Abstract
The notion of a “plurifunctional” nucleolus is now well established. However, molecular mechanisms underlying the biological processes occurring within this nuclear domain remain only partially understood. As a first step in elucidating these mechanisms we have carried out a proteomic analysis to draw up a list of proteins present within nucleoli of HeLa cells. This analysis allowed the identification of 213 different nucleolar proteins. This catalog complements that of the 271 proteins obtained recently by others, giving a total of ∼350 different nucleolar proteins. Functional classification of these proteins allowed outlining several biological processes taking place within nucleoli. Bioinformatic analyses permitted the assignment of hypothetical functions for 43 proteins for which no functional information is available. Notably, a role in ribosome biogenesis was proposed for 31 proteins. More generally, this functional classification reinforces the plurifunctional nature of nucleoli and provides convincing evidence that nucleoli may play a central role in the control of gene expression. Finally, this analysis supports the recent demonstration of a coupling of transcription and translation in higher eukaryotes.
INTRODUCTION
The highly complex organization of the cell nucleus reflects the intricate regulation of the various biological activities, including gene expression, that take place within this organelle (Lewis and Tollervey, 2000; Dundr and Misteli, 2001). In the interchromatin space, various molecular species constantly associate and dissociate to give rise to transient nuclear domains that structurally support DNA and RNA metabolism (Phair and Misteli, 2000; Misteli, 2001). At present, numerous nuclear domains have been pointed out, although the composition and function of most of them are not yet fully elucidated (Matera, 1999).
Nucleoli are dynamic nuclear domains that communicate constantly with the cytoplasm and with other nuclear domains (Chen and Huang, 2001). In the 1960s, it was demonstrated that nucleoli are the site of ribosome biogenesis. This function gives rise to their characteristic ultrastructural organization into three distinct regions visible by electron microscopy: the fibrillar center, the dense fibrillar component, and the granular component (Melese and Xue, 1995; Scheer and Hock, 1999; Olson et al., 2000). Ribosome biogenesis is a very complex process that involves the synthesis and assembly of four different mature rRNA molecules and of 80 different proteins. This process, which requires >150 different factors, is very efficient, because >14,000 ribosomal subunits can be synthesized every minute in an exponentially growing cell (Görlich and Mattaj, 1996). However, the molecular mechanisms governing the assembly of ribosomes and the majority of the nucleolar proteins involved in ribosome biogenesis are still ill defined. For instance, most of the ribonucleoproteins required for rRNA processing remain to be characterized (Weinstein and Steitz, 1999). Likewise, the mechanism by which mature ribosomal subunits are exported from nucleoli to the cytoplasm is not clearly elucidated (Aitchison and Rout, 2000; Kuersten et al., 2001). Furthermore, it was recently proposed that nucleoli may also play a crucial role in several other cellular processes such as the control of cell cycle, aging, and possibly mRNA export (Bond and Wold, 1993; Kadowaki et al., 1994b; Johnson et al., 1998; Pederson, 1998; Buonomo et al., 1999; Olson et al., 2000; Pederson and Politz, 2000; Visintin and Amon, 2000).
To gain insight into the multiple functions fulfilled by nucleoli, we have developed a proteomic analysis of purified nucleoli to determine their protein content. The recent advances in mass spectrometry (MS) techniques allowed us to identify 213 different proteins present within nucleoli purified from HeLa cells. About one-half of these proteins exhibit at least one known function. These proteins are involved in ribosome biogenesis, mRNA metabolism, and other cellular processes. Conversely, 48.8% of the identified proteins do not display well-characterized functions. However, sequence analyses, together with the finding that these proteins are indeed localized within nucleoli, allowed us to attribute hypothetical functions to some of them. A similar proteomic analysis has been reported recently (Andersen et al., 2002). Andersen and colleagues identified 271 proteins within purified human nucleoli. Interestingly, this allowed us to compare the data presented in these two independent studies; 133 proteins were common to both studies, 134 were unique to Andersen et al. (2002) study and 80 proteins were unique to this study. In conclusion, these two different but related proteomic analyses allowed identifying ∼350 different proteins that are localized within nucleoli of human cells. Preliminary functional analysis confirms the plurifunctional nature of nucleoli.
MATERIALS AND METHODS
Reagents for Proteomic Analysis
Unless stated otherwise, all reagents and chemicals were of the highest purity available. Water was purified using a MilliQ system (Millipore, Bedford, MA). Formic acid and acetic acid were from Fluka (Buchs, Switzerland). Hydrochloric acid, bromphenol blue, methanol, sodium chlorate, magnesium chlorate, and magnesium acetate were from Merck (Darmstadt, Germany). Acetonitrile (AcN) was from Biosolve (Volkenswaard, Holland), and trifluoroacetic acid (TFA), 1,4-dithiotreitol (DTT), 2,3-dihydroxybutane-1,4-dithiol (DTE), ammonium bicarbonate, iodoacetamide, glycerol, glycine, and porcine trypsin were from Sigma-Aldrich (St. Louis, MO). Phosphate-buffered saline was from BioWhittaker (East Rutherford, NJ), Tris and SDS were from Invitrogen (Carlsbad, CA), urea was from ICN Pharmaceuticals (Costa Mesa, CA), 3-[(3-cholamidopropyl)dimethylamino]-1-propane sulfonate (CHAPS) was from UBS (Cleveland, OH), and agarose was from Eurogentech (Liege, Belgium). Acrylamide:bis solution (37.5:1) and Biosafe Coomassie Blue were from Bio-Rad (Hercules, CA). Coomassie Brillant Blue R250 was from Amresco (Solon, OH). Proteins used as molecular mass references (low-molecular weight) and IPG Buffer 4-7 were from Amersham Biosciences (Piscataway, NJ).
