Abstract
CENP-A is a component of centromeric chromatin and defines active centromere regions by forming centromere-specific nucleosomes. We have isolated centromeric chromatin containing the CENP-A nucleosome, CENP-B, and CENP-C from HeLa cells using anti-CENP-A and/or anti-CENP-C antibodies and shown that the CENP-A/B/C complex is predominantly formed on α-satellite DNA that contains the CENP-B box (αI-type array). Mapping of hypersensitive sites for micrococcal nuclease (MNase) digestion indicated that CENP-A nucleosomes were phased on the αI-type array as a result of interactions between CENP-B and CENP-B boxes, implying a repetitive configuration for the CENP-B/CENP-A nucleosome complex. Molecular mass analysis by glycerol gradient sedimentation showed that MNase digestion released a CENP-A/B/C chromatin complex of three to four nucleosomes into the soluble fraction, suggesting that CENP-C is a component of the repetitive CENP-B/CENP-A nucleosome complex. Quantitative analysis by immunodepletion of CENP-A nucleosomes showed that most of the CENP-C and approximately half the CENP-B took part in formation of the CENP-A/B/C chromatin complex. A kinetic study of the solubilization of CENPs showed that MNase digestion first released the CENP-A/B/C chromatin complex into the soluble fraction, and later removed CENP-B and CENP-C from the complex. This result suggests that CENP-A nucleosomes form a complex with CENP-B and CENP-C through interaction with DNA. On the basis of these results, we propose that the CENP-A/B/C chromatin complex is selectively formed on the I-type α-satellite array and constitutes the prekinetochore in HeLa cells.
The centromere of higher eukaryotes was first defined as the primary constriction on mitotic chromosomes (39, 40) which is essential for faithful chromosome segregation during mitosis and meiosis (37). In vertebrate cells, the kinetochore, a three-layered disk-shaped structure composed of a dense inner plate, a lucent middle domain, and a dense outer plate, is formed at the centromere at mitosis and is the attachment site for spindle microtubules (5). Centromeric proteins known to date include constitutive proteins, such as CENP-A, -B, -C, and -H (37, 48), that are present at the centromere throughout the cell cycle, and transient proteins that appear after the onset of M-phase, such as CENP-E and -F, INCENP, Mad1, Mad2, Bub1, Bub2, and BubR1 (for a review, see reference 12). The constitutive proteins are detected as speckles (prekinetochores) in S-phase nuclei (55), and at least one of these proteins has been located in the inner kinetochore plate at M-phase (43).
The CENP-B gene codes for an 80-kDa protein (13) that binds to the 17-bp DNA motif known as the CENP-B box, which is present in human α-satellite and mouse minor satellite DNA (33), and causes nucleosome positioning around the CENP-B box region (62). The CENP-B gene seems to be nonessential, since knockout mice were viable without any apparent defect in growth and morphology (24, 30). Some functional homologues of CENP-B may remain to be found in mammals, since three different CENP-B homologues have been found in Schizosaccharomyces pombe (3).
The CENP-C gene is essential for chromosome segregation (17, 29), and its gene product (a 140-kDa protein) is a DNA-binding protein without apparent sequence specificity (49, 59). CENP-C has been detected at the inner kinetochore plate by electron microscopy (43), while CENP-B was reported to be located in the pairing domain (11). The CENP-A gene codes for a histone H3 variant: the C-terminal two-thirds of CENP-A is highly homologous to histone H3, but the remaining amino-terminal third is unique (36, 50). The histone fold domain located in the C-terminal region is essential for targeting CENP-A to the centromeric region (45, 50). The mouse CENP-A gene was shown to be essential by gene targeting (23).
In Saccharomyces cerevisiae, the CENP-A homologue, CSE4, has been isolated and is also essential for chromosome segregation (47). By mutational analysis, the functional domains of Cse4p were shown to be distributed across the histone fold domain, which is necessary for interaction with histone H4, and also in the amino-terminal 33 peptides (7, 18, 31). CENP-A homologues have been identified in Caenorhabditis elegans, Drosophila melanogaster, and S. pombe and are also essential for chromosome segregation (6, 21, 51).
In S. cerevisiae, the centromere is genetically defined within a 125-bp DNA region known as CEN (15, 16), and more than eight centromere-associated proteins, including Cse4p, are targeted to this cis-acting DNA region to form a higher-order chromatin complex (14, 25, 34) which is resistant to nuclease attack (4). In S. pombe, a much longer DNA sequence (40 to 120 kb), consisting of a 4- to 7-kb unique sequence (cc region) flanked by tens of kilobases of inverted repeat sequence, forms the centromere region (8, 19). It is speculated that some specific higher-order chromatin structure may be formed in the cc region, since in this region the nucleosome ladders produced by micrococcal nuclease (MNase) cleavage become smears when active centromeres are formed.
The centromeres of higher eukaryotes contain hundreds to thousands of kilobases of tandemly repeated DNA sequences, although their sequences and unit lengths are species specific (56). In humans, α-satellite (alphoid) arrays varying from 500 kb to 5 Mb are found at the centromeres (57). α-Satellite arrays consist of 171-bp monomer repeats that show chromosome-specific variation in sequence and higher-order repeat arrangement (1, 26, 28, 53). However, on rare occasions, stable neocentromeres have been found in euchromatic regions that lack repetitive DNA sequences, such as α-satellite (2, 44). Epigenetic establishment of centromeres has been reported in humans, D. melanogaster, and S. pombe (2, 44, 46, 58), and the newly formed centromeres in human cells contain all the centromere proteins studied except CENP-B (42). These results emphasize that the formation of DNA-protein complexes, especially formation of CENP-A nucleosomes, is critically important for formation of active centromeres.
Nucleosomes containing CENP-A may be what distinguish centromeric chromatin from euchromatin or noncentromeric heterochromatin, and may promote the formation of functional kinetochores. We have previously shown that CENP-A does indeed replace histone H3 in a nucleosomal reconstitution system in vitro and that the basic structure of the CENP-A nucleosome is the same as that of normal nucleosomes: an octamer of core histones (H3 or CENP-A, H4, H2A, and H2B) with DNA wrapped around it (63).
In the present work we have isolated the centromeric chromatin complexes from HeLa cells by chromatin immunoprecipitation (CHIP) using anti-CENP-A and/or anti-CENP-C antibodies. We have shown that CENP-A nucleosomes are formed predominantly on α-satellite DNA that contains CENP-B boxes (αI-type array) and that CENP-B and -C bind to the nucleosomal DNA to form the CENP-A/B/C prekinetochore chromatin complex. We therefore propose that centromeric chromatin complexes composed of CENP-A nucleosomes, CENP-B, and CENP-C constitute the prekinetochore complexes in HeLa cells.
MATERIALS AND METHODS
Cell growth and isolation of nuclei.
HeLa S3 cells were grown in spinner flasks to a concentration of 5 × 105 cells/ml in RPMI 1640 medium (Nissui Seiyaku, Tokyo, Japan) containing 5% calf serum. The nuclei were isolated as described previously (60).
ACA serum.
Anticentromere antibody (ACA)-positive serum (MI or AK) from scleroderma patients contained antibodies against the three major centromere autoantigens, CENP-A, CENP-B, and CENP-C (35, 60).
Preparation of monoclonal antibodies against CENP-A peptides and the amino-terminal half of CENP-B.
Monoclonal antibodies were prepared essentially as described previously (10). Human CENP-A peptides (amino acids 3 to 19 from the amino terminus) covalently linked to keyhole limpet hemocyanin (Pierce) were used as the CENP-A antigen. The amino-terminal half of CENP-B expressed in Escherichia coli cells and isolated as inclusion bodies was used as the CENP-B antigen. Mouse myeloma cells (P3-X63-Ag8.653) were used.
Preparation of anti-CENP-C antibodies.
The DNA sequence encoding the amino-terminal half of human CENP-C (amino acids 1 to 426) was amplified by PCR with a cDNA library of HeLa cells, and cloned into pFastBac HTa vector (Gibco-BRL) after digestion with NcoI and EcoRI. The primers used were 5′GGCCGGACCATGGCTGCGTCCGGTCTGGA and 5′ATAGAATCCTCTTCAGCTGGTTTAGCCAT. The isolated clones were sequenced, and a clone without amino acid replacement was selected. The amino-terminal half of CENP-C was expressed in SF9 cells using the Bac-to-Bac baculovirus expression system (Gibco-BRL) and purified on an Ni column. The polyclonal guinea pig antibodies against CENP-C were affinity purified with the same protein coupled to CNBr-activated Sepharose 4B beads (Amersham Pharmacia Biotech).
MNase digestion of HeLa nuclei.
The nuclear pellet (109 nuclei) was dissolved in 5 ml of ice-cold WB (20 mM HEPES [pH 8.0], 20 mM KCl, 0.5 mM EDTA, 0.5 mM dithiothreitol [DTT], 0.05 mM phenylmethylsulfonyl fluoride) containing 0.3 M NaCl. The nuclear suspension was digested with MNase (Roche Diagnostics) at 37°C after addition of CaCl2 to a final concentration of 2 mM. The reaction was stopped by the addition of EGTA to a final concentration of 5 mM with quick chilling. The digests were centrifuged with an R20A2 rotor (Hitachi) at 10,000 × g for 10 min at 4°C. The solubilized chromatin was recovered in the supernatant. The pellet was resuspended in 5 ml of the same buffer.
