Abstract
hnRNP A1 is a nucleocytoplasmic shuttling heterogeneous nuclear ribonucleoprotein that accompanies eukaryotic mRNAs from the active site of transcription to that of translation. Although the importance of hnRNP A1 as a regulator of nuclear pre-mRNA and mRNA processing and export is well established, it is unknown whether this is relevant for the control of proliferation, survival, and differentiation of normal and transformed cells. We show here that hnRNP A1 levels are increased in myeloid progenitor cells expressing the p210BCR/ABL oncoprotein, in mononuclear cells from chronic myelogenous leukemia (CML) blast crisis patients, and during disease progression. In addition, in myeloid progenitor 32Dcl3 cells, BCR/ABL stabilizes hnRNP A1 by preventing its ubiquitin/proteasome-dependent degradation. To assess the potential role of hnRNP A1 nucleocytoplasmic shuttling activity in normal and leukemic myelopoiesis, a mutant defective in nuclear export was ectopically expressed in parental and BCR/ABL-transformed myeloid precursor 32Dcl3 cells, in normal murine marrow cells, and in mononuclear cells from a CML patient in accelerated phase. In normal cells, expression of this mutant enhanced the susceptibility to apoptosis induced by interleukin-3 deprivation, suppressed granulocytic differentiation, and induced massive cell death of granulocyte colony-stimulating factor-treated cultures. In BCR/ABL-transformed cells, its expression was associated with suppression of colony formation and reduced tumorigenic potential in vivo. Moreover, interference with hnRNP A1 shuttling activity resulted in downmodulation of C/EBPα, the major regulator of granulocytic differentiation, and Bcl-XL, an important survival factor for hematopoietic cells. Together, these results suggest that the shuttling activity of hnRNP A1 is important for the nucleocytoplasmic trafficking of mRNAs that encode proteins influencing the phenotype of normal and BCR/ABL-transformed myeloid progenitors.
The leukemogenic potential of the BCR/ABL oncoproteins depends on their ability to transduce oncogenic signals leading to altered expression and/or function of critical regulators of hematopoietic cell proliferation, survival, and differentiation (21, 22, 29, 43, 53). We recently reported that expression and activity of the heterogeneous ribonucleoprotein (hnRNP) FUS are important for the tumorigenic potential, growth factor-independent proliferation, and altered differentiation of BCR/ABL-transformed myeloid progenitors (45). In these cells, BCR/ABL regulates FUS expression and activity by inducing a PKCβII-dependent phosphorylation that prevents the proteasome degradation of FUS (46). FUS proteolysis is mediated by the association with ubiquitinated hnRNP A1, which, in turn, undergoes proteasome-dependent degradation in cytokine-deprived myeloid precursors (46). FUS and hnRNP A1 are two associated RNA binding proteins that belong to the family of shuttling hnRNPs (31, 52, 60). hnRNPs are RNA polymerase II-associated proteins which control different cellular activities such as transcription, nuclear pre-mRNA processing, mRNA export, translation, and cytoplasmic mRNA stability (12, 31, 54).
The ubiquitously expressed hnRNP A1 is a well-characterized hnRNP, and its levels of expression are higher in proliferating and/or transformed cells than in differentiated tissues (3). hnRNP A1 has an important role in pre-mRNA and mRNA metabolism (16); it binds nascent pre-mRNA in a sequence-specific manner (7), promotes the annealing of cRNA strands (11, 26), and regulates splice site selection (8-10, 14, 36, 37), exon skipping or inclusion (5, 28), nuclear export of mature mRNAs (27), mRNA turnover (23, 24), and translation (57). Although primarily nuclear, hnRNP A1 shuttles continuously between the nucleus and the cytoplasm, where dissociates from its mRNA cargo and is rapidly reimported into the nucleus in a transportin 1-dependent manner (47, 49, 55). The nucleocytoplasmic shuttling activity of hnRNP A1 depends on ongoing RNA polymerase II transcription (47, 48) and on the integrity of the M9 domain, a 38-amino-acid sequence which controls both nuclear import and export (38) and serves as a specific sensor for transcription-dependent nuclear transport of hnRNP A1 (55). hnRNP A1 binds mRNA both in the nucleus and in the cytoplasm, and its involvement in the nucleocytoplasmic trafficking of mRNA molecules also depends on an intact M9 shuttling domain (27).
We show here that expression of hnRNP A1 is increased in BCR/ABL-expressing cells through a posttranslational mechanism that prevents its ubiquitin/proteasome-dependent degradation. Moreover, survival and differentiation of normal myeloid precursors, growth factor-independent proliferation and tumorigenic potential of BCR/ABL-expressing 32Dcl3 cells, and colony formation of primary CD34+ cells from a patient with chronic myelogenous leukemia (CML) in accelerated phase (CML-AP) were impaired by expression of a nuclear hnRNP A1 mutant deficient in nucleocytoplasmic shuttling.
MATERIALS AND METHODS
Cell cultures and primary cells.
The murine interleukin-3 (IL-3)-dependent 32Dcl3 myeloid precursor and its derivative cell lines were maintained in culture or induced to differentiate as described previously (2). Morphological differentiation was monitored by May-Grunwald and Giemsa staining of cytospin preparations. For assays requiring cell starvation, cells were washed four times in phosphate-buffered saline (PBS) and incubated for 8 to 12 h in RPMI supplemented with 10% fetal bovine serum and 2 mM l-glutamine. The 293T human embryonic kidney cell line transformed with adenovirus 5 DNA (American Type Culture Collection [ATCC], Manassas, Va.) and the amphotropic-packaging cell line Phoenix A (G. P. Nolan, Stanford University School of Medicine) (ATCC SD3444) were maintained in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum and 2 mM glutamine (GIBCO), grown for 16 to 18 h to 80% confluence, and transfected by the calcium phosphate-DNA precipitation method using the ProFection system (Promega). The empty pMT or LXSP plasmid was used to normalize amounts of transfected DNA.
The stable 32D-BCR/ABL cell line has been described previously (45), whereas BCR/ABL-expressing 32Dcl3 cells were obtained by retroviral infection with supernatants of Phoenix cells transfected with the pSRαWTp210BCR/ABL plasmid. 32Dcl3 cells transfected with empty vectors (LXSP, MSCVpuro, MIG-RI, and pSRαMSV-tkneo) were morphologically identical to parental cells.
Where indicated, parental and BCR/ABL-expressing 32Dcl3 cells were IL-3 starved (8 h) in the absence or in the presence of a 25 μM concentration of the proteasome inhibitor ALLN (Calbiochem). To inhibit BCR/ABL tyrosine kinase activity, 32D-BCR/ABL cells were cultured for 8 h in medium supplemented with the Abl-kinase inhibitor STI571 (1 μM) (Novartis). 293T cells were treated with actinomycin D as described (38, 47). To inhibit protein synthesis, parental and BCR/ABL-expressing 32Dcl3 cells were treated for the indicated times with cycloheximide at a concentration (20 μg/ml) equally tolerated by both cell lines.
Samples of mononuclear hematopoietic cells from bone marrow of patients with CML in chronic phase (CML-CP) and in myeloid blast crisis (CML-BC) (20) were Ficoll separated and directly lysed in Laemmli buffer (2 × 105 cells/20 μl) for Western blot analysis. CD34+ cells from leukophoresis of a CML-AP patient were purified by using the CD34 MultiSort kit (Miltenyi Biotec, Auburn, Calif.) and kept overnight in Iscove's modified Dulbecco medium supplemented with 20% FBS, 2 mM glutamine, and human recombinant IL-3 (20 ng/ml), IL-6 (20 ng/ml), Flt-3 ligand (100 ng/ml), and KL (100 ng/ml) (Stem Cell Technologies Inc., Vancouver, Canada). Normal murine hematopoietic marrow cells were obtained from the femurs of C57BL/6 mice after hypotonic lysis, Ficoll separation, and adherence to plastic. Mononuclear cells were kept for two days in complete Iscove's modified Dulbecco medium supplemented with murine recombinant IL-3 (2 ng/ml), IL-6 (1.2 ng/ml), and KL (10 ng/ml) and subjected to a second round of Ficoll separation. Primary (murine or human) hematopoietic cells (106) were infected with the indicated retroviral constructs and plated in methylcellulose for clonogenic assays.
Retroviral infection of 32Dcl3 cells and derivative cell lines, normal murine marrow cells, and CD34+ cells from a CML-AP patient.
