Abstract
In response to upstream signals, proteins in the Wiskott-Aldrich Syndrome protein (WASP) family regulate actin nucleation via the Arp2/3 complex. Despite intensive study of the function of WASP family proteins in nucleation, it is not yet understood how their distinct structural organization contributes to actin-based motility. Herein, we analyzed the activities of WASP and Scar1 truncation derivatives by using a bead-based motility assay. The minimal region of WASP sufficient to direct movement was the C-terminal WCA fragment, whereas the corresponding region of Scar1 was insufficient. In addition, the proline-rich regions of WASP and Scar1 and the Ena/VASP homology 1 (EVH1) domain of WASP independently enhanced motility rates. The contributions of these regions to motility could not be accounted for by their direct effects on actin nucleation with the Arp2/3 complex, suggesting that they stimulate motility by recruiting additional factors. We have identified profilin as one such factor. WASP- and Scar1-coated bead motility rates were significantly reduced by depletion of profilin and VASP and could be more efficiently rescued by a combination of VASP and wild-type profilin than by VASP and a mutant profilin that cannot bind proline-rich sequences. Moreover, motility of WASP WCA beads was not affected by the depletion or addback of VASP and profilin. Our results suggest that recruitment of factors, including profilin, by the proline-rich regions of WASP and Scar1 and the EVH1 domain of WASP stimulates cellular actin-based motility.
INTRODUCTION
For many cell types, the ability to move across a solid surface is fundamental to their biological function. Certain aspects of cell locomotion, such as the protrusion of the plasma membrane in lamellipodia and filopodia, are driven by the polymerization of actin filaments. To coordinate these behaviors, tight spatial and temporal control is exerted over several aspects of the polymerization cycle, including the nucleation of new actin filaments and the elongation of existing ones. Members of the Wiskott-Aldrich Syndrome protein (WASP) family, including WASP, N-WASP, and at least three variants of Scar/WAVE, seem to play a central role in regulating these processes. However, specifically how these different WASP family proteins contribute to various aspects of actin-based motility is not well understood.
WASP family proteins contain multiple regions (Figure 1), some of which bind to proteins involved in actin nucleation and elongation, and others to signaling molecules that regulate these activities. A conserved carboxy-terminal segment functions to stimulate the nucleation activity of the Arp2/3 complex (Higgs et al., 1999; Machesky et al., 1999; Rohatgi et al., 1999; Winter et al., 1999), an actin-nucleating factor in cells. This region, referred to as WCA, consists of a WASP-homology 2 domain (W) that binds to actin monomer (Machesky and Insall, 1998; Miki et al., 1998b), along with a basic connector sequence (C) and an acidic stretch (A) that interact with the Arp2/3 complex (Machesky and Insall, 1998; Marchand et al., 2001). Activation of the Arp2/3 complex by the WCA region leads to actin nucleation and cross-linking of newly formed filaments into branched arrays (Blanchoin et al., 2000).
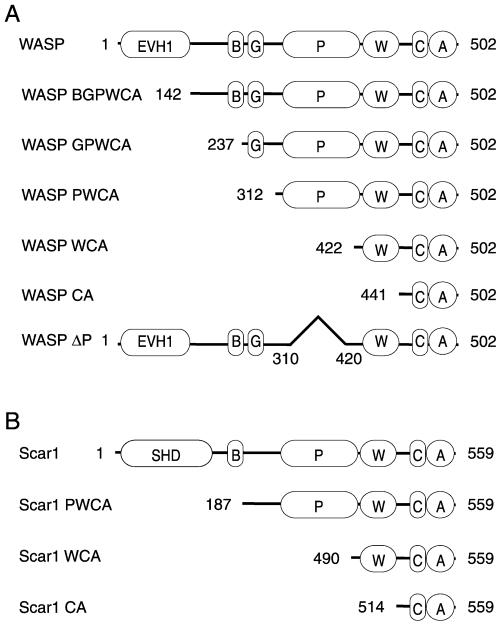
Figure 1.
N-terminal WASP and Scar1 truncation derivatives. (A) Schematic diagrams of WASP truncation and deletion derivatives. (B) Schematic diagrams of Scar1 truncation derivatives.
The activity of the WCA region of WASP family proteins is regulated by the central and amino-terminal regions. The central region consists of proline-rich sequences (P) that interact with the SH3 domains of adapter proteins (Grb2 and Nck), kinases, phospholipase Cγ, as well as WISH and IRSp53 (Bear et al., 2001; Takenawa and Miki, 2001). Binding of several of these proteins, including Grb2 (Carlier et al., 2000), Nck (Rohatgi et al., 2001), WISH (Fukuoka et al., 2001), and IRSp53 (Miki et al., 2000), enhances actin nucleation by WASP family proteins and the Arp2/3 complex. The P region of WASP family proteins also interacts with profilin (Miki et al., 1998b; Suetsugu et al., 1998) and VASP (Castellano et al., 2001; interaction only shown for WASP), actin-binding proteins that regulate actin dynamics. VASP influences actin dynamics by protecting the barbed end of filaments from capping, by inhibiting the branching activity of the Arp2/3 complex, and by contributing to nucleation with the Arp2/3 complex (Skoble et al., 2001; Bear et al., 2002). Profilin binds to actin monomer and promotes actin polymerization at the barbed ends of filaments, inhibits actin polymerization at the pointed ends, and catalyzes the exchange of ATP for ADP in actin monomers (Sun et al., 1995). Moreover, profilin potentiates actin nucleation by the Arp2/3 complex and N-WASP (Yang et al., 2000).
In contrast to the P and the WCA regions, the amino-terminal regions of WASP family proteins are more divergent. At their amino termini, WASP and N-WASP have an Ena/VASP homology 1 (EVH1) domain that binds to WASP-interacting protein (WIP) (Ramesh et al., 1997) and to phosphatidylinositol bisphosphate (PIP2) (Miki et al., 1996). They also contain a basic stretch (B) that interacts with PIP2 (Prehoda et al., 2000; Rohatgi et al., 2000) and a G protein-binding domain (G) that binds the Rho family GTPase Cdc42 (Symons et al., 1996). In their inactive, closed form, an intramolecular interaction between the G and the carboxy-terminal C region inhibits Arp2/3 complex-stimulating activity (Miki et al., 1998a; Rohatgi et al., 1999; Kim et al., 2000). Binding of Cdc42 and PIP2 disrupts this interaction and facilitates Arp2/3 complex activation (Rohatgi et al., 1999; Higgs and Pollard, 2000; Prehoda et al., 2000; Rohatgi et al., 2000). This mode of regulation is consistent with the cellular activities of WASP, which is involved in actin regulation downstream of Cdc42 (Symons et al., 1996). Unlike WASP and N-WASP, the amino terminus of Scar proteins consists of a unique Scar homology domain (SHD). The binding partners of the SHD are not yet well defined, although they may contribute to the observed regulation of Scar downstream of the Rho family GTPase Rac (Miki et al., 1998b, 2000).
Although WASP family proteins possess clear structural differences, how these variations contribute to their distinct functional and regulatory properties in cells is not understood. Moreover, despite numerous studies regarding the function of the regions of WASP family proteins in actin nucleation, the role of these regions in modulating actin-based motility has not been examined. To address these issues, we have devised a powerful in vitro motility system in which beads coated with WASP family proteins undergo actin-based motility in cytoplasmic extracts (Yarar et al., 1999). This bead-based assay recapitulates motile processes observed in cells and, due to the open nature of extracts, is amenable to experimental manipulation. Herein, we use this system together with a series of WASP and Scar1 truncation derivatives to define and differentiate the contributions of these protein domains to motility, and to test the role of interacting factors in this process.
MATERIALS AND METHODS
Generation of WASP and Scar1 Derivatives
DNA encoding WASP tagged at its N terminus with both Met Arg Gly Ser (MRGS) 6xHis and FLAG epitopes was amplified by polymerase chain reaction (PCR) from a human WASP cDNA (a generous gift of Arie Abo, PPD Discovery, Menlo Park, CA). The PCR product was subcloned into pFastBac1 (Invitrogen, Carlsbad, CA) and pGEX4T-1 (Amersham Biosciences, Piscataway, NJ) to generate the plasmids pDY10-1 and pDY7, respectively. DNAs encoding WASP truncation derivatives BGPWCA, GPWCA, PWCA, and WCA were also amplified by PCR and were subcloned into pDY10-1 to generate plasmids pDY10-2 (BPGWCA), pDY10-3 (GPWCA), pDY10-4 (PWCA), and pDY10-5 (WCA). DNA encoding WASP-CA was generated by PCR and subcloned into pDY7 to make plasmid pDY33. WASP ΔP was amplified by PCR from WASPΔP (a generous gift of Arie Abo) and subcloned into pDY10-1.