Immunoprecipitation.
Anti-CENP-A immunoglobulin G (IgG) (αCA) secreted by the monoclonal hybridoma cells and the IgG in ACA serum (MI) were purified on protein G-Sepharose columns (Pharmacia). Guinea pig anti-CENP-C IgG (gpαCC) was affinity purified on a CENP-C-linked Sepharose 4B column. The antibodies were eluted with 0.1 M glycine-HCl (pH 3.0). Each of these IgGs (≈10 mg each) was coupled to CNBr-activated Sepharose 4B (1 g of dry powder; Pharmacia). These IgG-linked Sepharose beads (50 μl) were added to the solubilized chromatin samples (1 ml) supplemented with 0.1% NP-40 and incubated for 6 to 12 h at 4°C with mild rotation. For CHIP with anti-CENP-B antibodies, anti-CENP-B IgG and protein G-Sepharose were added to the sample and incubated for several hours at 4°C with rotation. After incubation, the beads were washed three times with WB containing 0.3 M NaCl and 0.1% NP-40 and resuspended in 50 to 100 μl of SDS buffer (50 mM Tris-HCl [pH 8.0], 25 mM DTT, 1% sodium dodecyl sulfate [SDS], 15% glycerol). Proteins were eluted by incubation at 95°C for 5 min. The eluted proteins were quickly chilled with liquid nitrogen and stored at −80°C until needed.
SDS-PAGE and Western blotting.
The proteins were separated by SDS-12.5% or 7.5% polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membranes (Millipore) as previously described (52, 60). ACA serum (AK, 1:3,000 dilution) and goat anti-human IgG-horseradish peroxidase conjugate (Bio-Rad, 1:3,000 dilution) were used for immunodetection. All incubation was at 4°C overnight. Color development was carried out with Konica immunostain (Konica).
Glycerol gradient sedimentation.
A total of 3 ml of the solubilized chromatin sample was applied to 34 ml of a 5 to 20% glycerol gradient containing 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 0.1% NP-40, 2 mM DTT, and 0.15 M NaCl layered over 1 ml of 50% glycerol and centrifuged with an SW28 rotor (Beckman) at 22,000 rpm for 15.5 h at 4°C. Then 2-ml aliquots were fractionated from the bottom, and DNA and proteins were separated by 1% agarose gel electrophoresis and SDS-PAGE, respectively.
Cloning and sequencing of the DNA recovered by CHIP using anti-CENP-A, -B, or -C antibodies.
HeLa nuclei (2 × 108 nuclei/ml, 15 ml) were digested with 40 U of MNase (Boehringer) per ml for 10 min at 37°C. To remove short DNA fragments and free proteins, the soluble fraction of the digest was centrifuged through a 5 to 20% glycerol gradient, and fractions heavier than trinucleosomes were pooled and immunoprecipitated using anti-CENP-A, -B, or -C antibodies. After immunoprecipitation, the beads were treated with proteinase K, and the DNA was subsequently purified by phenol extraction. The DNA was cloned into 3′dT-tailed vector pCR2.1-TOPO using the TOPO TA cloning kit (Invitrogen) after treatment with Taq polymerase to add complementary 3′ A tails. The DNA sequences were determined using an ABI Prism 3700 DNA analyzer (Perkin Elmer Applied Biosystems) or an ABI Prism 377 DNA sequencer (Perkin Elmer Applied Biosystems). The sequences were compared to the database using the Blast server at the National Center for Biotechnology Information (NCBI).
Preparation of the A/B/C control mixture.
CENP-A, CENP-B, and CENP-C were expressed using the baculovirus Bac-to-Bac system (Gibco-BRL) and purified as follows. CENP-A and histone H4 with a 6x histidine tag at the amino terminus were coexpressed, and the CENP-A/his6-H4 complex was purified using an Ni column (63). CENP-B was purified with a Q-Sepharose column (60). The CENP-C gene encoding the full-size CENP-C protein was amplified by PCR with a cDNA library of HeLa cells (Clontech), and ligated to pFastBac vector. Full-size CENP-C was expressed in the baculovirus system and isolated as inclusion bodies; it was solubilized with 6 M urea and purified on a heparin-Sepharose column. The concentration of each protein was calculated from the band intensity after Coomassie brilliant blue (CBB) staining of SDS-PAGE gels, using bovine serum albumin (BSA) as a standard. To prevent loss during handling of the proteins, BSA and core histones were added.
PCR-based primer extension.
Primer extension procedures were performed as described previously (61, 62). Approximately 1 μg of HeLa DNA and the following primers, end labeled with [γ-32P]ATP, were used for each PCR primer extension reaction: CENP-B box l-primer, TCCCGTTTCCAACGAA; CENP-B box r-primer, TTTCGTTGGAAACGGG; mid-l primer, TTTTTATA/GC/GGAAGATA; and mid-r primer, TATCTTCC/GC/TATAAAAA. Methods for direct sequencing for position markers were as described previously (61). The samples were electrophoresed through 6% polyacrylamide-urea gels for 4 h or at 1,600 V. The gels were exposed to radiographic film (Kodak XAR5) with intensifying screens after drying.
RESULTS
Centromeric chromatin containing CENP-A is released into the soluble fraction by MNase digestion in the same way as bulk chromatin.
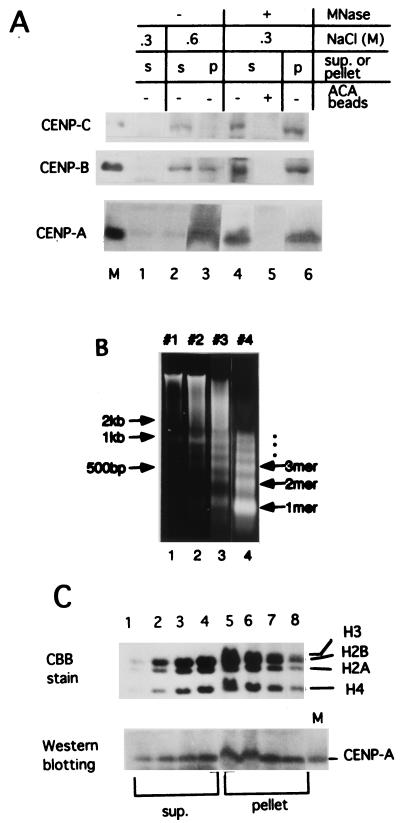
We examined the release of centromeric chromatin from HeLa nuclei into the soluble fraction by MNase digestion. Without MNase digestion, neither CENP-A, -B, nor -C was eluted into the soluble fraction by 0.3 M NaCl (Fig. 1A, lane 1); CENP-B and -C were eluted with 0.6 M NaCl (lane 2), while CENP-A was eluted with 2 M NaCl, like other core histones (data not shown). MNase digestion of the nuclei in 0.3 M NaCl released 30 to 50% of the centromeric proteins into the soluble fraction (lane 4, supernatant fraction; lane 6, pellet fraction). These bands were depleted by ACA serum, confirming that they were centromeric proteins (lane 5).
FIG. 1.
Bulk chromatin and centromeric chromatin were solubilized by MNase digestion of HeLa nuclei in 0.3 M NaCl. (A) Centromeric proteins CENP-A, -B, and -C were solubilized by MNase digestion. Isolated HeLa nuclei (2 × 108) were suspended with 1 ml of WB containing 0.3 M NaCl (sample a in lane 1 and sample c in lanes 4 to 6) or 0.6 M NaCl (sample b in lanes 2 and 3). Sample c was digested with 60 U of MNase per ml for 10 min at 37°C. Soluble and insoluble materials from each sample were separated by centrifugation. ACA beads were added to the supernatant of sample c and incubated overnight at 4°C. Pellets were resuspended in 1 ml of SDS buffer by extensive sonication and 5 μl of each sample was separated by SDS-7.5% (for CENP-B and CENP-C) or 12.5% (for CENP-A) PAGE, and centromeric proteins were detected by immunostaining with ACA serum. Lane 1, supernatant fraction of a; lane 2, supernatant fraction of b; lane 3, pellet fraction of b; lane 4, supernatant fraction of c before addition of ACA beads; lane 5, supernatant fraction of c after addition of ACA beads; lane 6, pellet fraction of c. Lane M, marker centromeric proteins, CENP-A, CENP-B, and CENP-C. (B) Size distribution of DNA fragments from bulk chromatin after MNase digestion. HeLa nuclei were digested with MNase to various extents. The fragmented DNA in the soluble fractions was extracted with phenol and electrophoresed through 1% agarose gel. DNA was detected with ethidium bromide staining. Lane 1, 20 U/ml for 2 min (40 U/ml × min, sample 1); lane 2, 20 U/ml for 4 min (80 U/ml × min, sample 2); lane 3, 40 U/ml for 5 min (200 U/ml × min, sample 3); lane 4, 80 U/ml for 45 min (3,600 U/ml × min, sample 4). Positions of the DNA size markers are indicated at the left. (C) Detection of core histones and CENP-A in each fraction. Soluble (sup.) and insoluble (pellet) fractions were subjected to SDS-12.5% PAGE, and the separated core histones were stained with Coomassie brilliant blue (upper panel). The proteins were transferred to a membrane and immunolabeled with ACA serum (AK) (lower panel). Lane M in the lower panel is a recombinant CENP-A marker protein. Lanes 1 to 4 correspond to samples 1 to 4 of the soluble (sup.) fractions, and lanes 5 to 8 to samples 1 to 4 of the pellet fractions.