32Dcl3 cell lines expressing wild-type (32D-WT-A1-HA and 32D-BCR/ABL-WT-A1-HA), mutant (32D-NLS-A1-HA and 32D-BCR/ABL/NLS-A1-HA) or both wild-type and mutant (32D-NLS-A1-HA/WT-A1-HA) hnRNP A1 or mutant hnRNP A1 and C/EBPα (32D-NLS-A1-HA/C/EBPα-HA) were generated by retroviral infection of parental and BCR/ABL-expressing 32Dcl3 cells. Transient expression of mutant hnRNP A1 in normal murine marrow cells and in CD34+ CML-AP cells was obtained by infection with the LXSP-NLS-A1-HA retrovirus. Infections were carried out as described previously (44). Briefly, infectious supernatants from transiently transfected Phoenix cells were collected at 48 h after transfection and used to infect normal or BCR/ABL-transformed (primary and 32Dcl3 derivative) cells; 24 h later, infected cells were either sorted for green fluorescent protein positivity or cultured in the presence of G418 (1 mg/ml) or puromycin (2.5 μg/ml) for clonal selection or clonogenic assays. Viral titers of infectious supernatants from Phoenix cells transfected with the LXSP and the LXSP-NLS-A1 retroviral constructs were determined as follows. NIH 3T3 cells were plated (70% confluent) in 60-mm-diameter dishes and infected with 1 ml of viral supernatant as described previously (44). After infection, cells were split at different dilutions and plated in the presence of puromycin (2.5 μg/ml); puromycin-resistant colonies were scored after 9 days. The CFU per milliliter of virus inoculum volume was calculated by multiplying the number of puromycin-resistant colonies by the split factor of 1/2 as recommended in the ATCC protocol. Comparable numbers of viral puromycin-resistant CFU per milliliter (1.64 × 106 to 1.82 × 106) were used for clonogenic assays of 32Dcl3 and CD34+ CML cells (see below). For Western blotting, cells were lysed either in Laemmli buffer (2 × 105 cells/20 μl) or in hypertonic buffer (10 mM HEPES [pH 7.5], 400 mM NaCl, 10% [vol/vol] glycerol, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 25 μg of aprotinin per ml, 10 μg of leupeptin per ml, 100 μg of pepstatin A per ml, 5 mM benzamidine, 1 mM Na3VO4, 50 mM NaF, 10 mM β-glycerolphosphate, and 1% [vol/vol] NP-40.
Plasmids.
The full-length hnRNP A1 cDNA was a kind gift of G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia). To construct plasmid pMT-A1-HA, wild-type hnRNP A1 cDNA was PCR amplified using an upstream primer containing a BamHI site and a downstream primer containing a mutation in the stop codon followed by the hemagglutinin (HA) epitope sequence and a HindIII restriction site. The PCR product was digested with BamHI and HindIII and subcloned into the cytomegalovirus-based vector pMT. The shuttling-deficient plasmid pMT-A1(G274A)-HA carrying the G274A mutation in the M9 domain (38) was generated by site-directed mutagenesis of pMT-A1-HA with the Quickeasy Mutagenesis system (Stratagene). To construct plasmid pMT-NLS-A1-HA, a double-stranded oligonucleotide containing the sequence of the bipartite-basic-type nuclear localization signal (NLS) (KRPAEDMEEEQAFKRSR) of hnRNP K (39) flanked at both ends by a BamHI site was subcloned in frame into plasmid pMT-A1(G274A)-HA previously digested with BamHI. Plasmids MSCVpuro-A1-HA and LXSP-NLS-A1-HA were generated by subcloning the Klenow-blunted A1-HA and NLS-A1-HA NotI/HindIII fragments into the HpaI site of MSCVpuro (Clontech) and LXSP (kind gift of A. Sacchi, Regina Elena Cancer Institute, Rome, Italy), respectively. Plasmid MIG-RI-WT-A1-HA was generated by subcloning A1-HA BamHI and HindIII-blunted fragment into the BglII- and HpaI-digested MIG-RI retroviral vector (44). To construct ΔuORF-C/EBPα-HA, rat C/EBPα cDNA was amplified by PCR from plasmid pC/EBPα (a kind gift of S. L McKnight, Tularik Inc., South San Francisco, Calif.) using a primer set in which the 5′ ends of the upstream and downstream primer start at the main ATG and at the stop codon of C/EBPα cDNA, respectively. The PCR product was used as a PCR template with a downstream primer that contains an EcoRI site at the 5′ end flanked by the HA tag sequence and by a mutated C/EBPα stop codon. The amplified product was directionally subcloned into the HpaI- and EcoRI-digested MIG-R1 vector. Each plasmid was sequenced to verify the presence of expected mutations and correct reading frames. Plasmids pSRαMSVtkneo, pSRαMSVtkneo-p210BCR/ABL, and LXSP-HA-FUS have been described previously (46). The mc/ebp-alpha-3′UTR plasmid containing part of the 3′ untranslated region of the murine c/EBPα cDNA was a kind gift of Daniel G. Tenen (Harvard Institute of Medicine, Boston, Mass.).
Western blot analysis.
Cells were harvested, washed twice with ice-cold PBS, and lysed (107 cells/100 μl of lysis buffer) in hypertonic buffer. Lysates were obtained and processed as described previously (46). Nuclear and cytoplasmic subcellular fractions were obtained as follows. Cells (107) were washed twice in ice-cold PBS and lysed in 1 ml of isotonic buffer (150 mM NaCl, 20 mM HEPES [pH 7.5]) supplemented with 0.2% NP-40 and with protease inhibitors (see above). After disruption of the cytoplasmic membrane, nuclei were collected by centrifugation (5 min, 500 × g, 4°C), lysed in isotonic buffer supplemented with 1% NP-40, and clarified by centrifugation. Cytoplasmic fractions were also further clarified by centrifugation (12,000 × g, 15 min, 4°C). For C/EBPα detection, cells (2 × 105 to 3 × 105) were washed twice with ice-cold PBS, lysed directly in 20 μl of Laemmli sodium dodecyl sulfate (SDS) sample buffer, denatured (10 min, 100°C) prior to fractionation by SDS-4 to 15% polyacrylamide gel electrophoresis, and processed for Western blotting as described previously (46). The antibodies used were as follows: monoclonal anti-hnRNP A1 (9H10) (38) and monoclonal anti-hnRNP C1/2 (4F4) (42) (kind gifts of G. Dreyfuss); monoclonal anti-HSP90; rabbit polyclonal anti-C/EBPα, anti-granulocyte colony-stimulating factor receptor (anti-G-CSFR), anti-Bcl-2, and anti-Bcl-XL (Santa Cruz Biotechnology, Santa Cruz, Calif.); monoclonal anti-Abl (Ab3; Oncogene Science); monoclonal anti-GRB2 and horseradish peroxidase-conjugated antiphosphotyrosine PY20 (Transduction Laboratories Inc.); and anti-HA (Babco, Berkeley, Calif.).
Pulse-chase.
32D-WT-A1-HA and 32D-BCR/ABL-WT-A1-HA cells were cultured for 90 min in RPMI 1640 without methionine and supplemented with 10% dialyzed FBS (Gibco BRL, Grand Island, N.Y.) and 2 ng of recombinant murine IL-3 (Gibco BRL) per ml at 106 cells/ml. Cells were washed and resuspended (5 × 106 cells/ml) in medium containing 250 μCi of [35S]methionine (NEN, Life Science Products, Boston, Mass.) per ml. After 1 h, cells were washed with methionine-containing RPMI and cultured (105 cells/ml) for 20 h in IL-3-containing medium supplemented with an excess of l-methionine (3 mg/ml) (Gibco BRL). At different times, cells were harvested and lysed in isotonic buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% NP-40) supplemented with protease and phosphatase inhibitors used at the indicated concentrations. Precleared extracts were incubated at 4°C for 2 h with Protein G Plus (Calbiochem)-coupled anti-HA antibody (Babco). Immunoprecipitated proteins were resolved by SDS-polyacrylamide gel electrophoresis, visualized by phosphorimaging (Molecular Dynamics) upon transfer onto a nitrocellulose membrane, and analyzed by densitometry. The half-life of the wild-type hnRNP A1 protein (t1/2) was calculated using the formula t1/2 = (0.693 × t)/ln (Nt/N0) as described previously (34).
Immunofluorescence microscopy.
293T cells were grown on a microscope glass slide and transfected with the HA-tagged wild-type, G274A, and NLS-A1 plasmids as described above. At 48 h after transfection, glass slides were washed in Hanks' balanced salt solution, and cells were fixed for 10 min in PBS containing 3.7% formaldehyde. Thereafter, cells were washed three times with PBS, permeabilized by incubation (10 min) in PBS-0.05% Triton X-100 (Sigma), rinsed again with PBS, and then blocked for 10 min in PBS-4% goat serum. Incubation with the anti-HA antibody (1:250 dilution) and with the fluorophore-labeled goat anti-mouse immunoglobulin G Alexa 488 A-11001 (1:200 dilution; Molecular Probes) were carried out at room temperature for 30 min. Slides were rinsed three times with PBS, treated with SlowFade Antifade reagent (Molecular Probes), and analyzed by confocal microscopy.
Northern blot analysis and RT-PCR with total and cytoplasmic RNAs.
Total RNA was extracted with Tri-Reagent (Sigma). Cytoplasmic RNA was prepared by adding 2 volumes of Tri-Reagent to the cytoplasmic fractions prepared as described above. For Northern blot analysis, RNA (15 μg) was fractionated onto denaturing 1% agarose-6.6% formaldehyde gels, transferred to a nylon membrane (Amersham), and hybridized to 32P-labeled hnRNP A1 cDNA (4) and to the murine C/EBPα 3′ untranslated region fragment (50). Reverse transcription-PCR (RT-PCR) was performed with cytoplasmic RNA (125 ng) reverse transcribed by using avian myeloblastosis virus reverse transcriptase (Roche, Inc.) and random examers (Pharmacia) as described previously (2). Bcl-XL levels were determined by PCR using a set of primers corresponding to nucleotides 100 to 120 and 920 to 945 of the reported cDNA sequence of the mouse Bcl-X gene. An internal BCL-XL primer (nucleotides 721 to 760) was used for Southern blot analysis to determine the specificity of the amplified PCR product. β-actin levels were monitored as a control for equal loading. Differences in Bcl-XL levels were detected after 23 to 28 PCR cycles. After 30 PCR cycles, levels of Bcl-XL were identical in cells expressing or not expressing the NLS-A1-HA hnRNP A1 mutant.