The DNA encoding Scar1 tagged at its N terminus with MRGS 6xHis and FLAG epitopes was amplified by PCR and subcloned into pDY10-1 to generate pTD9 and into pDY7 to generate pTD1. The Scar1 truncation derivative PWCA was generated by PCR and subcloned into pTD9 to make pTD10. The Scar1 WCA and CA derivatives were generated by PCR and subcloned into pTD1 to make pTD3 and pTD4.
Expression and Purification of Recombinant Proteins
Recombinant WASP and its truncation derivatives BGPWCA, GPWCA, PWCA, WCA, and WASP ΔP, and recombinant Scar1, its derivative PWCA, and VASP were expressed in Sf9 cells by using the baculovirus expression system. Baculovirus strains were generated and used for infections according to the Bac-to-Bac baculovirus expression system (Invitrogen). After 60 h of infection, cells were harvested by centrifugation at 500 × g for 10 min at 25°C, resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM KCl) with protease inhibitors and frozen in liquid N2. To prepare the lysate, cells were thawed and centrifuged at 200,000 × g.
To purify the recombinant proteins, Sf9 lysates were supplemented with 20 mM imidazole, incubated wtih Ni2+-NTA-agarose (QIAGEN, Valencia, CA) resin for 1 h at 4°C, washed with wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM KCl, 20 mM imidazole), and eluted with elution buffer (200 mM imidazole, 50 mM NaH2PO4, pH 8.0, 300 mM KCl, and protease inhibitors). Eluted proteins were further purified by gel filtration chromatography on a Superdex-200 column (Amersham Biosciences) equilibrated with control buffer [20 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.0, 100 mM KCl, 2 mM MgCl2, 5 mM EGTA, 1 mM EDTA, 0.5 mM dithiothreitol (DTT), 10% [vol/vol] glycerol].
Recombinant GST-WASP WCA, WASP-CA, SCAR-WCA, and SCAR-CA proteins were expressed as glutathione S-transferase (GST) fusions in Escherichia coli BL21-CodonPlus-RP cells (Stratagene, La Jolla, CA). Cells were grown to an OD600 of 0.5 and induced with 0.4 mM isopropyl β-d-thiogalactoside at 37°C for 3 h. Proteins were bound to glutathione-Sepharose (Amersham Biosciences), washed with phosphate-buffered saline, and eluted with 10 mM glutathione. Eluted proteins were further purified by gel filtration chromatography as described above. The GST tag was cleaved by incubation with thrombin (∼ 0.5 U/mg protein) for 5 min at 25°C, and the cleavage reaction was stopped by incubation with benzamidine Sepharose (Sigma-Aldrich, St. Louis, MO) for 20 min at 4°C. WASP and Scar1 derivatives were isolated from GST by Ni2+-NTA-agarose (QIAGEN) affinity chromatography as described above and transferred into control buffer by using Nap5 spin columns (Amersham Biosciences). All protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad, Hercules, CA) with bovine serum albumin as a standard.
Recombinant human GST-WT profilin and GST-H133S profilin (plasmids kindly provided by Changsong Yang and Sally Zigmond, University of Pennsylvania, Philadelphia, PA) were expressed in E. coli strain BL21 (DE), cleaved with thrombin, and purified as described previously (Yang et al., 2000). Recombinant GST-Cdc42 V12 (plasmid kindly provided by Arie Abo) was expressed in E. coli strain BL21 (DE). Protein was purified by glutathione affinity and gel filtration chromatography as described above and charged with guanosine-5′-O-(3-thio)triphosphate (GTPγS) as described previously (Ma et al., 1998). VASP was prepared as described previously (Skoble et al., 2001).
Pyrene-Actin Polymerization Assays
Human platelet Arp2/3 complex (Welch and Mitchison, 1998), rabbit skeletal muscle actin (Spudich and Watt, 1971), and pyrene-labeled actin (Kouyama and Mihashi, 1981) were prepared as described previously. Pyrene-actin polymerization assays were performed as described previously (Cooper et al., 1983) with the following modifications. Pyrene-actin and unlabeled actin were thawed and transferred into fresh G-buffer (2 mM Tris, pH 7.4, 0.2 mM CaCl2, 0.2 mM ATP, 0.2 mM DTT) by passing them over a 1-ml Sepharose G25 (Amersham Biosciences) spin column and were then mixed to generate 3 μM monomer solution with 10% pyrene actin. Arp2/3 complex (10 nM) or an equal volume of control buffer was mixed with initiation buffer (20 mM MgCl2, 10 mM EGTA, 5 mM ATP) and 20 nM WASP/Scar1 derivatives in control buffer. This solution and the G actin solution (final concentration 2 μM) were incubated at room temperature for 1 min and then mixed to initiate actin polymerization.
To measure the effects of GST Cdc42-V12 and PIP2 [PI(4,5)P2] on the activities of the WASP truncation derivatives, 10 nM Arp2/3 complex or control buffer was mixed with initiation buffer and 30 nM of each WASP derivative. To test the effect of PIP2 alone, the premix was added to 2 mM PIP2 (Calbiochem, San Diego, CA) and Cdc42 buffer (14 mM MOPS, pH 7.0, 68 mM KCl, 1.4 mM MgCl2, 22.4 mM EGTA, 0.14 mM ATP, 0.34 mM DTT, 7% [vol/vol] glycerol, 1 mM GTPγS, 20.7 mM MgCl2). To test the effect of Cdc42 alone, 10 μl of 2 μM GST Cdc42-V12 and 1 μl of control buffer were added to the premix. To test the effect of both PIP2 and Cdc42, 30 μM PIP2 and 300 nM GST Cdc42-V12 in Cdc42 buffer were added to the premix. The premix ± Cdc42-V12, ± PIP2 and 2 μM G actin were incubated at room temperature for 1 min and then mixed to initiate actin polymerization. Assembly kinetics was monitored using a Fluorolog 3 fluorometer (Instruments S.A.; excitation wavelength 365 nm, emission wavelength 407 nm) maintained at a temperature of 25°C.
Bead Motility Assays
For coating beads with protein, 1 μl of carboxylated polystyrene beads (0.5 μm in diameter, 2.68% solids; Polysciences, Warrington, PA) was incubated with various concentrations of purified proteins in 20 μl of final volume control buffer (20 mM MOPS, pH 7.0, 100 mM KCl, 2 mM MgCl2, 5 mM EGTA, 1 mM EDTA, 0.5 mM DTT, 10% [vol/vol] glycerol) for 1 h at room temperature, pelleted, and washed with XB (100 mM KCl, 0.1 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 10 mM K-HEPES, pH 7.7), and then resuspended in XB. The final amount of protein on the beads was determined by Western blotting with an anti-FLAG antibody by using known amounts of the FLAG-tagged proteins as a standard. For motility assays, beads were coated with a final concentration of 10–20 pmol of protein bound to 1 μl of bead slurry. For motility assays, beads were added to X. laevis egg extract (Theriot et al., 1994) supplemented with N-hydroxysuccinimidyl 5-carboxytetramethyl rhodamine-labeled actin (Kellogg et al., 1988) and 20× ATP-regenerating mix (150 mM creatine phosphate, 20 mM ATP, 2 mM EGTA, pH 7.7, 20 mM MgCl2). The extract was squashed between a slide and a coverslip and viewed after 5-min incubation at room temperature for WASP/Scar1 derivative-coated beads or 1-h incubation at room temperature for ActA-coated beads. To determine rates of movement, differential interference contrast or fluorescence images were recorded every 10 s, and rates of movement were determined by averaging the distance moved in a 60-s time interval by using MetaMorph software (Universal Imaging, Downington, PA).
Statistical analysis was performed using MINITAB software. Motility rates measured for the derivative-coated beads were compared using an analysis of variance test followed by a Tukey's multiple comparison test (set at 5%).
