visualized by confocal laser scanning microscopy (Zeiss, Oberkochen, Germany) using appropriate filters. To detect the localization of phospho-PLD1, after stimulation with 100 nM of EGF for 0.5 min, the cells were fixed in 4% (wt/vol) paraformaldehyde for 30 min at 37°C, washed with PBS, and incubated in blocking buffer (1% goat serum in PBS containing 0.2% Triton X-100) at 4°C for 1 h. Subsequently, the cells were incubated with anti-phospho-PLD1 antibody overnight at 4°C. After three washes with PBS containing 0.05% Triton X-100, the cells were incubated with rhodamine-conjugated anti-mouse antibody for 90 min. After three additional washes with PBS containing 0.05% Triton X-100, the cells were mounted and visualized by confocal laser scanning microscopy (Zeiss).
Measurement of In Vitro PLD Activity
PLD activity was measured by choline release from phosphatidylcholine, as described previously (Kim et al., 1999)
RESULTS
EGF Induces the Activation and Phosphorylation of PLD1
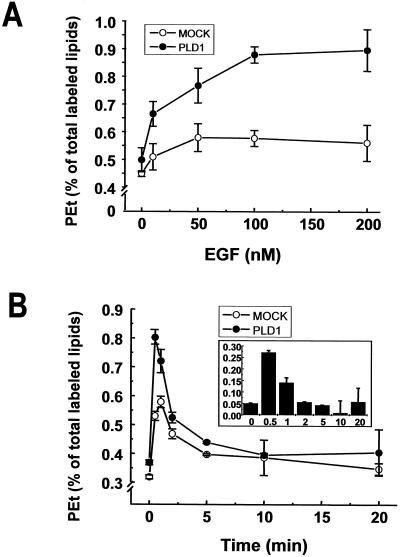
To investigate the molecular mechanism of PLD1 activation by EGF and especially the involvement of PKC, we used COS-7 cells transfected with rat PLD1. PLD activity was determined by use of the unique ability of PLD1 to produce phosphatidylethanol in the presence of ethanol. To access the dose response of EGF on PLD1, we monitored the formation of phosphatidylethanol at 1-min intervals after adding various doses of EGF. These treatments increased PLD1 activity at nanomolar EGF concentrations, and a saturated response was obtained with 100 nM of EGF (Figure 1A). To compare the kinetics of phosphorylation and activation, we monitored the formation of phosphatidylethanol at 1-min intervals by adding alcohol separately after EGF stimulation. The temporal response of PLD1 to EGF was very rapid: its activity peaked within only 30 s (with alcohol present from 0 to 1 min after the EGF stimulation), after which its activity decreased (Figure 1B). Recently, we reported that PLD1 undergoes phosphorylation at multiple sites, including serine2, threonine147, and serine561, on PMA stimulation (Kim et al., 1999). We also found that EGF induces multisite phosphorylation of PLD1, including serine2, threonine147, and serine561, and that the pattern of the PLD1 phosphopeptide map from EGF-stimulated cells was very similar to that obtained from PMA-stimulated cells (data not shown). Of the many sites phosphorylated, phosphorylation at threonine147 is one of the most easily monitored by use of the anti-phospho-threonine147 antibody (Kim Y et al., 2000). Maximal phosphorylation of threonine147 was observed at an EGF concentration of 100 nM (Figure 2A). The kinetics of the activation and the phosphorylation were very similar, because the phosphorylation also peaked within 30 s and then gradually diminished (Figure 2B). Thus, phosphorylation and activation seem to occur concurrently after EGF stimulation.
Figure 1.
Concentration and time-dependent activation of PLD1 by EGF in COS-7 cells. COS-7 cells (2.5 × 105) were transfected with 1 μg of vector (pCDNA3.1) alone or with rat PLD1. Twenty-four hours after transfection, the cells were starved for 24 h before being incubated with [ 3H]myristic acid for 3 h. (A) Cells were treated with various concentrations of EGF for 1 min in the presence of 1.5% ethanol. PLD activity was determined by measuring the formation of phosphatidylethanol (PEt) relative to total lipids. (B) To compare the kinetics of activation and phosphorylation, cells were stimulated with 100 nM of EGF, 1.5% ethanol was added at different times for 1-min intervals, and the lipids were then extracted for analysis. Time 0 min indicates incubation in the presence of 1.5% ethanol but no EGF. The other times mentioned indicate that the EGF was added at time 0 and that the ethanol was added 30 s before the time mentioned, i.e., 20 min means that the EGF was added at 0 min and that the cells were exposed to ethanol for 1 min between 19.5 min and 20.5 min. Inset, the PLD1 activity presented was calculated by subtracting the values obtained for cells transfected with vector alone (MOCK) from the values of cells overexpressing PLD1. Bars represent the range of duplicate determinations. The data shown are representative of three separate experiments.
Figure 2.
EGF-dependent phosphorylation of PLD1 in COS-7 cells. (A) COS-7 cells overexpressing PLD1 were treated with various concentrations of EGF for 1 min. PLD1 was immunoprecipitated from 1 mg of cell lysate with anti-PLD antibody and then subjected to immunoblot analysis with anti-phospho-PLD1 or anti-PLD antibody. (B) Quiescent COS-7 cells overexpressing PLD1 were stimulated with 100 nM of EGF for the indicated times, followed by immunoprecipitation and immunoblot analysis as described above. The data shown are representative of three independent experiments.
Phosphorylation of PLD1 Is Required for EGF-induced PLD1 Activation
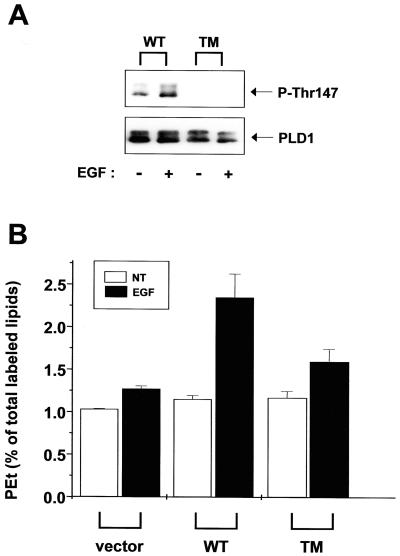
We previously identified serine2, threonine147, and serine561 as direct phosphorylation sites by PKC (21). Triple mutation (S2A/T147A/S561A) of these sites greatly attenuated PMA-induced PLD1 activity compared with WT PLD1. Therefore, we also examined the effect of triple mutation on EGF-induced PLD1 activity. As shown in Figure 3A, we confirmed that triple mutation completely blocked basal and EGF-induced threonine147 phosphorylation. The EGF-induced activity of triple-mutant PLD1 was significantly reduced compared with WT PLD1 (Figure 3B). This suggests that PKC-mediated phosphorylation is a major activation mechanism for EGF-induced PLD1 activity.
