Abstract
AFX-like Forkhead transcription factors, which are controlled by phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB) signaling, are involved in regulating cell cycle progression and cell death. Both cell cycle arrest and induction of apoptosis are mediated in part by transcriptional regulation of p27kip1. Here we show that the Forkheads AFX (FOXO4) and FKHR-L1 (FOXO3a) also directly control transcription of the retinoblastoma-like p130 protein and cause upregulation of p130 protein expression. Detailed analysis of p130 regulation demonstrates that following Forkhead-induced cell cycle arrest, cells enter G0 and become quiescent. This is shown by a change in phosphorylation of p130 to G0-specific forms and increased p130/E2F-4 complex formation. Most importantly, long-term Forkhead activation causes a sustained but reversible inhibition of proliferation without a marked increase in apoptosis. As for the activity of the Forkheads, we also show that protein levels of p130 are controlled by endogenous PI3K/PKB signaling upon cell cycle reentry. Surprisingly, not only nontransformed cells, but also cancer cells such as human colon carcinoma cells, are forced into quiescence by Forkhead activation. We therefore propose that Forkhead inactivation by PKB signaling in quiescent cells is a crucial step in cell cycle reentry and contributes to the processes of transformation and regeneration.
Mammalian cells require an extracellular proliferative signal directly after mitosis in order to keep on growing and dividing. When cells are faced with a lack of such a signal, they will either die or go into growth arrest in a postmitotic G1 phase. Two important intracellular signaling pathways that transduce such proliferative signals are the Ras and phosphatidylinositol 3-kinase (PI3K) pathways. Ras and PI3K can regulate various features of cell proliferation such as cytoskeletal rearrangements, gene transcription, DNA synthesis, and survival (reviewed in references 4 and 17). The proto-oncogene protein kinase B (PKB) is a major target of PI3K signaling in the control of cell proliferation (reviewed in reference 11), as it is involved in antiapoptotic signaling as well as cell cycle control. Recently, PKB was found to directly phosphorylate and inactivate a subfamily of Forkhead transcription factors consisting of AFX (FOXO4), FKHR (FOXO1), and FKHR-L1 (FOXO3a) (6, 29, 47). In addition, Ras, via the RalGEF/Ral pathway, cooperates with PKB in inhibiting AFX activity (29). Importantly, these two pathways are often found deregulated in tumor cells. Ras itself is mutated to an active form in 15% of all cancers, and the negative regulator of PI3K signaling, the tumor suppressor PTEN, has been shown to be mutated or deleted in a wide variety of tumors (reviewed in references 3 and 14).
Inactivation of the Forkhead transcription factors may play a major role in the control of cellular proliferation by the PI3K/PKB and Ras/Ral pathways. We and others have recently shown that all three Forkheads inhibit cell cycle progression at the G1/S transition, at least in part by controlling transcription of the gene for the p27kip1 cyclin-dependent kinase (cdk) inhibitor (7, 38, 42). Nevertheless, a p27kip1-independent mechanism for Forkhead-induced cell cycle arrest is likely to exist, since AFX was still able to partly reduce the activity of the cyclin E/cdk2 complex in the absence of p27kip1 (38).
The continuation of cell proliferation at various stages of the cell cycle involves inactivation of at least one of three members of the retinoblastoma family of nuclear pocket proteins. The general mechanism by which this family exerts its effects is the binding of different members of the E2F family of transcription factors; this binding actively represses genes required for cell cycle progression (reviewed in reference 21). The pRb/p105 protein is an essential component of the G1/S checkpoint. pRb is present at relatively constant levels throughout the cell cycle but is hyperphosphorylated by cyclin/cdk complexes and released from E2F-1 at the G1/S transition, allowing continuation through the cell cycle (reviewed in reference 50). Conversely, the p107 and pRb2/p130 proteins are regulated at the protein level as well as by phosphorylation. p107 protein levels are low during quiescence (commonly referred to as G0) and early G1 but high during the other stages of the cell cycle. p130 protein levels, on the other hand, are low in cycling cells but increase once cells exit the cell cycle (reviewed in reference 21). The rise in p130 protein levels at the G0 stage of the cell cycle is accompanied by a change in the phosphorylation of p130 from a hyperphosphorylated form (form 3) predominating in cycling cells to the hypophosphorylated forms in G0 cells (35, 36). The large amount of hypophosphorylated p130 in G0 cells binds to the E2F-4 transcription factor, which is thought to repress genes required for reentry into early G1 phase, thereby maintaining the quiescent state (51).
An addition to this well-documented function for p130 is the observation that p130 harbors a cyclin interaction motif (59) and may therefore in certain cases function as a “classical” cdk inhibitor. Indeed, a recent report (10) showed that serum-starved p27kip1−/− mouse embryo fibroblasts (MEFs) still display efficient inhibition of cyclin E/cdk2 activity. In these p27kip1−/− cells, p130 was found to bind to the cyclin E/cdk2 complexes, and inhibition of cdk2 activity within this complex was attributed to p130 binding. However, this finding is in apparent contrast to other reports showing that overexpression of exogenous p130 does not induce growth arrest (23). Thus, it is unclear whether and how endogenous p130 regulation can contribute to cell cycle arrest in the presence of other cell cycle regulators.
As stated, we have previously shown that Forkhead expression induces cell cycle arrest in G1. However, it remained to be established whether prolonged Forkhead activation would result in a definite (senescence) or reversible (quiescence) exit from the cell cycle into G0, or whether an initial G1 arrest is followed by apoptosis. The latter is suggested by experiments with pre-B cells in which Forkhead-regulated expression of p27kip1 resulted in cell death by apoptosis (16). Here we show that Forkheads can bind regulatory sequences within the p130 promoter region, increase p130 mRNA levels, and consequently regulate p130 protein levels. We provide evidence that the observed increase in p130 protein levels represents an entry of cells into a quiescent state. Furthermore, we show that endogenous PI3K signaling regulates p130 protein levels as well. Analysis of MEFs lacking various combinations of pRb family members shows that pRb family members are required for inhibition of bromodeoxyuridine (BrdU) incorporation by Forkheads but that p130 upregulation is not sufficient. Instead, p130 regulation is likely to be essential in establishing quiescence. These results show that the PI3K/PKB/Forkhead pathway is not only structurally but also functionally conserved throughout evolution, since both quiescence in mammalian cells and Dauer formation in Caenorhabditis elegans are reversible phenotypes. We also observe that tumor cells become quiescent following Forkhead expression, and these data suggest that for certain types of tumors (e.g., PTEN-negative tumors), PKB activation by autocrine growth factors or oncogenic mutations and consequent inhibition of Forkhead transcription factors are essential for enabling cell cycle reentry.
MATERIALS AND METHODS
Plasmids.
pBabe-FKHR-L1.A3 was created by ligating a Klenow fragment-blunted HindIII/BamHI fragment of pcDNA3-HA-FKHR-L1.A3 into Klenow fragment-blunted, BamHI-cut pBabe-puro. pcDNA3-HA-FKHR-L1.A3-ER has been described previously (16). pGL2-p130intron was created as follows. A 720-bp fragment of the first intron of the p130 gene was cloned from DLD-1 genomic DNA by PCR using the forward primer 5′-GAT GGC ACC ACT GAT ACA GAA G-3′ and the reverse primer 5′-CAA AGT GCT AGG AGA ATA TGT C-3′. This fragment was cloned into pGEMT and mutated at position +6 to create a KpnI restriction site. A KpnI-SalI fragment of pGEMT-p130intron was subsequently ligated into KpnI-XhoI-cut pGL2 to create pGL2-p130intron. pGL2-p130intron.DBEmut was created by site directed mutagenesis using the primer 5′-CAT AAA TAA ATA AGT AAA GAA ATA AAA GGA GGT ATT C-3′. All constructs were verified by automated sequencing. pBabe-HA-AFX has been described previously (38). pCMV-HA-p130 was a kind gift of D. Cobrinik.
Cell culture.
