Abstract
Histone acetyltransferases (HATs) such as CBP and p300 are regarded as key regulators of RNA polymerase II-mediated transcription, but the critical structural features of their HAT modules remain ill defined. The HAT domains of CBP and p300 are characterized by the presence of a highly conserved putative plant homeodomain (PHD) (C4HC3) type zinc finger, which is part of the functionally uncharacterized cysteine-histidine-rich region 2 (CH2). Here we show that this region conforms to the PHD type zinc finger consensus and that it is essential for in vitro acetylation of core histones and the basal transcription factor TFIIE34 as well as for CBP autoacetylation. PHD finger mutations also reduced the transcriptional activity of the full-length CBP protein when tested on transfected reporter genes. Importantly, similar results were obtained on integrated reporters, which reflect a more natural chromatinized state. Taken together, our results indicate that the PHD finger forms an integral part of the enzymatic core of the HAT domain of CBP.
Regulation at the level of transcription is the central mechanism by which cells respond to developmental and environmental cues. RNA polymerase II-mediated transcription in eukaryotes is to a large extent regulated at the level of chromatin, which forms a physical barrier against the binding of proteins to the promoter region of a target gene. The basic unit of chromatin is the nucleosome, which consists of an octamer of histone proteins around which the DNA is wrapped. The nucleosomes form an array that is further organized into higher-order chromatin structures. The two major enzymatic activities which make the DNA more accessible for the transcription machinery are the ATP-dependent chromatin remodeling complexes and histone acetyltransferases (HATs) (15, 32, 39). The prime targets of the HATs in chromatin are the N-terminal tails of the core histones H2A, H2B, H3, and H4. Acetylation of these tails results in neutralization of the positively charged lysines, thereby influencing DNA-histone and histone-histone contacts. Furthermore, posttranslational histone modifications including acetylation can create specific docking sites for regulatory proteins (64). This so-called histone code may also be involved in the establishment of epigenetic inheritance (62). In addition to their ability to acetylate histones, some HATs have also been shown to acetylate nonhistone proteins, including several sequence-specific transcription factors and the basal transcription factors TFIIE and TFIIF (reviewed in reference 60).
The acetyltransferases CREB binding protein (CBP) and the related protein p300 function as coactivators for a multitude of transcription factors (9, 23, 58, 63), including CREB (5, 16, 38). This coactivator function is not only due to their intrinsic HAT activity but is also the result of their ability to interact with other HAT proteins such as pCAF and SRC1 family members. Furthermore, CBP and p300 can stabilize the transcription complex by binding to several proteins simultaneously, thereby functioning as a scaffold or physical bridge. While CBP and p300 are essential coactivators for many different transcription factors, the relative importance of these different coactivator functions varies between transcription factors (34, 35, 37). Several lines of evidence underscore the importance of the CBP and p300 proteins in differentiation, growth control, and homeostasis. Firstly, CBP and p300 are required for embryonic development and viability, as observed in knockout mice (66). Secondly, recurrent chromosomal translocations involving CBP and p300 are found in human leukemias (30), and genetic alterations resulting in amino acid changes or protein truncations of p300 are found in solid tumors and tumor cell lines (21, 48). Finally, haploinsufficiency of CBP results in Rubinstein-Taybi syndrome (RTS) in humans, a developmental syndrome characterized by facial abnormalities, broad thumbs, big, broad toes, and mental retardation (54).
Comparison of the amino acid sequences of CBP and p300 from different species revealed the presence of numerous regions of near-identity, including the bromodomain, three cysteine-histidine-rich regions (CH1, -2, and -3), and the HAT domain, while other regions are poorly conserved (4) (see Fig. 1A). The bromodomain, which is found in many chromatin-associated proteins (31, 64), is thought to function as a histone binding motif (17, 29). While the CH1 and CH3 regions serve as binding sites for many different transcription factors and other proteins, the function of the CH2 region, which is partly located within the HAT domain, remains to be determined. Based on sequence homology, part of the CH2 region can be classified as a plant homeodomain (PHD) type zinc finger (1). This type of zinc finger, also named leukemia-associated-protein (LAP) finger (55) or trithorax consensus (TTC) finger (33), is characterized by a C4HC3 motif and is found predominantly in proteins that function at the chromatin level (1). Recently, the solution structures of the PHD fingers of the transcriptional repressor KAP-1 (13) and the Williams's syndrome transcription factor WSTF (52) were determined. In both proteins, two zinc atoms are coordinated by the cysteine and histidine residues in a cross-brace fashion, reminiscent of the zinc coordination found in the RING finger (10) (see Fig. 1B). Furthermore, a number of hydrophobic residues were shown to be involved in the stabilization of this structure. Although more than 300 (mainly nuclear) proteins contain one or more PHD fingers, relatively little is known about the function of this domain. Since many PHD finger-containing proteins reside in large multiprotein complexes, these zinc fingers have been proposed to be involved in protein-protein interactions (1).
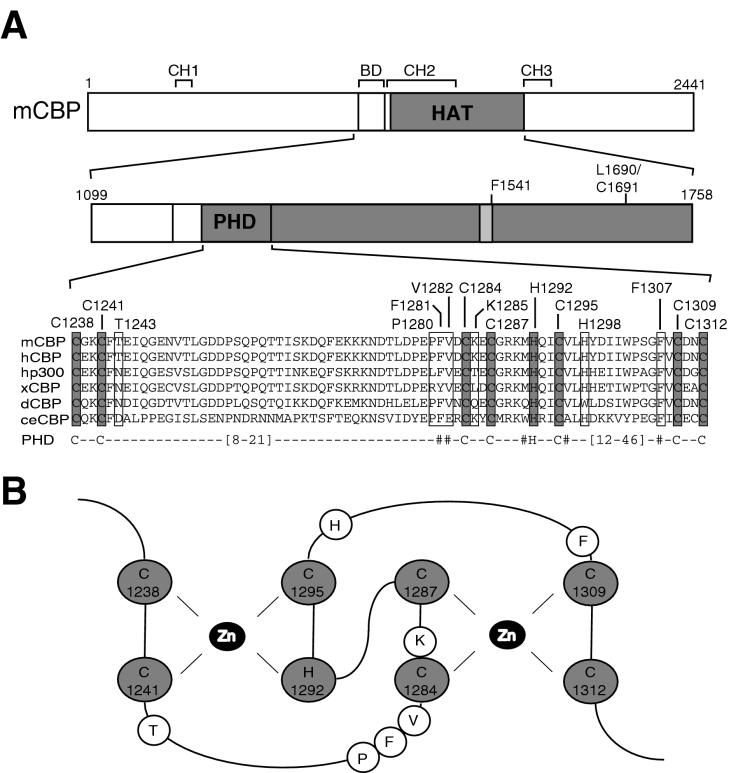
FIG. 1.