Figure 3.
EGF-induced PLD1 activity is reduced in cells carrying a triple mutation of phosphorylation sites. (A) COS-7 cells were transfected with WT or triple-mutant (TM) PLD1. Twenty-four hours after transfection, cells were starved for 24 h and then treated with 100 nM EGF for 0.5 min. PLD1 was immunoprecipitated from 2 mg of cell lysates. The proteins were separated by 8% SDS-PAGE and subjected to immunoblot analysis with anti-PLD or anti-phospho-PLD1 antibody. These data are representative of three independent experiments. (B) [3H]myristic acid–loaded COS-7 cells were treated with 100 nM EGF and 1.5% ethanol for 0.5 min. The PLD activity was determined by measuring the formation of phosphatidylethanol (PEt) relative to total lipids. The data are representative of three independent experiments.
Effect of Dominant Negative PKCα on EGF-induced PLD1 Phosphorylation and Activation
To investigate which isozyme of PKC is involved in EGF-induced PLD1 phosphorylation and activation and whether PLD1 is regulated in a phosphorylation-dependent manner, dominant negative (DN) types of PKCα and PKCδ were expressed with adenovirus expression vector. Infection of COS-7 cells with DN-PKCα or DN-PKCδ adenovirus resulted in dose-dependent increases in the amounts of PKCα or PKCδ as assessed by immunoblot analysis. The amounts of PKCα and PKCδ proteins in cells infected at a multiplicity of infection (MOI) (pfu/cell) of 20 were ∼20 times that of endogenous PKCα and PKCδ proteins, respectively (Figure 4A). The endogenous level of PKCδ protein was unchanged by infection with DN-PKCα adenovirus and vice versa. Interestingly, the expression of DN-PKCα inhibited the EGF-induced PLD1 phosphorylation (Figure 4B) and the EGF-induced PLD1 activation (Figure 4C), whereas the expression of DN-PKCδ potentiated the EGF-induced PLD1 phosphorylation (Figure 4B) and the EGF-induced PLD1 activation (Figure 4C). These data suggest that PKCα is a positive regulator of EGF-induced PLD1 activity and PKCδ is a negative regulator of the EGFR/PKCα/PLD1 pathway and that PKCα-mediated phosphorylation is a major activation mechanism of EGF-induced PLD1 signaling.
Figure 4.
Effect of DN-PKCα and DN-PKCδ on EGF-induced PLD1 phosphorylation and activation. (A) COS-7 cells were infected with DN-PKCα AdV or DN-PKCα AdV for 6 h at different MOIs, as indicated. Cell lysates (20 μg) were separated by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and analyzed by Western blot using anti-PKCα, anti-PKCδ, or anti-actin antibody. (B) COS-7 cells were transfected with WT PLD1. After 1 h, cells were infected with DN-PKCα AdV or DN-PKCδ AdV for 6 h at an MOI of 20. Twenty-four hours after infection, the cells were starved for 24 h and then treated with 100 nM EGF for 0.5 min. Cell lysates were immunoprecipitated with anti-PLD antibody, and the resulting precipitates were subjected to SDS-PAGE and quantified by use of anti-PLD or anti-phospho-PLD1 antibody. Data are representative of three separate experiments. (C) COS-7 cells were transfected with vector or WT PLD1. After 1 h, cells were infected with DN-PKCα AdV or DN-PKCδ AdV (MOI = 20 pfu/cell) for 6 h. After 43 h, cells were labeled with [3H]myristic acid (5 μCi/ml) for 3 h and then treated with 100 nM EGF for 0.5 min in the presence of 1.5% ethanol (vol/vol). The data shown represent mean ± SD values from three independent experiments performed in duplicate. PEt, phosphatidylethanol.
PLD1 in CEM Becomes Phosphorylated on EGF Treatment
The PMA-induced phosphorylation of PLD1 by PKC was previously observed to occur primarily in the caveolin-enriched, low-density membrane fraction, and therefore, the compartmentalization of PLD1 regulation was suggested (Kim Y et al., 2000). To pinpoint the site of EGFR-mediated PLD1 phosphorylation, we fractionated COS-7 cells and then monitored the phosphorylation of threonine147 in PLD1. A basal level of threonine147 phosphorylation was detected in the CEM, which was found to increase after EGF treatment (Figure 5A). Moreover, the EGF-induced phosphorylation of PLD1 in the CEM exhibited the same time course as that observed in whole cells, that is, the level of phosphorylation peaked within 30 s and then decreased gradually to the basal level (Figure 5B). We also observed the translocation of PKCα to the CEM in response to EGF treatment. PKCα was weakly detectable in the CEM before EGF treatment, but the amount of PKCα in the CEM clearly increased within 30 s of EGF treatment and then disappeared, suggesting that the temporal nature of PLD1 phosphorylation and PKCα translocation to the CEM are very similar. Interestingly, we also found the translocation of PKCδ to the CEM in response to EGF treatment (Figure 5A). Because the expression of DN-PKCδ potentiated EGF-induced PLD1 phosphorylation (Figure 4B) and EGF-induced PLD1 activation (Figure 4C), these results suggest that the translocation of PKCδ to the CEM induced by EGF treatment may contribute to the negative regulation of EGF-induced PLD1 activation.
Figure 5.
EGF-induced phosphorylation of PLD1 occurs in the CEM fraction. (A) After transfection with PLD1 and serum starvation, the COS-7 cells were incubated in the absence or presence of 100 nM EGF for 0.5 min. CEMs were prepared by sucrose density-gradient centrifugation. Twenty-five microliters of each fraction was subjected to 6–16% gradient SDS-PAGE followed by immunoblot analysis with anti-PLD, anti-phospho-PLD1, anti-PKCα, anti-PKCδ, and anti-caveolin-1 antibodies. (B) COS-7 cells overexpressing PLD1 were stimulated with 100 nM of EGF for the indicated times or with 100 nM of PMA as a control, and this was followed by sucrose density gradient centrifugation to prepare the CEM fraction. CEM lysates were immunoprecipitated (I.P.) with anti-PLD antibody, and the resulting precipitates were subjected to SDS-PAGE and quantified by use of anti-PLD or anti-phospho-PLD1 antibody. Equal volumes of fraction 5 were separated by 6–16% gradient SDS-PAGE, followed by immunoblot analysis with anti-PKCα or anti-caveolin-1 antibody. The data shown are representative of three separate experiments.