The DL23 cell line was created as follows. Linearized pcDNA3-HA-FKHR-L1.A3-ER (16) was transfected into DLD-1 human colon carcinoma cells by electroporation. Transfectants were selected for 2 weeks on 300 μg of Geneticin/ml. Subsequently, clones were isolated and analyzed for expression of the fusion protein. The DL23 subclone was chosen for further study. Primary MEFs derived from wild-type, p27kip1−/−, p130−/−, and p27kip1−/− p130−/− mice were a gift from B. Scheyen. MEFs, as well as A14, NIH 3T3, U87MG, and 3T3-PKB-ER (28) cells were grown on Dulbecco's modified Eagle medium with standard supplements. MCF7 cells were grown on DF12 medium. DL23 and DLD-1 cells were grown on RPMI-1640 supplemented similarly. 4-Hydroxy-tamoxifen (4OHT; Sigma) was diluted in RPMI 1640 and added to the cells at a final concentration of 500 nM (DL23 and DLD-1). LY294002 was dissolved in dimethyl sulfoxide and used at a final concentration of 10 μM.
Northern blotting.
Two micrograms of mRNA (polyA-Tract; Promega) purified from 1 mg of total RNA (RNAZol; TEL-TEST, Inc.) was run on a formaldehyde denaturing gel and blotted onto a GeneScreen-Plus nylon membrane (NEN). The blot was hybridized by using radiolabeled p130 (fragment of pCMV-HA-p130) and glyceraldehyde-3-phosphate dehydrogenase (NotI-linearized pUC19-GAPDH) probes.
Electrophoretic mobility shift assay.
Bandshift analysis of the human p130 gene using GST-AFX.DBD was performed as described previously (29). The oligonucleotides used were p130.DBEwt (5′-CTC ATA AAT AAA TAA GTA AAC AAA TAA AAG GAG GTA TTC-3′) and p130.DBEmut (5′-CTC ATA AAT AAA TAA GTA AAG AAA TAA AAG GAG GTA TTC-3′). Competition experiments were performed with 1:0, 1:10, 1:25, and 1:50 ratios of labeled to nonlabeled oligonucleotides. Bandshift analyses of NIH 3T3, DLD-1, DL23, and 3T3-PKB-ER cells were performed as described previously (55).
Antibodies, immunoprecipitations, and immunoblotting.
Anti-p130 (C-20), anti-E2F-4 (C-20), anti-ERα (MC-20), anti-cdk2 (M2), anti-cyclin D1 (H11), anti-cyclin E (C-19), and anti-actin (I-19) were from Santa Cruz. Anti-pRb was from PharMingen. Anti-p27kip1 was from Transduction Laboratories. Anti-Bim was from Affinity Bioreagents. Anti-phosphoT32-FKHR-L1 was a kind gift from A. Brunet. For immunoprecipitations, MEFs or DL23 cells were lysed in lysis buffer (50 mM Tris [pH 7.5], 1% Triton X-100, 100 mM NaCl, and 5 mM EDTA supplemented with NaF, aprotinin, leupeptin, benzamidine, and NaVO4), and the cleared supernatant was incubated at 4°C with 10% protein A-Sepharose and 3 μl of anti-cdk2 or anti-cyclin E for 3 h. Beads were washed three times and resuspended in 1× Laemmli sample buffer. For immunoblotting of total lysates, cells were lysed in 1× Laemmli sample buffer, electrophoresed, and blotted onto nitrocellulose membranes according to the standard protocol. Immunoblotting was performed by blocking the membranes in BLOTTO (2% nonfat dry milk-0.5% bovine serum albumin in PBS-Tween [phosphate-buffered saline plus 0.1% Tween 20]) for 1 h at room temperature. Primary and secondary antibody incubations were carried out in PBS-Tween overnight and for 2 h at 4°C, respectively, and the membrane was washed four times after each incubation. Proteins were visualized by standard enhanced chemiluminescence (Amersham) and autoradiography.
[35S]methionine pulse-labeling.
DLD-1 and DL23 cells were grown to subconfluency, labeled with 100 μM [35S]methionine for 2 h, and lysed in 50 mM Tris (pH 7.5)-1% Triton X-100. Samples were corrected for the amount of cells, and [35S]methionine incorporation was measured by scintillation counter.
BrdU incorporation, FACS analysis, and cyclin E-associated kinase assay.
BrdU incorporation, fluorescence-activated cell sorter (FACS) analysis, and cyclin E/cdk2 kinase assays were performed as described previously (38).
Retroviral infections.
Retroviral infections using pBabePuro, pBabe-HA-AFX, and pBabePuro-HA-FKHR-L1.A3 were performed as described previously (38).
Luciferase assay.
Luciferase measurements were performed as described previously (29).
RESULTS
Forkhead transcription factors cause upregulation of the p130 pocket protein.
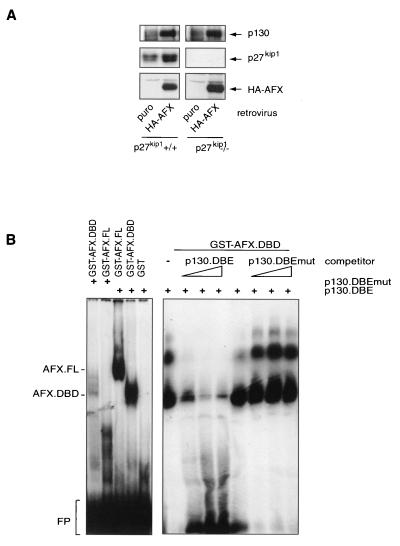
We have previously shown that infection of MEFs with a hemagglutinin (HA)-AFX-expressing retrovirus causes strong inhibition of cyclin E-associated kinase activity as well as of proliferation (38). While testing cell cycle regulators for their abilities to respond to Forkhead expression, we noticed an increase in protein expression of the pRb-like p130 protein. As shown in Fig. 1A, infection with an HA-AFX-expressing retrovirus increases p130 protein expression to similar levels in wild-type and p27kip1−/− cells compared to controls, indicating that this upregulation of p130 occurs independently of AFX-induced cell cycle arrest and/or the presence of p27kip1. We observed similar increases in p130 protein levels upon infection of the same cell lines with a retrovirus expressing HA-FKHR-L1.A3 (a mutant of FKHR-L1 that lacks all three inhibitory PKB phosphorylation sites) (data not shown) (6).
FIG. 1.
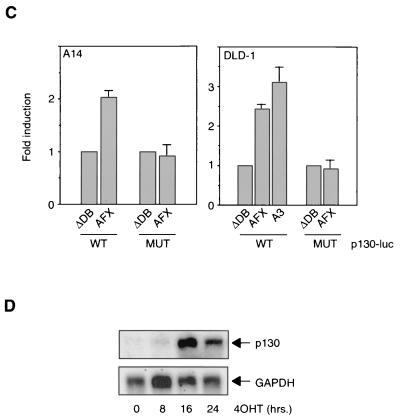
Forkhead transcription factors cause upregulation of the p130 pocket protein. (A) MEFs from wild-type (p27kip1+/+) or p27kip1 knockout (p27kip1−/−) mice were infected with either a control retrovirus or an HA-AFX-containing retrovirus. Total cellular lysates were subsequently analyzed for p130, p27kip1, and HA-AFX expression levels. (B) Purified GST or GST-DBD protein was incubated with a radiolabeled wild-type (p130.DBE) or mutant (p130.DBEmut) probe in the absence or presence of increasing amounts of a nonlabeled oligonucleotide. FL, full length; FP; free probe. (C) A14 or DLD-1 cells were transfected with pGL2-p130intron or pGL2-p130intron.DBEmut, with or without the indicated Forkhead constructs, and luciferase activity was measured. Data are averages from at least three experiments. (D) Two micrograms of RNA was electrophoresed, blotted onto a nylon membrane, and probed for the presence of p130 and GAPDH as a loading control by using radiolabeled probes.
Next, we investigated whether induction of p130 protein levels was mediated through transcriptional control of the p130 gene by these Forkhead transcription factors. Analysis of the human genome database revealed the presence of several suboptimal Forkhead binding sequences (DNA binding effect; TTGTTTAC [20]) in the 5′ flanking sequences of the p130 gene and one optimal inverse sequence in the first intron. To establish functionality, we tested whether the Forkheads can bind this optimal sequence. Purified GST-AFX and GST-DBD (respectively, full-length AFX and the AFX DNA-binding domain fused to glutathione S-transferase [29]) bound specifically to a radiolabeled oligonucleotide encompassing the DBE of the p130 gene but not to an oligonucleotide in which the DBE was mutated (Fig. 1B). Moreover, an unlabeled wild-type oligonucleotide, but not an unlabeled mutant oligonucleotide, could compete with the binding of the labeled wild-type probe to the Forkhead. Next, we cloned a 720-bp fragment of the first intron of the p130 gene and linked this to a luciferase reporter gene. Cotransfection of this reporter construct with Forkheads showed a clear increase in luciferase activity in A14 and DLD-1 cells (Fig. 1C). In keeping with the lack of Forkhead binding, the same construct now containing the mutant DBE did not respond to Forkhead expression. To further demonstrate transcriptional control, we used a cell line in which we could induce Forkhead activity by addition of 4OHT (for a complete description, see below). Activation of FKHR-L1 resulted in a clear increase in p130 mRNA levels (Fig. 1D). These data show that Forkheads can directly control transcription of the p130 gene.