Profilin Depletion/Addition
Control resin (glycine-Sepharose) or poly-l-proline (Sigma-Aldrich)-Sepharose resin was blocked for at least 2 h in 30 mg/ml bovine serum albumin and washed 3× with XB (100 mM KCl, 0.1 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 10 mM K-HEPES, pH 7.7). Resin was then added to X. laevis egg extract and incubated for 30 min at 4°C. The degree of depletion was determined by immunoblotting with an anti-Xenopus profilin antibody (see below) and comparing to known dilutions of control extract. Addback was done using 2 μM recombinant human profilin (WT or H133S) and/or 0.2 μM recombinant human VASP or an equivalent volume of buffer (20 mM MOPS, pH 7.0, 100 mM KCl, 2 mM MgCl2, 5 mM EGTA, 1 mM EDTA, 0.5 mM DTT, 10% [vol/vol] glycerol). After addition of profilin/VASP/buffer, the extract was incubated on ice for 20 min, and motility assays were performed as described above, except that slides were incubated for 20 min at room temperature before viewing. For poly-l-proline (PLP) addition experiments, soluble 10 μM PLP dissolved in XB, or XB alone was added to extract and incubated for 20 min. The extract was incubated on ice for 20 min and motility assays were performed as described above.
To determine rates of movement, differential interference contrast or fluorescence images were recorded every 5 s and rates of movement were determined by averaging the distance moved in 20-s time intervals by using MetaMorph software. For PLP depletion experiments, statistical analysis was performed as described above. For PLP addition, statistical significance was examined using a Student's t test.
WIP Pulldown
Carboxylated polystyrene beads (3 μl, 0.5 μm in diameter, 2.68% solids; Polysciences) were coated with 20 pmol of WASP or WASP GPWCA, and washed with control buffer (20 mM MOPS, pH 7.0, 100 mM KCl, 2 mM MgCl2, 5 mM EGTA, 1 mM EDTA, 0.5 mM DTT, 10% [vol/vol] glycerol). Coated beads were incubated with platelet extract, washed with control buffer, and proteins bound were determined by immunoblotting by using an anti-WIP antibody (kindly provided by Ines Anton and Narayanaswamy Ramesh, Harvard Medical School, Boston, MA).
Antibody Preparation
Profilin was purified from Xenopus egg extract by poly-l-proline affinity chromatography by using the procedures described in Janmey (1991). Purified profilin was used to immunize rabbits, and anti-Xenopus profilin antibodies were purified from serum as described previously (Harlow and Lane, 1999).
RESULTS
Regions of WASP Family Proteins That Function in Motility
To study the contribution of the regions of WASP family proteins to actin-based motility, we expressed and purified full-length WASP and Scar1, as well as a series of truncation and deletion derivatives of both proteins (Figure 1). We named each sequential truncation derivative for the regions remaining in the molecule. WASP derivatives included WASP-BGPWCA, WASP-GPWCA, WASP-PWCA, WASP-WCA, and WASP-CA, and Scar1 derivatives included Scar1-PWCA, Scar1-WCA, and Scar1-CA. We also generated a WASP deletion derivative missing the proline-rich region, which we called WASPΔP. The ability of each protein to direct motility was assessed using a bead-based assay. We showed previously that 1-μm-diameter beads coated with full-length WASP were motile in bovine brain extract and formed actin comet tails similar to those induced by the bacterial pathogen Listeria monocytogenes in infected cells (Yarar et al., 1999). In this study, we used X. laevis egg extracts, which have been shown to support the locomotion of L. monocytogenes (Theriot et al., 1994) and of beads coated with ActA, the L. monocytogenes surface protein that functions like WASP (Cameron et al., 1999). In addition, we used smaller diameter beads (0.5 μm), which were shown to exhibit a higher frequency of movement in Xenopus egg extract when coated with ActA (Cameron et al., 1999).
Initially, we compared the relative capacities of WASP, Scar1, and ActA to promote motility. Beads were coated with comparable concentrations of each protein (supplemental material, Figure S1), introduced into Xenopus egg extract, and monitored for their abilities to promote actin polymerization and movement (Figure 2). Motility rates were measured and the distributions of rates (Figure 3) were compared to determine whether they were statistically distinct at the 95% confidence level by using an analysis of variance test followed by a Tukey's multiple comparison test. Beads coated with full-length WASP moved at a rate of 8.3 ± 0.3 μm/min (mean ± 2 times SE; 95% confidence interval) in egg extracts, 42-fold faster than the rate for 1-μm beads in bovine brain extract (0.2 μm/min) (Yarar et al., 1999). This rate was significantly faster than for ActA- and Scar1-coated beads (6.5 ± 1.0 and 5.8 ± 0.4 μm/min, respectively), which were statistically indistinguishable from each other. Although both WASP beads and Scar1 beads initiated movement immediately after introduction into the extract, ActA-coated beads required a 20- to 30-min incubation before actin clouds were formed at the bead surfaces and 1 h before tail formation and motility were observed. This may be due to differences in the availability or function of factors that each protein recruits to stimulate actin polymerization.
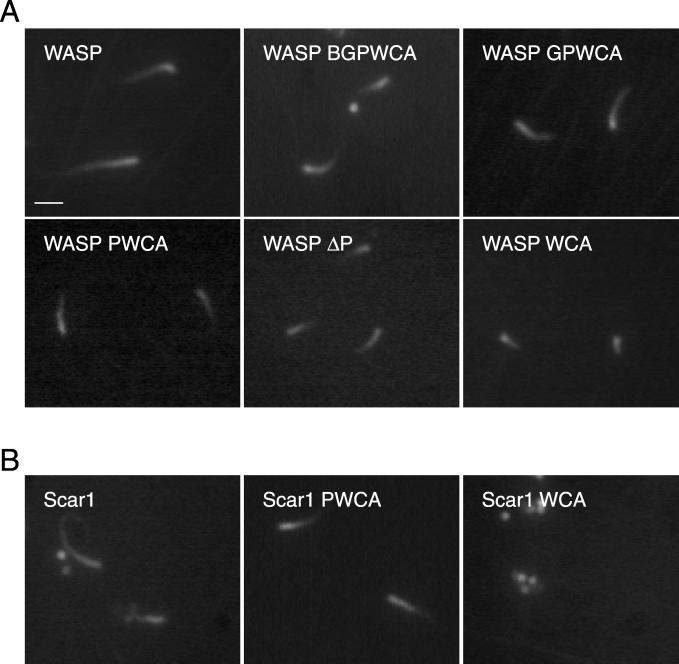
Figure 2.
Behavior of WASP/Scar1 derivative-coated beads in X. laevis egg extract, visualized by fluorescence microscopy. (A) Actin structures formed by WASP derivative-coated beads. (B) Actin structures formed by Scar1 derivative-coated beads. Images were taken after samples were incubated on the slide for 5 min. Bar, 2 μm.
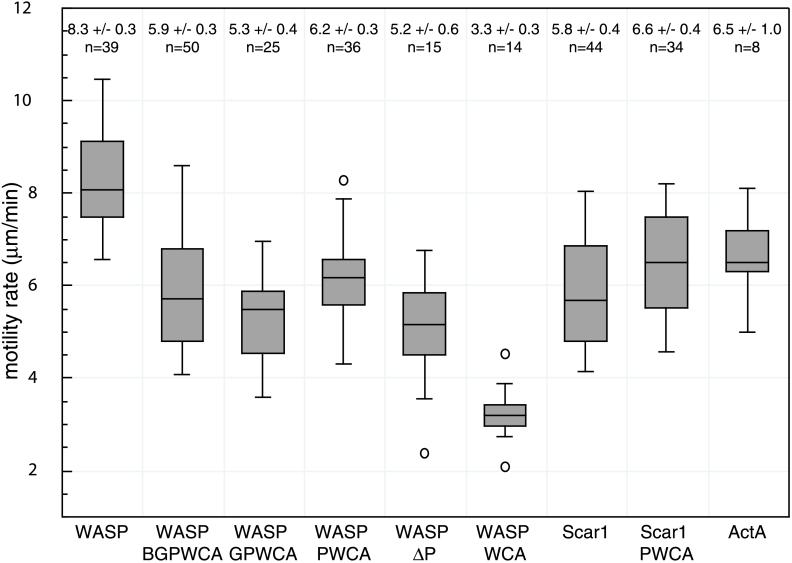
Figure 3.
Box plot distribution of motility rates of WASP/Scar1/ActA-coated beads in X. laevis egg extract. The average rates (± 2× SE; 95% confidence interval) and number of beads in each data set are presented above each distribution. The top line of each box represents the 3rd quartile, the middle line the median, and the bottom line the 1st quartile. The top and bottom whiskers indicate the maximum and minimum rates measured and the circles indicate outliers.