Clonogenic assay and tumorigenesis in SCID mice.
Methylcellulose colony formation assays were carried out as described previously (2). Where indicated, cells were plated in the presence of antibiotics (G418 at 1 mg/ml or puromycin at 1.25 μg/ml) and of different concentrations of IL-3 or G-CSF. Colonies (>125 μm) were scored 7 to 10 days later. 32D-BCR/ABL and 32D-BCR/ABL-NLS-A1-HA cells (5 × 106 cells/mouse) were injected subcutaneously into 5- to 7-week-old ICR SCID outbred mice (Taconic, Germantown, N.Y.). Before injection, cells were washed and resuspended (2.5 × 107 cells/ml) in PBS. Tumor growth was monitored every other day. Mice were sacrificed at 20 days postinjection, and the excised tumors were fixed in phosphate-buffered formalin.
RESULTS
BCR/ABL prevents ubiquitin/proteasome-dependent hnRNP A1 degradation.
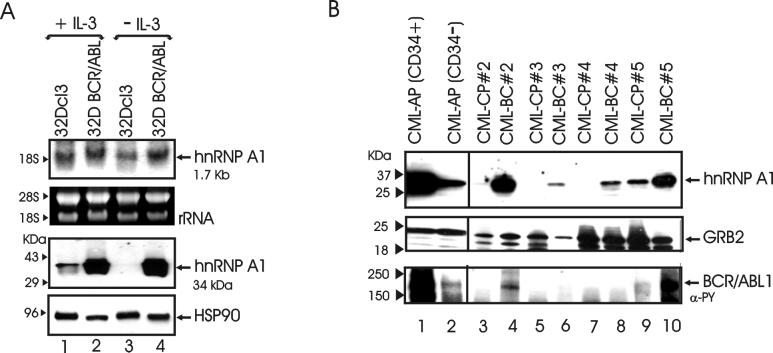
We recently reported that proteasome-mediated degradation of FUS requires association with hnRNP A1, which, in turn, undergoes ubiquitin/proteasome degradation in IL-3-deprived myeloid precursor 32Dcl3 cells (46). Since FUS expression correlates with that of BCR/ABL and its degradation is, in turn, suppressed by p210BCR/ABL (45, 46), we undertook experiments to determine whether levels of the FUS-associated hnRNP A1 are regulated by BCR/ABL in a manner similar to that of FUS. hnRNP A1 protein but not mRNA levels were markedly increased in BCR/ABL-expressing cells and were not influenced by the presence or absence of IL-3 (Fig. 1A). By contrast, hnRNP A1 protein was detectable at low levels in parental 32Dcl3 cells and downregulated upon cytokine deprivation (Fig. 1A, lanes 1 and 3). Likewise, hnRNP A1 levels were low or undetectable in 4 samples (representative of 10) of mononuclear marrows cells of CML-CP patients (Fig. 1B, lanes 3, 5, 7, and 9) but were clearly detectable upon transition into blast crisis (CML-BC patients) (Fig. 1B, lanes 4, 6, 8, and 10). Of note is that hnRNP A1 was more abundant in the CML-BC samples expressing detectable levels of BCR/ABL (Fig. 1B). hnRNP A1 expression in the CD34+ and CD34− fractions of mononuclear cells from a CML-AP patient, in which cytogenetic analysis revealed the presence of a double Philadelphia chromosome in approximately 20% metaphases, was also assessed. Of interest is that hnRNP A1 levels were higher in the CD34+ than in the CD34− fraction and correlated with those of BCR/ABL (Fig. 1B, lanes 1 and 2). Thus, it seems likely that hnRNP A1 expression correlates with that of BCR/ABL also in primary CML cells.
FIG. 1.
hnRNP A1 expression in normal and BCR/ABL-transformed cells. (A) Northern (top panel) and Western blot (bottom panel) analysis of hnRNP A1 expression in parental and BCR/ABL-expressing 32Dcl3 cells in the presence of IL-3 (lanes 1 and 3) or after IL-3 deprivation (12 h) (lanes 3 and 4). rRNA and HSP90 levels were used as controls for RNA and protein loading, respectively. (B) Western blot showing expression of hnRNP A1, BCR/ABL, and GRB2 in the CD34+ and CD34− fractions of mononuclear cells from a CML-AP patient (lanes 1 and 2) and in samples of mononuclear marrow cells from four CML-CP and four CML-BC patients (lanes 3 to 10).
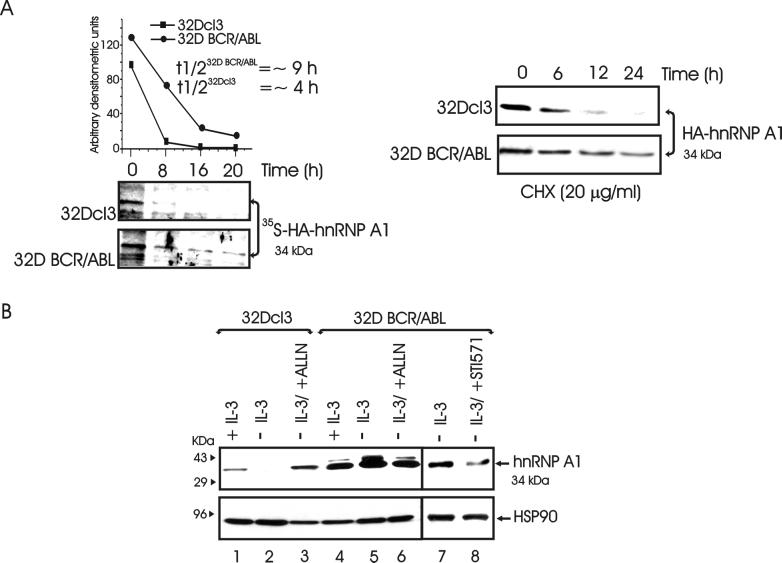
In parental 32Dcl3 cells, hnRNP A1 mRNA levels were similar to those in BCR/ABL-expressing cells regardless of the culture condition, suggesting that BCR/ABL induces posttranslational modifications that stabilize hnRNP A1 and prevent its proteasome-mediated degradation. Indeed, by pulse-chase experiments, the half-life of hnRNP A1 was longer in BCR/ABL-expressing cells (≈9 h) than in parental cells (≈4 h) (Fig. 2A, left panel). Consistent with these findings, treatment with the protein synthesis inhibitor cycloheximide, at a concentration (20 μg/ml) equally tolerated by parental and BCR/ABL-expressing 32Dcl3 cells during the time of exposure, resulted in downregulation of hnRNP A1 expression more rapidly in parental than in BCR/ABL-expressing 32Dcl3 cells (Fig. 2A, right panel). Thus, BCR/ABL expression appears to promote an increase in hnRNP A1 stability, possibly by preventing its proteasome-mediated degradation. Indeed, treatment with the proteasome inhibitor ALLN (25 μM) restored hnRNP A1 expression in IL-3-deprived parental 32Dcl3 cells (Fig. 2B, lanes 1 to 3), whereas it had no effect on hnRNP A1 levels in BCR/ABL-expressing cells (Fig. 2B, lanes 4 to 6). Of note is that hnRNP A1 levels were also decreased in BCR/ABL-expressing 32Dcl3 cells treated for 8 h with the specific ABL tyrosine kinase inhibitor STI571 (1 μM) (Fig. 2B, lanes 7 and 8), indicating that the enhanced hnRNP A1 expression is BCR/ABL tyrosine kinase dependent.
FIG. 2.
Role of BCR/ABL in the regulation of hnRNP A1 levels. (A) Left panel, stability of HA-tagged wild-type hnRNP A1 in exponentially growing parental and BCR/ABL-expressing 32Dcl3 cells. The half-life (t1/2) of hnRNP A1 was assessed by pulse-chase assay and quantitated by densitometry. Each point on the graph represents the relative amount of hnRNP A1 during the chase period; half-lives were calculated using the formula given in Materials and Methods. Right, levels of HA-tagged wild-type hnRNP A1 in parental and BCR/ABL-expressing cells treated with cycloheximide (CHX). (B) Effect of the proteasome inhibitor ALLN (lanes 1 to 6) and the ABL tyrosine kinase inhibitor STI571 (lanes 7 and 8) on endogenous hnRNP A1 levels in IL-3-deprived (8 h) parental and BCR/ABL-expressing cells. hnRNP A1 was detected with the 9H10 monoclonal antibody (38). HSP90 levels were monitored as a control for equal loading. Data are representative of those from three different experiments.
Generation of parental and BCR/ABL 32Dcl3 cell lines expressing a nucleus-localized shuttling-deficient hnRNP A1 mutant.