Forkhead expression induces a cell cycle exit into quiescence.
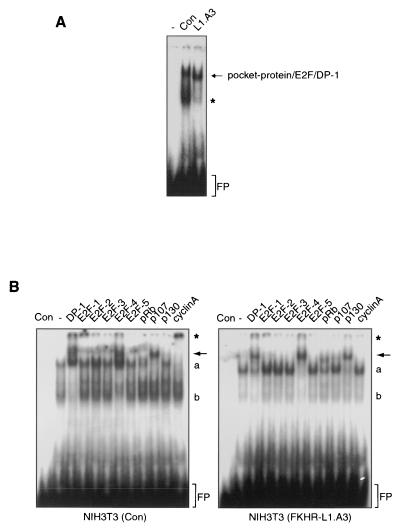
Increased expression of p130 protein is often associated with cell cycle exit and an entry into quiescence or senescence (21). We thus examined whether Forkhead-mediated p130 regulation may reflect cellular quiescence or senescence. Several general markers (e.g., reduction of general protein synthesis) for the G0 phase of the cell cycle have been described, but p130 appears to be the best-characterized marker. First, p130 protein is upregulated (51), as observed here after Forkhead activation (Fig. 1). Second, the p130 phosphorylation status changes from the hyperphosphorylated form 3 to the phosphorylated forms 1 and 2 (35, 36). Third, p130 associates with E2F-4, resulting in active repression of genes required for cell cycle reentry (51). To examine whether the Forkheads indeed cause cells to exit the cell cycle, we infected NIH 3T3 cells with a control virus or an HA-FKHR-L1.A3-expressing retrovirus and analyzed pocket-protein/E2F complexes by bandshift analysis using an oligonucleotide containing a consensus E2F family binding sequence (55). Normally growing NIH 3T3 cells infected with the control virus displayed moderate amounts of pocket protein/E2F complexes and relatively high levels of uncomplexed (free) E2F (Fig. 2A). Moreover, the major E2F complex consisted of p107, E2F-4, and cyclin A, as expected for cycling cells (Fig. 2B, left panel) (41). Proliferation of NIH 3T3 cells expressing the FKHR-L1.A3 protein, however, was strongly inhibited (data not shown) (38), and band supershift analysis using antibodies against the various E2F and pocket proteins showed that, compared to control cells, these cells displayed a complete shift in pocket protein/E2F complexes from mainly p107/E2F-4/cyclin A to p130/E2F-4 and a clear decrease in free E2F levels (Fig. 2B). Together with the increase in p130 protein levels, this indicates that Forkhead activity induces not only cell cycle arrest but also an exit from the cell cycle and entrance into a state of quiescence or senescence.
FIG. 2.
Elevated p130 protein levels are indicative of Forkhead-induced cell cycle exit. (A) Total cellular lysates from NIH 3T3 cells that were either left uninfected (−) or infected either with a control virus (Con) or with an FKHR-L1.A3-expressing retrovirus (L1.A3) were analyzed for the presence of E2F/DP-1/pocket protein complexes. Asterisk, free E2F; FP, free probe. (B) NIH 3T3 cells were infected and analyzed as for panel A. Supershifting antibodies are listed above the gel. Arrows indicate the disappearance of p107/E2F-4 complexes in the third lane from the right, and the appearance of p130/E2F-4 complexes in the second lane from the right, due to Forkhead activity. Asterisk, supershifted cyclin A-containing complexes; a, nonsupershifted E2F/pocket protein complexes; b, free E2F; FP, free probe.
Forkhead-mediated increase in p130 protein levels does not contribute to inhibition of cyclin E-associated kinase activity.
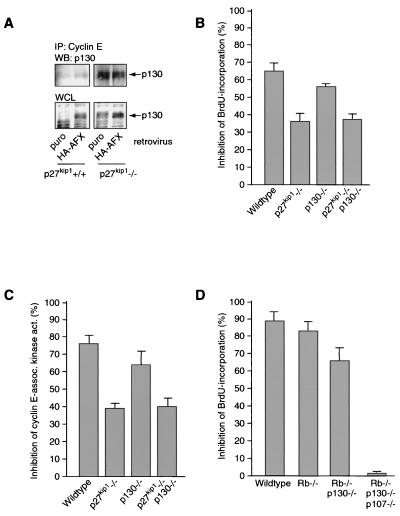
We have previously shown that Forkhead expression increases p27kip1 protein levels and inhibits cyclin E/cdk2 activity. It has been suggested that p130, like p27kip1, can function as a cdk inhibitor (10). In agreement with these recent observations, we observed some association of p130 with cyclin E/cdk2 complexes in wild-type cells, and this association was increased in p27kip1−/− cells (Fig. 3A). However, no further increase in the p130/cyclin E/cdk2 complex was observed upon introduction of HA-AFX either in wild-type or in p27kip1-deficient MEFs, although AFX did upregulate p130 in these experiments (Fig. 3A).
FIG. 3.
Forkhead-induced upregulation of p130 does not contribute to inhibition of cyclin E-associated kinase activity. (A) MEFs infected as for Fig. 1A were lysed, and the amount of p130 in cyclin E-containing complexes was analyzed by Western blotting (WB) of cyclin E immunoprecipitates (IP). WCL, whole-cell lysate. (B and C) Primary wild-type MEFs and MEFs from p27kip1, p130, or p27kip1 p130 knockout mice were infected with a control or an FKHR-L1.A3-encoding retrovirus. At 24 h postinfection, proliferation was measured by BrdU incorporation (B) and cyclin E-associated kinase activity (act.) (C). Graphs show relative inhibition of proliferation by FKHR-L1.A3, as measured by these two assays, compared to proliferation with the control. Data are averages from three independent experiments. (D) Primary wild-type MEFs and MEFs from pRb, pRb p130, and pRb p130 p107 knockout mice were treated with 10 nM LY249002 for 16 h, and proliferation was assayed by BrdU incorporation.
FKHR-L1.A3 induced strong inhibition of both BrdU incorporation and cyclinE/cdk2 activity in wild-type MEFs, similar to that observed upon overexpression of HA-AFX in the same cell lines (Fig. 3B and C, respectively) (38). These effects were greatly reduced, but not eliminated, in p27kip1−/− cells (Fig. 3B and C) (38). However, inhibition of BrdU incorporation or cyclin E-associated kinase activity upon Forkhead expression in MEFs lacking p130 differed only slightly from that for wild-type MEFs. Moreover, compared to that in p27kip1−/− MEFs, Forkhead expression in p130−/− p27kip1−/− cells showed no additional attenuation of cell proliferation arrest or decrease in cyclin E/cdk2 activity (Fig. 3B and C). These data suggest that p130 does not significantly contribute to the Forkhead-mediated decrease in cyclin E/cdk2 activity and in cellular proliferation. However, at this point we cannot exclude the possibility that either pRB or p107 or both act redundantly in the absence of p130. We therefore investigated the effect of Forkhead activation in MEFs lacking either pRb alone or combinations of the three pRb family members. Treatment of wild-type MEFs with LY294002 increased p130 expression (12) (data not shown) and inhibited BrdU incorporation in MEFs, as reported previously (Fig. 3D) (12, 38). LY294002 treatment inhibited BrdU incorporation in MEFs with a pRb gene deletion alone or in combination with a p130 gene deletion to the same extent as for wild-type MEFs. However, cells lacking all three pRb family members were no longer inhibited by LY294002. In keeping with this result, Forkhead expression in pRb−/− p107−/− p130−/− MEFs also no longer resulted in decreased BrdU incorporation (data not shown). However, in this experiment we could not control for the efficiency of infection by selection, since the selection markers used for creating pRb−/− p107−/− p130−/− MEFs precluded this. Taken together, these results suggest that indeed pRb or p107 or both can substitute for p130 in cell cycle arrest induced by the PI3K/PKB/Forkhead pathway.