Next, to evaluate the functions of the different regions of WASP in actin-based motility, each WASP derivative was tested using the bead assay. Beads coated with WASP-BGPWCA, WASP-GPWCA, WASP-PWCA, WASP-WCA, and WASPΔP were able to polymerize actin, form tails, and move immediately after introduction into the extract (Figure 2A). In contrast, WASP-CA–coated beads did not form tails, but were able to assemble a faint cloud of actin when coated with 20-fold more protein. Thus, WASP-WCA is the minimum region that is sufficient to direct motility in Xenopus egg extracts. The motility rates of these derivative-coated beads were also measured and compared with full-length WASP (Figure 3). In general, motility rates were proportional to the length of actin tails formed behind the moving beads, as was observed previously for moving L. monocytogenes in infected cells (Theriot et al., 1992). WASP-WCA, the minimal region required for motility, formed short tails and promoted slower movement (3.3 ± 0.3 μm/min) than any of the other derivatives tested. Derivatives containing the proline-rich region, including WASP-PWCA (6.2 ± 0.3 μm/min), WASP-GPWCA (5.3 ± 0.4 μm/min), and WASP-BGPWCA (5.9 ± 0.3 μm/min), formed longer tails and promoted faster movement. Similarly, WASPΔP (5.2 ± 0.6 μm/min), which lacks the P region but retains the WASP N terminus (EVH1-B-G), promoted faster motility than the WASP-WCA–coated beads. These four derivatives exhibited rate distributions that were statistically indistinguishable from one another and from ActA- and Scar1-coated beads. All of the WASP truncation derivatives promoted movement at significantly slower rates than did full-length WASP, which contains both the P region and the EVH1 domain. Taken together, these results indicate that the P region and the EVH1 domain of WASP make separate stimulatory contributions to motility.
In a similar analysis of Scar1 derivatives, only Scar1-PWCA was capable of directing actin polymerization, tail formation, and movement (Figure 2B). Beads coated with the smaller Scar1-WCA fragment polymerized actin but were not motile, and Scar CA-coated beads were only able to polymerize residual amounts of actin when coated with 20-fold excess protein. Thus, unlike WASP, the minimum fragment of Scar1 that was sufficient to direct motility was Scar1-PWCA. The rate of motility of Scar1-PWCA beads (6.6 ± 0.4 μm/min) was indistinguishable from the larger WASP derivative-coated beads but was statistically faster than beads coated with full-length Scar1 or WASP-WCA (Figure 3). These results indicate that the central P region of Scar1 plays an essential stimulatory role in motility, whereas the N-terminal SHD exhibits a slight inhibitory effect.
The observed differences in motility rates for WASP and Scar1 derivatives were not caused by small differences in the amount of protein coating the beads, as beads coated with amounts of each derivative varying over a ten-fold range moved at indistinguishable rates (Table 1). This is consistent with results found for beads coated with varying concentrations of ActA (Cameron et al., 1999). All derivatives were able to direct motility when coated onto the beads at a concentration of 1–2 pmol of protein per microliter of bead slurry, with the exception of WASP-WCA beads, which only generated tails when coated with 10–20 pmol of protein per microliter bead slurry (Table 1). Similarly, beads coated with GST-WASP WCA also required 10–20 pmol of protein bound per microliter of bead slurry to initiate motility, and moved at rates that were indistinguishable from WASP-WCA beads. These results support the conclusion that WASP-WCA is less potent than the larger derivatives, and thus is required at higher surface concentrations to initiate motility.
Table 1.
Dependence of bead motility on protein concentration
| 0.5 pmol | 1–2 pmol | 5 pmol | 10–20 pmol | |
|---|---|---|---|---|
| WASP | − | 7.5 ± 1.3 | 8.2 ± 1.0 | 7.8 ± 1.3 |
| WASP-BGPWCA | − | ± | 6.6 ± 0.9 | 6.4 ± 1.7 |
| WASP-GPWCA | − | ± | + | + |
| WASP-PWCA | − | 6.0 ± 0.9 | + | 6.4 ± 0.9 |
| WASP-WCA | − | − | − | 3.7 ± 0.7 |
| Scar1 | − | 6.4 ± 2.5 | 7.0 ± 1.4 | n.d. |
| Scar1-PWCA | − | 6.7 ± 1.0 | 6.2 ± 1.3 | 7.2 ± 0.9 |
Table showing the rates of motility (μm/min) at the indicated concentration of protein bound to 1 μl bead slurry. For rates that were not measured; +, tails formed; ±, tails do not persist and rates are difficult to measure; −, no tails formed; n.d., not determined.
Factors Other Than the Arp2/3 Complex Contribute to Motility
The EVH1 domain of WASP and the P regions of WASP and Scar could enhance motility by directly stimulating the actin-nucleating activity of the Arp2/3 complex, or by recruiting proteins that themselves stimulate actin polymerization. To distinguish between these possibilities, the activities of WASP and Scar1 derivatives were tested for Arp2/3 complex-stimulating activity by using the pyrene-actin polymerization assay (Figure 4). Full-length WASP (Yarar et al., 1999) and Scar1 exhibited comparably robust activities, although native WASP from bovine thymus has been shown to be autoinhibited in the absence of Cdc42 or PIP2 (Higgs and Pollard, 2000). The differences between recombinant and native WASP may reflect differences in posttranslational modification or folding upon expression in insect cells. However, we found that recombinant WASP was still able to bind to the interacting protein WIP (supplemental material, Figure S2), indicating that improper folding of the EVH1 domain is not the cause of activation. In contrast to full-length WASP, the WASP-BGPWCA and WASP-GPWCA derivatives were virtually inactive in actin nucleation, consistent with the previously described role of the B and G regions in autoinhibition (Rohatgi et al., 1999; Higgs and Pollard, 2000; Kim et al., 2000; Prehoda et al., 2000; Rohatgi et al., 2000). The Arp2/3 complex stimulatory activities of these truncations were dramatically enhanced by the addition of PIP2 and Cdc42-GTP (supplemental material, Figure S3), as reported for full-length native WASP (Higgs and Pollard, 2000) and N-WASP (Rohatgi et al., 1999; Higgs and Pollard, 2000; Kim et al., 2000; Prehoda et al., 2000; Rohatgi et al., 2000). Interestingly, although both truncations exhibited maximal stimulation with a combination of PIP2 and Cdc42-GTP, WASP-BGPWCA could be stimulated by either Cdc42 or PIP2 alone, whereas WASP-GPWCA could only be stimulated by Cdc42 in the presence of PIP2, as previously shown for the regulation of native full-length WASP (Higgs and Pollard, 2000). These results suggest that PIP2 can stimulate nucleation by binding to regions other than the previously identified binding site in the B region (Rohatgi et al., 1999; Higgs and Pollard, 2000; Kim et al., 2000; Prehoda et al., 2000; Rohatgi et al., 2000).
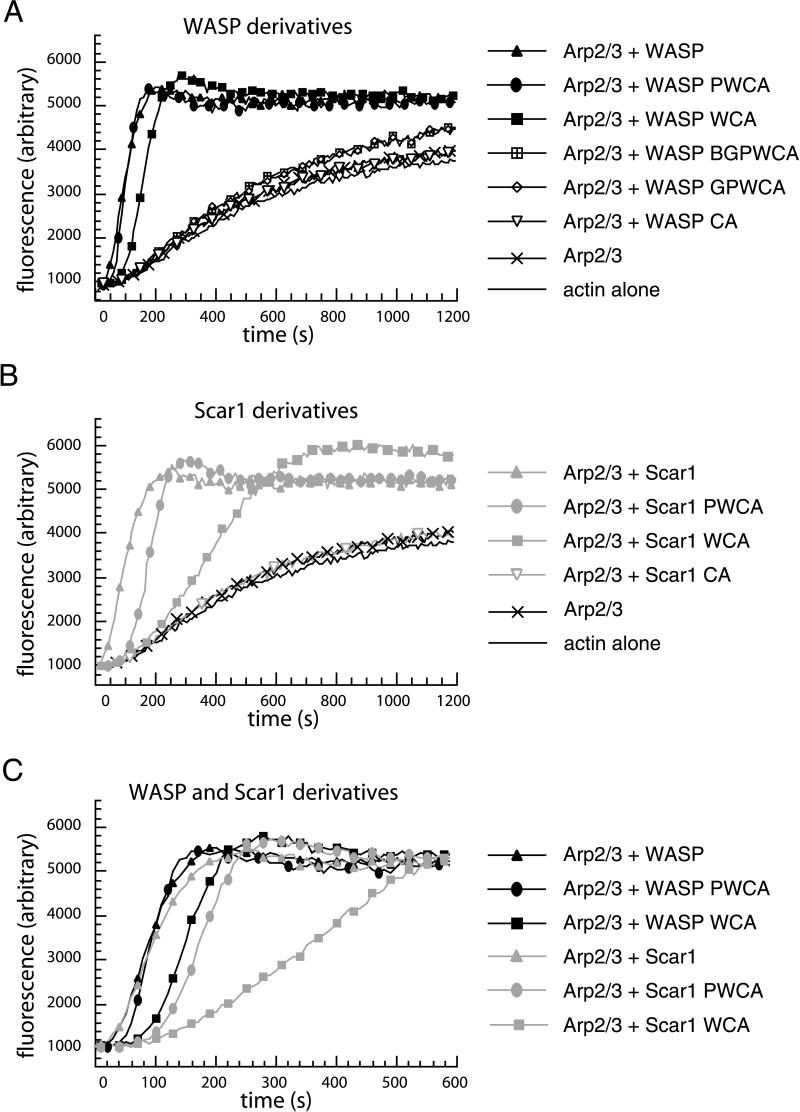
Figure 4.