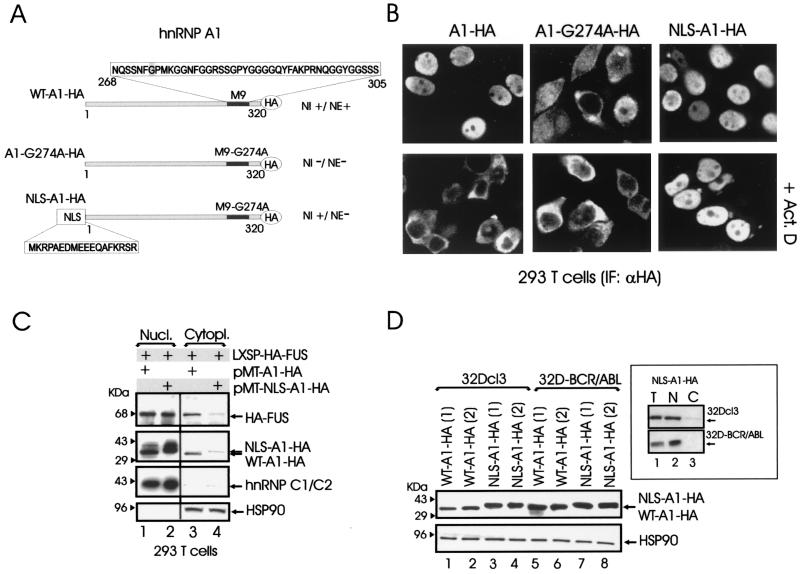
Several studies have shown that hnRNP A1 is a regulator of mRNA nuclear export (12, 54). The nucleocytoplasmic shuttling activity of hnRNP A1 depends on the integrity of a 38-amino-acid sequence, the M9 domain, which provides the signal for hnRNP A1 nuclear import and export (38). To determine the potential contribution of hnRNP A1 nucleocytoplasmic shuttling in the regulation of hematopoietic cell functions, we generated an hnRNP A1 mutant (NLS-A1-HA) expected to retain the hnRNP A1 nuclear localization (and perhaps nuclear function) while lacking nuclear export activity. The NLS-A1-HA construct (Fig. 3A) contains the bipartite-basic-type NLS of hnRNP K (39) fused in frame with the N terminus of an HA-tagged hnRNP A1 mutant (A1-G274A-HA) which lacked both nuclear import and export activities and inhibits hnRNP A1-dependent mRNA export when microinjected into nuclei of Xenopus laevis oocytes (27, 38). The subcellular localization of the NLS-A1-HA mutant was compared to that of HA-tagged wild-type hnRNP A1 (WT-A1-HA) and A1-G274A-HA mutant hnRNP A1 after transient transfection of 293T cells. As expected, NLS-A1-HA was localized only in the nucleus, while the G274A mutant accumulated in the cytoplasm (Fig. 3B). Moreover, treatment with actinomycin D, which induces the cytoplasmic relocation of wild-type hnRNP A1 (47), did not affect the subcellular localization of the NLS-A1-HA mutant, which remained entirely nuclear (Fig. 3B). Thus, the nucleus-localized NLS-A1-HA mutant has the potential to compete with wild-type hnRNP A1 for binding to, and nuclear export of, mRNAs that may be required for proliferation, survival, and differentiation of normal and leukemic myeloid progenitors.
FIG. 3.
Generation and expression of a nucleus-localized shuttling-deficient hnRNP A1 mutant. (A) Schematic representation of wild-type (WT-A1-HA) and mutant (A1-G274A-HA and NLS-A1-HA) hnRNP A1 constructs. Amino acid sequences of the hnRNP A1 M9 domain and of hnRNP K bipartite-basic NLS are boxed. NI, nuclear import; NE, nuclear export. (B) Anti-HA immunofluorescence shows the subcellular localization of WT-A1-HA, A1-G274A-HA, and NLS-A1-HA in transiently transfected 293T cells untreated or treated with actinomycin D (Act. D). (C) Effect of WT-A1-HA and NLS-A1-HA expression on nuclear (Nucl.) and cytoplasmic (Cytopl.) levels of HA-tagged FUS. Western blots show expression of HA-tagged FUS, HA-tagged wild-type (WT-A1-HA) and mutant (NLS-A1-HA) hnRNP A1, hnRNP C1/2, and HSP90 in nuclear and cytoplasmic fractions of 293T cells transiently transfected with the indicated plasmids. Expression of hnRNP C1/2 was used as a nuclear marker, while HSP90 was used as a cytoplasmic marker. Data are representative of those from three independent experiments. (D) Expression of wild-type and mutant hnRNP A1 in two clones of parental (lanes 1 to 5) and BCR/ABL-expressing (lanes 5 to 8) 32Dcl3 cells infected with the WT-A1-HA or the NLS-A1-HA retrovirus. The inset shows levels of NLS-A1-HA hnRNP A1 mutant in total lysates (lane T) and in nuclear (lane N) and cytoplasmic (lane C) fractions of parental and BCR/ABL-expressing 32Dcl3 cells.
By coimmunoprecipitation we found that FUS, an hnRNP whose altered expression affects differentiation and survival of normal and BCR/ABL-transformed myeloid progenitor cells (45), interacts with hnRNP A1 in the nucleus as well as in the cytoplasm (not shown). Since FUS is a shuttling protein that lacks known nuclear export or import signals (62), it is likely that its nuclear export is regulated in part by the association with hnRNP A1. Thus, we tested whether expression of the NLS-A1-HA mutant alters the subcellular distribution of ectopically expressed HA-FUS (LXSP-HA-FUS). Indeed, cytoplasmic levels of HA-FUS were decreased in 293T cells cotransfected with LXSP-HA-FUS and the cytomegalovirus-based pMT-NLS-A1-HA plasmid compared to those in cells coexpressing wild-type hnRNP A1 and HA-FUS (Fig. 3C, lanes 3 and 4).
Since the NLS-A1-HA mutant is likely to possess a dominant negative effect on the mRNA export activity of hnRNP A1, we generated parental and BCR/ABL 32Dcl3 cell lines ectopically expressing wild-type hnRNP A1 (32D-WT-A1-HA and 32D-BCR/ABL-WT-A1-HA) or the shuttling-deficient nucleus-localized mutant (32D-NLS-A1-HA and 32D-BCR/ABL-NLS-A1-HA) (Fig. 3D) and monitored proliferation, survival, and differentiation of these cell lines. As expected, in parental and BCR/ABL-expressing 32Dcl3 cells expression of the NLS-A1-HA mutant was readily detectable only in the nuclear compartment (Fig. 3D, inset).
Requirement of hnRNP A1 shuttling activity for survival and granulocytic differentiation of normal myeloid precursors.
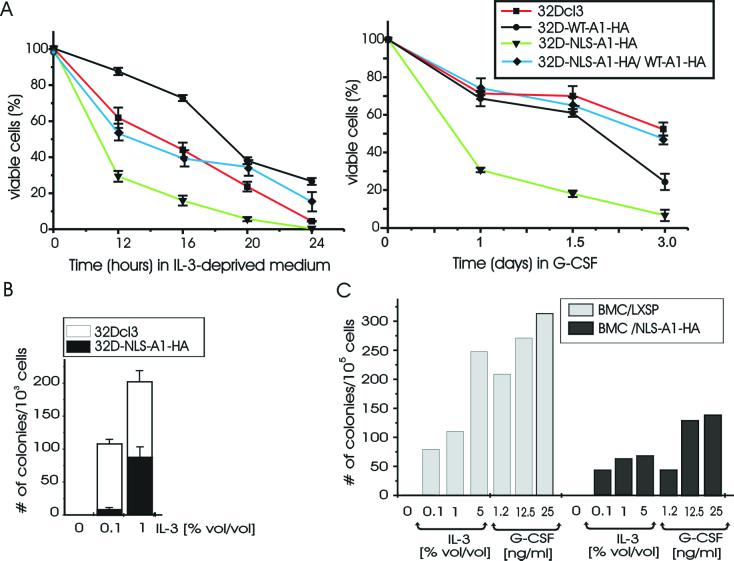
Parental and WT-A1-HA- and NLS-A1-HA-expressing myeloid precursor 32Dcl3 cells were either grown in the presence of IL-3, deprived of IL-3 for 12 to 24 h, or treated with G-CSF for 7 days. In IL-3-containing medium, 32Dcl3 cells expressing either the wild-type or the nucleus-localized shuttling-deficient hnRNP A1 proliferated like parental cells (not shown). At 12 h after IL-3 deprivation, dead cells were more frequent in 32D-NLS-A1-HA than in 32D-A1-HA cell cultures (≈70 versus ≈10%) (Fig. 4A, left panel); at 24 h, IL-3-deprived 32D-NLS-A1-HA cells were all dead, whereas ≈30% of wild-type hnRNP A1-expressing cells remained viable (Fig. 4A, left panel). Similarly, 32D-NLS-A1-HA cells were less clonogenic than parental cells when plated in methylcellulose in the presence of increasing concentrations of IL-3 (Fig. 4B). Although wild-type hnRNP A1-expressing cells were less prone then parental cells to cytokine deprivation-induced apoptosis, they did not become growth factor independent and were all dead after culture for 48 h in IL-3-deprived medium (not shown).
FIG. 4.