The isolated HeLa nuclei were digested with MNase to various extents, and the size distribution of the nucleosomal DNA in the soluble fraction was analyzed (Fig. 1B). Core histones in the soluble and insoluble fraction of each digest were separated by SDS-PAGE and detected by CBB staining (Fig. 1C, upper panel), and CENP-A was detected by immunostaining with ACA serum after blotting onto a membrane (Fig. 1C, lower panel). The results show that centromeric chromatin containing CENP-A was released into the soluble fraction by MNase digestion in the same way as bulk chromatin.
CENP-A, -B, and -C are coprecipitated by CHIP using anti-CENP-A or anti-CENP-C antibody.
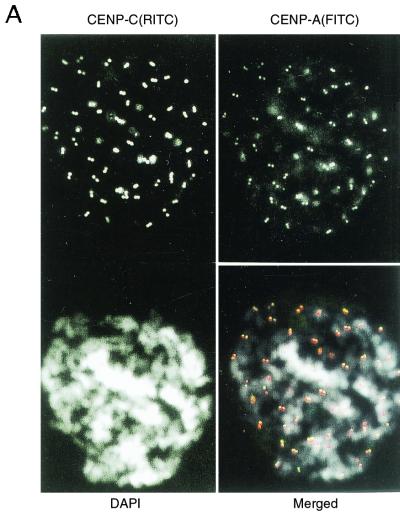
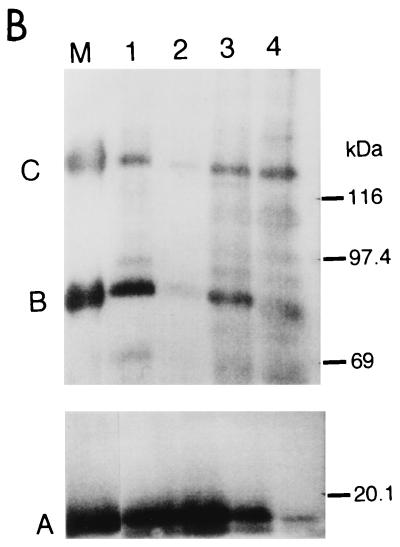
To investigate the structure and components of the centromeric chromatin that contains CENP-A nucleosomes by CHIP, we raised mouse monoclonal antibodies against amino-terminal polypeptides of human CENP-A and guinea pig antibody against human CENP-C, as described in Materials and Methods. These antibodies specifically recognize CENP-A and CENP-C in Western blots (data not shown). Figure 2A shows that the antibodies against CENP-A (top right) and CENP-C (top left) recognize the centromeric regions of HeLa chromosomes. The soluble fractions obtained by the weaker MNase digestion (40 U/ml, 5 min; sample 3 in Fig. 1B) and by the stronger MNase digestion (80 U/ml, 45 min; sample 4 in Fig. 1B) were subjected to CHIP analysis. When sample 3 was treated with anti-CENP-A antibody, CENP-B and CENP-C were coprecipitated with CENP-A (Fig. 2B, lane 1). CENP-A and -B were also coprecipitated with CENP-C with anti-CENP-C antibody (Fig. 2B, lane 3). But the amounts of these coprecipitates were greatly reduced by the stronger MNase digestion (Fig. 2B, lanes 2 and 4, sample 4). These results suggest that CENP-A nucleosomes form a complex with CENP-B and CENP-C, but that extensive cleavage of DNA breaks down the complex.
FIG. 2.
CENP-A, CENP-B, and CENP-C were coprecipitated by CHIP using anti-CENP-A and/or anti-CENP-C antibodies. (A) Indirect immunofluorescence microscopy using newly prepared anti-CENP-A (top right) and anti-CENP-C (top left) antibodies. Mitotic chromosomes from HeLa cells were stained with anti-CENP-A or anti-CENP-C antibodies. Second antibodies were anti-mouse IgG-fluorescein isothiocyanate (FITC) conjugate for CENP-A, and anti-guinea pig IgG-rhodamine isothiocyanate (RITC) conjugate for CENP-C. Chromosomes were stained with DAPI (4′,6′-diamidino-3-phenylindole, bottom left). The three panels were merged (bottom right). CENP-A, green; CENP-C, red; DNA, white. (B) CHIP of the solubilized chromatin. Isolated HeLa nuclei were digested with 40 U of MNase per ml for 5 min (lanes 1 and 3) or 80 U/ml for 45 min (lanes 2 and 4), and the soluble fractions were subjected to CHIP using anti-CENP-A (lanes 1 and 2) or anti-CENP-C (lanes 3 and 4) antibodies. The precipitated proteins were separated by SDS-7.5% (for CENP-B and -C) or 12.5% (for CENP-A) PAGE, and the centromere proteins were detected by Western blotting using ACA serum (AK). Lane M shows the positions of CENP-A, CENP-B, and CENP-C. Positions of molecular size markers are indicated at the right.
α-Satellite DNA containing CENP-B boxes (αI-type array) is concentrated by CHIP with either anti-CENP-A or anti-CENP-C antibodies.
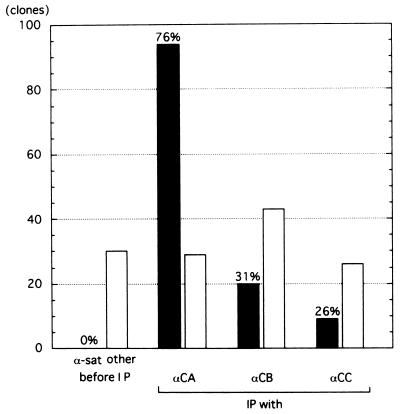
To examine the DNA component of the centromeric chromatin, the chromatin containing trinucleosomes and higher aggregates was recovered by 5 to 20% glycerol gradient sedimentation. The recovered chromatin was immunoprecipitated with the antibodies against CENP-A, -B, or -C. By slot blot analysis, α-satellite DNA was shown to be concentrated (data not shown). The precipitated DNA was cloned and sequenced. As summarized in Fig. 3, α-satellite DNA with CENP-B boxes was predominantly concentrated by CHIP using each of the three antibodies, while no α-satellite DNA was recovered from samples before immunoprecipitation (Fig. 3).
FIG. 3.
α-Satellite DNA fragments with CENP-B boxes were concentrated by CHIP using anti-CENP-A, anti-CENP-B, or anti-CENP-C antibodies. Before IP, clones prepared from the solubilized bulk chromatin before immunoprecipitation. IP, DNA fragments immunoprecipitated by anti-CENP-A (αCA), anti-CENP-B (αCB), or anti-CENP-C (αCC) antibodies were cloned into T vector and sequenced. More than 80% of the cloned fragments were longer than 500 bp. The black bars show the number of clones with α-satellite DNA that contained the CENP-B box. The open bars show the number of clones that were unique and contained no other repetitive sequence, not even α-satellite DNA without the CENP-B box.
It has already been shown that CENP-B preferentially binds to the CENP-B box. It was therefore to be expected that CHIP using anti-CENP-B antibody would precipitate α-satellite DNA with CENP-B boxes (20 clones, 31% of total clones), and the DNA from the other 44 clones (69%) was considered to be background, which may reflect the quality of this anti-CENP-B monoclonal antibody. CHIP using anti-CENP-C antibody significantly concentrated α-satellite DNA with CENP-B boxes (26%), while CHIP using anti-CENP-A antibody precipitated α-satellite DNA containing CENP-B boxes at a very high rate (76%). The isolated clones were all dimer repeats consisting of one α-satellite unit with a CENP-B box (box+) and the other without a CENP-B box (box−). Most of the clones could be classified into one of three families of type I α-satellite arrays; the results are summarized in Table 1. α-Satellite DNA fragments without CENP-B boxes and other repetitive sequences were not recovered with any of the CHIPs. These results show that centromeric chromatin containing CENP-A, -B, and -C is selectively formed on α-satellite DNA with CENP-B boxes (αI-type array) in HeLa cells.
TABLE 1.
Classification of the α-satellite DNA immunoprecipitated with anti-CENP-A antibodiesa
| Suprachromosomal family | Chromosome no. | Consensus sequence | No. of clones | % Homology |
|---|---|---|---|---|
| 1 | 1, 3, 5, 6, 7, 10, 12, 16, 19 | J2(+), J1(−) | 41 | 82-94 |
| 2 | 2, 4, 8, 9, 13, 14, 15, 18, 20, 21, 22 | D1(+), D2(−) | 43 | 82-92 |
| 3 | 11, 17, X | A(+), B(+), C(+), D(−), E(−) | 6 | 75-98 |
| 1, 2, or 3 | 4 | 76-80 |
α-Satellite clones recovered by immunoprecipitation with anti-CENP-A antibodies were classified into three suprachromosomal families (1). α-Satellite monomers with a CENP-B box are indicated by +, and monomers without the CENP-B box are marked by −. Each clone was highly homologous to one of the three families. Four clones could not be classified because they were equally homologous to all three families.