Effects of WASP/Scar1 truncations and the Arp2/3 complex on actin polymerization kinetics, measured using the pyrene-actin polymerization assay. (A) Actin (2 μM) in the presence of 10 nM Arp2/3 complex and 20 nM WASP derivatives. (B) Actin (2 μM) in the presence of 10 nM Arp2/3 complex and 20 nM Scar1 derivatives. (C) Comparison of a subset of WASP and Scar1 derivative curves in A and B.
The smaller derivatives, which lack the inhibitory regions, exhibited a range of activities. WASP-PWCA was as active as full-length WASP and Scar1, although minor differences between WASP and WASP-PWCA could be observed, suggesting that the EVH1 domain may have a subtle effect on actin polymerization. WASP-WCA and Scar1-PWCA were less active, but were comparable with one another. Scar1-WCA was significantly less active than the other derivatives, in agreement with previous findings (Machesky et al., 1999; Yamaguchi et al., 2000; Zalevsky et al., 2001), suggesting that the P region of Scar1 contributes to Arp2/3 complex activation. WASP-CA and Scar1-CA did not activate the Arp2/3 complex under the conditions tested, indicating that the W region is essential for activity.
When we compared these findings with the results from the bead motility assay (Table 2), we found that the direct effects of the regions of WASP and Scar1 on Arp2/3 complex stimulation could not entirely account for their influence on motility rates. For example, although the nucleating activity of WASP, WASP-PWCA, and Scar1 were similar to one another, WASP promoted significantly faster movement in the bead assay. This suggests that the EVH1 domain of WASP, which is absent in the other proteins, can stimulate motility through a mechanism that does not involve direct activation of the Arp2/3 complex. In addition, although Scar1-PWCA and WASP-WCA were comparable with one another in nucleation activity, Scar1-PWCA beads moved significantly faster than WASP-WCA beads. Thus, the P region of WASP family proteins also stimulates motility independently of its direct effect on Arp2/3 complex activity. Taken together, these observations suggest that the EVH1 and P regions of WASP and Scar1 recruit cytoplasmic factors other than the Arp2/3 complex that enhance motility.
Table 2.
Comparison of WASP/Scar1 derivatives; bead motility rates vs. Arp2/3 complex stimulation activity
| Bead motility rate (μm/min) | Arp2/3 stimulation | |
|---|---|---|
| WASP | 8.3 ± 0.3 | ++++ |
| WASP-BGPWCA | 5.8 ± 0.3 | +* |
| WASP-GPWCA | 5.3 ± 0.4 | +* |
| WASP-PWCA | 6.1 ± 0.3 | ++++ |
| WASP-WCA | 3.3 ± 0.3 | +++ |
| WASP-CA | 0 | − |
| Scar1 | 5.9 ± 0.4 | ++++ |
| Scar1-PWCA | 6.6 ± 0.4 | +++ |
| Scar1-WCA | 0 | ++ |
| Scar1-CA | 0 | − |
Arp2/3 complex stimulation activities are in the presence of purified WASP family derivatives, Arp2/3 complex, and actin (5% pyrene labeled). Asterisk (
) indicates derivatives that are autoinhibited.
Profilin Recruitment Enhances Motility Mediated by WASP Family Proteins
One appealing candidate for mediating the motility-promoting activity of the EVH1 and P regions of WASP family proteins is profilin, which interacts directly with the P regions of WASP and Scar1 (Miki et al., 1998b; Suetsugu et al., 1998) and indirectly with the EVH1 domain of WASP through WIP (Ramesh et al., 1997). A second candidate is VASP, an F-actin binding protein that binds directly to WASP and requires the P region for this interaction (Castellano et al., 2001). To investigate whether profilin and VASP play critical roles in WASP- and Scar1-bead motility, we first examined the capacity of WASP- and Scar1-coated beads to form actin tails and move in egg extracts depleted of profilin and VASP by using PLP. Treatment of egg extracts with PLP-Sepharose matrix resulted in the depletion of >90% of the profilin and VASP, but <5% of the total Arp2/3 complex and actin (our unpublished data). As a control, extracts were treated with N-terminally blocked Sepharose resin.
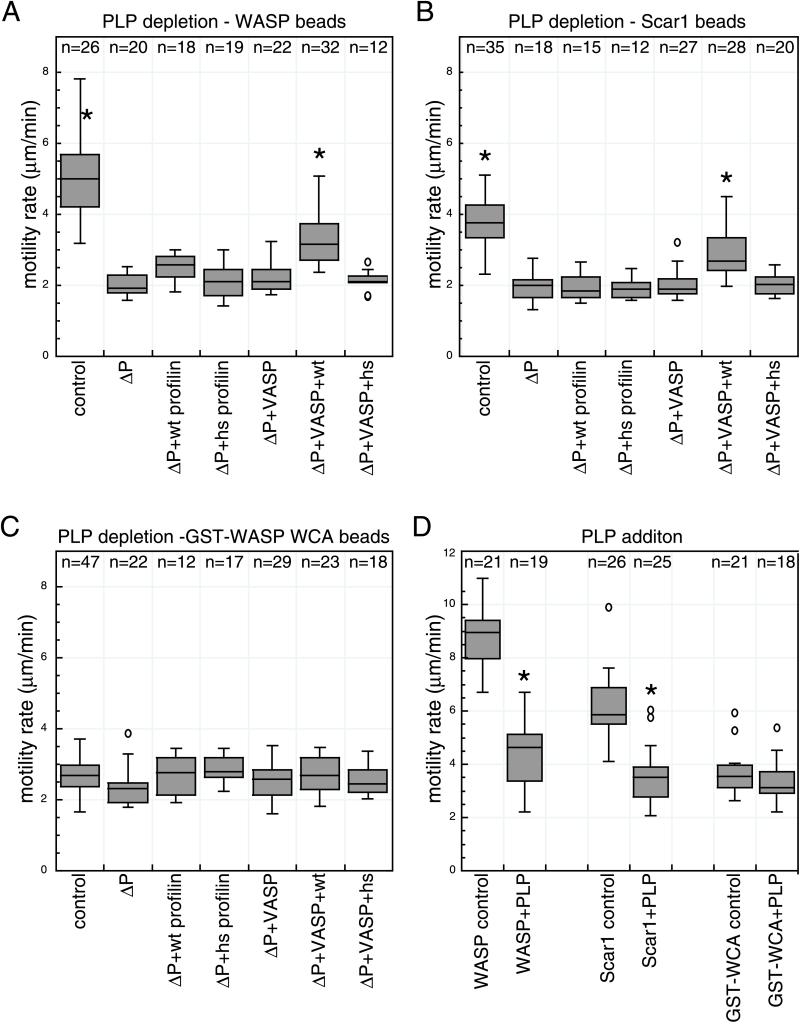
In the control extract, WASP- and Scar1-coated beads formed actin tails and moved through the extract (Figures 5 and 6), but at reduced rates compared with an untreated extract. In contrast, PLP depletion greatly compromised the motility of WASP- and Scar1-coated beads. Although these beads were able to form short tails, motility was reduced to rates observed for GST-WASP WCA-coated beads. Motility rates were also slowed upon the direct addition of 10 μM PLP to egg extracts (Figure 6D).
Figure 5.