Purification of Nucleoli from HeLa Cells
Nucleoli were purified using a procedure adapted from previously published protocols (Muramatsu and Onishi, 1978; Ochs, 1998). For a typical experiment, 3 × 108 HeLa cells were plated onto 245- × 245-mm Petri dishes in Eagle's minimum essential medium (Sigma-Aldrich, St. Louis, MO) containing 5% fetal calf serum (Eurobio, Les Ulis, France). Cells were incubated at 37°C under a 5% CO2 containing atmosphere. At ∼ 80% confluence, cells were washed with cold phosphate-buffered saline, pH 7.4, and scraped off while on ice. Cells were collected by centrifugation at 500 × g for 5 min. Cells were resuspended in 15 volumes of an hypotonic buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, and 1 mM MgCl2) and incubated on ice for 30 min. Cell lysis was performed by addition of a final concentration of 0.3% Nonidet P-40 (Roche Applied Science, Mannheim, Germany) and homogenization was performed using a 0.4-mm clearance Dounce homogenizer (Kimblr/Kontes, Owens, IL). Nuclei were collected by centrifugation at 1200 × g for 5 min and resuspended in 10 volumes of 0.25 M sucrose-containing 10 mM MgCl2. The supernatant containing the cytoplasmic fraction was harvested for further analyses. Nuclei were then purified by centrifugation at 1200 × g for 10 min through a 0.88 M sucrose cushion containing 0.05 mM MgCl2. Purified nuclei were resuspended in 10 volumes of 0.34 M sucrose containing 0.05 mM MgCl2 and sonicated on ice for several bursts of 30 s with 5-min intervals between them. Nucleoli were then purified from the resulting homogenate by centrifugation at 2000 × g for 20 min through a 0.88 M sucrose cushion containing 0.05 mM MgCl2. The supernatant containing the nucleoplasmic fraction devoid of nucleoli was harvested for further analyses. Purified nucleoli were resuspended in 0.34 M sucrose containing 0.05 mM MgCl2 for protein quantification (Bradford, 1976) and for further analyses.
Electron Microscopy Analyses
Fixation of nuclei and nucleoli pellets was performed by incubation for 10 min in 0.1 M cacodylate buffer, pH 7.4, containing 2% glutaraldehyde. Fixed pellets were washed 4 times with 0.2 M cacodylate buffer, pH 7.4. Postfixation was then performed by incubation for 30 min in 0.15 M cacodylate buffer, pH 7.4, containing 1% osmium tetroxide. Finally, pellets were dehydrated in ethanol and embedded in epoxy resin. Ultrathin sections were routinely contrasted with uranyl acetate and lead citrate and observed with a 1200 EX transmission electron microscope (JEOL, Tokyo, Japan) equipped with MegaView II high-resolution TEM camera and Analysis Soft Imaging System (Eloïse SARL, Roissy, France).
Electrophoresis and Western Blotting Procedures
For monodimensional SDS-PAGE, proteins from the total cells and from the different cellular fractions were solubilized in sample buffer (62.5 mM Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 0.1 M DTE, and traces of bromphenol blue) before separation on a 12.5% polyacrylamide gel (Laemmli, 1970). After electrophoresis, proteins were stained with Coomassie Brilliant Blue R250 or with Biosafe Coomassie Blue.
Western blot analysis of extracellular signal-regulated kinase (ERK)2 and nucleolin was carried out as described previously after separation by monodimensional SDS-PAGE (Diaz et al., 1993). Two micrograms of protein from each cellular fraction was separated by SDS-PAGE and transferred onto polyvinylidene difluoride membrane. Proteins were revealed using an anti-ERK2 polyclonal antibody diluted 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA) and an anti-nucleolin polyclonal antibody diluted 1:500 (a gift from P. Bouvet, ENS, Lyon, France).
For two-dimensional electrophoresis (2-DE), nucleolar proteins were first extracted from purified nucleoli using the acetic acid extraction method described previously (Waller and Harris, 1961; Madjar et al., 1979a). This method, originally developed to purify nucleic acid-free viral proteins (Fraenkel-Conrat, 1957), was used to remove remaining nucleic acids contained in the nucleolar fraction. The resulting nucleolar proteins were solubilized in isoelectric focusing (IEF) buffer containing urea. Briefly, nucleoli were resuspended in 0.34 M sucrose containing 0.05 mM MgCl2 and then magnesium acetate was added to a final concentration of 0.2 M before the addition of two volumes of glacial acetic acid. After 1 h at 4°C, the precipitated nucleic acids were removed by centrifugation. A second extraction of the remaining pellet was then performed. The two resulting supernatants were pooled and dialyzed against 500 volumes of 1 M acetic acid. Before electrophoresis, 500 μg of proteins was lyophilized and solubilized in IEF buffer containing 8 M urea, 2% CHAPS, 25 mM DTE, 0.5% IPG Buffer 4-7, and traces of bromphenol blue. IEF was carried out in the IPGPhor by using linear pH 4–7 IPG DryStrips (Amersham Biosciences) at 20°C with 40,000 Vh. On completion of the focusing time, proteins were reduced with DTE and alkylated with iodoacetamide. The second dimension was then performed under well-defined conditions with 650 Vh (Madjar et al., 1979b). Proteins were revealed with Biosafe Coomassie Blue.