Requirement of hnRNP A1 shuttling activity for survival and colony formation of myeloid precursor 32Dcl3 cells and primary murine marrow cells. (A) Effect of IL-3 deprivation (left) and G-CSF treatment (right) on the viability of parental and derivative cell lines ectopically expressing wild-type hnRNP A1 (32D-WT-A1-HA) or the nucleus-localized, shuttling-deficient hnRNP A1 mutant (32D-NLS-A1-HA) or coexpressing wild-type and shuttling-deficient hnRNP A1 (32D-NLS-A1-HA/WT-A1-HA). Each point represents the mean and standard deviation from three independent experiments. Cell death percentage was determined by trypan blue exclusion. (B) Methylcellulose colony formation, in the absence or in the presence of different concentrations of WEHI-3B conditioned medium used as a source of IL-3, from 32Dcl3 and 32D-NLS-A1-HA cells (103 cells/plate). Values are means and standard deviations for duplicate cultures from two independent experiments. (C) Clonogenic efficiency in the absence of growth factors or in the presence of increasing concentrations of WEHI conditioned medium or recombinant human G-CSF of murine mononuclear marrow cells (BMC) transduced with the empty LXSP or with the NLS-A1-HA retrovirus. After infection, cells (105 cells/plate) were plated in semisolid medium in the presence of 1.25 μg of puromycin per ml. The results are representative of those from two experiments performed in duplicate.
The importance of hnRNP A1 shuttling activity in normal myelopoiesis was assessed by investigating the effect of NLS-A1-HA expression on colony formation from primary murine mononuclear marrow cells. For this purpose, 105 primary murine mononuclear marrow cells, infected either with the LXSP-NLS-A1-HA or with the empty LXSP retrovirus, were plated in methylcellulose in the presence of puromycin (1.25 μg/ml) as a selectable marker and of increasing concentrations of WEHI conditioned medium as a source of IL-3 or recombinant human G-CSF. Compared to insert-less retrovirus-infected primary murine mononuclear marrow cells, expression of NLS-A1-HA induced 50 to 75% and 60 to 75% decreases in the numbers of IL-3- and G-CSF-derived colonies, respectively (Fig. 4C).
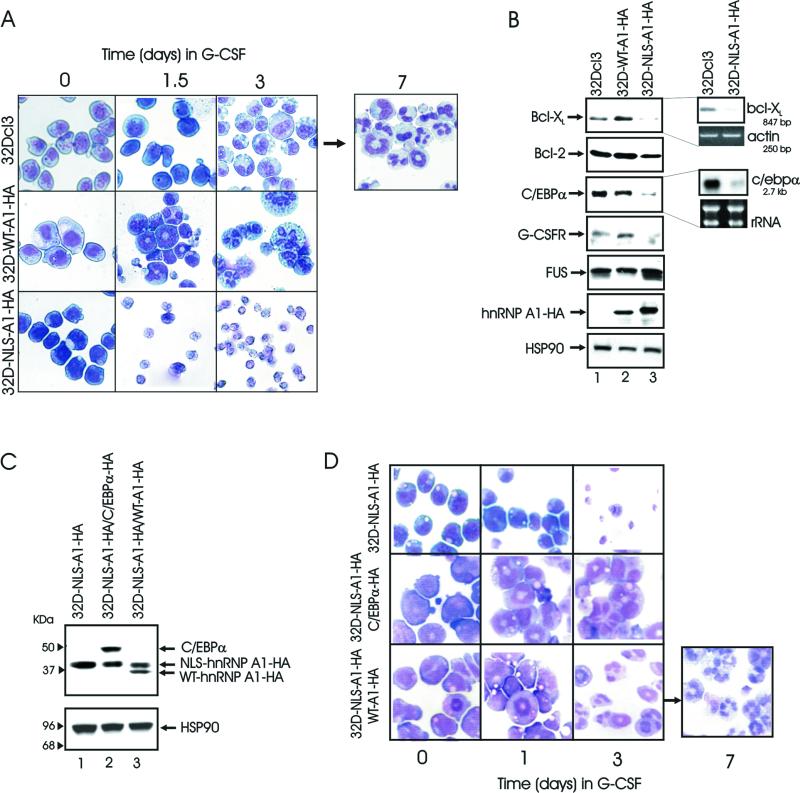
G-CSF-treated NLS-A1-HA-expressing 32Dcl3 cells showed morphological features of massive apoptosis (cytoplasmic shrinkage, nuclear condensation, and presence of apoptotic bodies) at day 1.5 (Fig. 5A, third row) and were all dead after 3 days (Fig. 4A, right panel, and 5A). Cultures of wild-type hnRNP A1-expressing cells revealed early signs of terminal differentiation as indicated by the presence of numerous polymorphonuclear cells at days 1.5 and 3 (Fig. 5A, second row) followed by death of the majority of cells at day 5 (not shown); parental 32Dcl3 cells remained viable and differentiated into neutrophils in 7 to 10 days (Fig. 5A, first row).
FIG. 5.
Requirement of hnRNP A1 shuttling activity for granulocytic differentiation of 32Dcl3 cells. (A) Representative microphotographs of May-Grunwald-Giemsa-stained cytospins of G-CSF-treated parental and 32Dcl3-derived cell lines. (B) Effect of WT-A1-HA and NLS-A1-HA expression on protein levels (left panels) of Bcl-2, Bcl-XL, C/EBPα, G-CSFR, and FUS and on mRNA levels (right panel) of Bcl-XL and c/ebpα. Bcl-XL cytoplasmic mRNA levels were detected by RT-PCR (see Materials and Methods); actin levels are shown as a control for equal loading. c/ebpα cytoplasmic mRNA levels were detected by Northern blotting using the murine 3′ untranslated region as a probe. rRNA levels are shown as a control for equal loading. The results are representative of those from three different experiments. (C) Western blot show expression of HA-tagged wild-type hnRNP A1 (lane 3) or C/EBPα (lane 2) in 32D-NLS-A1-HA cells. (D) G-CSF-stimulated granulocytic differentiation of 32D-NLS-A1-HA cells coexpressing WT-A1-HA or C/EBPα. Representative microphotographs of May-Grunwald-Giemsa-stained cytospins are shown.
To determine whether the effects of NLS-A1-HA expression on survival and differentiation of myeloid progenitor cells are due to altered hnRNP A1 function, 32D-NLS-A1-HA cells were transduced with the MIG-RI WT-A1-HA retrovirus (Fig. 5C, lane 3), sorted by the use of green fluorescent protein, and cultured in the absence of IL-3 or in the presence of G-CSF to monitor apoptosis susceptibility and ability to undergo granulocytic differentiation, respectively. In the absence of IL-3, 32D-NLS-A1-HA/WT-A1-HA cells were less prone than 32D-NLS-A1-HA cells to apoptosis induced by growth factor deprivation (Fig. 4A, left panel); in the presence of G-CSF, these cells were much more viable than the counterpart expressing NLS-A1-HA only (Fig. 4A, right panel) and underwent granulocytic differentiation (Fig. 5D, third row) with a kinetics similar to that of parental 32Dcl3 cells. Thus, it appears that in normal myeloid progenitors the hnRNP A1 shuttling-deficient mutant impairs normal hnRNP A1 functions.
To investigate potential mechanisms underlying both increased susceptibility to apoptosis and impaired differentiation of the NLS-A1-HA-expressing 32Dcl3 cells, steady-state mRNA and protein levels of the apoptosis suppressors Bcl-2 and Bcl-XL and of the regulator of granulocytic differentiation C/EBPα were assessed in parental and A1-WT-HA- and NLS-A1-HA-expressing 32Dcl3 cells. Compared to parental and 32D-WT-A1-HA cells, 32D-NLS-A1-HA cells showed reduced levels of Bcl-XL and C/EBPα (Fig. 5B). Expression of the C/EBPα-regulated G-CSFR was lower in 32D-NLS-A1-HA than in parental or 32D-WT-A1-HA cells (Fig. 5B), whereas levels of Bcl-2 or of the hnRNP A1-associated FUS protein were not significantly affected by expression of the NLS-A1-HA mutant hnRNP A1. Levels of c/EBPα and Bcl-XL (Fig. 5B) mRNAs were also reduced in 32D-NLS-A1-HA cells, in correlation with levels of the corresponding proteins. Thus, the altered response of 32D-NLS-A1-HA cells to IL-3 deprivation or G-CSF treatment might rest in the downregulation of Bcl-XL, C/EBPα, and G-CSFR expression, possibly reflecting defective nucleocytoplasmic trafficking of hnRNP A1-associated mRNAs. Ectopic expression of C/EBPα in 32D-NLS-A1-HA cells (Fig. 5C) restored G-CSF-dependent differentiation of 32D-NLA-A1-HA cells (Fig. 5D, second row).
Growth factor-independent proliferation and tumorigenesis of BCR/ABL-transformed cells is suppressed by the expression of the shuttling-deficient hnRNP A1 mutant.