Palmitoylation Is Critical for the Localization of PLD1 to the CEM
Lipid modifications of signaling molecules often facilitate their correct targeting to caveolae (Okamoto et al., 1998). Palmitoylation of PLD1 was recently observed, and the site of the fatty acylation was determined as cysteine240 and cysteine241 (Sugars et al., 1999). Consistent with this report, we found a lack of palmitoylation in the C240S/C241S mutant PLD1 (Figure 6A). To confirm whether palmitoylation of PLD1 is essential for the localization of PLD1 to the CEM, we examined the localization pattern of PLD1 in the C240S/C241S mutant. WT PLD1 was found to be located in the CEM and in the non-CEM fraction, but strikingly, most of the PLD1, which was expected to be CEM-located, remained in the non-CEM fraction in the mutant and was not cofractionated with the EGFR and caveolin-1 (Figure 6B). It has been reported previously that the EGFR is located in caveolae (Okamoto et al., 1998), and we also found that the EGFR is localized primarily in the CEM (Figure 6B). To exclude the possibility that the amount of caveolin-1 and EGFR in the CEM is changed by the transfection of the WT or the C240S/C241S mutant of PLD1, we examined the amounts of caveolin-1 and EGFR in the CEM fraction. However, no differences in the amounts of caveolin-1 or EGFR were found in the CEM of the WT- or C240S/C241S mutant-transfected cells (Figures 6B and 10). To determine whether palmitoylation of PLD1 is required for PLD1 localization to the CEM, GFP-PLD1 WT and GFP-PLD1 C240S/C241S were transiently coexpressed with RFP-caveolin-1 and RFP-EGFR in COS-7 cells. As shown in Figure 6C, GFP-PLD1 WT was generally present in the plasma membrane and on punctate structures in the cytoplasm. Similarly, caveolin-1 was localized in the plasma membrane and on punctate structures through the cytoplasm, indicating that PLD1 WT colocalized with caveolin-1. However, the GFP-C240S/C241S mutant showed a different distribution, because it was localized primarily to the plasma membrane and displayed a scattered distribution through the cytoplasm (Figure 6C). In addition, this mutant did not seem to colocalize with caveolin-1. Similarly, GFP-PLD1 WT colocalized with EGFR in the plasma membrane and on punctate structures inside cells, but the GFP-PLD1 C240S/C241S mutant did not seem to colocalize with EGFR (Figure 6D). These results suggest that palmitoylation of PLD1 is essentially required for PLD1 localization to the CEM and for colocalization with caveolin-1 and the EGFR. We also analyzed the distribution of PLD1 WT and C240S/C241S mutant by subcellular fractionation (Figure 6E). Although the GFP-C240S/C241S mutant displayed a scattered distribution through the cytoplasm, under conditions in which the majority of PLD1 WT was recovered with the membrane fraction, C240S/C241S mutant was also recovered with the membrane fraction, suggesting that C240S/C241S mutant was still localized to the membrane. From these results, we concluded that palmitoylation of PLD1 contributes to its localization to the CEM, but it cannot account completely for the localization on membranes in general.
Figure 6.
Palmitoylation is required for CEM localization of PLD1 and its colocalization with caveolin-1 and the EGFR. (A) COS-7 cells were transfected with control vector, WT PLD1, or C240S/C241S mutant. After 24 h, the cells were starved and loaded with [3H]palmitate (1 mCi/ml) for 5 h. Cell lysates (2 mg) were immunoprecipitated with anti-PLD antibody, and the resulting precipitates were subjected to 8% SDS-PAGE and then transferred to a nitrocellulose membrane. The PLD1 band was excised and its radioactivity monitored by Cerenkov counting. (B) CEMs were prepared from quiescent COS-7 cells expressing WT PLD1 or C240S/C241S mutant by sucrose density-gradient centrifugation. Twenty-five microliters of each fraction was subjected to 6–16% gradient SDS-PAGE followed by immunoblot analysis with anti-PLD, anti-EGFR, anti-caveolin-1, or anti-Bip/GRP78 antibody. The localizations and the amounts of the EGFRs and caveolin-1 in WT-PLD1–transfected cells and in mutant-transfected cells were identical. The blots shown are representative of three separate results. (C) COS-7 cells were cotransfected with GFP-PLD1 WT or GFP-PLD1 C240S/C241S and with RFP-caveolin-1. The cells were fixed, and GFP-PLD1 and RFP-caveolin-1 were examined under a confocal microscope. (D) COS-7 cells were cotransfected with GFP-PLD1 WT or GFP-PLD1 C240S/C241S and with RFP-EGFR. The cells were fixed, and GFP-PLD1 and RFP-EGFR were also examined by confocal microscopy. (E) COS-7 cells were transfected with vector, WT PLD1, or C240S/C241S mutant. After 48 h of expression, the cells were harvested, fractionated by hypotonic buffer, and ultracentrifuged to separate the cytosol and crude membrane fractions. The supernatant was designated cytosol (C) and kept, whereas the pellet containing the membrane fraction (M) was resuspended in buffer of volume equal to the supernatant. Equal volumes of the cytosol and membrane fractions were analyzed by immunoblot analysis with anti-PLD antibody.
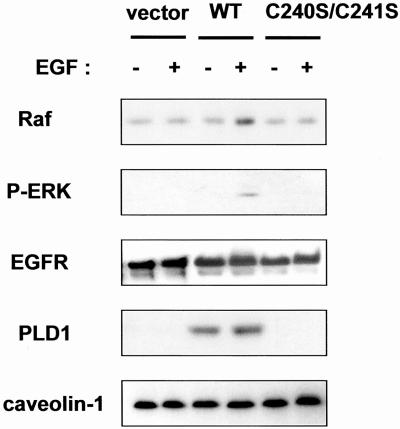
Figure 10.