CENP-A nucleosomes are phased on αI-type arrays as a result of CENP-B/CENP-B box interactions.
We have previously shown by in vitro reconstitution of the CENP-B/nucleosome complex that CENP-B/CENP-B box interactions cause nucleosome phasing around the CENP-B box region (62). As the present work has revealed that CENP-A nucleosomes are formed predominantly on αI-type arrays, we studied the question of whether the nucleosomes on αI-type arrays were phased in vivo as well as in vitro.
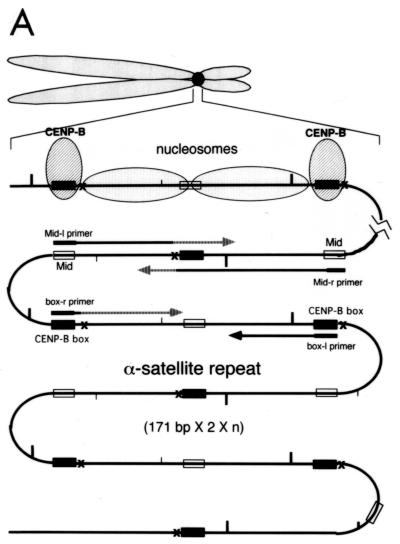
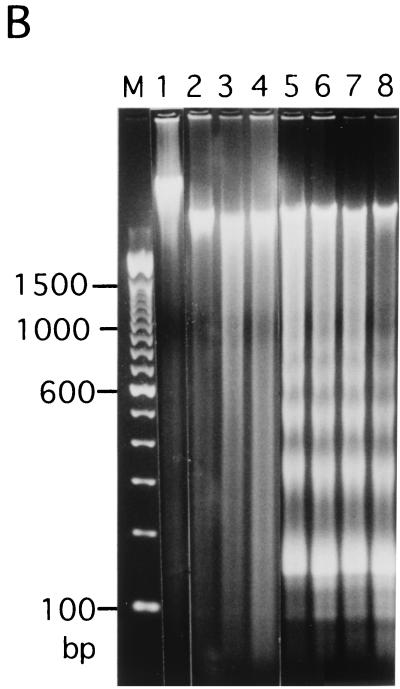
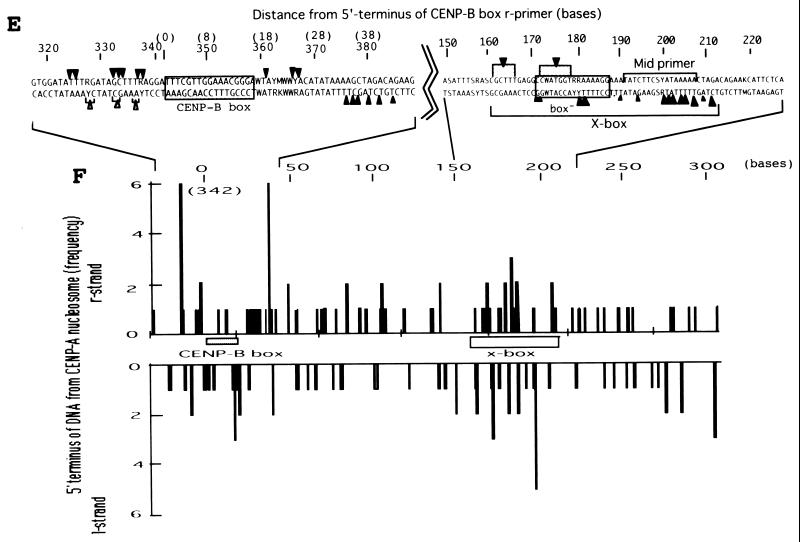
Figure 4A shows the strategy for in vivo MNase footprint analysis. The distance from the nearest CENP-B box of hypersensitive sites for MNase digestion was mapped by PCR-based primer extension analysis using the CENP-B box or mid-region (mid-primer) as primers. The mid-region is located approximately in the middle of adjacent CENP-B boxes (Fig. 4A) at the right side of CENP-B box− region (Fig. 4E, 191 to 208 bases from the 5′ terminus of the CENP-B box r-primer) (61). After MNase digestion of HeLa nuclei, the nuclear DNA was extracted and electrophoresed through an agarose gel (Fig. 4B). DNA ladders reflecting nucleosome structures were observed (Fig. 4B, lanes 5 to 8).
FIG. 4.
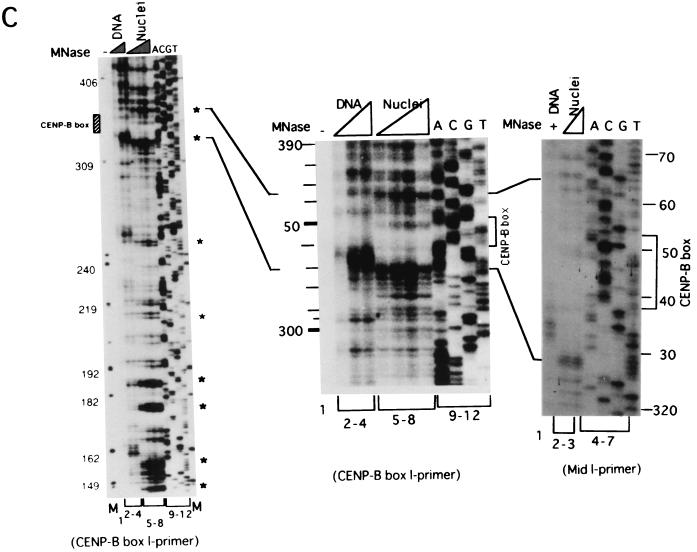
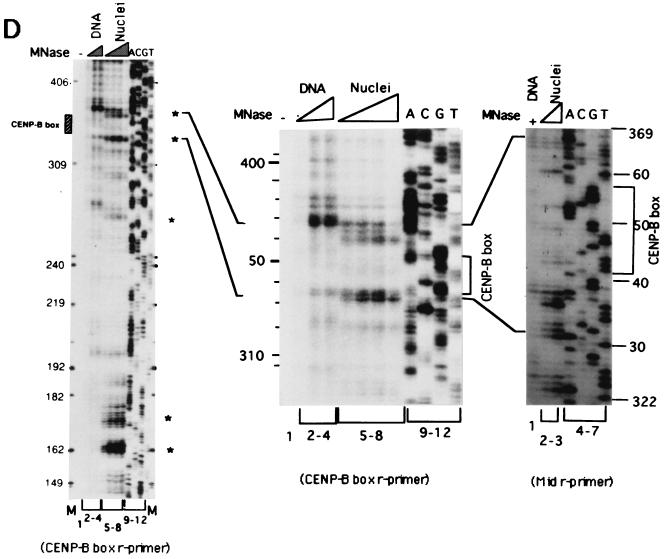
Mapping of hypersensitive sites for MNase digestion on αI-type arrays. (A) Strategy for in vivo mapping of cleavage sites by MNase digestion using CENP-B box primers or mid-primers. CENP-B boxes were located mainly in α-satellite dimer subfamilies. Dimer repeats are marked with thick vertical bars. CENP-B boxes and mid sequences are shown with solid and open boxes, respectively. The distances from the CENP-B box or the mid sequence to the hypersensitive sites for MNase digestion were measured by primer extension analyses using CENP-B boxes or mid sequences as primers. The primer for each sequence is indicated by a thick bar, and the direction of DNA synthesis is shown with arrows. Owing to the occasional deletion of 2 to 4 bases to the right of the CENP-B box (indicated by x in the figure), direct sequencing reactions were sometimes unreadable because of overlapping of the sequence ladder. (B) Agarose gel electrophoresis of HeLa DNA after MNase digestion. Purified DNA (20 μg, 0.1 ml; lanes 2 to 4) or nuclei (5 × 106, 0.1 ml; lanes 1 and 5 to 8) extracted from HeLa cells were digested with MNase (0.05 U for purified DNA and 15 U for nuclei) for 0 min (lane 1), 1 min (lane 2), 5 min (lane 3), 15 min (lane 4), 1 min (lane 5), 2 min (lane 6), 5 min (lane 7), and 20 min (lane 8). DNA from each sample was extracted with phenol and electrophoresed through a 1.7% agarose gel. Lane M shows a 100-bp DNA ladder. (C and D) PCR-primer extension analyses of the MNase digests with CENP-B box primers (left and middle panels) or mid primers (right panels). Left and middle panels, DNA (1 μg) from each of the digests shown in B was subjected to PCR-primer extension reactions using 32P-labeled CENP-B box l-primer (C) or r-primer (D), and the samples were electrophoresed for 4 h (left panels) or 6 h (middle panels). Lanes 1 to 8 correspond to the samples shown in lanes 1 to 8 of B. Lanes 9 to 12 show sequence ladders as position markers. Lane M is a 32P-labeled MspI digest of pBR322. The distance in bases from the CENP-B primer is indicated at the left. The area of the CENP-B box is marked with a shaded box, and hypersensitive bands are marked with asterisks (*). Right panels, higher-resolution analysis of the CENP-B box regions using mid l-primer (C) or r-primer (D). Lane 1 corresponds to lane 4 of B, and lanes 2 and 3 to lanes 6 and 7 of B. Lanes 4 to 7 show sequence ladders as position markers. The CENP-B box is indicated by a bracket at the right. The distance in bases from the CENP-B box is indicated at the right. (E) Mapping of MNase-hypersensitive sites around the CENP-B box and x-box regions. The DNA sequence obtained by direct sequencing of HeLa nuclei using the CENP-B box l-primer is shown (C, left, lanes 9 to 12) (61). The nucleotide position is indicated at the top as the distance in bases from the 5′ terminus of CENP-B box r-primer. Numbering around the CENP-B box region (left half of the figure) is counted from the adjacent CENP-B box. Position 342 corresponds to position 0, which is the start site for mapping around the x-box region (right half of the figure). Hypersensitive sites in the l-strand are indicated by upward-pointing arrowheads, and those in the r-strand by downward-pointing arrowheads. The upward-pointing open arrowheads with brackets at the left side of the CENP-B box indicate approximate loci (within brackets) for hypersensitive sites, because the DNA sequence of this region could not be read by direct sequencing. The CENP-B box is indicated by an open box (positions 1 to 17 or 343 to 359) and the corresponding area in the adjacent α-satellite monomer that has no affinity to CENP-B is shown as an open box (box) (positions 172 to 189). The mid-primer and x-box areas are indicated by brackets. R is A or G; Y is T or C; W is A or T; and S is G or C. (F) Mapping of 5′ termini of the α-satellite DNA fragments recovered by CHIP using anti-CENP-A antibody. 5′ termini of each strand from 94 clones of α-satellite DNA fragments (Fig. 3) were mapped on the α-satellite dimer repeat. The length of each vertical bar represents the frequency of the 5′ terminus mapping to each position on the sequence. The distance from the 5′ terminus of the CENP-B box r-strand is shown in bases at the top. Base 0 corresponds to base 342 from the adjacent CENP-B box.