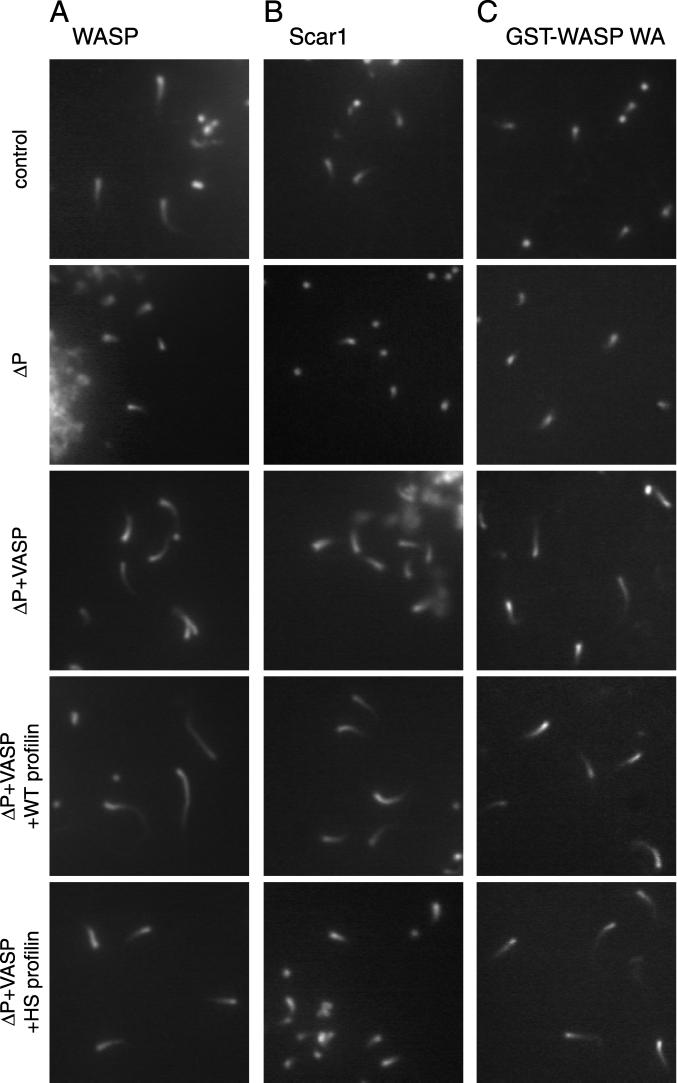
Effect of PLP depletion on WASP-, Scar1-, and GST-WASP WA bead motility. (A–C) Composite images of actin structures formed around WASP- (A), Scar1- (B), and GST-WASP WCA (C)-coated beads. Panels from top to bottom are: control depleted extract (control), poly-l-proline depleted extract (ΔP), ΔP extract supplemented with 0.2 μM VASP (Δp + VASP), ΔP extract supplemented with 0.2 μM VASP and 2 μM wild-type profilin (Δprofilin + VASP + wt profilin), and ΔP extract supplemented with 0.2 μM VASP and 2 μM H133S profilin (Δprofilin + VASP + HS profilin). Images were taken after samples were incubated on the slide for 30 min. Bar, 2 μm.
Figure 6.
Effect of PLP depletion/addition on WASP-, Scar1-, and GST-WASP WCA bead motility. (A–C) Box plot distribution of motility rates of WASP- (A), Scar1- (B), GST-WASP-WCA (C)-coated beads in conditions indicated: control depleted extract (control), poly-l-proline depleted extract (ΔP), ΔP extract supplemented with 0.2 μM VASP and/or 2 μM wild-type profilin (wt), and/or 2 μM H133S profilin (hs). Asterisks (*) denote population of beads that are significantly faster than beads moving in poly-l-proline–depleted extract. (D) Box plot distribution of motility rates of WASP/Scar1/GST-WASP WCA-coated beads in extract supplemented with 10 μM PLP or buffer (control). The number of beads in each data set is presented above each distribution. Asterisks (*) denote population of beads that move at rates significantly different than beads moving in control experiment.
The separate addition of either recombinant wild-type (wt) profilin (2 μM) or recombinant VASP (0.2 μM) to the depleted extract did not affect bead motility rates for WASP or Scar1, although VASP addition increased overall tail length (Figures 5 and 6). In contrast, the simultaneous addition of wt profilin and VASP to the PLP-depleted extract significantly increased motility rates of WASP and Scar1 beads (Figures 5 and 6). Even under these conditions, the rates were not fully restored to control levels, suggesting a technical limitation of the depletion experiment or a requirement for other factors that may have been removed during the depletion. The addition of a mutant form of profilin (R74E) that has a reduced affinity for actin (Korenbaum et al., 1998), alone or in combination with VASP, did not restore the bead motility rates (our unpublished data), indicating that the actin-binding activity of profilin is needed to enhance WASP and Scar1-bead motility rates. These results indicate that profilin and VASP, perhaps in combination with other factors, enhance WASP- and Scar1-bead motility in Xenopus egg extracts.
To examine whether the recruitment of profilin by the proline-rich sequences of WASP or Scar1 contributes to bead movement, we assessed the ability of another mutant form of profilin (H133S) to restore actin tail formation in depleted extracts. Profilin-H133S has a reduced affinity for polyproline sequences (Bjorkegren-Sjogren et al., 1997) such as those in WASP and Scar1, but has a normal affinity for actin (Egile et al., 1999; Yang et al., 2000). The addition of 2 μM profilin-H133S alone or in combination with VASP to depleted extracts did not increase the motility rates of either WASP- or Scar1-coated beads, indicating that binding of profilin to its polyproline ligands is important for its ability to stimulate WASP- and Scar1-bead motility.
To further examine whether profilin exerts its activity by interacting with the polyproline motifs within WASP and Scar1, or instead has a general effect on actin dynamics in the extract, we examined the behavior of GST-WASP WCA beads in the depleted extract. We reasoned that if profilin activity requires an interaction with WASP and Scar1, the motility rates of WASP-WCA beads should be unaffected by PLP depletion. Alternatively, if profilin exerts a global effect on actin dynamics, GST-WASP WCA bead movement should be compromised by PLP depletion. Strikingly, GST-WASP WCA bead motility was not affected by PLP depletion, PLP addition, or by the addback of VASP, wt profilin, or H133S profilin, separately or in combination (Figures 5 and 6). Thus, the effect of profilin on WASP- and Scar1-bead motility requires the EVH1 and P regions, which are absent from the WCA derivative. Interestingly, addition of VASP alone increased the length of actin tails formed behind the moving GST-WASP-WCA beads without affecting motility rates, indicating that VASP can exert an effect without directly interacting with WASP family proteins. In summary, our results strongly implicate the P region and EVH1 domain of WASP and Scar1 in enhancing actin-based motility by recruiting additional factors that include profilin.
DISCUSSION
A Threshold of Actin-nucleating Activity Is Required to Initiate Motility
The actin-nucleating and -branching activities of the Arp2/3 complex are critical for WASP-bead (Yarar et al., 1999) and L. monocytogenes motility (Loisel et al., 1999; May et al., 1999; Yarar et al., 1999). However, the precise relationship between actin nucleation and actin-based motility is not well defined. We have found that the smallest fragment of WASP that stimulates the nucleation activity of the Arp2/3 complex, WASP-WCA, is also the minimal region that is sufficient to support motility in egg extracts. Thus, it seems that the ability to stimulate the actin-nucleating and -branching activities of the Arp2/3 complex is sufficient to induce motility. In agreement with this conclusion, this region is also able to support actin-based bead motility in a reconstitution system consisting of purified proteins (Bernheim-Groswasser et al., 2002). Moreover, the corresponding N-terminal region of ActA, which contains similar functional regions (Skoble et al., 2000), also supports the movement of L. monocytogenes (Lasa et al., 1995). However, the N-WASP-WCA region is unable to support motility in bovine brain extract (Suetsugu et al., 2001b), most likely reflecting differences in the protein complement in brain and Xenopus eggs. Importantly, the WCA fragment of Scar1, which was less active than WASP-WCA in stimulating nucleation with the Arp2/3 complex (this report; Yamaguchi et al., 2000; Zalevsky et al., 2001), was unable to direct motility in Xenopus egg extract. This indicates that the WCA regions of different WASP family proteins are functionally distinct, and suggests that a threshold of nucleation activity must be achieved to initiate movement.
Enhancement of Motility by the Proline-rich Region and EVH1 Domain
Although the WASP-WCA fragment could direct movement, the presence of the P region and the EVH1 domain significantly enhanced motility rates. Moreover, the P region of Scar1 was required to initiate movement. Thus, the P regions and EVH1 domain play critical stimulatory roles. In agreement with the critical role of the P region, deletion of this region of N-WASP results in a reduction in the size of microspikes formed in response to EGF stimulation (Suetsugu et al., 1998) and an inhibition of actin polymerization by the bacterial pathogen Shigella flexneri (Mimuro et al., 2000), which recruits N-WASP to its surface to initiate motility in infected cells (Suzuki et al., 1998). Deletion of the N-WASP EVH1 domain has also been shown to reduce the rate of actin polymerization induced in bovine brain extract, as measured in a pyrene-actin assembly assay (Suetsugu et al., 2001a). Furthermore, missense mutations in the EVH1 domain of WASP result in >85% of all known cases of Wiskott-Aldrich syndrome (Schindelhauer et al., 1996).