In Gel Digestion
Fragments of the gel containing proteins of interest were cut out for digestion with trypsin by using the following procedure. Gel fragments obtained after one-dimensional electrophoresis (1-DE) were first destained by incubation in 100 μl of 50 mM ammonium bicarbonate and 30% AcN for 15 min at room temperature. Destaining solution was removed and fragments were then incubated for 35 min at 56°C in 25 μl of 10 mM DTT in 50 mM ammonium bicarbonate. DTT solution was then replaced by 25 μl of 55 mM iodoacetamide in 50 mM ammonium bicarbonate and the gel fragments were incubated for 45 min at room temperature in the dark. Gel pieces were then washed for 10 min with 100 μl of 50 mM ammonium bicarbonate and for 10 min with 100 μl of 50 mM ammonium bicarbonate and 30% AcN. Gel pieces obtained from 2-DE were only treated with 50 mM ammonium bicarbonate and 30% AcN for 20 min at room temperature because disulfide bond-containing proteins were already reduced and alkylated after IEF.
Gel pieces were then dried for 30 min in a Hetovac vacuum centrifuge (HETO, Allerod, Denmark). Dried pieces of gel were rehydrated for 45 min at 4°C in 5–20 μl of a solution of 50 mM ammonium bicarbonate containing trypsin at 6.25 ng/μl. After overnight incubation at 37°C, gel pieces were dried in high vacuum centrifuge before being rehydrated by the addition of 20 μl of H2O and finally dried again. Elution of the peptides was performed with 20 μl of 0.1% TFA for 20 min at room temperature with occasional shaking. The TFA solution containing the proteins was transferred to a polypropylene tube. A second elution of the peptides was performed with 20 μl of 0.1% TFA in 50% AcN for 20 min at room temperature with occasional shaking. The second TFA solution was pooled with the first one. The volume of the pooled extracts was reduced to 1–2 μl by evaporation under vacuum. Control extractions (blanks) were performed using pieces of gels devoid of proteins.
Protein Identification by Tandem Mass Spectrometry
Before nano-liquid chromatography separation, the volumes of peptide-containing solutions were adjusted to 7 μl by addition of a 0.1% formic acid solution. Samples were settled in a Triathlon autosampler (Spack, Emmen, Holland). For each experiment, 5 μl of peptide-containing solution was injected on a homemade C18 reverse phase column of 75 μm inner diameter (YMS-ODS-AQ200; Michrom Bioresource, Auburn, CA). Peptides were eluted with an AcN gradient in the presence of 0.1% formic acid, by using SunFlow pumps (SunChrom, Friderichsdorf, Germany). A flow splitter was used to decrease the flow rate from 200 to 0.4 μl/min. Peptides were analyzed with a Q-TOF mass spectrometer (Micromass, Wythenshawe, England). A 2700-V tension was applied on the nanoelectrospray capillary (New Objective, Woburn, MA). Argon was used as the collision gas. The collision energy was settled as a function of the precursor ion mass. MS/MS spectra were acquired by automatic switching between MS and MS/MS mode. Acquired MS/MS data were converted into a compatible format (DTA files) by ProteinLynx software (Micromass, Wythenshawe, England) and analyzed using the MASCOT search engine (http://www.matrixscience.com) against SWISS-PROT, TrEMBL, NCBInr, and EST databases. In cases of manual interpretation of MS/MS data, identification was performed by sequence only search by using the ProteinInfo search engine from PROWL (http://prowl.rockefeller.edu).
Protein Identification by Peptide Mass Fingerprinting
Before peptide mass fingerprinting, the volumes of peptide containing solutions were adjusted to 5 μl by addition of 0.1% TFA in 50% AcN. One microliter of each sample was deposited on a 2 × 96-well matrix-assisted laser desorption ionization target plate and dried in a vacuum container. Equal volumes of matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% AcN, 0.1% TFA) were added to the previously loaded digest. Samples were dried using a vacuum container. MS measurements were conducted with a matrix-assisted laser desorption ionization/time of flight mass spectrometer Voyager super STR (Applied Biosystems, Foster City, CA) equipped with a 337-nm nitrogen laser. The analyses were performed in the reflectron mode with an accelerating voltage of 20 kV, a delayed extraction parameter of 100–140 ns, and a low mass gate of 850 Da. Laser power was set slightly above threshold (10–15% higher than the threshold) for molecular ion production. Spectra were obtained by summation of 150 consecutive laser shots. Masses of the peaks were extracted from the spectra and used for protein identification using SmartIdent peptide mass fingerprint tool (Gras et al., 1999). The research was conducted against SWISS-PROT and TrEMBL databases.
RESULTS
Preparation of Highly Purified Nucleoli from HeLa Cells
Several cell fractionation procedures were tested to isolate nucleoli from HeLa cells (our unpublished data). From these experiments, a cell fractionation procedure adapted from Muramatsu and Onishi (1978) and Ochs (1998) was selected. This fractionation procedure generated highly purified nuclei as assessed by observation under light as well as electron microscopy (our unpublished data). The nuclei were disrupted by sonication and nucleoli were purified by differential centrifugation as indicated above in MATERIALS AND METHODS.