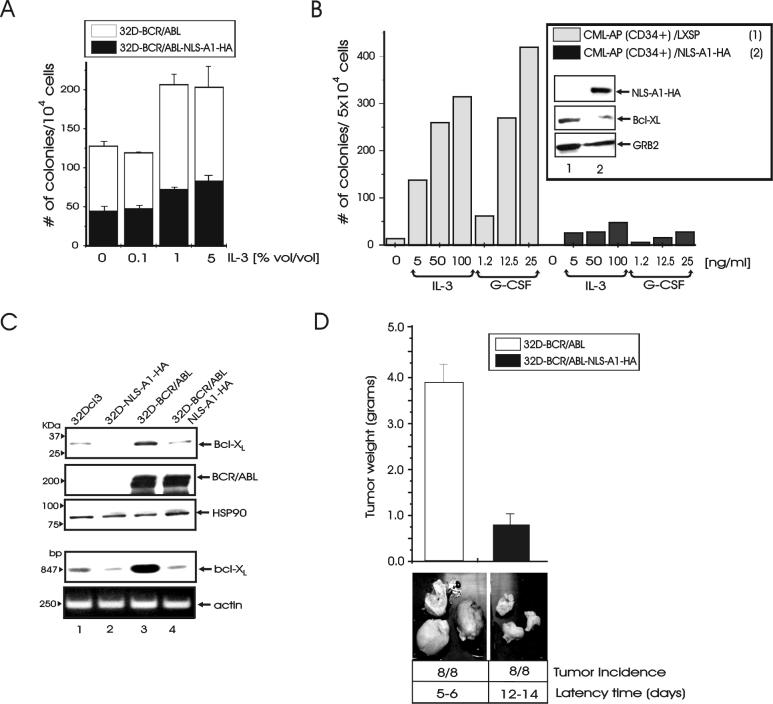
In IL-3-containing medium, proliferation of 32D-BCR/ABL-NLS-A1-HA cells was undistinguishable from that of 32D-BCR/ABL-WT-A1-HA or 32D-BCR/ABL cells (not shown). As expected, BCR/ABL- and BCR/ABL-WT-A1-HA-expressing 32Dcl3 cells were resistant to apoptosis induced by IL-3 deprivation. To determine whether expression of the shuttling-deficient hnRNP A1 mutant affects the phenotype of BCR/ABL-transformed cells, we assessed the effect of NLS-A1-HA on the colony-forming ability of BCR/ABL-expressing murine myeloid progenitor 32Dcl3 cells and primary CD34+ CML-AP (CML-APCD34+) cells. Thus, parental and NLS-A1-HA-expressing 32Dcl3 cells were infected with the pSRαMSVtkneo-p210BCR/ABL and pSRαMSVtkneo retrovirus and plated in methylcellulose (104 cells/plate) in the presence of G418 (1 mg/ml). Similarly, CML-APCD34+ cells were infected with the LXSP-NLS-A1-HA or with the LXSP retrovirus and plated in methylcellulose (5 × 104 cells/plate) in the presence of puromycin (1.25 μg/ml) as selectable marker. 32D-BCR/ABL cells formed a high number of colonies either in the absence or in the presence of increasing concentrations of IL-3-containing medium (Fig. 6A). By contrast, the colony-forming ability of freshly established 32D-BCR/ABL-NLS-A1-HA cells was markedly suppressed (≈60 to 65% inhibition) at each concentration of IL-3 in the semisolid culture (Fig. 6A). Likewise, the clonogenic efficiency of CML-APCD34+ cells was also dramatically reduced by expression of the NLS-A1-HA (≈85 to 95% inhibition), and the effect was essentially independent of the concentration of IL-3 or G-CSF in the semisolid medium (Fig. 6B). The reduced clonogenic efficiency of the NLS-A1-HA-expressing 32D-BCR/ABL and CML-APCD34+ cells was not due to reduced levels of BCR/ABL (Fig. 6C and 1B) but correlated with decreased expression of the antiapoptotic and BCR/ABL downstream effector Bcl-XL (Fig. 6C and inset of 6B).
FIG. 6.
Requirement of hnRNP A1 shuttling activity for colony formation and tumorigenesis of BCR/ABL-transformed cells. (A) Methylcellulose colony formation, in the absence or in the presence of different concentration of WEHI-3B conditioned medium used as a source of IL-3, from 32D-BCR/ABL and 32D-BCR/ABL-NLS-A1-HA cells (104 cells/plate). Values are means and standard deviations for duplicate cultures from two independent experiments. (B) Clonogenic efficiency in the absence of growth factors or in the presence of increasing concentrations of recombinant human IL-3 or G-CSF of primary CML-APCD34+ cells transduced with the empty LXSP or with the NLS-A1-HA retrovirus. After infection, cells (5 × 104 cells/plate) were plated in semisolid medium in the presence of 1.25 μg of puromycin per ml. Inset, Western blots show expression of NLS-A1-HA, Bcl-XL, and GRB2 in vector- and NLS-A1-HA-transduced CML-APCD34+ cells. (C) Expression of Bcl-XL protein (first panel) and mRNA (fourth panel) in 32Dcl3, 32D-NLS-A1-HA, 32D-BCR/ABL, and 32D-BCR/ABL-NLS-A1-HA cells. Levels of p210 BCR/ABL, HSP90, and actin were monitored as controls. Bcl-XL cytoplasmic mRNA levels were detected by RT-PCR (see Materials and Methods). (D) Subcutaneous tumors in SCID mice injected with 32D-BCR/ABL and 32D-NLS-A1-HA cells. The latency time (days) and tumor weight (means and standard deviations) were calculated; P < 0.01. The results are representative of those from two independent experiments.
To determine whether the shuttling activity of hnRNP A1 has a role in BCR/ABL-induced tumorigenesis, SCID mice (eight per group) were injected subcutaneously with 32Dcl3 cells expressing p210BCR/ABL alone or coexpressing p210BCR/ABL and the NLS-A1-HA mutant. 32D-BCR/ABL and 32D-BCR/ABL-NLS-A1-HA cells formed tumors in 5 to 6 and 12 to 14 days, respectively; at 20 days postinjection, tumors formed from 32D-BCR/ABL cells expressing the nuclear shuttling-deficient hnRNP A1 mutant showed an ≈80% decrease in weight compared to those formed from 32D-BCR/ABL cells (Fig. 6D).
DISCUSSION
We recently showed that FUS degradation by the 26S proteasome requires the formation of a multiprotein complex containing ubiquitinated hnRNP A1, which undergoes proteasome degradation in IL-3-deprived myeloid precursor cells (46). Since FUS proteolysis is suppressed by expression of the p210BCR/ABL oncoprotein (46), we asked whether hnRNP A1 levels are also regulated by BCR/ABL and whether interference with its nucleocytoplasmic shuttling function has an effect on the phenotypes of normal and BCR/ABL-transformed myeloid precursor cells. We show here that hnRNP A1 levels are more abundant in growth factor-independent 32D-BCR/ABL cells and in primary marrow cells from CML-BC patients than in parental 32Dcl3 and CML-CP cells. Moreover, treatment with the ABL tyrosine kinase inhibitor STI571 markedly reduced hnRNP A1 expression. Since enhanced hnRNP A1 expression correlates with high levels of BCR/ABL, which are more abundant during transition to blast crisis (18, 19), it is conceivable that upregulation of hnRNP A1 expression might contribute to the more aggressive phenotype of CML-BC marrow cells. Of note is that in primary CML cells, hnRNP A1 and BCR/ABL expression not only increased during disease progression but also were correlated with the percentage of blasts and resistance to STI571 treatment (20).
Mechanistically, the BCR/ABL-induced upregulation of hnRNP A1 expression reflects enhanced stability due to suppression of proteasome-dependent degradation. This is not unprecedented, since the deregulated kinase activity of BCR/ABL is required for transducing signals which regulate proteasome-dependent degradation of target proteins (13, 15, 46).
Preliminary evidence indicates that proteasome-mediated degradation of hnRNP A1, like that of FUS (46), was enhanced by c-Jun overexpression (not shown). Moreover, phosphomimetic mutation of hnRNP A1 serine 199 suppressed the degradation-promoting effect of c-Jun (not shown), suggesting that phosphorylation of hnRNP A1 on Ser 199 might prevent its proteasome-mediated degradation. The nucleocytoplasmic shuttling and RNA binding activities of hnRNP A1 are activated by the phosphatidylinositol 3-kinase- and BCR/ABL-regulated PKCζ (35, 41), which directly phosphorylates hnRNP A1 on serine 199 (40). Thus, BCR/ABL induction of PKCζ-dependent phosphorylation of hnRNP A1 may simultaneously suppress hnRNP A1 degradation and promote hnRNP A1-dependent nuclear export of mRNAs possibly required for BCR/ABL leukemogenic activity. It should be also noted that c-Jun is overexpressed in BCR/ABL-transformed cells and required for BCR/ABL-dependent leukemogenesis (51). Since c-Jun overexpression does not promote degradation of the S199D hnRNP A1 mutant (not shown), it seems likely that BCR/ABL-dependent phosphorylation of hnRNP A1 at serine 199 counteracts the degradation-promoting effects that c-Jun overexpression may have on hnRNP A1.