A conserved PHD finger in the acetyltransferase domain of CBP and p300. (A) Schematic representation of mouse CBP (mCBP), showing the position of the bromodomain (BD), the acetyltransferase domain (HAT), the PHD finger, and the CH1 through -3. Numbers refer to amino acid positions in mCBP. The putative CoA binding site is indicated by a lightly shaded box, and the positions of two HAT mutants, the F1541A (46) and L1690K C1691L (34) mutants, are shown. An alignment of the PHD fingers in the HAT domains of mCBP (GenBank accession number P45481), human CBP (hCBP) (Q92793), human p300 (hp300) (A54277), Xenopus CBP (xCBP) (20), Drosophila CBP (dCBP) (T13828), and Caenorhabditis elegans CBP (ceCBP) (P34545) is given. Putative zinc-coordinating cysteines and histidine are shaded; other amino acids that have been changed in this study are boxed. The PHD consensus sequence is shown below the alignment, with strongly conserved hydrophobic amino acids indicated (#). The lengths of the loops between the second and third and between the sixth and seventh zinc-coordinating residues vary between PHD fingers, as indicated. (B) Putative structure of the PHD finger in mCBP, based on the structures of the PHD fingers in KAP-1 (13) and WSTF (52). The zinc-coordinating cysteines and histidine and the zinc atoms (Zn) are indicated, as well as other amino acids that have been changed in this study.
Mammalian HAT enzymes can be divided into subfamilies (60, 61). However, it is currently difficult to classify a protein as a potential HAT enzyme on the basis of its amino acid sequence, since these subfamilies display no obvious similarity in their primary sequences or in the sizes of their HAT domains or the surrounding protein modules (36, 44, 50). The only region that is partly conserved between HAT subfamilies, either on the amino acid sequence level or the structural level or both (50, 65), is a small subdomain, first noticed in GCN5-related N-acetyltransferases, which encompasses the coenzyme A (CoA) binding site (45). As expected, mutation of critical residues in this motif results in loss of HAT function in, for example, CBP and p300 (35, 46). Additional amino acid sequences are required, however, since deletion of functionally uncharacterized regions outside this binding site also abolishes HAT activity (7, 51). The limited knowledge of the critical enzymatic and structural features of HAT modules prompted us to investigate the role of a putative PHD type zinc finger which is unique to the CBP and p300 HAT domains. Our data indicate that the PHD finger is an integral part of the enzymatic core of the acetyltransferase domain of CBP and that this domain is a key contributor to the transcriptional activity of the CBP protein.
MATERIALS AND METHODS
Plasmids.
All recombinant DNA work was performed according to standard procedures (6). The mammalian pcDNA3 expression vectors containing the DNA binding domain of the yeast transcription factor Gal4 (Gal4 DBD) and Gal4 DBD-CBP HAT (amino acids 1099 to 1758) (46), and the CBP HAT plasmid (amino acids 1099 to 1758) fused to glutathione S-transferase (GST) (7), were kind gifts from T. Kouzarides. RcRSV-CBPHA, originally a kind gift from R. H. Goodman, was digested with BamHI-XbaI or HindIII-NotI, and the full-length mouse CBP cDNA was cloned into the corresponding sites of pcDNA3-Gal4 DBD or pCDNA3.1, respectively. All point mutations were generated with the Quick Change mutagenesis kit (Stratagene) according to the manufacturer's instructions and were verified by sequencing. All reporter plasmids used were based on the pGL3 plasmid (Promega). The 5xGal-E1BTATA-Luc reporter (42) was a kind gift from M. Parker. This construct was cut with XbaI, and an XbaI fragment from ERE-TK-Luc (42), containing the herpes simplex virus (HSV) thymidine kinase (TK) promoter and luciferase sequences, was inserted to create 5xGal4-TK-Luc. The 5xGal-AdMLTATA-Luc reporter was generated by cloning the SacI-HindIII fragment of pGal4-M2-Luc, a kind gift from H.G. Stunnenberg, into pGL3 basic (Promega). GST-PCAF, generated by cloning a PCR fragment encoding amino acids 352 to 832 of human PCAF into pGEX2TK (Pharmacia), was a kind gift from F. Claessens. The plasmids containing His-tagged TFIIE34 and TFIIE56 (67) and the expression vectors for Gal4 DBD-CREB, its serine 133-to-alanine mutant (19), and the catalytic subunit of protein kinase A (PKA) (47) have been described previously.
Protein purifications.
Drosophila core histones were purified from embryo nuclear extracts essentially as described by Bulger and Kadonaga (12). Human His-tagged TFIIE34 was expressed in Escherichia coli (DE3), purified with nickel-nitrilotriacetic acid agarose resin (Qiagen), and further purified by SP Sepharose column chromatography as described elsewhere (67). The GST fusion protein of PCAF HAT was expressed in E. coli DH5α and purified according to standard procedures (6). The GST vectors containing wild-type and mutant CBP HAT were transformed into E. coli BL21-CodonPlus(DE3)-RIL (Stratagene). Bacteria were grown in Luria broth in the presence of 50 μM ZnCl2 at 25°C to an optical density at 600 nm of 0.5 and were induced with isopropyl-β-d-thiogalactopyranoside (0.15 mM) for 2 h at 25°C. Subsequently, bacteria were collected by centrifugation and resuspended in lysis buffer (25 mM HEPES [pH 7.6], 10% glycerol, 500 mM NaCl, 0.01% NP-40, 5 mM dithiothreitol, 2.5 mM MgCl2, 50 μM ZnCl2, and protease inhibitors). Lysozyme was added to a final concentration of 0.15 mg/ml, and the mixture was incubated for 30 min at 4°C with rotation. Lysates were freeze-thawed once, sonicated, cleared by centrifugation at 16,000 × g for 1 h, and stored at −70°C. GST proteins were purified on glutathione beads according to standard procedures (6).
Transient transfection experiments.