These DNA samples were subjected to primer extension analysis using 32P-labeled CENP-B box l-strand (Fig. 4C, left) or r-strand (Fig. 4D, left) as primers. MNase digestion of the nuclei produced several cleavage bands in very restricted regions (Fig. 4C and 4D, left, lanes 5 to 8) (indicated by * at the right of the figures), which were absent in the MNase digest of the purified DNA (lanes 2 to 4). Among them, the cleavage bands at the CENP-B box region in each strand were notable, because these bands were very similar to those obtained in the in vitro experiment (62).
To establish the sites of cleavage at the nucleotide level, the CENP-B box regions shown in Fig. 4C and 4D (left panels) were expanded by elongated electrophoresis (Fig. 4C and 4D, middle panels). Footprinting was also performed using mid-primers (Fig. 4C and 4D, right panels), and the cleavage bands were mapped on the DNA sequence (Fig. 4E). Very strong cleavage bands were also detected in the region of 145 to 190 bases from the primer sites (Fig. 4C and 4D, left panels), which corresponds approximately to the middle of the two adjacent CENP-B boxes (Fig. 4A). These bands were undetectable in the in vitro CENP-B/nucleosome reconstitution experiment (62). Therefore, the result suggests that some unknown factor(s) other than CENP-B might affect phasing of nucleosome formation in the region surrounded by CENP-B boxes. These cleavage sites were also mapped (Fig. 4E), and this region was termed the x-box. These results suggest that the nucleosomes are phased between the two CENP-B boxes, as shown by in vitro reconstitution experiments (62).
The 5′ termini of the α-satellite DNA fragments recovered by CHIP using anti-CENP-A antibody (Fig. 3) were also mapped on the α-satellite dimer repeat. The 5′ termini of each strand were clustered at two regions (Fig. 4F): one was at both edges of the CENP-B box, and the other was at the x-box. These 5′ termini at the CENP-B box and the x-box were compared with the results of MNase footprinting (Fig. 4E and 4F). These independently mapped cleavage sites for MNase digestion correspond well with each other. Therefore we conclude that CENP-B actually interacts with the CENP-B box in vivo and that CENP-A nucleosomes on αI-type arrays are phased as a result of CENP-B-CENP-B box interactions.
CENP-A/B/C chromatin can be trimmed to tri- and/or tetranucleosomes by MNase digestion.
The footprint analysis shows a repetitive configuration of the CENP-B/CENP-A nucleosome complex, reflecting the repetitive structure of the αI-type array, since CENP-B binds to the CENP-B box that is located mainly in alternate monomer units (61) and CENP-A nucleosomes are phased between the adjacent CENP-B boxes. Where then is CENP-C located? Does CENP-C associate with a large CENP-B-CENP-A chromatin complex or with an individual small repetitive complex?
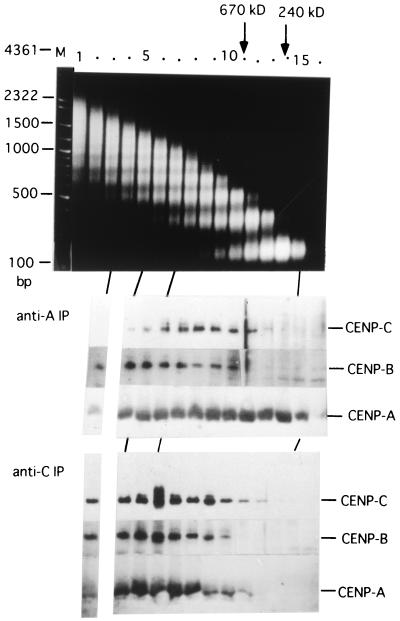
To examine molecular mass distribution of the CENP-A/B/C chromatin, the bulk chromatin solubilized with MNase digestion (60 U/ml, 15 min) was separated by 5 to 20% glycerol gradient sedimentation. Each fraction was subjected to CHIP analysis using anti-CENP-A (Fig. 5, middle panel) or anti-CENP-C (bottom panel) antibody, and CENP-A, -B, and -C were detected with ACA serum after SDS-PAGE. The top panel in Fig. 5 shows size distribution of nucleosomal DNA ladders in bulk chromatin of each fraction, which were used as molecular mass markers. The CHIPs with anti-CENP-C antibody (Fig. 5, bottom panel) show that the distribution pattern of the three CENPs is correlated, which suggests that CHIP with anti-CENP-C antibody mainly precipitates the CENP-A/B/C chromatin complex. Therefore, Fig. 5 (bottom panel) shows that MNase digestion of the nuclei (60 U/ml, 15 min) produced the CENP-A/B/C complex peaked at fraction 7 by glycerol gradient sedimentation. This fraction corresponds to the position of tetra- to pentanucleosomes in bulk chromatin, as shown in Fig. 5, top panel.
FIG. 5.
Size distributions of the centromeric chromatin separated by glycerol gradient sedimentation were examined by CHIP using anti-CENP-A or anti-CENP-C antibodies. HeLa nuclei (6 × 108, 3 ml) were digested with MNase for 15 min at 60 U/ml, and the solubilized chromatin fractions were centrifuged through a 5 to 20% glycerol density gradient. Aliquots of 2 ml were fractionated from the bottom. DNA was extracted from 10 μl of each fraction by phenol after proteinase K digestion and electrophoresed through a 1% agarose gel (top). Each 1.5-ml aliquot was subjected to CHIP analysis using anti-CENP-A (middle) or anti-CENP-C (bottom) antibodies. Distributions of CENP-A, CENP-B, and CENP-C were detected by Western blotting using ACA serum (AK) after SDS-7.5% (for CENP-B and CENP-C) and 12.5% (for CENP-A) PAGE. Marker proteins, catalase (240 kDa) and thyroglobulin (670 kDa) were centrifuged in parallel, and their positions are indicated at the top.
The molecular mass of the CENP-A/B/C complex is increased by the inclusion of some number of CENP-B and CENP-C (80 kDa + 140 kDa = 220 kDa), which roughly corresponds to one nucleosome unit, 240 kDa (Fig. 5, top panel). Therefore, the nucleosome number of the CENP-A/B/C chromatin should be less by one or two nucleosome units from the measured nucleosome number, depending on whether CENP-B and CENP-C are present as monomers or dimers in the CENP-A/B/C complex. Thus, the estimated number of nucleosomes contained in the peak CENP-A/B/C chromatin (fraction 7) would be three or four. These results support the possibility that CENP-C may associate rather uniformly with each of the repetitive CENP-B/CENP-A nucleosome complexes.
CHIP analysis with anti-CENP-A antibody (Fig. 5, middle panel) shows the presence of CENP-A mononucleosomes devoid of CENP-B and CENP-C (fractions 13 to 15). With further MNase digestion (60 U/ml, 45 min), the proportion of CENP-A mononucleosomes devoid of CENP-B and -C greatly increased to become the majority of the CENP-A-containing nucleosome population (data not shown). These results show that a secondary action of MNase digestion is to remove CENP-B and CENP-C from the CENP-A/B/C chromatin complex.