Despite extensive information on the function of hnRNP A1 in the control of pre-mRNA splicing (31), much less is known about the biological significance of hnRNP A1-dependent regulation of mRNA nucleocytoplasmic trafficking. Since nuclear export of hnRNP A1, and of the hnRNP A1-associated mRNA molecules, depends on the integrity of its M9 domain and on ongoing RNA polymerase II transcription (38, 55), we generated a nucleus-localized and shuttling-deficient hnRNP A1 mutant (NLS-A1-HA) harboring the G274A mutation in the M9 domain (38) and assessed its effect in normal and BCR/ABL-transformed 32Dcl3 myeloid precursor cells. In taking such an approach, we reasoned that expression of NLS-A1-HA would interfere with the nucleocytoplasmic shuttling activity of wild-type hnRNP A1. In this regard, microinjection of the G274A hnRNP A1 mutant into the nuclei of X. laevis oocytes specifically suppressed the nuclear export of radioactively labeled intronless mRNAs, most probably by saturating factors required for mRNA export (27). Likewise, mutational inactivation of the yeast Np13p, a functional homologue of hnRNP A1, also impaired the process of mRNA export (33). In our studies, expression of NLS-A1-HA was associated with inhibition of cytoplasmic localization of hnRNP A1-associated FUS (60), a protein that does not bear known nuclear import or export signals (61, 62), and decreased cytoplasmic levels of several mRNAs (not shown). Thus, it is likely that the NLS-A1-HA inhibits hnRNP A1-regulated mRNA trafficking also in hematopoietic cells.
In 32Dcl3 cells, expression of the NLS-A1-HA mutant markedly enhanced the susceptibility to apoptosis induced by IL-3-deprivation, reduced IL-3-dependent colony formation and suppressed G-CSF-stimulated granulocytic differentiation by promoting rapid cell death. Likewise, expression of NLS-A1-HA reduced the ability of primary mouse marrow cells to form IL-3- and G-CSF-derived colonies. Overexpression of wild-type hnRNP A1 in NLS-A1-HA-expressing 32Dcl3 cells decreased their susceptibility to apoptosis induced by IL-3 deprivation and restored G-CSF-stimulated granulocytic differentiation, strongly suggesting that the deleterious effects of NLS-A1-HA expression on myelopoiesis are indeed the consequence of impaired hnRNP A1 function.
Expression of the NLS-A1-HA mutant in BCR/ABL-transformed 32Dcl3 cells and primary CD34+ cells from a CML-AP patient reduced the methylcellulose colony-forming ability of both and impaired the leukemia-inducing effects of BCR/ABL-expressing 32Dcl3 cells, suggesting that enhanced hnRNP A1 shuttling activity favors BCR/ABL leukemogenesis. In a previous study (45) we showed that downregulation of the shuttling hnRNP FUS also correlated both in vitro and in vivo with reduced BCR/ABL leukemogenic potential. Since hnRNP A1 overexpression promotes FUS degradation in 293T cells (46) and might be required for FUS downmodulation during IL-3 starvation or G-CSF-induced differentiation of murine myeloid progenitor cells, we investigated FUS levels in wild-type and mutant hnRNP A1-expressing cells. In IL-3-cultured parental cells (Fig. 5) and BCR/ABL-expressing 32Dcl3 cells (not shown), FUS levels were apparently not affected by expression of wild-type or NLS-A1-HA hnRNP A1. This suggests that hnRNP A1 and FUS function independently in regulating survival and differentiation.
The effects of overexpressing the shuttling-deficient NLS-A1-HA mutant in parental and in BCR/ABL-expressing 32Dcl3 cells were markedly different from those of overexpressing wild-type hnRNP A1, which had no effect on BCR/ABL cells and accelerated differentiation of parental 32Dcl3 cells. Thus, the phenotype induced by ectopic expression of NLS-A1-HA most likely reflects the dominant negative effect of this mutant on hnRNP A1-mediated mRNA export and not the saturation of factors required for either hnRNP A1-dependent or -independent mRNA export. However, we cannot exclude the possibility that expression of the NLS-A1-HA mutant can interfere with the other nuclear functions of hnRNP A1.
In parental 32Dcl3 cells, expression of the NLS-A1-HA hnRNP A1 mutant was associated with a decrease in the cytosolic mRNA and protein levels of C/EBPα, the major regulator of granulocytic differentiation (50, 59), and Bcl-XL (6), a potent apoptosis suppressor in hematopoietic cells (1, 17, 25). Downregulation of the survival factor Bcl-XL was also noted in 32D-BCR/ABL cells and in primary CML-AP cells expressing the NLS-A1-HA hnRNP A1 mutant. Indeed, downregulation of C/EBPα and BCL-XL expression may account for the altered phenotype of NLS-A1-HA-expressing cells.
C/EBPα is required for granulocytic differentiation (59) most likely because it activates the transcription of many differentiation-related genes, including that encoding the G-CSFR (56, 58). Indeed, G-CSFR levels were downmodulated in NLS-A1-HA-expressing 32Dcl3 cells, suggesting that reduced levels of G-CSF-dependent signals might cause impaired differentiation and massive apoptosis of G-CSF-treated NLS-A1-HA-expressing cells. Consistent with this hypothesis, expression of C/EBPα in NLS-A1-HA-expressing 32Dcl3 cells restored G-CSF-induced granulocytic differentiation.
Since hnRNP A1 binds intronless pre-mRNAs (27, 30) and c/ebpα pre-mRNA does not contain introns (32), it is conceivable that hnRNP A1 may negatively control the export of c/ebpα mRNA. Alternatively, the effect of NLS-A1-HA on c/ebpα mRNA expression may not be direct but rather may be mediated by other factors influencing c/ebpα transcription, mRNA stability, or mRNA export. For example, in exponentially growing 32Dcl3 cells, overexpression of degradation-resistant S256D FUS, but not of degradation-prone S256A FUS, leads to downregulation of C/EBPα (not shown). Suppression of C/EBPα expression by the constitutively active S256D FUS mutant might depend on the increased affinity of S256D FUS for hnRNP A1 (unpublished observation); thus, formation of this complex may inhibit hnRNP A1 activity, causing nuclear retention of C/EBPα mRNA with a consequent decrease in the levels of translatable cytoplasmic C/EBPα mRNA. Expression of the NLS-A1-HA hnRNP A1 mutant markedly downregulates Bcl-XL expression in parental and BCR/ABL-expressing cells and in primary cells from a CML-AP patient. Consistent with the importance of Bcl-XL for the survival of growth factor-dependent normal and BCR/ABL-transformed hematopoietic cells (1, 6, 17, 25), the increased propensity to apoptosis and the diminished leukemogenic potential of NLS-A1-HA-expressing normal and BCR/ABL-transformed cells, respectively, might depend on the downregulation of the antiapoptotic Bcl-XL. Suppression of Bcl-XL mRNA expression by the mutant hnRNP A1 might be the direct consequence of reduced Bcl-XL mRNA export or may depend on altered expression or function of factors, e.g., STAT-5 (25), that regulate its transcription.
In conclusion, we have provided evidence for a novel function of hnRNP A1 as a regulator of normal hematopoiesis and BCR/ABL leukemogenesis. The role of hnRNP A1 in hematopoiesis is probably dependent on the effects on nucleocytoplasmic trafficking of mRNA molecules that encode factors (e.g., Bcl-XL and C/EBPα) essential for survival and differentiation and are abnormally regulated upon BCR/ABL-dependent transformation of myeloid progenitors.
Acknowledgments
A.I. and G.S. contributed equally to this work.
We thank G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia) for hnRNP A1 cDNA and antibody, N. Flomenberg (Bone Marrow Transplant Unit, Thomas Jefferson University, Philadelphia, Pa.) for providing samples of CML-AP cells, H. Radomska (Harvard Institute of Medicine, Boston, Mass.) for helpful scientific discussion, and Cathy Franzeo for editorial assistance in preparation of the manuscript.
R. Trotta is supported by NIH training grant T32-CA09662. G. Santilli and C. Guerzoni were supported in part by a fellowship from the A. Serra Foundation for Cancer Research and Therapy. C. Gambacorti-Passerini is supported in part by the Italian Association for Cancer Research (AIRC) and by a grant from the Ministero della Sanità, Italy. This work was supported in part by NIH grants to B. Calabretta.