The localization of PLD1 to the CEM is required for EGF-induced Raf-1 translocation to the CEM and ERK phosphorylation. COS-7 cells transfected with control vector, WT, or C240S/C241S mutant of PLD1 were starved and then stimulated with 100 nM of EGF for 0.5 min. CEM fractions were prepared, and equal amounts of proteins (5 μg) were separated by gel electrophoresis before immunoblotting with anti-Raf-1, anti-phospho-ERK, anti-EGFR, anti-PLD, and anti-caveolin-1 antibody. The blots shown are representative of three separate results.
Complex Formation between PLD1 and Caveolin-1 and the EGFR within the CEM
As is shown by Figure 7, the EGFR and caveolin-1 were coimmunoprecipitated with PLD1 WT but not with PLD1 C240S/C241S in the CEM fraction, which suggests complex formation between PLD1, caveolin-1, and the EGFR within the CEM. The amount of each molecule in the immune complexes was not substantially changed by EGF treatment. The observation that PLD1 is coimmunoprecipitated with caveolin-1 and the EGFR can also be explained by the fact that these proteins are colocalized to membrane domains that are insoluble in nonionic detergent rather than by the formation of a complex between these proteins. To exclude this possibility, we used Nonidet P-40, deoxycholate, and β-octylglucopyranoside to extract the CEM proteins. Before these detergents were used, caveolin-1, PLD1, and the EGFR were detected in the 150,000 × g pellet of the CEM fraction, but after these detergents were used, these proteins were detected in the 150,000 × g supernatant of the CEM fraction (data not shown). This result suggests that palmitoylation of PLD1 is essentially required for its localization to the CEM, in which PLD1 forms a molecular complex with the EGFR and caveolin-1.
Figure 7.
Complex formation of PLD1 with caveolin-1 and with the EGFR in the CEM. COS-7 cells transfected with WT or C240S/C241S mutant of PLD1 were incubated in the presence or absence of 100 nM of EGF for 0.5 min. The CEM fraction was prepared, and the proteins were solubilized in an extraction buffer (10 mM Tris-HCl, pH 7.4, 10 mM EDTA, 1% Nonidet P-40, 0.4% deoxycholate, 60 mM β-octylglucopyranoside, 0.5 mM phenylmethylsulfonylfluoride, 1 μg/ml leupeptin, and 5 μg/ml aprotinin). The sample was centrifuged at 150,000 × g for 1 h, and the solubilized supernatant was incubated with anti-PLD antibody precoupled to protein A agarose resin. The immune complexes were subjected to 6–16% gradient SDS-PAGE followed by immunoblot analysis with anti-PLD, anti-EGFR, or anti-caveolin-1 antibody. The blots shown are representative of three separate results. I.P., immunoprecipitate.
Palmitoylation of PLD1 Is Critical for Its Phosphorylation and Activation by EGF
The palmitoylation-deficient mutant proved to be a useful tool in the study of the importance of the localization of PLD1 to the CEM. EGF-induced phosphorylation of C240S/C241S was remarkably reduced compared with the WT (Figure 8A), and EGF-induced PLD1 activity was also attenuated in this mutant (Figure 8B). To eliminate the possibility that the observed decreased phosphorylation and activation were somehow caused by a conformational change induced by the point mutation itself, we examined the in vitro phosphorylation status and the ARF-dependent or PKC-dependent activity of the palmitoylation-deficient PLD1 mutant versus WT PLD1. As shown in Figure 8, C and D, the C240S/C24 1S mutant was phosphorylated in vitro by PKCα to the same extent as WT PLD1, and no differences were detected in the ARF-dependent or PKCα-dependent activity of this mutant versus the WT. These results suggest that the localization of PLD1 to the CEM is critical for the phosphorylation and activation of PLD1 by PKCα.
Figure 8.
Palmitoylation of PLD1 is required for its phosphorylation and activation by EGF. (A) COS-7 cells transfected with WT or C240S/C241S mutant PLD1 were incubated in the absence or presence of 100 nM of EGF for 0.5 min. PLD1 was immunoprecipitated, and the immune complex was subjected to immunoblot analysis with anti-phospho-PLD1 or anti-PLD antibody. (B) COS-7 cells (2.5 × 105) were transfected with vector alone, WT PLD1, or C240S/C241S mutant PLD1. The cells were stimulated with 100 nM of EGF in the presence of 1.5% ethanol for 0.5 min, and the lipids were extracted to determine PLD activity. The numbers indicate the amount of phosphatidylethanol (PEt) formed after EGF stimulation. Data represent mean ± SD values of three independent experiments performed in duplicate. (C) Lysates from WT or mutant PLD1-transfected COS-7 cells were immunoprecipitated (I.P.) with anti-PLD1 antibody, and the immune complex was phosphorylated in vitro with PKCα in the presence of PMA and ATP at 37°C for 15 min and then immunoblotted with anti-phospho-PLD1 antibody. (D) In vitro PLD activity was measured with mixed lipid vesicles. Cell homogenates (0.5 μg) were assayed for PLD activity in the presence or absence of 1 μM ARF/10 μMGTPγS or 10 nM PKCα/100 nM PMA. The data shown are representative of three separate experiments.
EGF-Induced Phosphorylation of PLD1 Occurs Primarily in the Plasma Membrane of COS-7 Cells
Caveolin-1 is the defining protein component of caveolae within the plasma membrane, but significant amounts of caveolin-1 are known to exist in intracellular compartments, such as the ER and the trans-Golgi network (Smart et al., 1999). Liquid-ordered domains like caveolae can also form within the Golgi apparatus and may play a role in the biosynthetic trafficking of cholesterol from the ER to the plasma membrane (Smart et al., 1999). Therefore, the CEM fraction in Figures 5 and 6 may also contain membranes derived from cytoplasmic organelles. To visualize the localization of PLD1 phosphorylation in intact cells, we stained phosphorylated PLD1 with phospho-PLD1 mAb after EGF simulation. The GFP-PLD1 WT overexpressed in COS-7 cells was generally present in the plasma membrane and in cytoplasmic vesicular structures, and after treatment with EGF, phosphorylated PLD1 was found primarily in the plasma membrane (Figure 9). Conversely, in C240S/C241S mutant-transfected COS7 cells, phosphorylated PLD1 was not detected in the plasma membrane. These results therefore indicate that PLD1 phosphorylation may occur in the caveolae-like domains of the plasma membrane and not in those of the cytoplasmic endomembranes and furthermore, that palmitoylation of PLD1 contributes to EGF-dependent phosphorylation of PLD1 in the plasma membrane.