Forkhead-induced cell cycle exit is mediated by the PI3K/PKB pathway.
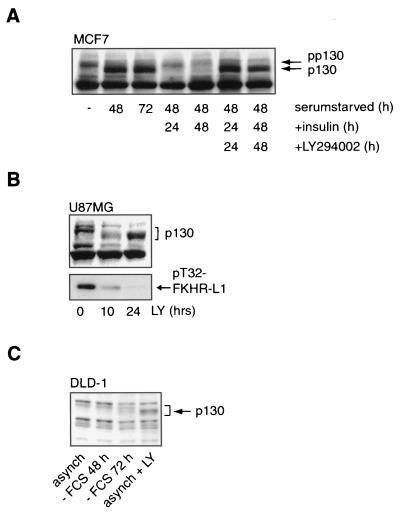
To examine whether the endogenous PI3K/PKB/Forkhead pathway is involved in p130 regulation, we investigated whether the increase in p130 protein levels upon serum starvation can be reversed through activation of the PI3K/PKB signaling pathway. Indeed, serum deprivation of a variety of cell lines, including the insulin-responsive cell line MCF7, resulted in an increase in p130 protein levels, which could be reversed by addition of insulin (Fig. 4A). However, preincubation with the PI3K inhibitor LY294002 completely blocked this, showing the PI3K dependence of p130 regulation in these cells (Fig. 4A). Similarly, treatment of the PTEN-negative glioblastoma cell line U87MG with LY294002 for 24 h resulted in Forkhead dephosphorylation and thus activation (6, 29), with a concomitant increase in p130 protein levels (Fig. 4B). Importantly, treatment of the human colon carcinoma cell line DLD-1 with LY294002 resulted in an increase in p130 levels, whereas serum starvation for as long as 72 h could not accomplish this (Fig. 4C). We attribute this differential response to LY294002 treatment and serum starvation to autocrine growth factor production by this tumor cell line. Taken together, these data show that the PI3K/PKB/Forkhead pathway is involved in the regulation of p130 protein levels.
FIG. 4.
Forkhead-induced cell cycle exit is mediated by PI3K signaling. (A) MCF7 cells were serum starved for 48 or 72 h and subsequently treated with insulin for 24 or 48 h, with or without a 30-min preincubation with LY294002. p130 protein levels were then analyzed by Western blotting. pp130, hyperphosphorylated p130. (B) U87MG cells were treated with 10 μM LY294002 (LY) for 0, 10, or 24 h. Total cellular lysates were immunoblotted for the presence of p130 protein and T32-phosphorylated FKHR-L1. (C) p130 protein levels of asynchronous (asynch) DLD-1 cells were compared to those of DLD-1 cells that were either serum starved (− FCS) for 48 or 72 h or treated with LY294002 for 16 h.
Conditional activation of FKHR-L1 causes growth arrest and increased p130 protein levels in human colon carcinoma cells.
Previous work has shown that activation of PI3K is sufficient to drive serum-deprived cells back into the cell cycle (27). Because Forkhead activation causes cells to withdraw from the cell cycle through regulation of p27kip1 and p130, we wanted to address the possibility that inactivation of Forkheads as a consequence of PI3K signaling is essential for cell cycle reentry. Therefore, we created an inducible Forkhead system and investigated, first, whether specific Forkhead activation in such cells is responsible for the control of p130 protein levels and, second, whether Forkhead activation can induce reversible cell cycle exit and entry, a hallmark of quiescence. For these experiments, we chose the DLD-1 cell line, in which cell cycle exit (as measured by p130 protein levels) cannot be induced by 72 h of serum starvation, in contrast to 24 h of treatment with LY294002 (Fig. 4C). We created the DL23 cell line, a DLD-1 subclone that stably expresses a fusion of HA-FKHR-L1.A3 with a modified form of the estrogen receptor (ER) hormone-binding domain (HA-FKHR-L1.A3-ER) (16). ER fusion proteins are usually inactive until the cells are presented with the ligand for the modified ER 4OHT (32). 4OHT is thought to activate the fusion protein by releasing repressors, probably heat shock proteins, bound to the ER moiety of the fusion protein. Indeed, HA-FKHR-L1.A3-ER is localized in the cytosol and thus is likely to be inactive unless cells are treated with 4-OHT, which results in translocation to the nucleus (data not shown).
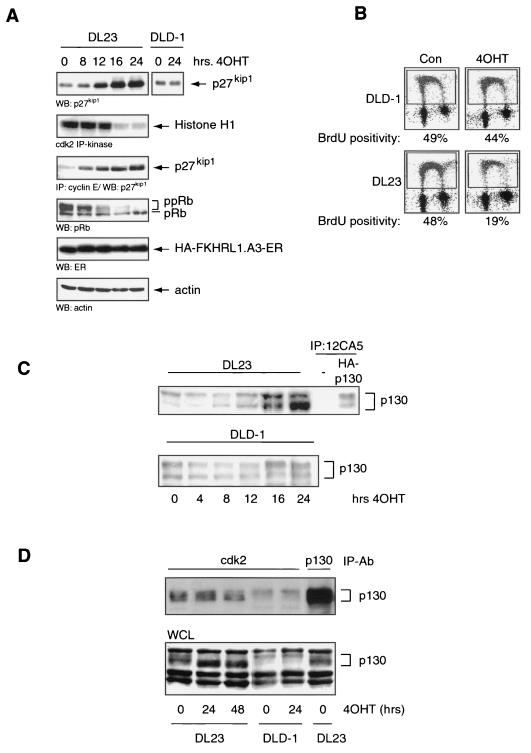
Specific activation of the Forkhead transcription factor by 4OHT in the DL23 cell line resulted in upregulation of p27kip1 protein, hypophosphorylation of pRb, inhibition of cdk2 activity, and subsequent cell cycle arrest (Fig. 5A and B), consistent with data obtained for NIH 3T3 cells (38). These cell cycle-inhibitory events are specific for Forkhead activity, since the DLD-1 cell line did not display such effects upon 4OHT addition (Fig. 5A and B; also data not shown). Next, by Western blotting, we analyzed p130 protein levels upon specific Forkhead activation. As seen in Fig. 5C, treatment of the DL23 cell line with 4OHT for 16 to 24 h resulted in a strong increase in p130 protein levels, indicating that indeed the Forkheads may be mediators of the effect of LY294002 treatment on p130 levels observed in Fig. 4C. As with the MEFs for which data are shown in Fig. 1 and 3, the Forkhead-induced increase in p130 levels in DL23 cells does not seem to contribute to the inhibition of cdk2 activity, since the presence of p130 in cdk2-containing complexes does not significantly change upon 4OHT treatment (Fig. 5D).
FIG. 5.
Conditional activation of FKHR-L1 causes growth arrest and increased p130 protein levels in human colon carcinoma cells. (A) DL23 and DLD-1 cells were left untreated (0 h) or treated with 500 nM 4OHT for 8, 12, 16, or 24 h. Total cellular lysates were electrophoresed and blotted for the presence of p27kip1, pRb, HA-FKHR-L1.A3-ER,or actin. Cell lysates were analyzed for cdk2 kinase activity by cdk2 immunoprecipitation (IP) and in vitro kinase reaction using histone H1 as a substrate. The cdk2 kinase reaction products were blotted, autoradiographed, and probed for the presence of p27kip1. WB, Western blot. (B) BrdU incorporation of DL23 and DLD-1 cells left untreated (Con) or treated with 500 nM 4OHT for 16 h. (C) Total cellular lysates from DLD-1 and DL23 cells treated without (0 h) or with 500 nM 4OHT for 4, 8, 12, 16, or 24 h were analyzed by Western blotting for the presence of the p130 retinoblastoma-like protein. Anti-HA immunoprecipitations of A14 cells transiently transfected with empty vector or pCMV-HA-p130 served as controls. (D) DL23 and DLD-1 cells, left untreated (0 h) or treated with 4OHT for 24 or 48 h, were lysed, and cdk2 was immunoprecipitated. The cdk2 immunoprecipitates were subsequently analyzed for the presence of p130 by Western blotting. WCL, whole-cell lysate; IP-Ab, immunoprecipitating antibody.