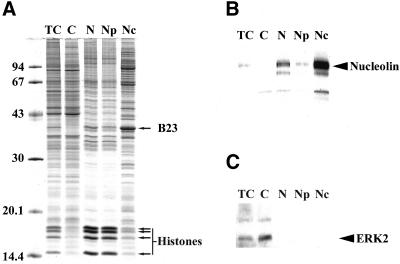
To verify the quality of the selected fractionation procedure, 10 μg of each cellular fraction was separated by 1-DE (Figure 1A). The band patterns obtained after separation of proteins extracted from total cells and from the cytoplasmic fraction are very similar. However, the four bands (indicated by arrows) whose molecular masses are between 14.4 and 20.1 kDa are abundant in the total cell extract, whereas they are only very slightly detected in the cytoplasmic fraction. These bands were identified by MS as histones. The band patterns obtained after separation of nuclear and nucleoplasmic fractions are very similar to each other, both of them contain four bands strongly stained (indicated by arrows). Remarkably, the band pattern obtained after separation of the nucleolar fraction is very different from those obtained after separation of all the other cellular fractions and from that obtained after separation of total cell extract. One of the more intense bands of the nucleolar fraction contains B23 as identified by MS (Figure 1A). An identical conclusion was drawn after a similar analysis which was performed in the same experimental conditions but after staining of the proteins with silver nitrate, a procedure known to be more sensitive although less quantitative than that using Coomassie Blue (our unpublished data). These analyses indicate that the nucleolar fraction is highly enriched in proteins specific to this fraction.
Figure 1.
Analysis of the cellular fractions obtained during nucleoli purification. (A) 1-DE separation of proteins extracted from total cells (TC) and from subcellular fractions indicated on the top of the gel (C, cytoplasm; N, nuclei; Np, nucleoplasm; Nc, nucleoli). For each fraction, 10 μg protein was separated on a 12.5% polyacrylamide gel. Proteins were stained with Coomassie brilliant blue R250. Positions of B23 and histones are indicated by arrows on the right of the panel. Sizes of the molecular weight markers are indicated in kilodaltons on the left of the panel. (B, C) Western blot analyses of the different cellular fractions described in A using an anti-ERK2 antibody (B) and an anti-nucleolin antibody (C). The position of ERK2 and nucleolin is indicated by an arrow on the right of panels B and C, respectively.
The efficiency of this specific enrichment was confirmed first by identification of B23 by MS (Figure 1A) and second by Western blot analysis of the different protein extracts by using an anti-nucleolin antibody (Figure 1B). As expected, nucleolin is not detected in the cytoplasmic fraction, whereas it is present in the nuclear fraction, barely detected in the nucleoplasmic fraction, and very abundant in the nucleolar fraction. Finally, to verify that nuclear fractions were not contaminated with cytoplasmic proteins, a Western blot analysis of the different protein extracts was performed using an anti-ERK2 antibody (Figure 1C). ERK2 was detected in total cell extract and was highly enriched in the cytoplasmic fraction. ERK2 was not detected in the nuclear, nucleoplasmic, or nucleolar fractions, demonstrating that under our experimental conditions ERK2 was predominantly cytoplasmic and the nuclear fractions were free of at least one protein that is known to migrate from the cytoplasm to the nucleus under specific conditions (Lenormand et al., 1993).
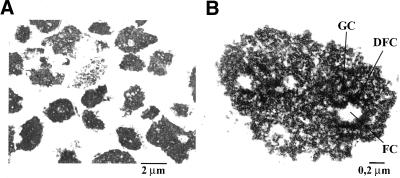
Electron microscope analyses revealed that the nuclear fraction was devoid of contamination with any cytoplasmic organelles (our unpublished data). Furthermore, electron microscope analyses of the nucleolar fraction showed that the only recognizable structures in this fraction were nucleoli (Figure 2A). A higher magnification of purified nucleoli revealed that they have conserved their characteristic ultrastructure composed of three distinct compartments: the fibrillar center, the dense fibrillar component, and the granular component (Figure 2B).
Figure 2.
Transmission electron micrographs of purified nucleoli. The nucleolar fraction was obtained from HeLa cells, as described in MATERIALS AND METHODS and submitted to electron microscopy analyses (A) Nucleoloar fraction at 4000× magnification. Nucleoli are the main structures observed in the last fraction of the cellular fractionation procedure. (B) Nucleolar fraction at 20,000× magnification. Purified nucleoli have conserved their characteristic ultrastructure in three main compartments: FC, fibrillar center; DFC, dense fibrillar component; and GC, granular component.
Sequential Proteomic Analysis of Nucleolar Proteins Separated by SDS-PAGE
To identify the largest number of nucleolar proteins, we have performed a proteomic analysis with purified nucleoli. For this, an amount of nucleoli corresponding to 15 μg of protein was separated by one-dimensional SDS-PAGE and the separated proteins stained with Biosafe Coomassie Blue (Figure 3). This gel was sequentially cut into 108 fragments, with the positions indicated to the right of the figure. Proteins contained in each fragment were in gel digested with trypsin. Peptides were extracted from gel fragments and analyzed by nano-liquid chromatography-electrospray ionization-Q-q-time of flight mass spectrometry. The high sensitivity of direct coupling of nano-high-performance liquid chromatography and tandem mass spectrometry allowed identification of low-abundance proteins in a complex mixture and is therefore particularly well adapted for the identification of proteins separated by one-dimensional SDS-PAGE. Peptides were separated on a homemade reverse phase microcolumn before ionization by electrospray and analysis in a tandem mass spectrometer. Using this technique, up to 15 different proteins were found in one single gel fragment (fragment 12 in Supplemental Table, supplementary data).