Figure 9.
EGF-induced phosphorylation of PLD1 occurs specifically in the plasma membrane. COS-7 cells transfected with GFP-WT PLD1 were starved and then stimulated with 100 nM of EGF for 0.5 min. The cells were fixed and stained with anti-phospho-PLD1 antibody, and rhodamine-conjugated anti-mouse antibody was used as a secondary antibody. GFP-PLD1 and phosphorylated PLD1 (Phospho-PLD1) were examined under a confocal microscope. The data shown are representative of four separate experiments.
The Localization of PLD1 to the CEM Is Required for EGF-induced Raf-1 Translocation and ERK Phosphorylation in the CEM
We next addressed the question of whether the palmitoylation and localization of PLD1 to the CEM are necessary for the Raf-ERK pathway. COS-7 cells were transfected with control vector, WT, or C240S/C241S mutant PLD1 and stimulated with EGF for 0.5 min, and each CEM fraction in the vector, WT-transfected, and C240S/C241S mutant-transfected cells was obtained. Interestingly, the amount of Raf-1 in the CEM was rapidly increased by EGF treatment in the WT-transfected cells but not in the vector or C240S/C241S mutant-transfected cells (Figure 10). In addition, ERK phosphorylation in the CEM was also elevated by EGF treatment only in the WT-transfected cells. These results demonstrate that the palmitoylation-dependent localization of PLD1 to the CEM contributes to the localized formation of PA and that this has a critical function in the activation of the MAPK cascade through Raf-1 translocation to the CEM.
DISCUSSION
PLD1 has been found to be expressed at various subcellular locations, but the functions of and the regulatory mechanisms involving PLD1 at each cellular location are not well understood. Previously, we reported that PLD1 cofractionated with caveolin-1, a marker for CEM (Kim et al., 1999). However, the knowledge of the functional importance of the correct CEM localization of PLD1 with respect to PLD1 regulation and its signaling pathway are limited. In this study, for the first time, we present experimental evidence that proves that the correct localization of PLD1 to the CEM via palmitoylation is critical for PLD1 regulation and EGF signaling.
The PKC-dependent regulatory mechanism of PLD has been a central issue in PLD regulation studies (Exton, 1999). However, the nature of the receptor-mediated stimulations associated with the phosphorylation of PLD1 and the issue of whether phosphorylation is essential for the PLD1 activity induced by receptor stimulation remained to be determined. Previously, we reported that PKC-mediated phosphorylation is required for PMA-induced PLD1 activation (Kim et al., 1999). We also observed that PKCα-mediated PLD1 phosphorylation is required for EGF-induced activation, because DN-PKCα, which encodes a kinase-defective mutant of PKCα (K368R), inhibited EGF-induced PLD1 phosphorylation and activation (Figure 3, B and C). Surprisingly, DN-PKCδ, which encodes a kinase-defective mutant of PKCδ (K376R), enhanced EGF-induced PLD1 phosphorylation and activation (Figure 3B and 3C). Previously, PKC isoforms α and δ were reported to have antagonistic effects on PLD activity (Hornia et al., 1999). Thus, the translocation of PKCδ to the CEM induced by EGF treatment may contribute to the negative regulation of EGF-induced PLD1 activation. Although the molecular mechanism of the involvement of PKCδ on the regulation of EGF-induced PLD1 activation is still unknown, in this report, we show that PKCα and PKCδ, which are activated via EGFR activation, antagonistically regulate PLD1 by phosphorylation-dependent mechanisms.
The activation of PLD1 by EGF stimulation seems to be composed of several intricate mechanisms. PLD1 can be regulated by various activators, such as PKC, ARF, and Rho family proteins (Exton, 1999). Although we have suggested the involvement of phosphorylation in the EGF-induced activation of PLD1 within the CEM, the precise role of the phosphorylation in the activation process remains unknown. The phosphorylation itself is not likely to increase the catalytic activity directly, because the catalytic activity of PLD1, activated by PKCα in vitro, did not require ATP (Hammond et al., 1997). Previously, Rac1 or Ras/Ral signaling was reported to be required for the activation of PLD after EGF stimulation (Hess et al., 1997; Voss et al., 1999), and Rac1 and Ral A are known to stimulate PLD1 by direct interaction (Hammond et al., 1997; Kim et al., 1998). Therefore, the possibility exists that PKCα phosphorylation facilitates the binding of PLD1 stimulators, such as the low-molecular-weight G proteins, to PLD1.
Compartmentalization of PLD1 in the CEM has been reported recently, but the important domain or motif required for correct localization is not known. Lipid modification is required or, at least, greatly facilitates the targeting of a protein to the caveolae (Smart et al., 1999). The C-terminal domain of caveolin-1 also undergoes palmitoylation at three residues (Uittenbogaard and Smart, 2000). PLD1 has been reported to be palmitoylated at cysteine240 and cysteine241, which reside in the PH domain (Sugars et al., 1999). In the present study, WT PLD1 is enriched in the CEM fraction (Figure 5), colocalized with caveolin-1 (Figure 6C), and coimmunoprecipitated with caveolin-1 (Figure 7). Previously, we also reported the direct interaction of PLD1 with caveolin-1 (Kim et al., 1999). Caveolin-1 is the structural component of caveolae within the plasma membrane, but significant amounts of caveolin-1 are known to exist in intracellular compartments, such as the ER and the trans-Golgi network (Smart et al., 1999) (Figure 6C). In Figure 9, PKCα-phosphorylated PLD1 was found primarily in the plasma membrane, suggesting that WT PLD1 phosphorylation may occur in the caveolae-like domains of the plasma membrane and not in those of the cytoplasmic endomembranes. Conversely, in C240S/C241S mutant-transfected cells, phosphorylated PLD1 was not detected in the plasma membrane. The palmitoylation-deficient mutant of PLD1 (C240S/C241S) was excluded from the CEM fraction (Figure 6B), and the C240S/C241S mutant could not be coimmunoprecipitated with caveolin-1 in the CEM, although WT was coimmunoprecipitated with caveolin-1. Therefore, palmitoylation of PLD1 contributes to EGF-dependent phosphorylation of PLD1 in the plasma membrane and may be a targeting signal to direct PLD1 to a specific plasma membrane domain, the caveolae.