The human osteosarcoma cell line U-2 OS and the adenovirus type 5-transformed human embryo retina cell line 911 (18) and its derivatives were routinely maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Gibco Life Technologies), 100 μg of penicillin/ml, and 100 μg of streptomycin/ml. For luciferase reporter assays, cells were plated into 24-well microtiter plates and transfected by using the calcium phosphate coprecipitation method with Gal4 DBD expression plasmids at 200 ng/well, 150 ng of pcDNA-LacZ plasmid (Invitrogen) as an internal control, and 1 μg of a reporter plasmid [or pBluescript SK(−) in the case of the 911 reporter cells]. Transfections with CREB in 911 reporter cells contained 100 ng of Gal4 DBD-CREB, 100 ng of PKA expression vector, 100 ng of pcDNA-CBP, and 1 μg of pBluescript SK(−). After 24 h, cells were washed with 1× HEPES-buffered saline and then maintained in medium for another 24 h. Cells were then washed with phosphate-buffered saline (PBS) and harvested in lysis buffer (25 mM Tris phosphate [pH 7.8], 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100). Extracts were assayed for luciferase activity according to the manufacturer's protocol (Promega) and for β-galactosidase activity (6) as a control. For immunoprecipitation experiments, cells were plated in 9-cm dishes and transfected with Fugene (Roche) with 10 μg of expression plasmid.
Isolation of reporter cell lines.
911 cells were transfected with the 5xGal-E1BTATA-Luc reporter or the 5xGal-AdMLTATA-Luc reporter together with a CMV-neo plasmid by the calcium phosphate coprecipitation method as described above. After 24 h, cells were replated into 5-cm dishes, and Geneticin (G418; 600 μg/ml) was added. Resistant clones were isolated after 6 days and cultured under continuous G418 selection.
Immunoprecipitations.
Cells, which had previously been transfected, were washed in ice-cold PBS, collected in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.1% NP-40, and protease inhibitors), and incubated for 30 min at 4°C. Lysates were freeze-thawed once and cleared by centrifugation. Protein concentrations were determined by the Bradford method. Cell lysates containing equal amounts of protein (600 μg) were incubated for 3 to 4 h at 4°C under constant rotation with a polyclonal antibody against the Gal4 DBD (SC-570; Santa Cruz) which had been precoupled to protein A-Sepharose beads. Immunocomplexes were washed four times in lysis buffer, and one-sixth of the amount (corresponding to 100 μg of cell lysate) was taken for Western blot analysis, while the remainder was used in acetyltransferase assays.
Acetyltransferase assays.
Acetyltransferases expressed in mammalian cells or bacteria were isolated by immunoprecipitation or GST purification, respectively, as described above. In some experiments the acetyltransferases were preincubated with the chelating agent 1,10-phenanthroline (5 or 10 mM; Sigma) for 30 min at 4°C. Acetyltransferases were incubated directly with Drosophila core histones (5 μg) or purified TFIIE34 (2 μg) in AIPH buffer (20 mM Tris-HCl [pH 8.0], 60 mM NaCl, 2 mM EDTA, 0.2% NP-40, 40 μM phenylmethylsulfonyl fluoride) containing [14C]acetyl-CoA (0.05 μCi) for 40 min at 30°C. Reactions were stopped by addition of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were separated on SDS-PAGE gels with 15% (for histones) or 10% (for TFIIE34) polyacrylamide, fixed, and stained with Coomassie brilliant blue. Subsequently, gels were enhanced with Amplify (Amersham) and dried, and labeled proteins were visualized by fluorography.
Western blotting.
Proteins, separated by SDS-PAGE, were transferred onto Immobilon membranes (Millipore). Blots were blocked in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 5% nonfat dried milk powder and subsequently incubated with 1:1,000 dilutions of a monoclonal antibody against the Gal4 DBD (SC-510; Santa Cruz) or a polyclonal antibody against acetylated lysine residues (06-933; Upstate Biotechnology). After a wash, blots were incubated with peroxidase-conjugated antibodies (1:10,000; Jackson ImmunoResearch Laboratories). Blots were washed again, and immunoreactive bands were visualized by enhanced chemiluminescence.
RESULTS
A conserved PHD finger is essential for the in vitro HAT activity of CBP.
Deletion analyses to determine the minimal HAT domains of CBP (amino acids 1232 to 1709) and p300 (amino acids 1195 to 1673) have indicated that regions N- and C-terminal of the putative CoA binding site (amino acids 1459 to 1541 in mouse CBP) are essential for acetylation of histones (27, 60). The precise function of these domains is unknown, but the N terminus of the HAT domain of CBP harbors a region (amino acids 1238 to 1312) that conforms to the C4HC3 consensus of PHD type zinc fingers (1) (Fig. 1A). Alignment of CBP and p300 proteins from different metazoans (Fig. 1A) and the recently isolated orthologs from the plant Aradopsis thaliana (11) (data not shown) revealed that all the putative zinc-coordinating cysteines (C1238, C1241, C1284, C1287, C1295, C1309, and C1312) and histidine (H1292) are strictly conserved. Extrapolation of the solution structures of the PHD fingers from KAP-1 (13) and WSTF (52) to the putative PHD finger in CBP suggests that the first zinc atom would be coordinated by C1238, C1241, H1292, and C1295, while C1284, C1287, C1309, and C1312 would coordinate the second zinc atom (Fig. 1B). To investigate the role of this PHD finger motif in CBP HAT function, the putative zinc-coordinating residues were mutated to alanines. In addition, we also targeted H1298, a residue not predicted to be part of a PHD type zinc ligation scheme (Fig. 1B), as well as several other nonconserved residues (T1243, P1280, V1282, and K1285) for mutation. Finally, we mutated the phenylalanines at positions 1281 and 1307, since all PHD finger proteins have hydrophobic residues, which are often aromatic (1), at this position. As controls, the known F1541A (46) and L1690K/C1691L (34) HAT mutants, each harboring a mutation C-terminal of the PHD finger (Fig. 1A), were included.