Figure 3.
Sequential analysis of nucleolar proteins separated by SDS-PAGE. Nucleolar proteins (15 μg) were separated by SDS-PAGE on a 12.5% polyacrylamide gel and stained with Bio-Safe coomassie blue. This gel was then sequentially cut into 108 fragments. Each cut was numbered. Their position within the gel and their number are indicated by dashes on the right side of the gel. Molecular masses of known proteins separated in the same gel are indicated on the left of the figure.
After fragmentation of the peptides, all obtained collision-induced dissociation (CID) spectra were converted in a mascot compatible format (DTA files) before searching against SWISS-PROT, TrEMBL, NCBInr, and/or EST databases. Matching results with at least three peptides per protein were selected in a first round of screening. CID spectra matching with a significant score, but with only one or two peptides per protein, were manually confirmed (de novo sequencing) and searched in sequence-only mode against the same databases. A similar procedure was applied with interpretable CID spectra that did not return a protein entry with the mascot software. After this manual and computer analysis of MS/MS data, 306 polypeptides were found in the 108 fragments, corresponding indeed to 190 different proteins. The number of the gel fragment analyzed, the name of the proteins identified, their database accession numbers, their molecular masses (MW), and their isoelectric points (pI) are reported in Supplemental Table. Sequences of the peptides obtained from 25 proteins did not permit discrimination between several entries of the databases. The entries that correspond to proteins very close in sequence are indicated in Supplemental Table.
Separation of Nucleolar Proteins by Two-dimensional Electrophoresis and Identification by Mass Spectrometry
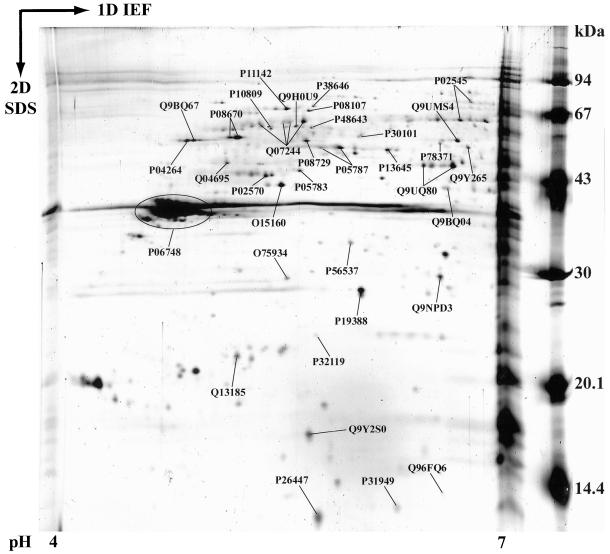
Nucleolar proteins with a pI below 7 were poorly represented in the first set of proteins identified after separation by 1-DE (Supplemental Table, 30% of the 190 different identified proteins). To enrich the nucleolar protein catalog in protein with a pI below 7 and to obtain an annotated 2-DE map of acidic nucleolar proteins, a 2-DE separation of proteins extracted from purified nucleoli was undertaken in the 4–7 pI range before their identification by MS. Identification of the proteins contained in the more intense spots observed after 2-DE and staining with Biosafe Coomassie Blue was performed. Identity of these proteins was indicated on an equivalent 2-DE gel performed with the same sample under the same experimental conditions but whose proteins where stained with silver nitrate (Figure 4). The proteins contained in 46 different spots were identified by either peptide mass fingerprinting or tandem mass spectrometry or both and were found to represent 35 different nucleolar proteins (Supplemental Table).
Figure 4.
Annotated 2-DE map of acidic nucleolar proteins. Nucleoli from HeLa cells were purified as described in MATERIALS AND METHODS. Nucleolar proteins were extracted with acetic acid before separation by 2-DE. Proteins were separated by IEF on immobilized pH gradients 4–7 in the first dimension. They were then separated by SDS-PAGE in the second dimension. Finally, proteins were identified by mass spectrometry. The image presented here is a representative gel stained with silver nitrate. The 35 identified proteins are labeled with their SWISS-PROT or TrEMBL accession numbers.
Functional Classification of 213 Proteins
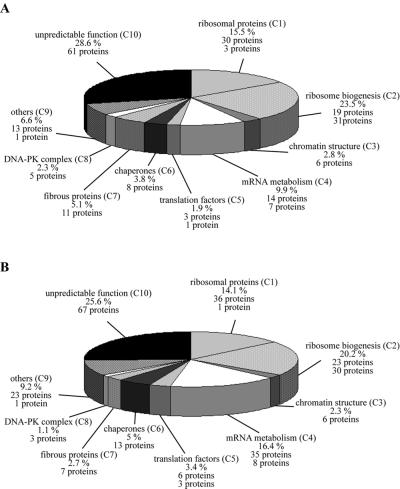
A total of 213 different proteins were identified within the nucleolar fraction; 190 proteins and 23 additional proteins were identified after separation by 1-DE and 2-DE, respectively (Supplemental Table). An extensive bibliographic analysis was carried out for each of the 213 proteins. From this, it seemed that 109 (51.2%) of the 213 proteins exhibit at least one known biological function, whereas the 104 (48.8%) remaining ones did not display a well-defined biological function. Therefore, sequence homology searches were carried out between each of these 104 proteins and those of the SWISS-PROT and TrEMBL databases. This analysis was performed using the BLAST network service (www.expasy.org). A strong homology was found between some of these 104 proteins and proteins with known functions present in the databases explored. This finding allowed the attribution of hypothetical functions to 43 proteins, whereas no specific function could be predicted for the 61 remaining. Altogether, these analyses permitted the description of functional classes (C1–C10) and the grouping of the identified nucleolar proteins according to their known or hypothetical biological functions into these functional classes (Supplemental Table). The different classes deduced from these analyses and the number of proteins contained in each class are presented in Figure 5A. An identical analysis was carried out for the proteins identified by Andersen et al. (2002) (Figure 5B).