Most of the solubilized CENP-C and approximately half the solubilized CENP-B form complexes with CENP-A nucleosomes.
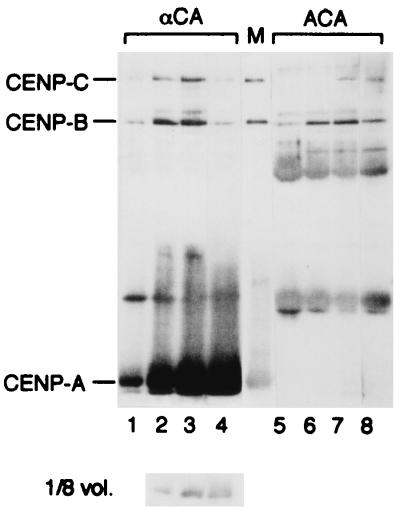
It is difficult to detect CENP-B and CENP-C with Western blotting directly in bulk chromatin fractions because of high background. The newly prepared anti-CENP-A antibody could quantitatively precipitate CENP-A chromatin (Fig. 6, lanes 5 to 8). Therefore, we measured the relative amount of CENP-B or CENP-C that interacted with CENP-A nucleosomes by two-step immunoprecipitation using anti-CENP-A antibody and ACA serum. Each of the four samples described in Fig. 1B was immunoprecipitated with anti-CENP-A antibody, and the supernatants were then precipitated with ACA serum. Both sets of precipitates (αCA and ACA) were analyzed by Western blotting with ACA.
FIG. 6.
Quantitative analysis of CENP-A, -B, and -C in CENP-A/B/C chromatin. The four MNase digests shown in Fig. 1B were immunoprecipitated with anti-CENP-A antibodies (lanes 1 to 4; 12-μl samples of αCA). The supernatants were subsequently precipitated with ACA serum (lanes 5 to 8; 12-μl samples of ACA) to isolate and detect the residual CENPs. The precipitated proteins were resolved by SDS-12.5% PAGE and immunoblotted using ACA serum. Because of overloading of CENP-A in the αCA samples (lanes 2 to 4), 1.5-μl samples were also immunoblotted and are shown below. The diffuse bands between CENP-A and CENP-B in both samples were IgGs released from the antibody-Sepharose beads. Lane M shows a CENP-A/B/C marker mixture containing CENP-A (61 fmol/μl), CENP-B (23 fmol/μl), and CENP-C (25 fmol/μl), A/B/C molar ratio = 2.7:1:1.
As shown in Fig. 6 (lanes 1 to 4), CENP-B and CENP-C were coprecipitated with CENP-A. Figure 6 (lanes 5 to 8) indicates that the CENP-A chromatin is depleted from the supernatant solutions, and CENP-C is also largely depleted even at early stages of digestion (Fig. 6, lanes 5 to 6). In contrast, about half of the solubilized CENP-B remains in the supernatant (Fig. 6, lanes 5 to 6). When the precipitate shown in lane 3 was further digested with MNase or treated with 0.6 M NaCl, CENP-B and CENP-C were released into the supernatant (data not shown). These results suggest that most of the solubilized CENP-C and approximately half the solubilized CENP-B formed complexes with CENP-A nucleosomes through interaction with DNA.
The amounts of CENP-B and CENP-C increased (Fig. 6, lanes 1 to 3), reached a maximum (lane 3), and then decreased (lane 4) as DNA cleavage progressed, while the amount of CENP-A continued to increase. As the present work is highly dependent on MNase digestion, the condition of MNase digestion in each experiment is summarized in Table 2 so that the data from each figure can be compared. The data from Fig. 5 and Fig. 6 are complementary, although they were from different digestions, in the point that MNase digestion produced CENP-A mononucleosomes devoid of CENP-B and CENP-C as well as the CENP-A/B/C chromatin in Fig. 5 and the ratio of CENP-B (and/or CENP-C)/CENP-A in the immunoprecipitates decreased at stronger MNase digestion in Fig. 6.
TABLE 2.
Summary of the conditions of MNase digestion in each experiment
These results show that MNase digestion of HeLa nuclei caused two events. First, it released mainly CENP-A/B/C chromatin complexes into the soluble fraction in the early stages of digestion, and second, it removed CENP-B and/or CENP-C from the complexes after more extensive cleavage.
DISCUSSION
By CHIP analyses using anti-CENP-A and/or anti-CENP-C antibodies, we have shown in this paper that the CENP-A nucleosome forms a complex with CENP-B and CENP-C in HeLa cells (Fig. 2) and that CENP-A/B/C complexes are selectively formed on α-satellite DNA containing CENP-B boxes (αI-type array) (Fig. 3). Mapping of the hypersensitive sites for MNase digestion around the CENP-B box regions showed that CENP-A nucleosomes are phased on the αI-type array as a result of CENP-B/CENP-B box interactions (Fig. 4). This implies that the CENP-B/CENP-A nucleosome complex forms a repetitive configuration reflecting the repetitive DNA sequence.
Molecular mass analyses by glycerol gradient sedimentation showed that MNase digestion of the nuclei released a CENP-A/B/C chromatin complex into the soluble fraction consisting of tri- or tetranucleosomes (Fig. 5). Immunodepletion of the prekinetochore chromatin using anti-CENP-A antibody showed that most of the CENP-C and approximately half the CENP-B took part in complex formation (Fig. 6). The kinetic study shown in Fig. 6 indicated that MNase digestion of the nuclei first released the CENP-A/B/C complexes into the soluble fraction and later removed CENP-B and CENP-C from the released complexes.
We could not detect CENP-H in the isolated CENP-A/B/C chromatin complex (data not shown). It might be released during purification of the complex because of the high salt concentration (0.3 M NaCl). The analyses with the autoantiserum EJ, which was expected to contain anti-CENP-G antibody (20), detected CENP-B and CENP-C but not CENP-G in the isolated chromatin complex (data not shown). Our analyses with the serum by immunoblotting and indirect immunofluorescence microscopy using several mammalian cultured cell lines revealed that the EJ antiserum currently available contained no antibodies against CENP-G, but contains anti-CENP-B and anti-CENP-C antibodies (data not shown).
Solubilization of centromeric chromatin by MNase digestion of HeLa nuclei.
We tried to solubilize the centromeric chromatin by MNase digestion of the nuclei, since the materials that are solubilized by DNA cleavage may be candidates for chromatin components. We confirmed that CENP-A, -B, and -C were all solubilized by MNase digestion (Fig. 1A). Up to ≈70% of CENP-A nucleosomes could be solubilized (Fig. 1C), but curiously, the solubility of CENP-B and -C did not increase beyond that shown in Fig. 1A, and instead that of CENP-B decreased during further MNase digestion (data not shown). The solubility of the chromatin proteins sharply decreased with lower salt concentration (0.15, 0.2, or 0.25 M NaCl), as a result of nonspecific interaction with insoluble materials (data not shown). A decrease in the amount of CENP-B or -C after extensive MNase digestion was also greater at the lower salt concentration (data not shown).
In this study, we found that MNase digestion excised CENP-B and CENP-C from the CENP-A/B/C chromatin, as well as solubilizing the CENP-A/B/C complex. We noticed that the decrease in solubility of CENP-B and -C after extensive MNase digestion correlated with the excision of CENP-B and -C from the CENP-A/B/C complex; the free CENP-B and -C molecules may have been titrated out of the soluble fraction by nonspecific binding to insoluble materials at 0.3 M NaCl. With weaker MNase digestion, solubilization of the CENP-A/B/C chromatin dominates over the liberation of free CENP-A nucleosomes and free CENP-B or -C molecules, and therefore CENP-A, -B, and -C were reproducibly coprecipitated by CHIP with anti-CENP-A or -C antibodies. We confirmed that ratios of CENP-A, -B, and -C in the coprecipitates were constant at lower salt concentration with weaker MNase digestion, although amounts of the coprecipitates were highly dependent on salt concentration (0.15 to 0.3 M; data not shown).
Prekinetochore chromatin containing CENP-A nucleosomes is selectively formed on I-type α-satellite arrays.
Vafa and Sullivan (54) reported that hemagglutinin (HA)-tagged CENP-A, exogenously expressed in HeLa cells, was mainly located on α-satellite DNA. In this work we isolated chromatin containing endogenous CENP-A and DNA longer than α-satellite trimer (510 bp) by using monoclonal antibodies raised against CENP-A. As shown in Fig. 3, the precipitated chromatin DNA fragments were predominantly α-satellite (76% of the total clones), and surprisingly, all the α-satellite DNA belonged to the subfamilies containing CENP-B boxes.