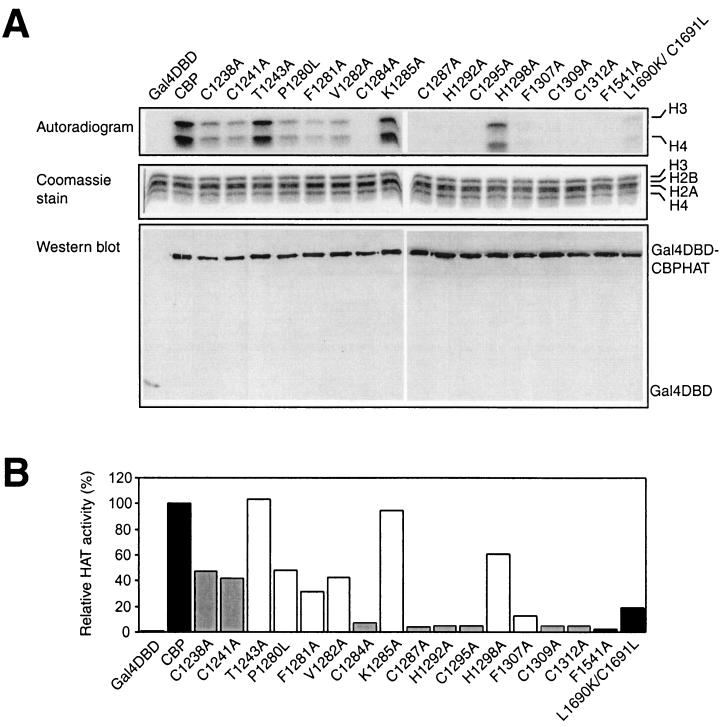
To analyze the in vitro HAT activity of wild-type CBP and the mutants described above, we first overexpressed the region of CBP encompassing the HAT domain (amino acids 1099 to 1758) fused to Gal4 DBD in U-2 OS cells. Subsequently, these proteins were isolated by immunoprecipitation with an anti-Gal4 DBD antibody and tested for the ability to acetylate purified Drosophila core histones in vitro. As observed previously (35, 51), Gal4 DBD-CBP HAT acetylated core histones in vitro, with a preference for histones H3 and H4 (Fig. 2). Mutation of the six most C-terminal zinc-coordinating residues (C1284, C1287, H1292, C1295, C1309, and C1312) and F1307, which is part of the hydrophobic core of PHD proteins (13), resulted in an almost-complete loss of HAT activity. A second class of mutants, including those with mutations of the N-terminal cysteines C1238 and C1241, the conserved hydrophobic F1281, and the nonconserved P1280, V1282, and H1298, displayed reduced activity. The third group of mutants, encompassing the nonconserved residues T1243 and K1285, showed no significant change in HAT activity from that for wild-type CBP. As expected, the known F1541A and L1690K/C1691L HAT mutants showed severely reduced capacities to acetylate histones (34, 46). As a control, Western blot analysis was performed; it showed that comparable amounts of the Gal4 DBD fusions were immunoprecipitated (Fig. 2A). These findings suggest that the CBP HAT domain contains an essential PHD type zinc finger, since mutation of amino acids that are part of the PHD finger signature results in a partial or complete loss of HAT activity.
FIG. 2.
The PHD finger is essential for the in vitro HAT activity of the CBP HAT domain. (A) Gal4 DBD fusions of wild-type CBP HAT or various mutants, overexpressed in U-2 OS cells and immunoprecipitated with an antibody against the Gal4 DBD, were incubated with purified Drosophila core histones in the presence of [14C]acetyl-CoA. Reaction products were separated by SDS-PAGE and stained with Coomassie brilliant blue to verify the presence of equal amounts of histones in each lane. Gels were subsequently dried, and labeled proteins were detected by fluorography. An aliquot from the same immunoprecipitation was used for Western blot analysis with an antibody against the Gal4 DBD. (B) Quantification of the data presented in panel A, as determined by PhosphorImager analysis. Data are presented relative to the activity of wild-type CBP (taken as 100%). Solid bars, wild-type CBP and HAT mutants F1541A and L1690K/C1691L; shaded bars, zinc-coordinating PHD finger mutants; open bars, other PHD finger mutants.
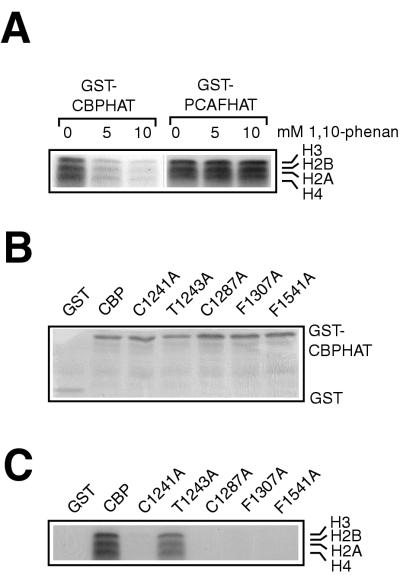
To substantiate the importance of zinc coordination in the CBP HAT domain, we tested whether the zinc-chelating agent 1,10-phenanthroline affected in vitro HAT activity. As shown in Fig. 3A, increasing concentrations of 1,10-phenanthroline clearly inhibited the ability of bacterially expressed CBP HAT to acetylate histones. The HAT domain of PCAF, which does not contain (putative) zinc fingers, also efficiently acetylated histones in vitro. Histone acetylation by PCAF HAT, however, was not sensitive to the presence of 1,10-phenanthroline (Fig. 3A). These results show that the inhibitory effect of 1,10-phenanthroline is specific for CBP, and they support the mutational analysis, which indicates that zinc coordination within the PHD finger motif is important for the HAT activity of CBP (Fig. 2).
FIG. 3.
Zinc ligation and coordination are essential for the in vitro HAT activity of CBP. (A) GST fusions of the HAT domains of CBP and PCAF were preincubated with 1,10-phenanthroline (5 or 10 mM) and subsequently incubated with purified Drosophila core histones in the presence of [14C]acetyl-CoA. Reaction products were processed as described in the legend to Fig. 2. (B) GST fusions of the CBP HAT domain isolated on glutathione-Sepharose beads were separated by SDS-PAGE and stained with Coomassie brilliant blue. (C) GST-CBP HAT fusion proteins were incubated with purified Drosophila core histones in the presence of [14C]acetyl-CoA. Reaction products were processed as described above.
To obtain further evidence for the functional significance of the PHD finger, we performed in vitro acetylation assays with bacterially expressed CBP proteins. Equal amounts of GST-CBP fusion proteins were purified on glutathione beads (Fig. 3B) and tested for the ability to acetylate core histones in vitro. The effects of the different amino acid substitutions on HAT activity (Fig. 3C) were similar to those observed with the Gal4 DBD fusions (Fig. 2), indicating that the importance of the PHD finger is an intrinsic property of the CBP protein. Interestingly, the reduction in the HAT activity of the C1241A mutant was more pronounced when it was expressed in bacteria, suggesting that the phenotype of this mutant might be somewhat compensated for by posttranslational modifications such as phosphorylation (2) or through interactions with other cellular proteins (14, 26, 53, 59).
Taken together, these results strongly support the notion that the region between amino acids 1238 and 1312 of CBP forms a PHD type (C4HC3) zinc finger which is essential for the ability to acetylate histones in vitro.
The PHD finger of CBP is also essential for the acetylation of nonhistone proteins.