Figure 5.
Functional classes for nucleolar proteins identified in the two independent proteomic analyses of purified human nucleoli. Functional classes were deduced for the 213 nucleolar proteins identified in this study and listed in the supplemental table, online. (A) and for 262 nucleolar proteins identified in the study of Andersen et al. (2002) (B). The name of the class with its corresponding abbreviation used in the supplemental table (C1–C10) is given. The percentage of total proteins found within each class is indicated. In addition, for each class, the number of proteins with a demonstrated involvement in the biological process is given, followed by the number of proteins with hypothetical involvement in this biological process in italics.
Completion of the Human Nucleolar Database
The two similar high-throughput proteomic analyses of human nucleoli available at present (Andersen et al., 2002; this study) should allow to initiate the construction of a comprehensive annotated human nucleolar database. For this, we have carried out a fine comparison of the results obtained in both studies to determine whether some of the proteins identified in this study were identified in Andersen et al. (2002) study. Each of the 213 proteins listed in Supplemental Table were submitted to a BLASTp search (www.expasy.org) against the deduced proteins obtained from the Unigene, GenBank, or IPI entries provided by Andersen et al. (2002). Several homologous proteins were detected between both studies. Using the PeptideMass program (www.expasy.org), each homologous protein identified in Andersen's study was digested and treated in silico with parameters simulating our experimental conditions. The peptides obtained were then compared with those obtained experimentally for our homologous protein. If all the peptides were identical, the probability that the two proteins represent the same protein was therefore high. This was the case for 133 proteins. Without more information from Andersen et al. (2002) study, we have decided to consider that these proteins are identical. These proteins are indicated in Supplemental Table. Conversely, if all the peptides of the two homologous proteins were not identical, we concluded that the two proteins were different. This was the case for 80 of the proteins of Supplemental Table.
DISCUSSION
The notion of a “plurifunctional nucleolus” was proposed previously (Pederson, 1998). However, at present, a comprehensive view of the biological processes that might occur within this subnuclear structure is still missing. Unquestionably, identification of proteins contained within nucleoli represents one of the first indispensable steps to fulfill this tremendous task. For this reason, a proteomic analysis was developed and reported herein to obtain a catalog of proteins contained within nucleoli of human cells. This catalog of 213 proteins seemed complementary to that of the 271 proteins obtained recently by others (Andersen et al., 2002) and allowed the establishment of a list of 350 different nucleolar proteins. However, establishing a list of nucleolar proteins is not sufficient per se to point out the different biological processes that might occur within nucleoli. This is why, when possible, the biological functions of the 350 proteins found in nucleoli were determined and gathered together according to several functional classes corresponding to well-defined major biological processes.
The first step of our proteomic analysis of human nucleoli was to purify these membraneless nuclear structures from HeLa cells and to verify their purity. While performing the fractionation procedure, integrity and purity of nuclei as well as enrichment of the nucleolar fraction with nucleoli were assessed by analysis of each fraction under light and electron microscopes. This analysis showed clearly that nuclei were the only organelles present in the nuclear fraction and that the nucleolar fraction contained a very high concentration of nucleoli, which conserved their ultrastructural integrity.
The enrichment of proteins specific to nucleolar and/or other fractions was evaluated by separation of the different fractions by 1-DE followed by Coomassie and silver nitrate staining as well as by Western blot. These analyses showed clearly that numerous proteins, such as histones were present almost exclusively in the fractions from nuclear origin, confirming the high quality of the nucleocytoplasmic separation procedure. In addition, the finding that in our experimental conditions, ERK2 was found by Western blot analysis exclusively in the cytoplasmic fraction suggested that extremely low contamination of the nuclear fractions, if any, with cytoplasmic proteins was present. More importantly, these analyses demonstrated that the majority of the proteins visible in the nucleolar fraction were not detected in the other fractions. In particular, the nucleolar fraction was highly enriched in two proteins that have been previously shown to be among the most abundant proteins of nucleoli, B23 and nucleolin (Bugler et al., 1982; Roussel and Hernandez-Verdun, 1994).
Because all of these results indicated very strongly that our nucleolar fraction was highly enriched in nucleoli and in proteins specific for this fraction, the next step of our proteomic analysis was to perform an extensive identification of the proteins contained within this fraction. For this, proteins from this fraction were separated by 1-DE and 2-DE through polyacrylamide gels. The separated proteins were stained with Coomassie Blue. Proteins of interest were in gel digested with trypsin and resulting peptides analyzed by mass spectrometry and for most of them by nano-liquid chromatography-electrospray ionization-Q-q-time of flight mass spectrometry to obtain sequence information. These data permitted the identification of 190 different proteins after separation by 1-DE and 23 supplementary proteins after separation by 2-DE.