The PLD1 was found to be regulated by its interaction with phosphatidylinositol 4,5-bisphosphate (PIP)2-containing membranes (Hammond et al., 1995). Therefore, there are two possibilities for the localization signal in the PLD1. One involves a hydrophobic interaction between the palmitate of PLD1 and the lipid components of CEM, and the other an interaction between the PLD1 and membrane PIP2. In the present study, interestingly, the double mutant (C240S/C241S) of the palmitoylation sites on PLD1 mislocalized from the CEM to the non-CEM fraction, although the binding property of C240S/C241S mutant to PIP2 seemed to be unchanged versus the WT protein (Figure 8D). However, a substantial portion of C240S/C241S mutant was recovered in the membrane fraction (Sugars et al., 1999) (Figure 6E). Thus, these results raise the possibility that the key factor for CEM localization of PLD1 is palmitoylation modification and that the general factor of membrane localization is not palmitoylation modification but other actions, such as interactions with membrane lipid or proteins.
Because the palmitoylation/depalmitoylation process is readily reversed by enzymatic machinery within the cells (Camp and Hofmann, 1993), palmitoylation may represent a means by which the function of PLD1 can be regulated. Our results demonstrate the presence of PLD1 in two different cellular compartments, i.e., the CEM and the non-CEM. WT PLD1 in the non-CEM seemed to be a nonpalmitoylated form and was not regulated by PKCα. The molecular mechanisms of PLD1 palmitoylation/depalmitoylation, caused by a putative palmitoyl transferase and a unknown cysteine palmitoyl thioesterase, respectively, are unknown. Thus, studies on the palmitoylation/depalmitoylation processes in cells will contribute to the understanding of the regulatory mechanism of PLD1.
Different results have been reported regarding localization of EGFR in cells. On the basis of detergent-free membrane fractionation experiments, as much as 40–60% of the total pool of EGFR at the plasma membrane has been reported to localize to caveolae in nonstimulated cells not overexpressing the EGFR (Mineo et al., 1999). In A431 cells, which overexpress the EGFR, similar detergent-free fractionation experiments have shown that the majority of the EGFR is localized within caveolin-enriched low-buoyant-density membrane domains (Couet et al., 1997; Pike and Miller, 1998; Waugh et al., 1999). Recently, Ringerike et al. (2002), using immuno-electron microscopy, reported that 7% of the total number of EGFRs at the plasma membrane are within caveolae and 40% of EGFRs localizing to the plasma membrane are within anti-PLAP patched rafts in nonstimulated A431 cells. Therefore, it seems that the EGFR is within both caveolae and raft. Couet et al. (1997) reported the coimmunoprecipitation of EGFR using anti-caveolin IgG and the direct interaction of caveolin-1 with EGFR. Park et al. (2000) also reported that EGFR was coimmunoprecipitated with caveolin-1. In this study, we showed that PLD1 is cofractionated with caveolin-1 and EGFR (Figure 6B), coimmunoprecipitated with caveolin-1 and EGFR (Figure 7), and colocalized with caveolin-1 and EGFR (Figure 6, C and D). However, the palmitoylation-deficient mutant remained in the non-CEM and could not form a molecular complex with the EGFR and caveolin-1. Consequently, it was not receptive to EGFR signaling and could not be phosphorylated or activated by PKCα. This finding demonstrates that the CEM localization of PLD1 is required for complex formation with the EGFR and caveolin-1, as well as for the regulation of PLD1 by PKCα.
Whereas the natures of the events occurring at the caveolae within the plasma membrane remain unresolved, it is clear that the activation of Raf-1 by growth factors requires the translocation of Raf-1 from the cytoplasm to the plasma membrane, where it is activated by interacting with GTP-bound Ras (Mineo et al., 1997; Roy et al., 1997), by PKC phosphorylation (Kolch et al., 1993; Carrol and May, 1994) and by tyrosine kinases (Fabian et al., 1993; Marais et al., 1995). It has also been reported that Raf-1 interacts with phosphatidylserine and PA in vitro (Ghosh et al., 1996), and it has been shown that mutations that disrupt Raf-PA interactions prevent the recruitment of Raf-1 to membranes and insulin-dependent MAPK phosphorylation (Rizzo et al., 2000). We also found that the overexpression of PLD1 WT but not of C240S/C241S mutant concurrently induced transient EGF-dependent Raf-1 translocation to the CEM and ERK phosphorylation. These results demonstrate that transient translocation of Raf to the CEM and ERK phosphorylation by EGF are dependent on the localized regulation of PLD1 within the CEM.
In conclusion, in this study, we demonstrate the molecular mechanism and the importance of the CEM localization of PLD1 in the EGF signaling pathway. A focus on the components of caveolae will be a useful strategy in the search to identify the molecular networks of PLD1 in signaling pathways.
Abbreviations used:
- CEM
caveolin-enriched membrane
- EGF
epidermal growth factor
- GFP
green fluorescent protein
- PA
phosphatidic acid
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PKC
protein kinase C
- PLC-γ1
phospholipase C-γ1
- PLD
phospholipase D
- PMA
phorbol 12-myristate 13-acetate
- RFP
red fluorescent protein
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–02–0100. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–02–0100.
This work was supported in part by the POSTECH Research Fund, the National Research Laboratory of the Ministry of Science and Technology, and the Center for Cell Signaling Research in the Republic of Korea.