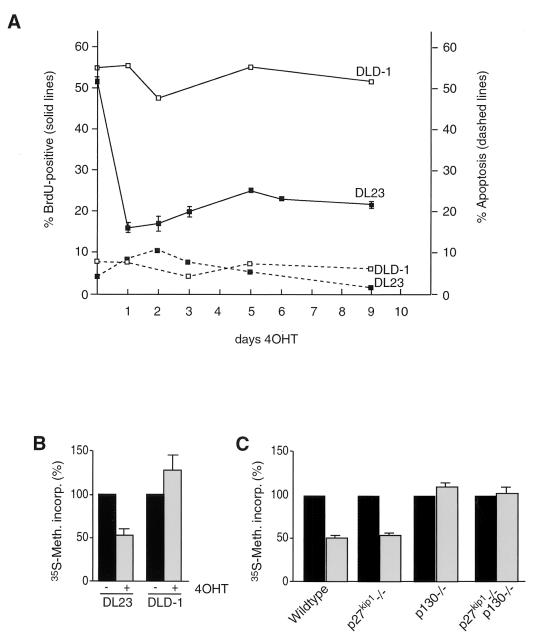
To study more closely the possible role of the Forkheads in the regulation of cell cycle exit and entry in DL23 cells, we first chose to determine the effect of prolonged Forkhead activity on cellular proliferation. To this end, we added 4OHT to DL23 cells for as long as 9 days and measured sub-G1 DNA content and BrdU incorporation to determine apoptosis and proliferative status, respectively. As seen in Fig. 6A, activation of FKHR-L1.A3 by 4OHT results in rapid, drastic, and continued inhibition of BrdU incorporation, but no great increase in the relative amount of cells in apoptosis was observed, even after 9 days. Finally, We measured general protein synthesis activity as another marker of quiescence. As shown in Fig. 6B, protein synthesis was clearly slowed down in DL23 cells treated with 4OHT for 48 h, as measured by [35S]methionine pulse-labeling, indicating that by this criterion as well, cells had entered G0. The results presented in Fig. 3 suggest that regulation of p130 by itself is not sufficient to inhibit cyclinE/cdk2 activity and cell proliferation. We therefore investigated whether, instead, the Forkhead-induced decrease in general protein synthesis is dependent on p130. To this end MEFs were infected with control and AFX retroviruses, and protein synthesis was measured by [35S]methionine pulse-labeling. Interestingly, protein synthesis was not reduced either in p130−/− or in p27−/− p130−/− MEFs following Forkhead expression. Although general protein synthesis can be influenced in many ways, this result clearly suggests that p130 regulation is essential for establishing quiescence.
FIG. 6.
Long-term activation of FKHR-L1 causes continued arrest.(A) DLD-1 and DL23 cells were treated with 500 nM 4OHT for 9 days. Cells were seeded to approximately 5% confluency. Cells were given fresh 4OHT-containing medium every day and were split whenever cell density reached 70 to 80%. Typically, during the whole experiment, only the DLD-1 control cells were split and seeded again to 5% confluency (usually at day 5). Samples were taken at various time points to measure proliferation by BrdU incorporation (solid lines) and cell death by FACS analysis of sub-G1 population (dashed lines). (B) DLD-1 and DL23 cells were left untreated (−) or treated with 500 nM 4OHT for 48 h (+) and pulse-labeled with [35S]methionine to measure protein synthesis. (C) Primary wild-type MEFs and MEFs from p27kip1, p130, or p27kip1 p130 knockout mice were infected with control virus (pBabe-puro) (solid bars) or FKHR-L1.A3-encoding retrovirus (shaded bars) and pulse-labeled with [35S]methionine at 24 h postinfection to measure protein synthesis.
Human colon carcinoma cells are forced to exit the cell cycle by specific activation of FKHR-L1.
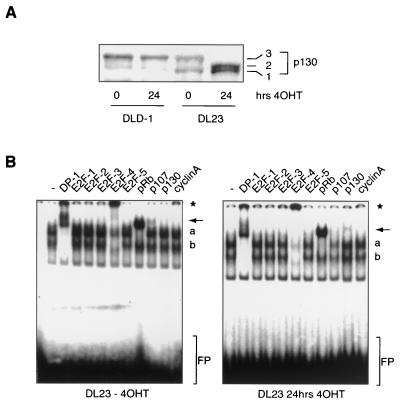
To determine whether the observed increase in p130 protein levels along with the sustained arrest is indicative of Forkhead-induced quiescence or senescence in DL23 cells, we analyzed the G0 markers mentioned above. First we examined the phosphorylation status of p130 protein upon 4OHT treatment of DL23 cells. Normal cycling DL23 cells show most p130 protein hyperphosphorylated to form 3, as described for other cycling cells (Fig. 7A) (35, 36). Twenty-four hours of 4OHT treatment, however, resulted in an increase in the relative amount of p130 protein phosphorylated to forms 1 and 2 (Fig. 7A). We next examined the functional interaction between p130 and E2F-4 using band supershift analysis. The interaction of the p130/E2F-4 complex with E2F binding sequences was increased in cells containing the activated Forkhead, whereas p107-containing complexes were lost (Fig. 7B). Furthermore, as with NIH 3T3 cells, by using an antibody against cyclin A, we observed a decrease in levels of p107/cyclin A/E2F-4 complexes. At this point, it should be noted that the most abundant pocket protein/E2F complex observed in DL23 cells proved to be pRb/E2F-4; however, this appears not to be uncommon for human cells (19, 24, 39, 57). Taken together, these data suggest that activation of Forkhead transcription factors induces cells to exit the cell cycle and enter G0. Moreover, the data imply that the effect of the Forkheads on proliferation is sufficiently strong to cause human tumor cells to enter quiescence or senescence.
FIG. 7.
Specific activation of FKHR-L1 causes human carcinoma cells to exit the cell cycle. (A) Total cellular lysates from DLD-1 and DL23 cells that were left untreated (0 h) or treated with 500 nM 4OHT for 24 h were electrophoresed on a 6% polyacrylamide gel and immunoblotted for the presence of p130. The three phosphorylated forms of the p130 protein are indicated as 1, 2, and 3. (B) Total cellular lysates from DL23 cells left untreated (left) or treated with 500 nM 4OHT for 48 h (right) were analyzed for the presence of E2F/DP-1/pocket protein complexes. Supershifting antibodies are listed above the gel. Arrows indicate the disappearance of p107/E2F-4 complexes in the third lane from the right and the appearance of p130/E2F-4 complexes in the second lane from the right upon 4OHT treatment. Asterisk, supershifted E2F-4- or cyclin A-containing complexes; a, nonsupershifted E2F/pocket protein complexes; b, free E2F; FP, free probe.
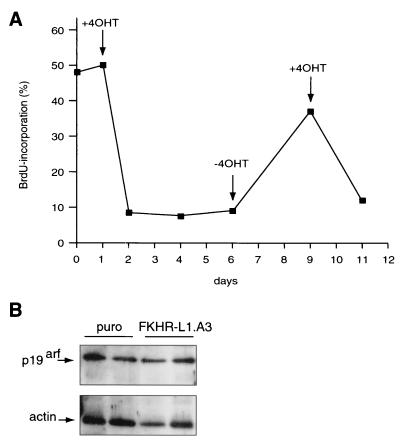
Importantly, cellular quiescence is a reversible phenotype, and this clearly distinguishes quiescence from senescence, which is irreversible. The inducible Forkhead system enabled us to directly address whether the Forkhead-induced phenotype is indeed reversible. To this end, we treated DL23 cells for as long as 5 days with 4OHT, after which cells were grown for 3 days in medium without 4OHT. BrdU incorporation after 5 days of 4OHT treatment was diminished from 50 to 9%, but this phenotype reverted almost completely upon withdrawal of 4OHT for 3 subsequent days (Fig. 8). All cells had reentered the cell cycle, since more than 95% of the population stained BrdU positive when given BrdU for 40 h (data not shown). The cells that reentered the cell cycle upon removal of 4OHT were efficiently arrested again upon readdition of 4OHT, reflecting true reversibility (Fig. 8). The latter result also demonstrates that we did not select a non-4OHT-responsive subpopulation during the initial prolonged arrest.
FIG. 8.