The final step of our proteomic analysis was to outline the biological processes in which the 213 proteins could be involved. A bibliographic analysis was performed to obtain functional data for each of the 213 proteins. From this first analysis, it seemed that several functional studies were available for 109 out of the 213 proteins, whereas no functional studies were available for the 104 remaining proteins. This allowed us to determine unambiguously the biological mechanisms in which these 109 proteins are involved. Further analyses were performed to gain insights into the functions of the 104 proteins for which no functional studies were available. A search within protein databases was made to find proteins homologous to each of these 104 proteins. This search allowed the identification of a homologous protein with a well-defined function for 43 proteins of the 104, permitting the assignment of a hypothetical function. No function could be attributed for the remaining 61 proteins representing 28.6% of the 213 proteins.
As expected, numerous proteins found within nucleoli are either ribosomal proteins (15.5%) or proteins involved in ribosome biogenesis (23.5%). Interestingly, this analysis allowed us to propose clearly that 31 proteins of this latter category, which are from human origin, participate to ribosome biogenesis due to their homology with previously characterized proteins, which are for most of them from yeast origin. Moreover, the high proportion in our nucleolar fractions of proteins involved in well-established nucleolar functions strongly indicates that our nucleolar fraction is highly enriched in nucleoli. Several other proteins found within nucleoli in the current analysis are involved in the ultrastructural organization of the nucleus: fibrous proteins (5.1%) and structural proteins of the chromatin (2.8%) (Verheijen et al., 1986; Gerner et al., 1998; Belmont et al., 1999; Jung et al., 2000; Bergquist et al., 2001). Five proteins (2.3%) belong to the DNA-dependent protein kinase system, a multifunctional complex shown to be involved in many biological processes such as DNA repair and telomere maintenance (Dynan and Yoo, 1998; Featherstone and Jackson, 1999; d'Adda di Fagagna et al., 2001). Several proteins (6.6%) were arbitrarily regrouped into the class named “others” because they are involved in different biological processes, and each of them would require the construction of a novel specific functional class. For example, importin α-2 subunit, casein kinase II, ubiquitin, poly[ADP-ribose] polymerase-1, or S100 proteins are proteins from this category (Allende and Allende, 1995; Pickart, 2001; Tong et al., 2001; Weis et al., 1995; Donato, 2001). One of the features emerging from our classification is the surprising finding that ∼12% of the 213 proteins are proteins involved in the regulation of every step of mRNA metabolism, i.e., their synthesis, splicing, editing, nuclear export, and also their translation and degradation. As illustrated in Table 1, most of these proteins participate in several steps of mRNA metabolism. Indeed, many of these proteins have never been shown to be localized within nucleoli by using other techniques. Therefore, at this stage of the study, we cannot exclude that some of these proteins are found artifactually within the nucleolar fraction. Conversely, we cannot exclude also that these proteins are found in small amount and very transiently within nucleoli rendering them difficult to visualize using other techniques. Many more experiments would be required to address this question.
Table 1.
. Nucleolar proteins involved in the regulation of mRNA metabolism
The main steps of mRNA metabolism are indcated and underlined. The 25 proteins belonging to classes C4 and C5 (see Figure 5A) are classified according to their demonstrated implication in mRNA metabolism. Proteins in bold are involved in several steps. Proteins in italics and regrouped at the end of each section are proteins with hypothetical functions. Each protein is followed by one or several numbers that refer to articles listed at the bottom of the table. This functional classification is based on these articles.
Analysis of the biological functions of the nucleolar proteins identified previously by Andersen et al. (2002) demonstrated that these proteins could be classified according to the same functional classes elaborated in this study. Therefore, this study, by allowing the classification of a total of ∼350 nucleolar proteins according to the biological processes in which they are involved, firmly confirms the plurifunctional nature of nucleoli and outlines biological processes taking place within these nuclear domains (Andersen et al., 2002; this study). In particular, the present analysis shows clearly that translational regulators, chaperones, and also proteins involved in mRNA processing are found within nucleoli, in addition to other components of the translation machinery such as ribosomes (Leary and Huang, 2001), tRNAs (Bertrand et al., 1998; Pederson and Politz, 2000), signal recognition particle (Politz et al., 2000; this study), and even mRNAs (Kalland et al., 1991; Bond and Wold, 1993; Kadowaki et al., 1994a). This provides molecular evidence to the recent demonstration that translation can occur within nuclei of human cells (Iborra et al., 2001) and suggests that nucleoli themselves could play a central role in the control of this process. Indeed, one may suppose that, depending on the physiological or pathological state of the cell, highly specialized translation machines preassemble within nucleoli before being either exported to the cytoplasm or directly used in nuclei to translate certain classes of mRNAs. Alternatively, these preassembled translation machines may participate to the mRNA quality control (Hentze and Kulozik, 1999).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Simone Peyrol (CeCIL, Lyon, France) for generous advice for electron microscope experiments. We thank Dr. Philippe Bouvet (ENS) who kindly provided the anti-nucleolin antibody. We thank Drs. Jackie Lavigne and Frédéric Catez for critical reading of the manuscript. Electron microscope experiments were performed using the Center Commun d'Imagerie Laennec facilities. This work was supported by the Institut National de la Santé et de la Recherche Médicale and by the Swiss National Fund for Scientific Research (grant 31-59095.99). Y.C. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
Online version of this article contains supplementary material. Online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0271. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0271.