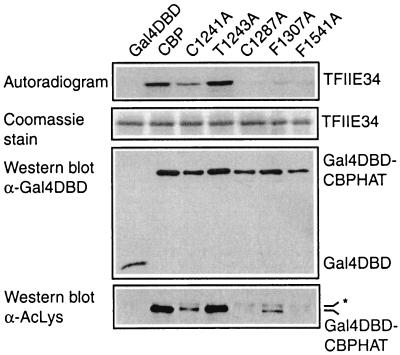
Since histones are not the only in vitro substrates of the acetyltransferase activity of CBP, we next examined wild-type CBP and a selection of mutants for their abilities to acetylate nonhistone proteins. For this, the small (TFIIE34) and large (TFIIE56) subunits of the basal transcription factor TFIIE were purified from bacteria and incubated with immunopurified wild-type and mutant Gal4 DBD-CBP HAT proteins. As shown previously (28), wild-type CBP acetylated the small subunit of TFIIE (TFIIE34) but not the larger TFIIE56 subunit (Fig. 4 and data not shown). Acetylation of TFIIE34 by the T1243A mutant was comparable to that by wild-type CBP, whereas the C1287A and F1541A mutants failed to acetylate TFIIE34 and the F1307A and C1241A mutants showed reduced activity. Similar results were obtained with TFIIF (data not shown), another substrate for in vitro acetylation by CBP (28). Furthermore, Western blot analysis of the Gal4 DBD-CBP HAT proteins showed a pattern for CBP autoacetylation, detected with an antibody against acetylated lysine residues, similar to that for TFIIE34 acetylation (Fig. 4). Thus, our mutational analysis of CBP yielded similar results for core histones (Fig. 2 and 3), TFIIE (Fig. 4), CBP itself (Fig. 4), and TFIIF (data not shown). This suggests that the PHD finger plays a general role in the acetylation reaction and is not involved in determining substrate specificity.
FIG. 4.
The PHD finger is essential for acetylation of nonhistone proteins. Gal4 DBD fusions of wild-type CBP HAT or various mutants, isolated as described in the legend to Fig. 2, were incubated with bacterially expressed TFIIE34 in the presence of [14C]acetyl-CoA. Reaction products were processed as described in the legend to Fig. 2. An aliquot from the same immunoprecipitation was used for Western blot analysis with antibodies against the Gal4 DBD or acetylated lysine residues (AcLys), as indicated. The asterisk on the right indicates aspecific binding of the anti-acetyl antibody.
Role of the PHD finger in the transcriptional activity of CBP.
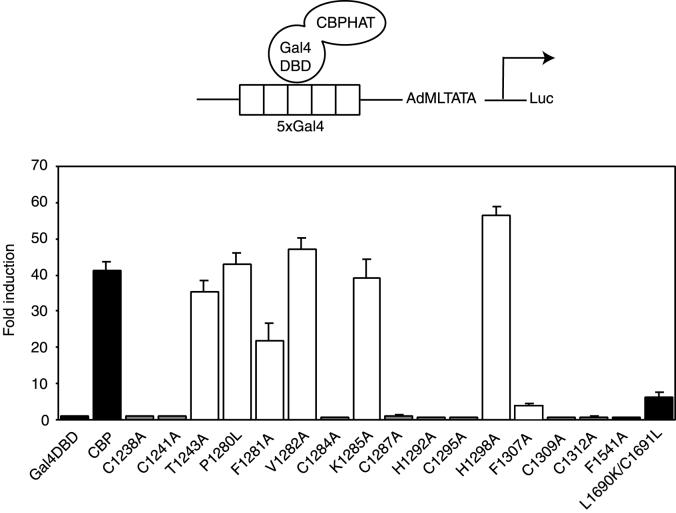
Having established that the PHD finger is essential for in vitro acetyltransferase activity, we wanted to examine its relevance for the transcriptional activity of the CBP protein. Previously, Gal4 DBD-CBP HAT was shown to stimulate transcription from a reporter containing five Gal4 binding sites in front of the adenovirus major late TATA box (5xGal4-AdMLTATA-Luc) in a HAT-dependent fashion (46). In agreement with this, we found that the Gal4 DBD-CBP HAT fusion stimulated transcription from this reporter approximately 40 times more strongly than the Gal4 DBD alone in U-2 OS cells (Fig. 5). The transcriptional activity of the CBP HAT domain depended on the integrity of the PHD finger, since mutation of any of the conserved putative zinc-ligating residues and F1307, which is part of the hydrophobic core of PHD fingers (13), resulted in an almost-complete loss of activity. Altering the conserved F1281 reduced the activity of the fusion protein. In contrast, the nonconserved amino acids T1243, P1280, V1282, K1285, and H1298 are not critical for the transcriptional activity of the fusion protein, since mutating them had no significant effect. As expected, the F1541A and L1690K C1691L HAT mutants showed dramatic reductions in activity, confirming that activation of the reporter depends on the integrity of the HAT function (46).
FIG. 5.
Mutations in the PHD finger disrupt the transcriptional activity of the CBP HAT domain. U-2 OS cells were transfected with expression vectors for Gal4 DBD fusions of the HAT domain of CBP and a 5xGal4-AdMLTATA-Luc reporter, schematically represented at the top. Activation of the luciferase reporter, normalized for β-galactosidase activity, is shown as fold induction over that with the Gal4 DBD alone. Solid bars, wild-type CBP and the HAT mutants F1541A and L1690K/C1691L; shaded bars, zinc-coordinating PHD finger mutants; open bars, other PHD finger mutants. Results are averages of a minimum of three independent experiments assayed in duplicate ± standard errors of the means.
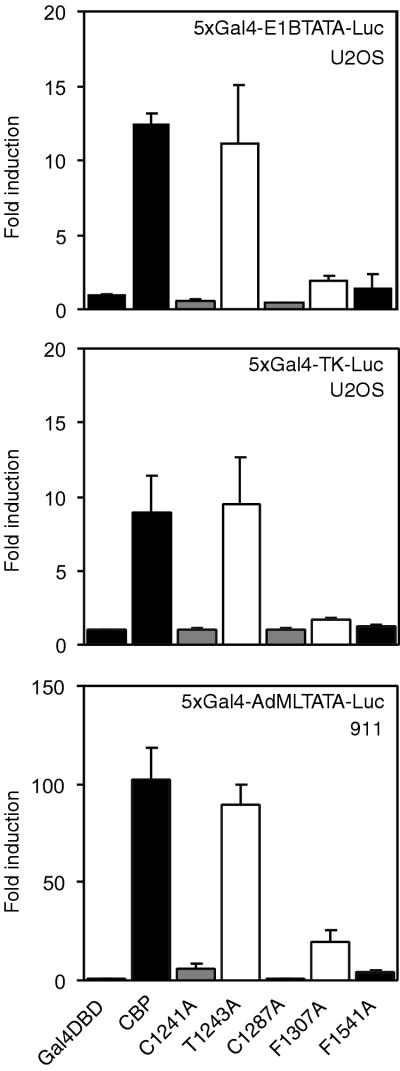
To investigate whether these results depended on the promoter context, two reporters containing five Gal4 binding sites in front of either the adenovirus E1B TATA box (5xGal4-E1BTATA-Luc) or the HSV TK promoter (5xGal4-TK-Luc) were used. As shown in Fig. 6, wild-type CBP and a selection of mutants gave a pattern of activity similar to that with the 5xGal4-AdML-Luc reporter (Fig. 5). In addition, the same constructs were tested for the ability to activate the 5xGal-AdMLTATA-Luc reporter in a different cell line. In 911 cells the activity of the wild-type CBP HAT domain was higher than that observed in U-2 OS cells (Fig. 5), but the effects of the amino acid substitutions were similar for the two cell lines. These results show that the phenotypes of the different mutants are independent of the promoter context or cellular background.