REFERENCES
- Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- Brown FD, Thompson N, Saqib KM, Clark JM, Powner D, Thompson NT, Solari R, Wakelam MJ. Phospholipase D1 localizes to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Curr Biol. 1998;8:835–838. doi: 10.1016/s0960-9822(98)70326-4. [DOI] [PubMed] [Google Scholar]
- Camp LA, Hofmann SL. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-ras. J Biol Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- Carpenter G, Cohen S. Epidermal growth factor. J Biol Chem. 1990;265:7709–7712. [PubMed] [Google Scholar]
- Carrol MP, May WS. Protein kinase C-mediated serine phosphorylation directly activates Raf-1 in murine hematopoietic cells. J Biol Chem. 1994;269:1249–1256. [PubMed] [Google Scholar]
- Cockcroft S. G-protein-regulated phospholipases C, D and A2-mediated signaling in neutrophils. Biochim Biophys Acta. 1992;1113:135–160. [PubMed] [Google Scholar]
- Colley WC, Sung T-C, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Cook SJ, Wakelam MJ. Epidermal growth factor increases sn-1,2-diacylglycerol levels and activates phospholipase D-catalyzed phosphatidylcholine breakdown in Swiss 3T3 cells in the absence of inositol-lipid hydrolysis. Biochem J. 1992;285:247–253. doi: 10.1042/bj2850247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couet J, Sargiacomo M, Lisanti MP. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins: caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J Biol Chem. 1997;272:30429–30438. doi: 10.1074/jbc.272.48.30429. [DOI] [PubMed] [Google Scholar]
- English D, Cui Y, Siddiqui RA. Messenger functions of phosphatidic acid. Chem Phys Lipids. 1996;80:117–132. doi: 10.1016/0009-3084(96)02549-2. [DOI] [PubMed] [Google Scholar]
- Exton JH. Regulation of phospholipase D. Biochim Biophys Acta. 1999;1439:121–133. doi: 10.1016/s1388-1981(99)00089-x. [DOI] [PubMed] [Google Scholar]
- Fabian JR, Daar IO, Morrison DK. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami K, Takenawa T. Phosphatidic acid that accumulates in platelet-derived growth factor-stimulated Balb/c 3T3 cells is a potential mitogenic signal. J Biol Chem. 1992;267:10988–10993. [PubMed] [Google Scholar]
- Ghosh S, Strum JC, Sciorra VA, Daniel L, Bell RM. Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid: phosphatidic acid regulates the translocation of Raf-1 in 12-O-tetradecanoyl phorbol-13-acetate-stimulated Madin-Darby canine kidney cells. J Biol Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- Ha KS, Exton JH. Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts. J Cell Biol. 1993;123:1789–1796. doi: 10.1083/jcb.123.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, Nozawa Y, Prestwich GD, Frohman MA, Morris AJ. Characterization of two alternately spliced forms of phospholipase D1: activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J Biol Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- Hess JA, Ross AH, Qiu RG, Symons M, Exton JH. Role of Rho family proteins in phospholipase D activation by growth factors. J Biol Chem. 1997;272:1615–1620. doi: 10.1074/jbc.272.3.1615. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Honda A, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Hornia A, Lu Z, Sukezane T, Zhong M, Joseph T, Frankel P, Foster DA. Antagonistic effects of protein kinase C α and δ on both transformation and phospholipase D activity mediated by the epidermal growth factor receptor. Mol Cell Biol. 1999;19:7672–7680. doi: 10.1128/mcb.19.11.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Morgan C, Cockcroft S. Phospholipase D and membrane traffic: potential roles in regulated exocytosis, membrane delivery and vesicle budding. Biochim Biophys Acta. 1999;1439:229–244. doi: 10.1016/s1388-1981(99)00097-9. [DOI] [PubMed] [Google Scholar]
- Kaszkin M, Seidler L, Kast R, Kinzel V. Epidermal growth factor-induced production of phosphatidylalcohols by HeLa cells and A431 cells through activation of phospholipase D. Biochem J. 1992;287:51–57. doi: 10.1042/bj2870051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Han JM, Lee SM, Kim Y, Lee TG, Park JB, Lee SD, Suh P-G, Ryu SH. Phospholipase D1 in caveolae: regulation by protein kinase Calpha and caveolin-1. Biochemistry. 1999;38:3763–3769. doi: 10.1021/bi982478+. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SD, Han JM, Lee TG, Kim Y, Park JB, Lambeth JD, Suh PG, Ryu SH. Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA. FEBS Lett. 1998;430:231–235. doi: 10.1016/s0014-5793(98)00661-9. [DOI] [PubMed] [Google Scholar]
- Kim Y, Han JM, Han BR, Lee KA, Kim JH, Lee BD, Jang IH, Suh PG, Ryu SH. Phospholipase D1 is phosphorylated and activated by protein kinase C in caveolin-enriched microdomains within the plasma membrane. J Biol Chem. 2000;275:13621–13627. doi: 10.1074/jbc.275.18.13621. [DOI] [PubMed] [Google Scholar]
- Kim Y, et al. Phosphorylation and activation of phospholipase D1 by protein kinase C in vivo: determination of multiple phosphorylation sites. Biochemistry. 1999a;38:10344–10351. doi: 10.1021/bi990579h. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim JE, Lee SD, Lee TG, Kim JH, Park JB, Han JM, Jang SK, Suh PG, Ryu SH. Phospholipase D1 is located and activated by protein kinase C alpha in the plasma membrane in 3Y1 fibroblast cell. Biochim Biophys Acta. 1999b;1436:319–330. doi: 10.1016/s0005-2760(98)00120-9. [DOI] [PubMed] [Google Scholar]
- Kolch W, Heidecker G, Kochs G, Hummel R, Vahidi H, Mischak H, Finkenzeller G, Marme D, Rapp UR. Protein kinase C alpha activates RAF-1 by direct phosphorylation. Nature. 1993;364:249–252. doi: 10.1038/364249a0. [DOI] [PubMed] [Google Scholar]
- Kuroki T, Kashiwagi M, Ishino K, Huh N, H, Ohba M. Adenovirus-mediated gene transfer to keratinocytes. J Invest Dermatol Symp Proc. 1999;4:153–157. doi: 10.1038/sj.jidsp.5640200. [DOI] [PubMed] [Google Scholar]
- Lee TG, et al. Phorbol myristate acetate-dependent association of protein kinase C alpha with phospholipase D1 in intact cells. Biochim Biophys Acta. 1997;1347:199–204. doi: 10.1016/s0005-2760(97)00083-0. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kim HS, Pai J-K, Ryu SH, Suh P-G. Activation of phospholipase D induced by platelet-derived growth factor is dependent upon the level of phospholipase C-gamma 1. J Biol Chem. 1994;269:26842–26847. [PubMed] [Google Scholar]