Our results indicate that the direct effects of the WASP EVH1 domain and the P regions of WASP and Scar1 on actin nucleation with the Arp2/3 complex is not sufficient to account for the increased motility rates conferred by these regions in cell extracts. This suggests that the enhancement of motility by the P region and EVH1 domain is most likely mediated by proteins that bind to these regions. A number of binding partners for the P regions in WASP and Scar1 have been identified, including a variety of SH3 domain-containing proteins, such as Grb2 (Carlier et al., 2000), IRSp53 (Miki et al., 2000), WISH (Fukuoka et al., 2001), and Nck (Rohatgi et al., 2001). These proteins have been shown to increase the Arp2/3 complex-stimulating activity of WASP family proteins, suggesting one possible mechanism for enhancing motility. The P region also binds to profilin (for WASP and Scar1) and to VASP (binding only examined for WASP), actin-binding proteins whose functional properties are discussed below (Miki et al., 1998b; Suetsugu et al., 1998; Castellano et al., 2001). To date, the only identified EVH1 domain-binding protein is WIP (Ramesh et al., 1997). Although the biochemical activity of WIP remains unclear, it acts in concert with N-WASP in cells to promote the formation of filopodia (Martinez-Quiles et al., 2001), making it a candidate stimulatory factor. Moreover, its binding partners Nck (Anton et al., 1998; Rohatgi et al., 2001) and profilin (Yang et al., 2000) enhance actin nucleation and filament polymerization and may mediate the stimulatory effect of the EVH1 domain on motility. Alternatively, factors other than WIP may be recruited to the EVH1 domain to enhance motility rates.
Unlike the EVH1 domain, the SHD of Scar1 had only a slight effect on motility, suggesting that its primary function may be in regulating Scar1 activity or localization. This observation highlights the functional distinctions between the EVH1 and SHD domains and suggests that they serve as determinants for the different activities of WASP and Scar1 in cells. However, because the binding partners of the SHD are unknown, it is not yet clear how this domain contributes to Scar1 function.
Similarly, the WASP B and G regions did not affect motility in our assay. Although the G region allows truncations of WASP to be autoinhibited in solution and prevents them from stimulating the Arp2/3 complex, beads coated with WASP-BGPWCA or WASP-GPWCA polymerized purified actin in the presence of purified Arp2/3 complex, indicating that autoinhibition is relieved by binding to the beads (D.Y. and M.D.W., unpublished observations). This may explain our observation that activation or inhibition of G proteins in egg extracts by the addition of GTPγS or of dominant negative forms of Cdc42 and Rac had no effect on the formation of actin clouds or tails or on motility rates (D.Y. and M.D.W., unpublished observations). This also suggests that G proteins do not enhance motility of the coated beads once movement has been initiated.
Role of Profilin and VASP in Motility Mediated by WASP Family Proteins
We found that removal of >90% of both profilin and VASP by treatment of Xenopus egg extracts with PLP dramatically slowed the motility of both WASP and Scar1 beads. Faster motility rates could be substantially restored by the addition of wild-type profilin together with VASP, but not with the addition of a mutant form of profilin that does not bind to proline-rich sequences. Importantly, the motility of GST-WASP-WCA beads was not affected either by PLP depletion or by the addback of VASP and profilin. This demonstrates that profilin exerts its effects by interacting with proline-rich sequences in WASP family proteins either directly, or indirectly through VASP or an unidentified factor.
Consistent with the critical role of profilin demonstrated herein, several studies have shown that profilin enhances the motility rates of the bacterial pathogens L. monocytogenes and Shigella flexneri in cell extracts and in a reconstitution system consisting of purified proteins (Theriot et al., 1994; Marchand et al., 1995; Egile et al., 1999; Loisel et al., 1999; Mimuro et al., 2000). Although profilin binds directly to the P regions of WASP family proteins (Miki et al., 1998b; Suetsugu et al., 1998) and indirectly with the EVH1 domain of WASP via WIP (Ramesh et al., 1997), the functional importance of this binding interaction has been debated. Although some suggest that profilin binding to the polyproline ligands is not required to enhance Shigella actin-based motility (Egile et al., 1999), others have reported that the direct binding of profilin to polyproline ligands is important for its stimulatory role (Mimuro et al., 2000). Our results strongly suggest that an interaction between profilin and the P regions of WASP and Scar1 is vital for profilin to enhance actin-based motility. In support of a role for profilin binding to proline-rich sequences, this activity is important for its ability to stimulate Arp2/3 complex and WASP protein-mediated actin nucleation in neutrophil extracts in response to activated Cdc42 (Yang et al., 2000). However, we were unable to detect an effect of profilin on actin nucleation by purified WASP/Scar1 and Arp2/3 complex (D.Y. and M.D.W., unpublished observations), suggesting that, if it affects actin nucleation, it only does so in the presence of other cytosolic factors. In addition to polyproline binding, we and others have found that the ability of profilin to bind to actin is also required for its role in actin-based motility (Egile et al., 1999; Mimuro et al., 2000; this study). Recruitment of profilin may function in motility by enhancing actin nucleation with Arp2/3 complex, or by increasing the local concentration of polymerization competent actin through desequestration of actin from thymosin β4 (Pantaloni and Carlier, 1993).
Although profilin can bind directly to the P region of WASP and Scar1, it also binds to the proline-rich motif in VASP (Reinhard et al., 1995). Therefore, VASP may function in part by recruiting profilin to WASP and Scar1. Interestingly, Listeria move at reduced rates in cells expressing a derivative of VASP missing the profilin binding site (Geese et al., 2002), suggesting a role for the interaction between Ena/VASP proteins and profilin in actin-based motility. VASP may also play a role in WASP and Scar1-bead motility that is independent of its interaction with profilin. This idea is supported by our observation that the addition VASP increases the length of actin tails formed by WASP- and Scar1-coated beads, and by the finding that VASP can enhance Listeria and WASP-bead motility in the absence of profilin in a reconstitution system consisting of purified proteins (Loisel et al., 1999; Castellano et al., 2001). This effect may result from the ability of VASP to bundle F-actin (Bachmann et al., 1999; Laurent et al., 1999), which may stabilize actin tails and protect them from depolymerization. VASP also enhances the nucleating activity of ActA and the Arp2/3 complex, reduces the branching frequency of the Arp2/3 complex, and protects the barbed end of actin filaments from capping proteins (Skoble et al., 2001; Bear et al., 2002). Further experimentation will be required to clarify the contributions of each activity of VASP in actin-based bead motility mediated by WASP-family proteins and to determine whether VASP exerts its effect on actin-based motility by binding to WASP and Scar1.
Although our results indicate that profilin and VASP enhance the rates of motility of WASP- and Scar1-coated beads, it is notable that bead motility rates in PLP-treated extracts were not completely restored by the addition of purified profilin and VASP. This suggests that other polyproline binding factors, for example SH3 or WW domain containing proteins, may also contribute to motility.
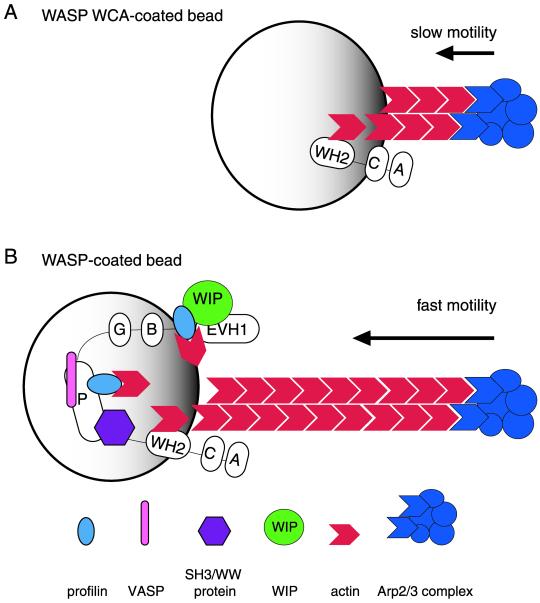
Based on our findings, we propose a model for the function of the regions of WASP-family proteins in actin-based motility (illustrated for WASP in Figure 7). The WASP WCA region, the minimum region that stimulates actin nucleation with the Arp2/3 complex, is sufficient to promote slow rates of actin-based bead motility. Full-length WASP and Scar1, which possess additional functional regions, recruit other factors that promote more rapid rates of bead motility. Regions of WASP and Scar1 that enhance motility include the P region, which recruits profilin, VASP (only demonstrated for WASP), and SH3-domain containing proteins, and the EVH1 domain of WASP, which recruits WIP and profilin. These factors may enhance motility by stimulating the nucleating activity of the Arp2/3 complex, or by stabilizing or promoting the elongation of newly formed filaments. Hence, the acceleration of both filament nucleation and elongation by factors recruited by the P regions of WASP and Scar1 and the EVH1 domain of WASP may regulate cellular actin-based motility.