FIG. 6.
Transcriptional activity of the CBP HAT domain on various promoters and cell lines. U-2 OS (top and center panels) and 911 (bottom panel) cells were transfected with expression vectors for Gal4 DBD fusions of the HAT domain of CBP and a 5xGal4-E1BTATA-Luc (top), a 5xGal4-TK-Luc (center), or a 5xGal4-AdMLTATA-Luc (bottom) reporter. Data are presented as described in the legend to Fig. 5.
In summary, there is a good correlation between in vitro HAT activity (Fig. 2 and 3) and transcriptional activity (Fig. 4 and 5) for the majority of mutants. Importantly, all the PHD mutants that cannot acetylate core histones in vitro also fail to activate transcription. We conclude, therefore, that the integrity of the PHD finger in the CBP HAT domain is a prerequisite for HAT-dependent transcription, in agreement with its essential role in in vitro acetyltransferase activity.
Effect of PHD mutations on full-length CBP.
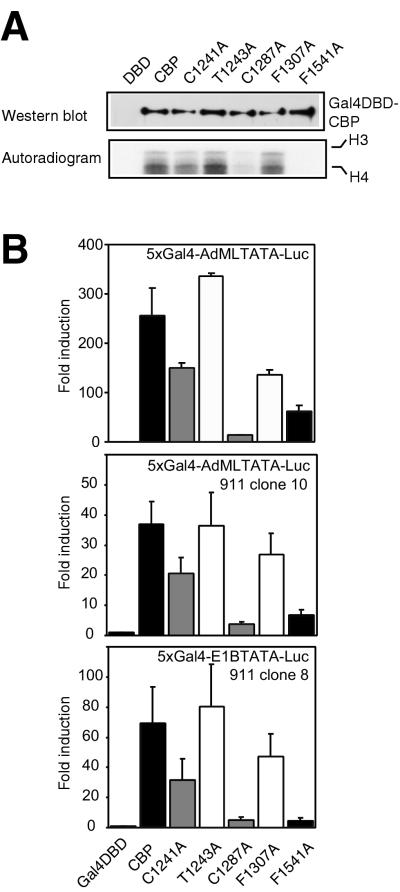
Since regions other than the HAT domain are also important for CBP to function as a transcriptional coactivator (34, 35, 37), we next investigated the importance of the PHD finger for the HAT function and for transcriptional activity in the context of the full-length protein. First, Gal4 DBD fusions of full-length CBP and a selection of mutants were overexpressed in cells, isolated by immunoprecipitation, and tested for their abilities to acetylate core histones. As shown in Fig. 7A, wild-type CBP and the T1243A mutant acetylated histones to similar extents, while the C1287A and F1541A mutants showed dramatic reductions in activity. Mutation of C1241 or F1307 resulted in a partial reduction in activity, most clearly in the former mutant.
FIG. 7.
Effects of PHD mutations on the HAT activity and transcriptional activity of full-length CBP. (A) Gal4 DBD fusions of full-length wild-type CBP HAT or various mutants, overexpressed in 911 cells, were incubated with purified Drosophila core histones in the presence of [14C]acetyl-CoA. Reaction products were processed as described in the legend to Fig. 2. (B) 911 cells (top panel) were transiently transfected with expression vectors for Gal4 DBD fusions of full-length CBP and a 5xGal4-AdMLTATA-Luc reporter. Similar experiments were performed on 911 cells stably transfected with the 5xGal4-AdMLTATA-Luc reporter (clone 10) (center panel) or the 5xGal4-E1BTATA-Luc reporter (clone 8) (bottom panel). Data are presented as described in the legend to Fig. 5.
Next, the transcriptional activity of these constructs was investigated in transient transfection assays. Wild-type CBP and the T1243A mutant activated the 5xGal4-AdMLTATA-Luc reporter in 911 cells to similar extents, while mutation of F1541 or C1287 resulted in a clear reduction of transcriptional activity (Fig. 7B, top panel). The C1241A and F1307A mutants showed partial losses of activity compared to wild-type CBP. Similar results were obtained over a range of concentrations of CBP (50 to 400 ng/well) (data not shown). These data show that the transcriptional phenotypes of the mutants parallel their in vitro HAT activities (Fig. 7A). Interestingly, for some mutants (e.g., C1241A), the correlation between these activities is greater in the full-length protein than in the isolated HAT domain (Fig. 2 and 5).
To examine the role of the PHD finger on promoters within a natural chromatin environment, we isolated individual clones of 911 cells with Gal4 reporters stably integrated into the genome. Introduction of the full-length CBP protein and the mutant versions fused to the Gal4 DBD into cells stably transfected with the 5xGal4-AdMLTATA-Luc reporter (clone 10) resulted in a pattern of activation similar to that observed when a transiently transfected reporter was used (Fig. 7B, center panel). Similar results were obtained with several other independent clones (data not shown), indicating that these results are not due to specific integration events. Furthermore, when cells with an integrated 5xGal4-E1BTATA-Luc reporter (clone 8) were used, the transcriptional-activity pattern of wild-type CBP and the mutant versions was again similar (Fig. 7B, bottom panel).
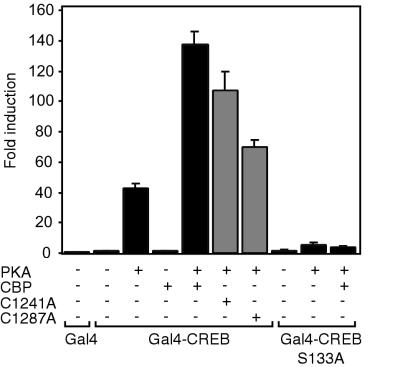
Next we investigated the abilities of PHD mutants to potentiate transcription when they are not tethered directly to a promoter via the Gal4 DBD. For this, cells with an integrated 5xGal4-E1BTATA-Luc reporter (clone 8) were transfected with a Gal4-DBD fusion of CREB, a transcription factor which requires CBP (5, 34), in the absence or presence of CBP and the C1241A and C1287A PHD mutants. Recruitment of CBP by CREB requires PKA-mediated phosphorylation of CREB at serine 133 (5, 16, 38). As shown in Fig. 8, CBP potentiated PKA-activated CREB, while the C1241A and C1287A PHD mutants, particularly the latter, showed reduced activity. No effect of CBP was observed in the absence of PKA or when the PKA phosphorylation site in CREB was mutated to alanine (S133A), indicating that the stimulatory effect of CBP depends strictly on its recruitment by activated CREB.