REFERENCES
- 1.Amarante-Mendes, G. P., A. J. McGahon, W. K. Nishioka, D. E. Afar, O. N. Witte, and D. R. Green. 1998. Bcl-2-independent Bcr-Abl-mediated resistance to apoptosis: protection is correlated with up regulation of Bcl-xL. Oncogene 16:1383-1390. [DOI] [PubMed] [Google Scholar]
- 2.Bellon, T., D. Perrotti, and B. Calabretta. 1997. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood 90:1828-1839. [PubMed] [Google Scholar]
- 3.Biamonti, G., M. T. Bassi, L. Cartegni, F. Mechta, M. Buvoli, F. Cobianchi, and S. Riva. 1993. Human hnRNP protein A1 gene expression. Structural and functional characterization of the promoter. J. Mol. Biol. 230:77-89. [DOI] [PubMed] [Google Scholar]
- 4.Biamonti, G., M. Buvoli, M. T. Bassi, C. Morandi, F. Cobianchi, and S. Riva. 1989. Isolation of an active gene encoding human hnRNP protein A1. Evidence for alternative splicing. J. Mol. Biol. 207:491-503. [DOI] [PubMed] [Google Scholar]
- 5.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boise, L. H., M. Gonzalez-Garcia, C. E. Postema, L. Ding, T. Lindsten, L. A. Turka, X. Mao, G. Nunez, and C. B. Thompson. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597-608. [DOI] [PubMed] [Google Scholar]
- 7.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buvoli, M., F. Cobianchi, and S. Riva. 1992. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 20:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartegni, L., M. Maconi, E. Morandi, F. Cobianchi, S. Riva, and G. Biamonti. 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259:337-348. [DOI] [PubMed] [Google Scholar]
- 10.Chabot, B., M. Blanchette, I. Lapierre, and H. La Branche. 1997. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 17:1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobianchi, F., C. Calvio, M. Stoppini, M. Buvoli, and S. Riva. 1993. Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res. 21:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 2000. Connections between the processing and nuclear export of mRNA: evidence for an export license? Proc. Natl. Acad. Sci. USA 97:4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai, Z., R. C. Quackenbush, K. D. Courtney, M. Grove, D. Cortez, G. W. Reuther, and A. M. Pendergast. 1998. Oncogenic Abl and Src tyrosine kinases elicit the ubiquitin-dependent degradation of target proteins through a Ras-independent pathway. Genes Dev. 12:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deutsch, E., A. Dugray, B. AbdulKarim, E. Marangoni, L. Maggiorella, S. Vaganay, R. M'Kacher, S. Douc Rasy, F. Eschwege, W. Vainchenker, A. G. Turhan, and J. Bourhis. 2001. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood 97:2084-2090. [DOI] [PubMed] [Google Scholar]
- 16.Dreyfuss, G., M. J. Matunis, S. Pinol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 17.Dumon, S., S. C. Santos, F. Debierre-Grockiego, V. Gouilleux-Gruart, L. Cocault, C. Boucheron, P. Mollat, S. Gisselbrecht, and F. Gouilleux. 1999. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene 18:4191-4199. [DOI] [PubMed] [Google Scholar]
- 18.Elmaagacli, A. H., D. W. Beelen, B. Opalka, S. Seeber, and U. W. Schaefer. 2000. The amount of BCR-ABL fusion transcripts detected by the real-time quantitative polymerase chain reaction method in patients with Philadelphia chromosome positive chronic myeloid leukemia correlates with the disease stage. Ann. Hematol. 79:424-431. [DOI] [PubMed] [Google Scholar]
- 19.Gaiger, A., T. Henn, E. Horth, K. Geissler, G. Mitterbauer, T. Maier-Dobersberger, H. Greinix, C. Mannhalter, O. A. Haas, K. Lechner, et al. 1995. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood 86:2371-2378. [PubMed] [Google Scholar]
- 20.Gambacorti-Passerini, C., R. Barni, E. Marchesi, M. Verga, M. Rossi, F. Rossi, P. Pioltelli, E. Pogliani, and M. Corneo. 2001. Sensitivity of the abl inhibitor STI571 in fresh leukaemic cells obtained from chronic myelogenous leukaemia patients in different stages of the disease. Br. J. Haematol. 112:972-974. [DOI] [PubMed] [Google Scholar]
- 21.Ghaffari, S., G. Q. Daley, and H. F. Lodish. 1999. Growth factor independence and BCR/ABL transformation: promise and pitfalls of murine model systems and assays. Leukemia 13:1200-1206. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, M. Y. 1999. Biological consequences of the BCR/ABL fusion gene in humans and mice. J. Clin. Pathol. 52:719-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton, B. J., E. Nagy, J. S. Malter, B. A. Arrick, and W. F. Rigby. 1993. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 268:8881-8887. [PubMed] [Google Scholar]
- 24.Henics, T., A. Sanfridson, B. J. Hamilton, E. Nagy, and W. F. Rigby. 1994. Enhanced stability of interleukin-2 mRNA in MLA 144 cells. Possible role of cytoplasmic AU-rich sequence-binding proteins. J. Biol. Chem. 269:5377-5383. [PubMed] [Google Scholar]
- 25.Horita, M., E. J. Andreu, A. Benito, C. Arbona, C. Sanz, I. Benet, F. Prosper, and J. L. Fernandez-Luna. 2000. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of Bcl-xL. J. Exp. Med 191:977-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Idriss, H., A. Kumar, J. R. Casas-Finet, H. Guo, Z. Damuni, and S. H. Wilson. 1994. Regulation of in vitro nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein protein A1 by reversible phosphorylation. Biochemistry 33:11382-11390. [DOI] [PubMed] [Google Scholar]
- 27.Izaurralde, E., A. Jarmolowski, C. Beisel, I. W. Mattaj, G. Dreyfuss, and U. Fischer. 1997. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J. Cell Biol. 137:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, Z. H., W. J. Zhang, Y. Rao, and J. Y. Wu. 1998. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. USA 95:9155-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kantarjian, H. M., A. Deisseroth, R. Kurzrock, Z. Estrov, and M. Talpaz. 1993. Chronic myelogenous leukemia: a concise update. Blood 82:691-703. [PubMed] [Google Scholar]
- 30.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673-682. [DOI] [PubMed] [Google Scholar]
- 31.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 32.Landschulz, W. H., P. F. Johnson, E. Y. Adashi, B. J. Graves, and S. L. McKnight. 1988. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 2:786-800. [DOI] [PubMed] [Google Scholar]
- 33.Lee, M. S., M. Henry, and P. A. Silver. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 10:1233-1246. [DOI] [PubMed] [Google Scholar]
- 34.Luscher, B., and R. N. Eisenman. 1988. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol. Cell. Biol. 8:2504-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majewski, M., M. Nieborowska-Skorska, P. Salomoni, A. Slupianek, K. Reiss, R. Trotta, B. Calabretta, and T. Skorski. 1999. Activation of mitochondrial Raf-1 is involved in the antiapoptotic effects of Akt. Cancer Res. 59:2815-2819. [PubMed] [Google Scholar]
- 36.Matter, N., M. Marx, S. Weg-Remers, H. Ponta, P. Herrlich, and H. Konig. 2000. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 275:35353-35360. [DOI] [PubMed] [Google Scholar]
- 37.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 38.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 39.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Municio, M. M., J. Lozano, P. Sanchez, J. Moscat, and M. T. Diaz-Meco. 1995. Identification of heterogeneous ribonucleoprotein A1 as a novel substrate for protein kinase C zeta. J. Biol. Chem. 270:15884-15891. [DOI] [PubMed] [Google Scholar]
- 41.Nakanishi, H., K. A. Brewer, and J. H. Exton. 1993. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 268:13-16. [PubMed] [Google Scholar]
- 42.Nakielny, S., and G. Dreyfuss. 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 134:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osarogiagbon, U. R., and P. B. McGlave. 1999. Chronic myelogenous leukemia. Curr. Opin. Hematol. 6:241-246. [DOI] [PubMed] [Google Scholar]
- 44.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrotti, D., S. Bonatti, R. Trotta, R. Martinez, T. Skorski, P. Salomoni, E. Grassilli, R. V. Lozzo, D. R. Cooper, and B. Calabretta. 1998. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 17:4442-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrotti, D., A. Iervolino, V. Cesi, M. Cirinna, S. Lombardini, E. Grassilli, S. Bonatti, P. P. Claudio, and B. Calabretta. 2000. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CbetaII-dependent pathway. Mol. Cell. Biol. 20:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 48.Pinol-Roma, S., and G. Dreyfuss. 1991. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science 253:312-314. [DOI] [PubMed] [Google Scholar]
- 49.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 50.Radomska, H. S., C. S. Huettner, P. Zhang, T. Cheng, D. T. Scadden, and D. G. Tenen. 1998. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18:4301-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raitano, A. B., J. R. Halpern, T. M. Hambuch, and C. L. Sawyers. 1995. The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc. Natl. Acad. Sci. USA 92:11746-11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ron, D. 1997. TLS-CHOP and the role of RNA-binding proteins in oncogenic transformation. Curr. Top. Microbiol. Immunol. 220:131-142. [DOI] [PubMed] [Google Scholar]
- 53.Sawyers, C. L. 1999. Chronic myeloid leukemia. N. Engl. J. Med. 340:1330-1340. [DOI] [PubMed] [Google Scholar]
- 54.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 55.Siomi, M. C., P. S. Eder, N. Kataoka, L. Wan, Q. Liu, and G. Dreyfuss. 1997. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138:1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, L. T., S. Hohaus, D. A. Gonzalez, S. E. Dziennis, and D. G. Tenen. 1996. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood 88:1234-1247. [PubMed] [Google Scholar]
- 57.Svitkin, Y. V., L. P. Ovchinnikov, G. Dreyfuss, and N. Sonenberg. 1996. General RNA binding proteins render translation cap dependent. EMBO J. 15:7147-7155. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, X., E. Scott, C. L. Sawyers, and A. D. Friedman. 1999. C/EBPalpha bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 59.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zinszner, H., R. Albalat, and D. Ron. 1994. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 8:2513-2526. [DOI] [PubMed] [Google Scholar]
- 61.Zinszner, H., D. Immanuel, Y. Yin, F. X. Liang, and D. Ron. 1997. A topogenic role for the oncogenic N-terminus of TLS: nucleolar localization when transcription is inhibited. Oncogene 14:451-461. [DOI] [PubMed] [Google Scholar]
- 62.Zinszner, H., J. Sok, D. Immanuel, Y. Yin, and D. Ron. 1997. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci. 110:1741-1750. [DOI] [PubMed] [Google Scholar]