REFERENCES
- Aitchison JD, Rout MP. The road to ribosomes. Filling potholes in the export pathway. J Cell Biol. 2000;151:F23–F26. doi: 10.1083/jcb.151.5.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende JE, Allende CC. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Dietzel S, Nye AC, Strukov YG, Tumbar T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Bergquist J, Gobom J, Blomberg A, Roepstorff P, Ekman R. Identification of nuclei associated proteins by 2D-gel electrophoresis and mass spectrometry. J Neurosci Methods. 2001;109:3–11. doi: 10.1016/s0165-0270(01)00395-8. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond VC, Wold B. Nucleolar localization of myc transcripts. Mol Cell Biol. 1993;13:3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur J Biochem. 1982;128:475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Michienzi A, De Angelis FG, Bozzoni I. The Rev protein is able to transport to the cytoplasm small nucleolar RNAs containing a Rev binding element. RNA. 1999;5:993–1002. doi: 10.1017/s1355838299990064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- Diaz J-J, Simonin D, Massé T, Deviller P, Kindbeiter K, Denoroy L, Madjar J-J. The herpes simplex virus type 1 Us11 gene product is a phosphorylated protein found to be non-specifically associated with both ribosomal subunits. J Gen Virol. 1993;74:397–406. doi: 10.1099/0022-1317-74-3-397. [DOI] [PubMed] [Google Scholar]
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H. Degradation of tobacco virus with acetic acid. Virology. 1957;4:1–4. doi: 10.1016/0042-6822(57)90038-7. [DOI] [PubMed] [Google Scholar]
- Gerner C, Holzmann K, Grimm R, Sauermann G. Similarity between nuclear matrix proteins of various cells revealed by an improved isolation method. J Cell Biochem. 1998;71:363–374. doi: 10.1002/(sici)1097-4644(19981201)71:3<363::aid-jcb5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic Transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gras R, Muller M, Gasteiger E, Gay S, Binz PA, Bienvenut W, Hoogland C, Sanchez JC, Bairoch A, Hochstrasser DF, Appel RD. Improving protein identification from peptide mass fingerprinting through a parameterized multi-level scoring algorithm and an optimized peak detection. Electrophoresis. 1999;20:3535–3550. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3535::AID-ELPS3535>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- Johnson FB, Marciniak RA, Guarente L. Telomeres, the nucleolus and aging. Curr Opin Cell Biol. 1998;10:332–338. doi: 10.1016/s0955-0674(98)80008-2. [DOI] [PubMed] [Google Scholar]
- Jung E, Hoogland C, Chiappe D, Sanchez JC, Hochstrasser DF. The establishment of a human liver nuclei two-dimensional electrophoresis reference map. Electrophoresis. 2000;21:3483–3487. doi: 10.1002/1522-2683(20001001)21:16<3483::AID-ELPS3483>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants [published erratum in J. Cell Biol. (1994) 6, 1627] J Cell Biol. 1994a;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Hitomi M, Chen S, Tartakoff AM. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol Biol Cell. 1994b;5:1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalland KH, Langhoff E, Bos HJ, Gottlinger H, Haseltine WA. Rex-dependent nucleolar accumulation of HTLV-I mRNAs. New Biol. 1991;3:389–397. [PubMed] [Google Scholar]
- Kuersten S, Ohno M, Mattaj IW. Nucleocytoplasmic transport. Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leary DJ, Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–150. doi: 10.1016/s0014-5793(01)03143-x. [DOI] [PubMed] [Google Scholar]
- Lenormand P, Sardet C, Pages G, L'Allemain G, Brunet A, Pouyssegur J. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Madjar J-J, Arpin M, Buisson M, Reboud JP. Spot position of rat liver ribosomal proteins by four different two-dimensional electrophoreses in polyacrylamide gel. Mol Gen Genet. 1979a;171:121–134. doi: 10.1007/BF00269998. [DOI] [PubMed] [Google Scholar]
- Madjar J-J, Michel S, Cozzone AJ, Reboud JP. A method to identify individual proteins in four different two-dimensional gel electrophoresis systems: application to Escherischia coli ribosomal proteins. Anal Biochem. 1979b;92:174–182. doi: 10.1016/0003-2697(79)90641-9. [DOI] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Melese T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Onishi T. Isolation and purification of nucleoli and nucleolar chromatin from mammalian cells. Methods Cell Biol. 1978;17:141–210. doi: 10.1016/s0091-679x(08)61142-5. [DOI] [PubMed] [Google Scholar]
- Ochs RL. Methods used to study structure and function of the nucleolus. Methods Cell Biol. 1998;53:303–321. doi: 10.1016/s0091-679x(08)60884-5. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Politz JC. The nucleolus and the four ribonucleoproteins of translation. J Cell Biol. 2000;148:1091–1095. doi: 10.1083/jcb.148.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, Hernandez-Verdun D. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp Cell Res. 1994;214:465–472. doi: 10.1006/excr.1994.1283. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Tong WM, Cortes U, Wang ZQ. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim Biophys Acta. 2001;1552:27–37. doi: 10.1016/s0304-419x(01)00035-x. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers H, Vooijs P, van Venrooij W, Ramaekers F. Protein composition of nuclear matrix preparations from HeLa cells: an immunochemical approach. J Cell Sci. 1986;80:103–122. doi: 10.1242/jcs.80.1.103. [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A. The nucleolus: the magician's hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:372–377. doi: 10.1016/s0955-0674(00)00102-2. [DOI] [PubMed] [Google Scholar]
- Waller JR, Harris JI. Studies on the composition of the protein from Escherischia coli ribosomes. Proc Natl Acad Sci USA. 1961;47:18–24. doi: 10.1073/pnas.47.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Weis K, Mattaj IW, Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.