FIG. 8.
PHD mutations reduce the ability of CBP to coactivate CREB. 911 cells stably transfected with a 5xGal4-E1BTATA-Luc reporter (clone 8) were transfected with either Gal4 DBD alone, Gal4 DBD-CREB, or the S133A mutant, with or without the catalytic subunit of PKA, and with or without an expression vector containing full-length CBP or the C1241A or C1287A PHD mutant, as indicated. Data are presented as described in the legend to Fig. 5.
In summary, these results show that mutation of the PHD finger in the context of the full-length protein affects the HAT activity and transcriptional activity of the CBP protein. Furthermore, given the dramatic reduction in transcriptional activity observed when HAT activity is abolished, we conclude that under these circumstances the HAT domain is a key contributor to the transcriptional activity of CBP.
DISCUSSION
Although acetyltransferases are generally recognized as key regulators of transcription, the critical enzymatic and structural features of HAT modules remain ill defined. In the present study we show that a small region in the CBP HAT domain which is part of the uncharacterized CH2 region conforms to the PHD type zinc finger consensus and that it is essential for the acetyltransferase activity of CBP. In addition, disruption of the PHD finger also reduced the transcriptional activity of the CBP protein, both on transfected and on integrated, chromatinized reporter genes.
Interestingly, several lines of evidence strongly link amino acid substitutions in PHD fingers to human disease. First, mutations in the PHD finger of the ATRX protein, resulting in conversion of the first and sixth zinc-coordinating cysteines into arginine and phenylalanine, respectively, predispose individuals to α-thalassemia (22). Second, two point mutations in the PHD finger of the AIRE-1 gene, which is linked to autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), have been described; one of these alters the third cysteine into a tyrosine residue (8, 56). Third, a number of point mutations have been found in the ING1 gene, a candidate tumor suppressor gene, one of which changes the second cysteine of the PHD finger into a serine residue (24). A single amino acid change (R1379P) in CBP, resulting in complete loss of HAT activity, was recently identified in a patient suffering from RTS (49). Based on our results demonstrating that single amino acid changes in the PHD finger of CBP result in partial or complete losses of HAT activity, it will be of interest to examine whether such mutations are present in RTS patients.
The presence of a bona fide PHD finger is unique to the HAT domain of the CBP/p300 subfamily. When PHD fingers were first identified, they were suggested to be involved in protein-protein interactions (1). Indeed, PHD fingers have been shown to be essential for the interactions of KAP-1 with Mi-2α (57), ZMOX1a with 14-3-3 proteins (25), and SPBP with RNF4 (41). Furthermore, a special type of zinc finger, the extended PHD finger, was shown to be responsible for oligomerization of the AF10 protein (40). Given the location of the PHD finger within the HAT domain, it is possible that in CBP this zinc finger is involved in substrate binding, as shown for the unrelated C2HC zinc finger in the MYST family member MOF (3). The fact that the PHD mutations affected the acetylation of all substrates tested to similar extents implies that in CBP the PHD finger would function as a relatively aspecific docking site for acetylation substrates. This might explain the promiscuous enzymatic behavior of CBP/p300 compared to that of several other HATs (60). Furthermore, such a low-specificity binding function of the PHD finger could also contribute to the interaction of CBP/p300 with chromatin, since the bromodomain of p300 was recently shown to be essential but not sufficient for this interaction (43). Alternatively, the PHD finger might not function primarily in intermolecular protein interactions but mainly serve in maintaining the structural integrity of the CBP/p300 HAT domain. In either case, however, it is clear that the PHD finger cannot be functionally separated from the HAT domain and should therefore be viewed as an integral part of the enzymatic core of the CBP protein.
Acknowledgments
We thank F. Claessens, R. H. Goodman, N. C. Jones, T. Kouzarides, M. G. Parker, and H. G. Stunnenberg for plasmid constructs. We are grateful to G. Chalkley for Drosophila core histone preparations, R. G. J. Vries and J. H. A. Martens for helpful discussions and N. Little and T. Mahmoudi for critical reading of the manuscript.
E.K. and H.T were supported by the Dutch Cancer Society (KWF).
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396:184-186. [DOI] [PubMed] [Google Scholar]
- 3.Akhtar, A., and P. B. Becker. 2001. The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep. 2:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arany, Z., W. R. Sellers, D. M. Livingston, and R. Eckner. 1994. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 77:799-800. [DOI] [PubMed] [Google Scholar]
- 5.Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-229. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. Kingston, D. Moore, J. J. Seidman, J. Smith, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 7.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 8.Bjorses, P., M. Halonen, J. J. Palvimo, M. Kolmer, J. Aaltonen, P. Ellonen, J. Perheentupa, I. Ulmanen, and L. Peltonen. 2000. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am. J. Hum. Genet. 66:378-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blobel, G. A. 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 10.Borden, K. L., and P. S. Freemont. 1996. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6:395-401. [DOI] [PubMed] [Google Scholar]
- 11.Bordoli, L., M. Netsch, U. Luthi, W. Lutz, and R. Eckner. 2001. Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res. 29:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulger, M., and J. T. Kadonaga. 1994. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5:241-262.