Forkhead-induced G0 is reversible. (A) DL23 cells were seeded to approximately 5% confluency. Confluency was monitored during the experiment, as described for Fig. 6A, but none of the dishes at any of the indicated time points required splitting. Cells were given fresh medium without 4OHT (−4OHT) or with 500 nM 4OHT (+4OHT) every day. When 4OHT was removed (at day 6), cells were washed twice with medium without 4OHT before the fresh medium was added. Similarly, when cells were again given 4OHT (at day 9), they were washed twice with medium containing 4OHT before the fresh medium was added. Samples were taken at days 0, 1, 2, 3, 4, 6, 9, and 11, and BrdU incorporation was measured. (B) Samples of primary wild-type MEFs from two independent infections with either a control virus (puro) or an FKHR-L1.A3-encoding retrovirus were analyzed for p19arf expression by immunoblotting.
Finally, we investigated the effect of Forkhead activation on parameters of senescence. No LacZ staining could be observed in DL23 cells even after prolonged treatment with 4OHT (data not shown). In addition, in DLD-1, cells the p14arf gene is extensively methylated (58), and consequently we could not observe any p14arf protein either before or after 4OHT treatment (data not shown). We therefore also analyzed p19arf expression in MEFs infected with AFX retrovirus. No change in p19arf expression could be observed following Forkhead expression (Fig. 8B). Taken together, these results demonstrate that the Forkhead-mediated cell cycle exit indeed represents entry into quiescence (G0).
DISCUSSION
In this study we show that signaling-independent activation of the Forkhead transcription factors AFX and FKHR-L1, which are normally controlled by the PI3K/PKB signaling pathway (6, 29), results in an increase in protein expression of the retinoblastoma-like protein p130. Expression of p130 is controlled directly at the transcriptional level via an inverse DBE located in the first intron, not an uncommon position for regulatory elements in transcriptional control (25, 49). Previously we have shown that Forkheads induce cell cycle arrest through regulation of p27kip1 (38). Here we show that cells not only arrest but in addition exit from the cell cycle and enter quiescence (G0). Entrance into G0 is indicated not only by the upregulation of p130 protein itself but also by the change in the p130 phosphorylation pattern and the increase in E2F-4/p130 complex formation. These changes have been described as characteristic for quiescent cells (36, 51) and indicate that activation of AFX-like Forkhead transcription factors is sufficient to force cells out of the cell cycle. The report of others that activation of PI3K, and thereby inactivation of Forkheads, is sufficient to drive serum-deprived cells into the cell cycle (27) suggested that Forkhead activation or inactivation is sufficient to regulate, respectively, cell cycle exit and entry. Indeed, we could show that prolonged growth factor-independent activation of FKHR-L1 results in sustained but reversible growth arrest. In addition, this result provides evidence that cells do not enter senescence, and this conclusion is further corroborated by a lack of p19arf regulation by Forkhead activation.
In this respect, our data are remarkably consistent with the established role of these Forkhead transcription factors in C. elegans. In this nematode, the AFX/FKHR/FKHR-L1-like DAF-16 Forkhead transcription factor is regulated by a PI3K/PKB-like pathway and induces longevity and Dauer formation (31, 44, 45). The latter effect is an exit from development at the second larval stage in adverse situations such as lack of nutrients. The Dauer phenotype is reversible, allowing the worm to reenter the developmental program when conditions change for the better (reviewed in reference 54). Thus, the regulation of Dauer in C. elegans by the PI3K/PKB/Forkhead pathway in many ways resembles quiescence rather than senescence or apoptosis.
Although p130 is generally considered to be a marker of quiescence, it is at present unclear what its role can be in the actual establishment of quiescence. One possibility is that if cells are arrested by any other mechanisms (e.g., an increase in p27kip1 levels), p130 acts as a block to prevent cell cycle reentry, for example, by preventing the expression of certain genes. Alternatively, p130 may be actively involved in establishing the arrest. The observations that LY249002 treatment no longer inhibits proliferation of pRb−/− p107−/− p130−/− MEFs (Fig. 3D) and that these MEFs do not arrest upon serum deprivation (13) indicate that expression of one of the pRb family members is essential to establish cell cycle arrest in the absence of PI3K signaling. The mechanism, however, remains unclear. A Forkhead-induced increase in p130 levels does not result in inhibition of cyclin E-dependent cdk2 activity either in wild-type or in p27kip1−/− MEFs (Fig. 3C), suggesting a cyclinE/cdk2-independent mechanism. In contrast to a role in driving or establishing cell cycle arrest, our observation that in p130−/− and p27−/− p130−/− MEFs, Forkheads no longer repress general protein synthesis suggests that p130 regulation may indeed be of importance for establishing quiescence. In keeping with this, 4OHT treatment resulted in induction of p130 expression, which was delayed for several hours compared to p27kip1 expression (compare Fig. 5A and 5C; also data not shown).
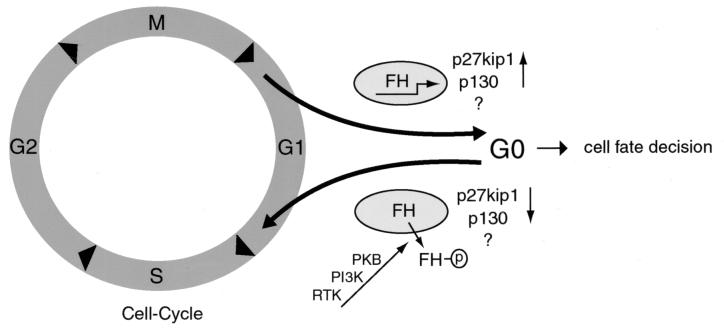
Taken together, the data presented here suggest an elegant model in which the PI3K/PKB signaling pathway regulates cell cycle exit and entry through (in)activation of the AFX-like Forkhead transcription factors (Fig. 9). In this model the absence of PI3K signaling results in Forkhead activation and increased expression of p27kip1 and p130. We interpret the results on p130 regulation presented here as indicating that p27kip1 regulation is required for inducing cell cycle arrest and that p130 regulation is required for establishing cell cycle exit and entry into quiescence. However, it is clear that other cell cycle regulators in addition to p27kip1 and p130 must be affected by the Forkheads, since we find that in MEFs deficient in both p130 and p27kip1, Forkheads can still cause a decrease in cdk2 activity (Fig. 3C). If PI3K becomes activated by growth factor treatment, inactivation of Forkhead results in decreased p27kip1 and p130 expression, and this allows quiescent cells to reenter the cell cycle. Consistent with this model are the observations that disruption of p27kip1 expression or specific activation of PI3K can drive quiescent cells into G1 (27, 30, 34, 37, 48).
FIG. 9.
Model for cell cycle regulation by the PI3K/PKB/Forkhead pathway. When cells do not receive growth factors in early G1, PKB is not activated and Forkheads (FH) remain active. This results in upregulation of the genes for p27kip1 and p130 along with yet unidentified target genes, leading to cell cycle exit in nonhematopoietic cells and, with the additional regulation of the Bim and FasL gene products, to apoptosis in hematopoietic cells. Subsequent activation of PKB through receptor tyrosine kinases (RTK) leads to phosphorylation of FH (indicated by the circled p) and inactivation of Forkheads by nuclear exclusion and therefore to cell cycle reentry via downregulation of the Forkhead cell cycle-regulatory target genes. See the text for further details.
It has been proposed that the presence of p130/E2F-4 repressor complexes on DNA has the effect of conferring reversibility on the cell cycle exit program (21). This is potentially important in the context of oncogenic transformation and regeneration. Conceivably, quiescent tissue is forced back into the cell cycle by activation of the PI3K/PKB pathway. Indeed, ligand-independent activation of PKB in quiescent cells reduces the amount of p130 protein (data not shown), indicating a return to the G1 phase of the cell cycle, and PI3K activated in a similar manner (PI3K-ER) also drives cells out of quiescence (27). This cell cycle reentry function of PKB might contribute to the oncogenic effect of mutations or deletions in the PTEN tumor suppressor (46, 52), especially in normally quiescent tissue such as the brain, where PTEN genetic alterations are quite common (26, 43). Once inactivated, PTEN no longer inhibits PKB-dependent inactivation of the Forkheads, and thus cells reenter the cell cycle.