Alexandrov et al. (1) classified α-satellite subfamilies containing CENP-B boxes into three suprachromosomal families, which include all the human centromeres except the Y. We have previously shown by direct sequencing of HeLa DNA using CENP-B box primers that most CENP-B boxes were located on α-satellite dimer subfamilies (61). The precipitated α-satellite DNA was classified into one of the three families on the basis of homology of each sequence (Table 1), suggesting that the precipitated DNA fragments cover the centromeres of all chromosomes in HeLa cells. The centromeric regions of human chromosome 21 consist of two contiguous long α-satellite arrays. One is α21-I of ≈1.3 Mb, which belongs to suprachromosomal family 2 and consists mainly of the D1-D2 dimer repeat, so that the CENP-B boxes will normally occur in alternate monomer units (26). The other is α21-II of ≈1.9 Mb, which contains very few CENP-B boxes, and homologies between different monomer units are low (26). Similar DNA structures were found in human chromosomes 13 and 14 (53). The present results demonstrate that CENP-A/B/C chromatin complexes are selectively formed on the α-I type array.
By indirect immunofluorescence microscopy of elongated human centromeres, antigens for ACA serum were shown to be localized to the αI-type array (26). Consistent with this, the introduction of yeast artificial chromosomal DNA with an 80-kbp αI-type array into a human cultured cell line, HT1080, resulted in the efficient formation of artificial minichromosomes, but introduction of an αII-type array did not (27). These results suggest that the αI-type array is sufficient to establish active centromeres de novo.
One of the characteristics of this repetitive sequence is the presence of CENP-B boxes. As shown in this paper, CENP-B/CENP-B box interactions are involved in the formation of prekinetochore chromatin complexes. Another factor is the presence of the x-box region (Fig. 4). This region is highly conserved (41), and MNase footprint analyses in vivo (Fig. 4) and in vitro (62) suggested that some unknown factor(s) might be involved in the chromatin formation around this region. Interestingly, α-satellite DNA from the human Y chromosome and African green monkey chromosomes contains a sequence homologous to the x-box.
Where is CENP-C located on the large CENP-B/CENP-A nucleosome complex on the αI-type array?
It has already been confirmed that the CENP-B/CENP-B box interaction was necessary and sufficient for CENP-B to localize at the centromeric region (38). In this paper we showed that CENP-A nucleosomes were phased on αI-type arrays. It is therefore highly probable that the CENP-B/CENP-A nucleosome complex mainly forms a repetitive configuration that reflects the repetitiveness of the αI-type array. The next important question is where the CENP-C is located on the CENP-B/CENP-A nucleosome complex. There are two possibilities: (i) CENP-C interacts with a large CENP-B/CENP-A nucleosome complex, or (ii) a CENP-C monomer or dimer interacts with each of the repetitive units of the CENP-B/CENP-A nucleosome complex. These possibilities could be experimentally distinguished. Figure 5 showed that the CENP-A/B/C chromatin could be reduced to a three- to four-nucleosome complex without losing CENP-B and CENP-C. The results clearly excluded the former possibility and supported the latter.
CENP-A/B/C chromatin complexes may become components of the inner kinetochore plate during M-phase.
The immunodepletion study shown in Fig. 6 indicated that approximately half the released CENP-B remained in the soluble fraction after immunodepletion of chromatin containing CENP-A. At an early stage of MNase digestion (samples 1 and 2 in Fig. 6), free CENP-B molecules should not yet have been excised from the chromatin complexes, since two simultaneous cleavages by MNase at the CENP-B box region were needed to excise a CENP-B molecule from the chromatin complex. So the CENP-B left in the soluble fractions at early stages of MNase digestion after immunodepletion of the CENP-A chromatin (Fig. 6, lanes 5 to 6) would still bind to chromatin that did not contain CENP-A. We have previously shown that the CENP-B/nucleosome complex could be assembled in vitro equally well using either normal (62) or CENP-A nucleosomes (63), and formed a specific DNA structure at the CENP-B box region which became hypersensitive to MNase (62).
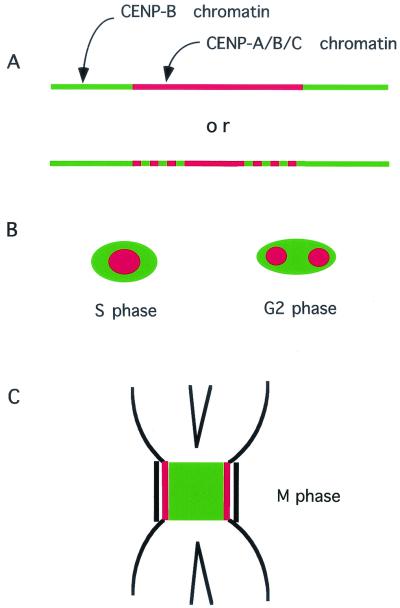
These results suggest that half the CENP-B molecules on αI-type arrays form a complex with normal nucleosomes, as illustrated in Fig. 7A. CENP-A and CENP-C, observed by indirect immunofluorescence microscopy, were seen as round dots and completely overlapped at S- and M-phases (55), while CENP-B was sometimes seen as a diffuse patch that included a dot (or double dots) of CENP-A (55). These observations agree well with the present results that most of the solubilized CENP-C and about half the solubilized CENP-B formed a complex with CENP-A nucleosomes (Fig. 7B). By immunoelectron microscopy, CENP-C was reported to be localized to the inner kinetochore plate (43). CENP-A was proposed to be a component of the inner kinetochore plate on the grounds of colocalization of CENP-A and -C by optical microscopy (55). The present work revealed that CENP-A and -B form a chromatin complex with CENP-C, and therefore CENP-A and -B must also be the components of the inner kinetochore plate (Fig. 7C).
FIG. 7.
Proposed model for the distribution of CENPs. (A) The A/B/C chromatin complex (CENP-A nucleosome/CENP-B/CENP-C complex; red) occupies 50% of the αI-type array, and the normal nucleosome/CENP-B complex (green) occupies the remaining 50%. (B) The distribution of CENP-A, -B, and -C in S- and G2-phases as seen by light microscopy. Red represents the prekinetochore, which contains the A/B/C chromatin complex. Green represents the centromeric heterochromatin, which consists of the normal nucleosome/CENP-B complex. (C) The figure represents the results of electron microscopy observations and the present results in M-phase. The CENP-A/B/C chromatin complex (red) constitutes the inner kinetochore domain, and the CENP-B/normal nucleosome complex (green) constitutes the pairing domain.
Model structure for the prekinetochore chromatin in HeLa cells.
We have previously proposed a model structure for the CENP-B/nucleosome complex (62). The present results suggest that this model would be applicable to the CENP-B/CENP-A nucleosome complex only if normal nucleosomes were replaced by CENP-A nucleosomes. As the CENP-A/B/C complex can be trimmed to a small chromatin complex containing 3 to 4 nucleosomes (Fig. 5), CENP-C might bind distributively to the repetitive CENP-B/CENP-A nucleosome complex. As CENP-C is a DNA-binding protein with no apparent sequence specificity (49, 59), it remains to be shown where and how it binds on the prekinetochore complex. The present study may provide a molecular basis for the repeat subunit model of Zinkowsky et al. (64) and is basically similar to the molecular organization of the human centromere proposed by Choo (9).
Acknowledgments
We thank K. Todokoro for anti-CENP-H antibody, B. R. Brinkley for autoantiserum EJ, G. Tamiya and M. Tomizawa for DNA sequencing, and M. Yanagida, K. H. A. Choo, H. Masukata, and K. Sullivan for helpful comments.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas, a Grant-in-Aid for Scientific Research (c), a grant from the Ministry of Agriculture, Forestry and Fisheries for Establishment of Transgenic Agro-biofarm Systems, a Grant-in-Aid for JSPS Fellows to H.Y., and the Sasakawa Scientific Research Grant from the Japan Science Society and Research Fellowship of JSPS for Young Scientists to S.A.
REFERENCES
- 1.Alexandrov, I. A., T. D. Mashkova, T. A. Akopian, L. I. Medledev, L. L. Kisselev, S. P. Mitkevich, and Y. B. Yurov. 1991. Chromosome-specific alpha satellites: two distinct families on human chromosome 18. Genomics 11:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Barry, A., E. Howmann, M. Cancilla, S. R., and K. Choo. 1999. Sequence analysis of an 80 kb human neocentromere. Hum. Mol. Genet. 8:217-227. [DOI] [PubMed] [Google Scholar]
- 3.Baum, M., and L. Clark. 2000. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol. Cell. Biol. 8:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom, K. S., and J. Carbon. 1982. Yeast centromere DNA is a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29:305-317. [DOI] [PubMed] [Google Scholar]
- 5.Brinkley, B. R., M. M. Valdivia, A. Tousson, and R. D. Balczon. 1989. Mitosis: molecules and mechanisms. Academic Press, Orlando, Fla.