- 13.Capili, A. D., D. C. Schultz, F. J. Rauscher III, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C. J., Z. Deng, A. Y. Kim, G. A. Blobel, and P. M. Lieberman. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung, W. L., S. D. Briggs, and C. D. Allis. 2000. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 12:326-333. [DOI] [PubMed] [Google Scholar]
- 16.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 17.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 18.Fallaux, F. J., O. Kranenburg, S. J. Cramer, A. Houweling, H. Van Ormondt, R. C. Hoeben, and A. J. Van Der Eb. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215-222. [DOI] [PubMed] [Google Scholar]
- 19.Flint, K. J., and N. C. Jones. 1991. Differential regulation of three members of the ATF/CREB family of DNA-binding proteins. Oncogene 6:2019-2026. [PubMed] [Google Scholar]
- 20.Fujii, G., R. Tsuchiya, Y. Itoh, K. Tashiro, and S. Hirohashi. 1998. Molecular cloning and expression of Xenopus p300/CBP. Biochim. Biophys. Acta 1443:41-54. [DOI] [PubMed] [Google Scholar]
- 21.Gayther, S. A., S. J. Batley, L. Linger, A. Bannister, K. Thorpe, S. F. Chin, Y. Daigo, P. Russell, A. Wilson, H. M. Sowter, J. D. Delhanty, B. A. Ponder, T. Kouzarides, and C. Caldas. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 24:300-303. [DOI] [PubMed] [Google Scholar]
- 22.Gibbons, R. J., S. Bachoo, D. J. Picketts, S. Aftimos, B. Asenbauer, J. Bergoffen, S. A. Berry, N. Dahl, A. Fryer, K. Keppler, K. Kurosawa, M. L. Levin, M. Masuno, G. Neri, M. E. Pierpont, S. F. Slaney, and D. R. Higgs. 1997. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 17:146-148. [DOI] [PubMed] [Google Scholar]
- 23.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 24.Gunduz, M., M. Ouchida, K. Fukushima, H. Hanafusa, T. Etani, S. Nishioka, K. Nishizaki, and K. Shimizu. 2000. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 60:3143-3146. [PubMed] [Google Scholar]
- 25.Halbach, T., N. Scheer, and W. Werr. 2000. Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res. 28:3542-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamamori, Y., V. Sartorelli, V. Ogryzko, P. L. Puri, H. Y. Wu, J. Y. Wang, Y. Nakatani, and L. Kedes. 1999. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96:405-413. [DOI] [PubMed] [Google Scholar]
- 27.Henttu, P. M., E. Kalkhoven, and M. G. Parker. 1997. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol. Cell. Biol. 17:1832-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson, S., and L. Pillus. 1999. Modifying chromatin and concepts of cancer. Curr. Opin. Genet. Dev. 9:175-184. [DOI] [PubMed] [Google Scholar]
- 31.Jeanmougin, F., J. M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson. 1997. The bromodomain revisited. Trends Biochem. Sci. 22:151-153. [DOI] [PubMed] [Google Scholar]
- 32.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 33.Koken, M. H., A. Saib, and H. de The. 1995. A C4HC3 zinc finger motif. C. R. Acad. Sci. Ser. III 318:733-739. [PubMed] [Google Scholar]
- 34.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 35.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 37.Kurokawa, R., D. Kalafus, M. H. Ogliastro, C. Kioussi, L. Xu, J. Torchia, M. G. Rosenfeld, and C. K. Glass. 1998. Differential use of CREB binding protein-coactivator complexes. Science 279:700-703. [DOI] [PubMed] [Google Scholar]
- 38.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 39.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 40.Linder, B., R. Newman, L. K. Jones, S. Debernardi, B. D. Young, P. Freemont, C. P. Verrijzer, and V. Saha. 2000. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J. Mol. Biol. 299:369-378. [DOI] [PubMed] [Google Scholar]
- 41.Lyngso, C., G. Bouteiller, C. K. Damgaard, D. Ryom, S. Sanchez-Munoz, P. L. Norby, B. J. Bonven, and P. Jorgensen. 2000. Interaction between the transcription factor SPBP and the positive cofactor RNF4. An interplay between protein binding zinc fingers. J. Biol. Chem. 275:26144-26149. [DOI] [PubMed] [Google Scholar]
- 42.Mak, H. Y., S. Hoare, P. M. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marmorstein, R. 2001. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2:422-432. [DOI] [PubMed] [Google Scholar]
- 45.Marmorstein, R., and S. Y. Roth. 2001. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 11:155-161. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Balbas, M. A., A. J. Bannister, K. Martin, P. Haus-Seuffert, M. Meisterernst, and T. Kouzarides. 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellon, P. L., C. H. Clegg, L. A. Correll, and G. S. McKnight. 1989. Regulation of transcription by cyclic AMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 86:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muraoka, M., M. Konishi, R. Kikuchi-Yanoshita, K. Tanaka, N. Shitara, J. M. Chong, T. Iwama, and M. Miyaki. 1996. p300 gene alterations in colorectal and gastric carcinomas. Oncogene 12:1565-1569. [PubMed] [Google Scholar]
- 49.Murata, T., R. Kurokawa, A. Krones, K. Tatsumi, M. Ishii, T. Taki, M. Masuno, H. Ohashi, M. Yanagisawa, M. G. Rosenfeld, C. K. Glass, and Y. Hayashi. 2001. Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 10:1071-1076. [DOI] [PubMed] [Google Scholar]
- 50.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 51.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 52.Pascual, J., M. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J. Mol. Biol. 304:723-729. [DOI] [PubMed] [Google Scholar]
- 53.Perissi, V., J. S. Dasen, R. Kurokawa, Z. Wang, E. Korzus, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1999. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl. Acad. Sci. USA 96:3652-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrij, F., R. H. Giles, H. G. Dauwerse, J. J. Saris, R. C. Hennekam, M. Masuno, N. Tommerup, G. J. van Ommen, R. H. Goodman, D. J. Peters, et al. 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376:348-351. [DOI] [PubMed] [Google Scholar]
- 55.Saha, V., T. Chaplin, A. Gregorini, P. Ayton, and B. D. Young. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. USA 92:9737-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saugier-Veber, P., N. Drouot, L. M. Wolf, J. M. Kuhn, T. Frebourg, and H. Lefebvre. 2001. Identification of a novel mutation in the autoimmune regulator (AIRE-1) gene in a French family with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Eur. J. Endocrinol. 144:347-351. [DOI] [PubMed] [Google Scholar]
- 57.Schultz, D. C., J. R. Friedman, and F. J. Rauscher III. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15:428-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shikama, N., J. Lyon, and N. B. La Thangue. 2000. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230-236. [DOI] [PubMed] [Google Scholar]
- 59.Soutoglou, E., B. Viollet, M. Vaxillaire, M. Yaniv, M. Pontoglio, and I. Talianidis. 2001. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 20:1984-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, S. 2001. One HAT size fits all? Nat. Struct. Biol. 8:8-10. [DOI] [PubMed] [Google Scholar]
- 62.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 63.Vo, N., and R. H. Goodman. 2001. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276:13505-13508. [DOI] [PubMed] [Google Scholar]
- 64.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 65.Yan, Y., N. A. Barlev, R. H. Haley, S. L. Berger, and R. Marmorstein. 2000. Crystal structure of yeast esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6:1195-1205. [DOI] [PubMed] [Google Scholar]
- 66.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 67.Yokomori, K., C. P. Verrijzer, and R. Tjian. 1998. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc. Natl. Acad. Sci. USA 95:6722-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]