Certain differentiated tissues, such as liver, smooth and skeletal muscle, pancreatic, and auditory sensory epithelial tissues, have the ability to regenerate. Recent data indicate that both p27kip1 and p130 are involved in defining the exact pool of skeletal muscle and auditory sensory epithelial reserve cells that confer this capacity to regenerate (8, 34). In addition, high p27kip1 protein levels have been implicated in maintenance of the quiescent state of germinal center/memory B cells (56), and p130 or p27kip1 has been implicated in liver, pancreas, and smooth muscle regeneration (1, 5, 9, 40). Based on the data presented in this report, it is tempting to speculate that in the cells of tissues such as liver, pancreas, and muscle, the PI3K/PKB/Forkhead pathway contributes to cell cycle reentry.
The data presented here show that specific activation of FKHR-L1 alone is sufficient to induce human tumor cells to exit the cell cycle. Not only can Forkheads inhibit proliferation of human colon carcinoma cells; they also have the ability to arrest human leukemia cells, human glioblastoma cells, human renal carcinoma cells, and human osteosarcoma cells (38). This potent capacity of the Forkheads may indicate that proliferation during the process of oncogenic transformation cannot occur in the presence of active Forkheads. Possibly, during this transformation process, endogenous Forkhead activities have been diminished in such tumor cells. Indeed, human U87MG glioblastoma cells (this study) and human Jurkat leukemia cells (unpublished data) have high levels of phosphorylated and thus inactivated FKHR-L1. Furthermore, exogenous FKHR is located in the cytoplasm, and thus kept inactive, in human renal and prostate cancer cells (42). The putative importance of this pathway in oncogenesis is further supported by the findings that in many tumors p27kip1 (reviewed in reference 33) or p130 expression (2, 53) is low.
Forced activation of the Forkheads that are regulated by PKB either causes cells to exit the cell cycle, as described here, or to go into apoptosis. It is conceivable that in the latter case, cell cycle arrest and/or exit in the absence of PI3K/PKB signaling is deleterious because of a lack of survival signals and/or activation of default death pathways. The ability of Forkheads to induce proapoptotic genes such as those encoding Bim and FasL (6, 15) may constitute part of this default death pathway. Conditional activation of FKHR-L1 in the DL23 cell line used in this study does not result in upregulation of Bim (unpublished data), whereas activation of the same construct in Ba/F3 cells (pre-B cells) does (15). Furthermore, the DLD-1 cell line, which is the parental cell line for the DL23 clone, is not sensitive to FasL-induced apoptosis despite high levels of CD95 (18, 22). We do not yet understand the mechanism by which the default death pathways are silenced in these cells, but it will be interesting to examine what other cellular factors determine why Bim is not Forkhead responsive in nonhematopoietic cells and why the presence of CD95 does not necessarily mean FasL responsiveness.
In conclusion, we have identified a novel function for the Forkhead transcription factors AFX, FKHR, and FKHR-L1 in cell cycle control. We have shown that these Forkheads induce cells to exit the cell cycle and enter a state of quiescence, as demonstrated by upregulation of protein levels of the retinoblastoma family member p130, an increase in levels of p130/E2F-4 complexes, and the reversibility of cell cycle arrest. The last observation further emphasizes the importance of activation of the PI3K/PKB pathway in processes such as oncogenic transformation, in that it may relieve the inhibitory constraints on cell cycle progression imposed by Forkhead-mediated regulation of p27kip1 and p130 levels, resulting in reentry into the cell cycle.
Acknowledgments
We thank D. Cobrinik, H. Ariga, A. Brunet, B. Scheyen, and J. H. Dannenberg for providing useful reagents. We thank Johannes L. Bos for ongoing discussions, support, and critical reading of the manuscript. We also thank R. Watson and C. Sardet for critical reading of the manuscript.
G.J.P.L.K. was supported by a grant from Chemical Sciences (NWO-CW). E.W.-F.L. and J.G. are supported by the Leukemia Research Fund of the United Kingdom.
G. J. P. L. Kops and R. H. Medema contributed equally to this work.
REFERENCES
- 1.Albrecht, J. H., R. Y. Poon, C. L. Ahonen, B. M. Rieland, C. Deng, and G. S. Crary. 1998. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene 16:2141-2150. [DOI] [PubMed] [Google Scholar]
- 2.Baldi, A., V. Esposito, A. De Luca, C. M. Howard, G. Mazzarella, F. Baldi, M. Caputi, and A. Giordano. 1996. Differential expression of the retinoblastoma gene family members pRb/p105, p107, and pRb2/p130 in lung cancer. Clin. Cancer Res. 2:1239-1245. [PubMed] [Google Scholar]
- 3.Bos, J. L. 1998. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 17:6776-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos, J. L. 1995. p21ras: an oncoprotein functioning in growth factor-induced signal transduction. Eur. J. Cancer 31:1051-1054. [DOI] [PubMed] [Google Scholar]
- 5.Bouzahzah, B., M. Fu, A. Iavarone, V. M. Factor, S. S. Thorgeirsson, and R. G. Pestell. 2000. Transforming growth factor-β1 recruits histone deacetylase 1 to a p130 repressor complex in transgenic mice in vivo. Cancer Res. 60:4531-4537. [PubMed] [Google Scholar]
- 6.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 7.Brunet, A., J. Park, H. Tran, L. S. Hu, B. A. Hemmings, and M. E. Greenberg. 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 21:952-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carnac, G., L. Fajas, A. L'Honore, C. Sardet, N. J. Lamb, and A. Fernandez. 2000. The retinoblastoma-like protein p130 is involved in the determination of reserve cells in differentiating myoblasts. Curr. Biol. 10:543-546. [DOI] [PubMed] [Google Scholar]
- 9.Claudio, P. P., L. Fratta, F. Farina, C. M. Howard, G. Stassi, S. Numata, C. Pacilio, A. Davis, M. Lavitrano, M. Volpe, J. M. Wilson, B. Trimarco, A. Giordano, and G. Condorelli. 1999. Adenoviral RB2/p130 gene transfer inhibits smooth muscle cell proliferation and prevents restenosis after angioplasty. Circ. Res. 85:1032-1039. [DOI] [PubMed] [Google Scholar]
- 10.Coats, S., P. Whyte, M. L. Fero, S. Lacy, G. Chung, E. Randel, E. Firpo, and J. M. Roberts. 1999. A new pathway for mitogen-dependent cdk2 regulation uncovered in p27Kip1-deficient cells. Curr. Biol. 9:163-173. [DOI] [PubMed] [Google Scholar]
- 11.Coffer, P. J., J. Jin, and J. R. Woodgett. 1998. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J. 335:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collado, M., R. H. Medema, I. Garcia-Cao, M. L. Dubuisson, M. Barradas, J. Glassford, C. Rivas, B. M. Burgering, M. Serrano, and E. W. Lam. 2000. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J. Biol. Chem. 275:21960-21968. [DOI] [PubMed] [Google Scholar]
- 13.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cristofano, A., and P. P. Pandolfi. 2000. The multiple roles of PTEN in tumor suppression. Cell 100:387-390. [DOI] [PubMed] [Google Scholar]
- 15.Dijkers, P. F., R. H. Medema, J. J. Lammers, L. Koenderman, and P. J. Coffer. 2000. Expression of the pro-apoptotic bcl-2 family member bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10:1201-1204. [DOI] [PubMed] [Google Scholar]
- 16.Dijkers, P. F., R. H. Medema, C. Pals, L. Banerji, N. S. Thomas, E. W. Lam, B. M. Burgering, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27KIP1. Mol. Cell. Biol. 20:9138-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481-507. [DOI] [PubMed] [Google Scholar]
- 18.Fukazawa, T., T. Fujiwara, Y. Morimoto, J. Shao, M. Nishizaki, Y. Kadowaki, A. Hizuta, L. B. Owen-Schaub, J. A. Roth, and N. Tanaka. 1999. Differential involvement of the CD95 (Fas/APO-1) receptor/ligand system on apoptosis induced by the wild-type p53 gene transfer in human cancer cells. Oncogene 18:2189-2199. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa, Y., S. Iwase, J. Kikuchi, M. Nakamura, H. Yamada, and M. Matsuda. 1999. Transcriptional repression of the E2F-1 gene by interferon-alpha is mediated through induction of E2F-4/pRB and E2F-4/p130 complexes. Oncogene 18:2003-2014. [DOI] [PubMed] [Google Scholar]