- Liu P, Ying Y, Anderson RG. Platelet-derived growth factor activates mitogen-activated protein kinase in isolated caveolae. Proc Natl Acad Sci USA. 1997;94:13666–13670. doi: 10.1073/pnas.94.25.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez I, Arnold RS, Lambeth JD. Cloning and initial characterization of a human phospholipase D2 (hPLD2): ADP-ribosylation factor regulates hPLD2. J Biol Chem. 1998;273:12846–12852. doi: 10.1074/jbc.273.21.12846. [DOI] [PubMed] [Google Scholar]
- Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo C, Anderson RGW, White MA. Physical association with Ras enhances activation of membrane-bound Raf (RafCAAX) J Biol Chem. 1997;272:10345–10348. doi: 10.1074/jbc.272.16.10345. [DOI] [PubMed] [Google Scholar]
- Mineo C, Gill GN, Anderson RGW. Regulated migration of epidermal growth factor receptor from caveolae. J Biol Chem. 1999;274:30636–30643. doi: 10.1074/jbc.274.43.30636. [DOI] [PubMed] [Google Scholar]
- Mineo C, James GL, Smart EJ, Anderson RGW. Localization of epidermal growth factor-stimulated Ras/Raf-1 interaction to caveolae membrane. J Biol Chem. 1996;271:11930–11935. doi: 10.1074/jbc.271.20.11930. [DOI] [PubMed] [Google Scholar]
- Mineo C, Ying Y-S, Chapline C, Jaken S, Anderson RGW. Targeting of protein kinase Cα to caveolae. J Cell Biol. 1998;141:601–610. doi: 10.1083/jcb.141.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M, Ishino K, Kashiwagi M, Kawabe S, Chida K, Huh NH, Kuroki T. Induction of differentiation in normal human keratinocytes by adenovirus-mediated introduction of the eta and delta isoforms of protein kinase C. Mol Cell Biol. 1998;18:5199–5207. doi: 10.1128/mcb.18.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- Park S-K, Provost JJ, Bae CD, Ho W-T, Exton JH. Cloning and characterization of phospholipase D from rat brain. J Biol Chem. 1997;272:29263–29271. doi: 10.1074/jbc.272.46.29263. [DOI] [PubMed] [Google Scholar]
- Park WY, Park JS, Cho KA, Kim DI, Ko YG, Seo JS, Park SC. Up-regulation of caveolin attenuates epidermal growth factor signaling in senescent cells. J Biol Chem. 2000;275:20847–20852. doi: 10.1074/jbc.M908162199. [DOI] [PubMed] [Google Scholar]
- Pike LJ, Miller JM. Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J Biol Chem. 1998;273:22298–22304. doi: 10.1074/jbc.273.35.22298. [DOI] [PubMed] [Google Scholar]
- Ringerike T, Blystad FD, Levy FO, Madshus IH, Stang E. Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J Cell Sci. 2002;115:1331–1340. doi: 10.1242/jcs.115.6.1331. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Watkins SC, Romeo G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid, and is independent of association with Ras. J Biol Chem. 2000;275:23911–23918. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- Roth MG, Bi K, Ktistakis NT, Yu S. Phospholipase D as an effector for ADP-ribosylation factor in the regulation of vesicular traffic. Chem Phys Lipids. 1999;98:141–152. doi: 10.1016/s0009-3084(99)00026-2. [DOI] [PubMed] [Google Scholar]
- Roy S, Lane A, Yan J, McPherson R, Hancock JF. Activity of plasma membrane-recruited Raf-1 is regulated by Ras via the Raf zinc finger. J Biol Chem. 1997;272:20139–20145. doi: 10.1074/jbc.272.32.20139. [DOI] [PubMed] [Google Scholar]
- Shen Y, Xu L, Foster DA. Role for phospholipase D in receptor-mediated endocytosis. Mol Cell Biol. 2001;21:595–602. doi: 10.1128/MCB.21.2.595-602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaaby R, Jensen T, Hansen HS, Frohman MA, Seedorf K. PLD2 complexes with the EGF receptor and undergoes tyrosine phosphorylation at a single site upon agonist stimulation. J Biol Chem. 1998;273:33722–33727. doi: 10.1074/jbc.273.50.33722. [DOI] [PubMed] [Google Scholar]
- Smart EJ, Graf GA, McNiven MA, Sessa WC, Engelman JA, Scherer PE, Okamoto T, Lisanti MP. Caveolins, liquid-ordered domains, and signal transduction. Mol Cell Biol. 1999;19:7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugars JM, Cellek S, Manifava M, Coadwell J, Ktistakis NT. Fatty acylation of phospholipase D1 on cysteine residues 240 and 241 determines localization on intracellular membranes. J Biol Chem. 1999;274:30023–30027. doi: 10.1074/jbc.274.42.30023. [DOI] [PubMed] [Google Scholar]
- Toda K, Nogami M, Murakami K, Kanaho Y, Nakayama K. Colocalization of phospholipase D1 and GTP-binding-defective mutant of ADP-ribosylation factor 6 to endosomes and lysosomes. FEBS Lett. 1999;442:221–225. doi: 10.1016/s0014-5793(98)01646-9. [DOI] [PubMed] [Google Scholar]
- Uittenbogaard A, Smart EJ. Palmitoylation of caveolin-1 is required for cholesterol binding, chaperone complex formation, and rapid transport of cholesterol to caveolae. J Biol Chem. 2000;275:25595–25599. doi: 10.1074/jbc.M003401200. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Voss M, Weernink PA, Haupenthal S, Moller U, Cool RH, Bauer B, Camonis JH, Jakobs KH, Schmidt M. Phospholipase D stimulation by receptor tyrosine kinases mediated by protein kinase C and a Ras/Ral signaling cascade. J Biol Chem. 1999;274:34691–34698. doi: 10.1074/jbc.274.49.34691. [DOI] [PubMed] [Google Scholar]
- Waugh MG, Lawson D, Hsuan JJ. Epidermal growth factor receptor activation is localized within low-buoyant density, non-caveolar membrane domains. Biochem J. 1999;337:591–597. [PMC free article] [PubMed] [Google Scholar]
- Way G, O'Luanaigh N, Cockcroft S. Activation of exocytosis by cross-linking of the IgE receptor is dependent on ADP-ribosylation factor 1-regulated phospholipase D in RBL-2H3 mast cells: evidence that the mechanism of activation is via regulation of phosphatidylinositol 4,5-bisphosphate synthesis. Biochem J. 2000;346:63–70. [PMC free article] [PubMed] [Google Scholar]
- Yeo E-J, Exton JH. Stimulation of phospholipase D by epidermal growth factor requires protein kinase C activation in Swiss 3T3 cells. J Biol Chem. 1995;270:3980–3988. doi: 10.1074/jbc.270.8.3980. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Altshuller YM, Hammond SM, Hayes F, Morris AJ, Frohman MA. Loss of receptor regulation by a phospholipase D1 mutant unresponsive to protein kinase C. EMBO J. 1999;18:6339–6348. doi: 10.1093/emboj/18.22.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]