- 6.Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth, and S. Henikoff. 1999. A histone-H3-like protein in C. elegans. Nature 401:547-548. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., R. E. Baker, K. C. Keith, K. Harris, A. Stoler, and M. Fitzgerald-Hayes. 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20:7037-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chikashige, Y., N. Kinoshita, Y. Nakaseko, T. Matsumoto, S. Murakami, O. Niwa, and M. Yanagida. 1989. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57:739-751. [DOI] [PubMed] [Google Scholar]
- 9.Choo, K. H. A. 2000. Centromerization. Trends Cell Biol. 10:182-188. [DOI] [PubMed]
- 10.Coligan, J. E., a. m. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober. 1991. Current protocols in immunology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 11.Cooke, C. A., R. L. Bernat, and W. C. Earnshaw. 1990. CENP-B: A major human centromere protein located beneath the kinetochore. J. Cell Biol. 110:1475-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobie, K. W., K. L. Hari, K. A. Maggert, and G. H. Karpen. 1999. Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev. 9:206-217. [DOI] [PubMed] [Google Scholar]
- 13.Earnshaw, W. C., K. F. Sullivan, P. S. Machlin, C. A. Cooke, D. A. Kaiser, T. D. Pollard, N. F. Rothfield, and D. W. Cleveland. 1987. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104:817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espelin, C. W., K. B. Kaplan, and P. K. Sorger. 1997. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 139:1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 29:235-244. [DOI] [PubMed] [Google Scholar]
- 16.Fleig, U., J. D. Beinhauer, and J. H. Hegemann. 1995. Functional selection for the centromere DNA from yeast chromosome VII. Nucleic Acids Res. 23:922-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukagawa, T., and W. R. A. Brown. 1997. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 6:2301-2308. [DOI] [PubMed] [Google Scholar]
- 18.Glowczewski, L., P. Yang, T. Kalashnikova, S. M. Santisteban, and M. M. Smith. 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahnenberger, K. M., M. P. Baum, C. M. Polizzi, J. Carbon, and L. Clarke. 1989. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, D., Z. C., K. Woods, X. L., D. Turner, R. K. Busch, B. R. Brinkley, and H. Busch. 1998. CENP-G: a new centromeric protein that is associated with the α-1 satellite DNA subfamily. Chromosoma 107:189-197. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff, S., K. Ahmad, J. S. Platero, and B. V. Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz, R. A., D. A. Agard, J. W. Sedat, and C. L. Woodcock. 1994. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J. Cell Biol. 125:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. A. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenp A) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson, D. F., K. J. Fowler, E. Earle, R. Saffery, P. Kalitsis, H. Trowell, J. Hill, N. G. Wreford, D. M. deKrester, M. R. Cancilla, E. Howman, L. Hii, S. M. Cutts, D. V. Irvine, and K. H. A. Choo. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyland, K. M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeno, M., H. Masumoto, and T. Okazaki. 1994. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range α-satellite DNA arrays of human chromosome 21. Hum. Mol. Genet. 3:1245-1257. [DOI] [PubMed] [Google Scholar]
- 27.Ikeno, M., B. Grimes, T. Okazaki, M. Nakano, K. Saitoh, H. Hoshino, N. McGill, H. Cooke, and H. Masumoto. 1998. Construction of YAC based mammalian artificial chromosomes. Nat. Biotechnol. 16:431-439. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen, A. L., C. J. Bostock, and A. L. Bak. 1987. Homologous subfamilies of human alphoid repetitive DNA on different nucleolus organizing chromosomes. Proc. Natl. Acad. Sci. USA 84:1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalitsis, P., K. J. Fowler, E. Earle, J. Hill, and K. H. A. Choo. 1998. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and embryo death. Proc. Natl. Acad. Sci. USA 95:1136-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapoor, M., R. Montes, G. Liu, G. Lozano, C. Cummings, M. Muncini, I. Ouspenski, B. R. Brinkley, and G. S. May. 1998. The cenpB gene is not essential in mice. Chromosoma 107:570-576. [DOI] [PubMed] [Google Scholar]
- 31.Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant cse4p from histone H3. Mol. Cell. Biol. 19:6130-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa, K., H. Masumoto, M. Ikeda, and T. Okazaki. 1995. Analysis of protein-DNA and protein-protein interactions of centromere protein B (CENP-B) and properties of the DNA-CENP-B complex in the cell cycle. Mol. Cell. Biol. 15:1602-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masumoto, H., H. Masukata, Y. Muro, N. Nozaki, and T. Okazaki. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meluh, P. B., P. Yang, P. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:6.07-613. [DOI] [PubMed]
- 35.Muro, Y., H. Masumoto, K. Yoda, N. Nozaki, M. Ohashi, and T. Okazaki. 1992. Centromere protein B assembles human centromeric α-satellite DNA at the 17 bp sequence, CENP-B box. J. Cell Biol. 116:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer, D. K., K. O'Day, H. L. Trong, H. Charbonneau, and R. L. Margolis. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluta, A. F., A. M. Mackay, A. M. Ainszein, I. G. Goldberg, and W. C. Earnshaw. 1995. The centromere: hub of chromosomal activities. Science 270:1591-1594. [DOI] [PubMed] [Google Scholar]
- 38.Pluta, A. F., N. Saitoh, I. Goldberg, and W. C. Earnshaw. 1992. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J. Cell Biol. 116:1081-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rattner, J. R. 1987. The organization of the mammalian kinetochore: a scanning electron microscope study. Chromosoma 95:175-181. [DOI] [PubMed] [Google Scholar]
- 40.Ris, H., and P. L. Witt. 1981. Structure of the mammalian kinetochore. Chromosoma 82:153-170. [DOI] [PubMed] [Google Scholar]
- 41.Romanova, L. Y., G. V. Derigain, T. D. Mashkova, I. G. Tumeneva, A. R. Mushegian, L. l. Kisselev, and I. A. Alexandrov. 1996. Evidence for selection in evolution of alpha satellite DNA: the central role of CENP-B/pJα binding region. J. Mol. Biol. 261:334-340. [DOI] [PubMed] [Google Scholar]
- 42.Saffery, R., D. V. Irvine, B. Griffiths, P. Kalitsis, L. Wordeman, and K. H. Choo. 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 9:175-185. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh, H., J. Tomkiel, C. A. Cooke, H. Ratrie, I. I. I., M. Maurer, N. F. Rothfield, and W. C. Earnshaw. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70:115-125. [DOI] [PubMed] [Google Scholar]
- 44.Sart, D., M. R. Cancilla, E. Earle, J. Mao, R. Saffery, K. M. Tainton, P. Kalitsis, J. Martyn, A. E. Barry, and A. Choo. 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of nonα-Satellite DNA. Nat. Genet. 16:144-153. [DOI] [PubMed] [Google Scholar]
- 45.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner, N. C., and L. Clarke. 1994. A novel epigenetic effect can alter centromere function in fission yeast. Cell 79:865-874. [DOI] [PubMed] [Google Scholar]
- 47.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 48.Sugata, N., S. Li, W. C. Earnshaw, T. J. Yen, E. Munekata, K. Yoda, H. Masumoto, P. E. Warurton, and K. Todokoro. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere-kinetochore complexes. Hum. Mol. Genet. 9:2919-2926. [DOI] [PubMed] [Google Scholar]
- 49.Sugimoto, K., K. K., A. Shibata, and M. Himeno. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5:132-141. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, K. F., M. Hechenberger, and M. Khaled. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, K., E. S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288:2215-2219. [DOI] [PubMed] [Google Scholar]
- 52.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trowell, H. E., A. Nagy, B. Vissel, and K. H. A. Choo. 1993. Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum. Mol. Genet. 2:1639-1649. [DOI] [PubMed] [Google Scholar]
- 54.Vafa, O., and K. Sullivan. 1997. Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7:897-900. [DOI] [PubMed] [Google Scholar]
- 55.Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K. F. Sullivan, G. G. Poirier, and W. C. Earnshaw. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromere. Curr. Biol. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 56.Willard, H. F. 1990. Centromeres of mammalian chromosomes. Trends Genet. 6:410-416. [DOI] [PubMed] [Google Scholar]
- 57.Willard, H. F. 1998. Centromeres: the missing link in the development of human artificial chromosomes. Curr. Opin. Genet. Dev. 8:219-225. [DOI] [PubMed] [Google Scholar]
- 58.Williams, B. C., T. Murphy, M. Goldberg, and G. Karpen. 1998. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 18:30-37. [DOI] [PubMed] [Google Scholar]
- 59.Yang, C. H., J. Tomkiel, H. Saitoh, D. H. Johnson, and W. C. Earnshaw. 1996. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 16:3576-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoda, K., K. Kitagawa, H. Masumoto, Y. Muro, and T. Okazaki. 1992. A human centromere protein, CENP-B, has a DNA binding domain containing four potential α-helices at the NH2 terminus, which is separable from dimerizing activity. J. Cell Biol. 119:1413-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoda, K., and T. Okazaki. 1997. Site specific base deletions in human α-satellite monomer DNAs are associated with regularly distributed CENP-B boxes. Chromosome Res. 5:207-211. [DOI] [PubMed] [Google Scholar]
- 62.Yoda, K., S. Ando, A. Okuda, A. Kikuchi, and T. Okazaki. 1998. In vitro assembly of CENP-B/α-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells 3:533-548. [DOI] [PubMed] [Google Scholar]
- 63.Yoda, K., S. Ando, S. Morishita, K. Houmura, K. Hashimoto, K. Takeyasu, and T. Okazaki. 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97:7266-7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinkowsky, R. P., J. Meyne, and B. R. Brinkley. 1991. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]