- 20.Furuyama, T., T. Nakazawa, I. Nakano, and N. Mori. 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grana, X., J. Garriga, and X. Mayol. 1998. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 17:3365-3383. [DOI] [PubMed] [Google Scholar]
- 22.Han, S. Y., S. Y. Choung, I. S. Paik, H. J. Kang, Y. H. Choi, S. J. Kim, and M. O. Lee. 2000. Activation of NF-κB determines the sensitivity of human colon cancer cells to TNFα-induced apoptosis. Biol. Pharm. Bull. 23:420-426. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, K., T. Farkas, J. Lukas, K. Holm, L. Ronnstrand, and J. Bartek. 2001. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 20:422-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humbert, P. O., C. Rogers, S. Ganiatsas, R. L. Landsberg, J. M. Trimarchi, S. Dandapani, C. Brugnara, S. Erdman, M. Schrenzel, R. T. Bronson, and J. A. Lees. 2000. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell 6:281-291. [DOI] [PubMed] [Google Scholar]
- 25.Jin, S., F. Fan, W. Fan, H. Zhao, T. Tong, P. Blanck, I. Alomo, B. Rajasekaran, and Q. Zhan. 2001. Transcription factors Oct-1 and NF-YA regulate the p53-independent induction of the GADD45 following DNA damage. Oncogene 20:2683-2690. [DOI] [PubMed] [Google Scholar]
- 26.Kleihues, P., and H. Ohgaki. 2000. Phenotype vs. genotype in the evolution of astrocytic brain tumors. Toxicol. Pathol. 28:164-170. [DOI] [PubMed] [Google Scholar]
- 27.Klippel, A., M. A. Escobedo, M. S. Wachowicz, G. Apell, T. W. Brown, M. A. Giedlin, W. M. Kavanaugh, and L. T. Williams. 1998. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol. Cell. Biol. 18:5699-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohn, A. D., A. Barthel, K. S. Kovacina, A. Boge, B. Wallach, S. A. Summers, M. J. Birnbaum, P. H. Scott, J. C. Lawrence, Jr., and R. A. Roth. 1998. Construction and characterization of a conditionally active version of the serine/threonine kinase Akt. J. Biol. Chem. 273:11937-11943. [DOI] [PubMed] [Google Scholar]
- 29.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630-634. [DOI] [PubMed] [Google Scholar]
- 30.Ladha, M. H., K. Y. Lee, T. M. Upton, M. F. Reed, and M. E. Ewen. 1998. Regulation of exit from quiescence by p27 and cyclin D1-CDK4. Mol. Cell. Biol. 18:6605-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, K., J. B. Dorman, A. Rodan, and C. Kenyon. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319-1322. [DOI] [PubMed] [Google Scholar]
- 32.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lloyd, R. V., L. A. Erickson, L. Jin, E. Kulig, X. Qian, J. C. Cheville, and B. W. Scheithauer. 1999. p27kip1: a multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am. J. Pathol. 154:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowenheim, H., D. N. Furness, J. Kil, C. Zinn, K. Gultig, M. L. Fero, D. Frost, A. W. Gummer, J. M. Roberts, E. W. Rubel, C. M. Hackney, and H. P. Zenner. 1999. Gene disruption of p27Kip1 allows cell proliferation in the postnatal and adult organ of corti. Proc. Natl. Acad. Sci. USA 96:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayol, X., J. Garriga, and X. Grana. 1995. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene 11:801-808. [PubMed] [Google Scholar]
- 36.Mayol, X., J. Garriga, and X. Grana. 1996. G1 cyclin/CDK-independent phosphorylation and accumulation of p130 during the transition from G1 to G0 lead to its association with E2F-4. Oncogene 13:237-246. [PubMed] [Google Scholar]
- 37.McIlroy, J., D. Chen, C. Wjasow, T. Michaeli, and J. M. Backer. 1997. Specific activation of p85-p110 phosphatidylinositol 3′-kinase stimulates DNA synthesis by ras- and p70 S6 kinase-dependent pathways. Mol. Cell. Biol. 17:248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 39.Moberg, K., M. A. Starz, and J. A. Lees. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16:1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morisset, J., J. C. Aliaga, E. L. Calvo, J. Bourassa, and N. Rivard. 1999. Expression and modulation of p42/p44 MAPKs and cell cycle regulatory proteins in rat pancreas regeneration. Am. J. Physiol. 277:G953-G959. [DOI] [PubMed]
- 41.Mudryj, M., S. H. Devoto, S. W. Hiebert, T. Hunter, J. Pines, and J. R. Nevins. 1991. Cell cycle regulation of the E2F transcription factor involves an interaction with cyclin A. Cell 65:1243-1253. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, N., S. Ramaswamy, F. Vazquez, S. Signoretti, M. Loda, and W. R. Sellers. 2000. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 20:8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng, H. K., and P. Y. Lam. 1998. The molecular genetics of central nervous system tumors. Pathology 30:196-202. [DOI] [PubMed] [Google Scholar]
- 44.Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee, H. A. Tissenbaum, and G. Ruvkun. 1997. The Forkhead transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994-999. [DOI] [PubMed] [Google Scholar]
- 45.Paradis, S., and G. Ruvkun. 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12:2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramaswamy, S., N. Nakamura, F. Vazquez, D. B. Batt, S. Perera, T. M. Roberts, and W. R. Sellers. 1999. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA 96:2110-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rena, G., S. Guo, S. C. Cichy, T. G. Unterman, and P. Cohen. 1999. Phosphorylation of the transcription factor Forkhead family member FKHR by protein kinase B. J. Biol. Chem. 274:17179-17183. [DOI] [PubMed] [Google Scholar]
- 48.Rivard, N., G. L'Allemain, J. Bartek, and J. Pouyssegur. 1996. Abrogation of p27Kip1 by cDNA antisense suppresses quiescence (G0 state) in fibroblasts. J. Biol. Chem. 271:18337-18341. [DOI] [PubMed] [Google Scholar]
- 49.Rowntree, R. K., G. Vassaux, T. L. McDowell, S. Howe, A. McGuigan, M. Phylactides, C. Huxley, and A. Harris. 2001. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 10:1455-1464. [DOI] [PubMed] [Google Scholar]
- 50.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 51.Smith, E. J., G. Leone, J. DeGregori, L. Jakoi, and J. R. Nevins. 1996. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol. Cell. Biol. 16:6965-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stambolic, V., A. Suzuki, J. L. de la Pompa, G. M. Brothers, C. Mirtsos, T. Sasaki, J. Ruland, J. M. Penninger, D. P. Siderovski, and T. W. Mak. 1998. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95:29-39. [DOI] [PubMed] [Google Scholar]
- 53.Susini, T., F. Baldi, C. M. Howard, A. Baldi, G. Taddei, D. Massi, S. Rapi, L. Savino, G. Massi, and A. Giordano. 1998. Expression of the retinoblastoma-related gene Rb2/p130 correlates with clinical outcome in endometrial cancer. J. Clin. Oncol. 16:1085-1093. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, J. H. 1993. Chemosensory regulation of development in C. elegans. Bioessays 15:791-797. [DOI] [PubMed] [Google Scholar]
- 55.van der Sman, J., N. S. Thomas, and E. W. Lam. 1999. Modulation of E2F complexes during G0 to S phase transition in human primary B-lymphocytes. J. Biol. Chem. 274:12009-12016. [DOI] [PubMed] [Google Scholar]
- 56.Wagner, E. F., M. Hleb, N. Hanna, and S. Sharma. 1998. A pivotal role of cyclin D3 and cyclin-dependent kinase inhibitor p27 in the regulation of IL-2-, IL-4-, or IL-10-mediated human B cell proliferation. J. Immunol. 161:1123-1131. [PubMed] [Google Scholar]
- 57.Wu, L., E. C. Goodwin, L. K. Naeger, E. Vigo, K. Galaktionov, K. Helin, and D. DiMaio. 2000. E2F-Rb complexes assemble and inhibit cdc25A transcription in cervical carcinoma cells following repression of human papillomavirus oncogene expression. Mol. Cell. Biol. 20:7059-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng, S., P. Chen, A. McMillan, A. Lafuente, M. J. Lafuente, A. Ballesta, M. Trias, and J. K. Wiencke. 2000. Correlations of partial and extensive methylation at the p14ARF locus with reduced mRNA expression in colorectal cancer cell lines and clinicopathological features in primary tumors. Carcinogenesis 21:2057-2064. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, L., E. Harlow, and B. D. Dynlacht. 1995. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 9:1740-1752. [DOI] [PubMed] [Google Scholar]