Figure 7.
Model explaining the contributions of WASP/Scar1 regions to actin-based motility. (A) WCA derivative of WASP stimulates the nucleation activity of the Arp2/3 complex and is the minimal region that is sufficient to promote actin-based motility. (B) In full-length WASP, the EVH1 domain and P region recruit cytosolic factors, including profilin, that increase the rate of motility by enhancing actin nucleation with the Arp2/3 complex and/or facilitating actin polymerization.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Changsong Yang and Sally Zigmond for the gift of GST-profilin plasmids, Ines Anton and Narayanaswamy Ramesh for the gift of anti-WIP antibody, and Arie Abo for the gift of the WASPΔP plasmid. We thank Lisa Cameron, Matthew Footer, and Julie Theriot for helpful advice. We thank Justin Skoble, Rebecca Heald, Erin Goley, and especially Iain Cheeseman for critical reading of the manuscript and the Welch laboratory, Iain Cheeseman, and Justin Skoble for stimulating conversations during the course of this work. This work was supported by grants from the University of California Cancer Research Coordinating Committee, the Hellman Family Faculty Fund, and National Institutes of Health grant GM-59609 (to M.W.).
Footnotes
Online version of this article contains supplemental figures. Online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0294. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0294.
REFERENCES
- Anton IM, Lu W, Mayer BJ, Ramesh N, Geha RS. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adaptor protein Nck. J Biol Chem. 1998;273:20992–20995. doi: 10.1074/jbc.273.33.20992. [DOI] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Bear JE, Krause M, Gertler FB. Regulating cellular actin assembly. Curr Opin Cell Biol. 2001;13:158–166. doi: 10.1016/s0955-0674(00)00193-9. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IM, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–524. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bernheim-Groswasser A, Wiesner S, Golsteyn RM, Carlier MF, Sykes C. The dynamics of actin-based motility depend on surface parameters. Nature. 2002;417:308–311. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- Bjorkegren-Sjogren C, Korenbaum E, Nordberg P, Lindberg U, Karlsson R. Isolation and characterization of two mutants of human profilin I that do not bind poly-l-proline. FEBS Lett. 1997;418:258–264. doi: 10.1016/s0014-5793(97)01376-8. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci USA. 1999;94:1408–1413. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Nioche P, Broutin-L'Hermite I, Boujemaa R, Le Clainche C, Egile C, Garbay C, Ducruix A, Sansonetti P, Pantaloni D. GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex. J Biol Chem. 2000;275:21946–21952. doi: 10.1074/jbc.M000687200. [DOI] [PubMed] [Google Scholar]
- Castellano F, Le Clainche C, Patin D, Carlier MF, Chavrier P. A WASp-VASP complex regulates actin polymerization at the plasma membrane. EMBO J. 2001;20:5603–5614. doi: 10.1093/emboj/20.20.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T. A novel neural Wiskott-Aldrich Syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. J Cell Biol. 2001;152:471–482. doi: 10.1083/jcb.152.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geese M, Loureiro JJ, Bear JE, Wehland J, Gertler FB, Sechi AS. The contribution of Ena/VASP proteins to the intracellular motility of Listeria requires phosphorylation and the proline-rich core but not F-actin binding or multimerization. Mol Biol Cell. 2002;13:2383–2396. doi: 10.1091/mbc.E02-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA. Polyproline affinity method for purification of platelet profilin and modification with pyrene-maleimide. Methods Enzymol. 1991;196:92–99. doi: 10.1016/0076-6879(91)96011-f. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies; A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Mitchison TJ, Alberts BM. Behavior of microtubules and actin filaments in living Drosophila embryos. Development. 1988;103:675–686. doi: 10.1242/dev.103.4.675. [DOI] [PubMed] [Google Scholar]
- Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Korenbaum E, Nordberg P, Bjorkegren-Sjogren C, Schutt CE, Lindberg U, Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–9283. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labeled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Lasa I, David V, Gouin E, Marchand JB, Cossart P. The amino-terminal part of ActA is critical for the actin-based motility of Listeria monocytogenes; the central proline-rich region acts as a stimulator. Mol Microbiol. 1995;18:425–436. doi: 10.1111/j.1365-2958.1995.mmi_18030425.x. [DOI] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehman A, Grobe L, Jockusch BM, Wehland J, Gertler FB, Carlier MF. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Marchand JB, Moreau P, Paoletti A, Cossart P, Carlier MF, Pantaloni D. Actin-based movement of Listeria monocytogenes: actin assembly results from the local maintenance of uncapped filament barbed ends at the bacterium surface. J Cell Biol. 1995;130:331–343. doi: 10.1083/jcb.130.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Quiles N, Rohatgi R, Anton IM, Medina M, Saville SP, Miki H, Yamaguchi H, Takenawa T, Hartwig JH, Geha RS, Ramesh N. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat Cell Biol. 2001;3:484–491. doi: 10.1038/35074551. [DOI] [PubMed] [Google Scholar]
- May RC, Hall ME, Higgs HN, Pollard TD, Chakraborty T, Wehland J, Machesky LM, Sechi AS. The Arp2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr Biol. 1999;9:759–762. doi: 10.1016/s0960-9822(99)80337-6. [DOI] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998a;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998b;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- Mimuro H, Suzuki T, Suetsugu S, Miki H, Takenawa T, Sasakawa C. Profilin is required for sustaining efficient intra- and intercellular spreading of Shigella flexneri. J Biol Chem. 2000;275:28893–28901. doi: 10.1074/jbc.M003882200. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Dyche Mullins R, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with Wiskott-Aldrich Syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Nollau P, Ho HY, Kirschner MW, Mayer BJ. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J Biol Chem. 2001;276:26448–26452. doi: 10.1074/jbc.M103856200. [DOI] [PubMed] [Google Scholar]
- Schindelhauer D, Weiss M, Hellebrand H, Golla A, Hergersberg M, Seger R, Belohradsky BH, Meindl A. Wiskott-Aldrich syndrome: no strict genotype-phenotype correlations but clustering of missense mutations in the amino-terminal part of the WASP gene product. Hum Genet. 1996;98:68–76. doi: 10.1007/s004390050162. [DOI] [PubMed] [Google Scholar]
- Skoble J, Auerbuch V, Goley ED, Welch MD, Portnoy DA. Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J Cell Biol. 2001;155:89–100. doi: 10.1083/jcb.200106061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. Identification of another actin-related protein (Arp) 2/3 complex binding site in neural Wiskott-Aldrich syndrome protein (N-WASP) that complements actin polymerization induced by the Arp2/3 complex activating (VCA) domain of N-WASP. J Biol Chem. 2001a;276:33175–33180. doi: 10.1074/jbc.M102866200. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Yamaguchi H, Takenawa T. Requirement of the basic region of N-WASP/WAVE2 for actin-based motility. Biochem Biophys Res Commun. 2001b;282:739–744. doi: 10.1006/bbrc.2001.4619. [DOI] [PubMed] [Google Scholar]
- Sun HQ, Kwiatkowska K, Yin HL. Actin monomer binding proteins. Curr Opin Cell Biol. 1995;7:102–110. doi: 10.1016/0955-0674(95)80051-4. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miki H, Takenawa T, Sasakawa C. Neural Wiskott-Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 1998;17:2767–2776. doi: 10.1093/emboj/17.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Rosenblatt J, Portnoy DA, Goldschmidt-Clermont PJ, Mitchison TJ. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76:505–517. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Welch MD, Mitchison TJ. Purification and assay of the platelet Arp2/3 complex. Methods Enzymol. 1998;298:52–61. doi: 10.1016/s0076-6879(98)98008-9. [DOI] [PubMed] [Google Scholar]
- Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Miki H, Suetsugu S, Ma L, Kirschner MW, Takenawa T. Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proc Natl Acad Sci USA. 2000;97:12631–12636. doi: 10.1073/pnas.190351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Huang M, DeBiasio J, Pring M, Joyce M, Miki H, Takenawa T, Zigmond SH. Profilin enhances Cdc42-induced nucleation of actin polymerization. J Cell Biol. 2000;150:1001–1012. doi: 10.1083/jcb.150.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch MD. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11:1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.