Abstract
Cleavage by the V(D)J recombinase at a pair of recombination signal sequences creates two coding ends and two signal ends. The RAG proteins can integrate these signal ends, without sequence specificity, into an unrelated target DNA molecule. Here we demonstrate that such transposition events are greatly stimulated by—and specifically targeted to—hairpins and other distorted DNA structures. The mechanism of target selection by the RAG proteins thus appears to involve recognition of distorted DNA. These data also suggest a novel mechanism for the formation of alternative recombination products termed hybrid joints, in which a signal end is joined to a hairpin coding end. We suggest that hybrid joints may arise by transposition in vivo and propose a new model to account for some recurrent chromosome translocations found in human lymphomas. According to this model, transposition can join antigen receptor loci to partner sites that lack recombination signal sequence elements but bear particular structural features. The RAG proteins are capable of mediating all necessary breakage and joining events on both partner chromosomes; thus, the V(D)J recombinase may be far more culpable for oncogenic translocations than has been suspected.
The immune system's vast repertoire of B- and T-cell receptors results from a site-specific gene rearrangement process known as V(D)J recombination. Assorted variable (V), diversity (D), and joining (J) coding gene segments are recombined in developing lymphocyte precursors to form the variable region genes that encode antigen-binding sites of immunoglobulin and T-cell receptor molecules (40). The DNA is first cleaved at specific recombination signal sequences (RSS) located adjacent to the coding segments. RSS consist of conserved heptamer and nonamer sequences, separated by either 12 or 23 nucleotides of spacer DNA (referred to as 12 RSS and 23 RSS). The RAG1 and RAG2 recombinase proteins, assisted by either of the nonspecific DNA-bending proteins HMG1 and HMG2 (41), generate a nick between the RSS and the adjacent coding segment by hydrolysis. The newly formed 3′ OH group then attacks the phosphodiester bond connecting the coding segment to the RSS on the opposite strand. This transesterification reaction forms a hairpin coding end (which contains the coding segment) and a blunt signal end (which contains the RSS) (19). These ends are then joined to form the rearranged products, the coding and signal joints.
The mechanisms responsible for guiding the broken ends to ensure their proper joining remain poorly understood. In the test tube, the RAG proteins remain tightly associated with the signal ends in a postcleavage complex, which includes the coding ends (2, 8). Recent analysis of RAG mutants bearing specific defects in signal and coding joint formation indicates that the RAG proteins indeed play critical roles in both coding and signal joint formation and supports the notion that the broken ends remain associated with the RAG proteins in a postcleavage complex (30, 37). The joining steps of V(D)J recombination require several double-strand break repair factors in addition to the RAG proteins (reviewed in reference 6). Coding joint formation is necessarily preceded by the endonucleolytic opening of the hairpin coding ends. Although the identity of the nuclease that performs this function has not been established, it is known that the RAG proteins are capable of opening hairpins in vitro by hydrolysis (3, 38). We recently identified RAG mutants that display severe defects in hairpin opening in vitro and in coding joint formation in vivo, thus providing evidence that the RAG proteins play a role in hairpin opening in vivo (30, 37).
The signal ends may adopt a fate other than forming signal joints: the RAG proteins can join signal ends to unrelated target DNA molecules by transesterification, inserting either a single RSS or a 12 RSS-23 RSS pair into the target and forming single- or double-ended integration products, respectively (1, 9). In short, the RAG proteins can serve as a transposase. Because RAG-mediated transposition could prove to be a potent source of genomic instability in lymphocytes (9, 34), its mechanism and regulation are of both scientific interest and biological import.
It is reasonable to suppose that transposition might be regulated by target selection, a step that would not affect V(D)J recombination itself but could serve as a critical control point in the transposition reaction. The nature of the target sites chosen by the RAG transposase could have profound biological ramifications. For example, target “hot spots,” if they exist, could induce oncogenic chromosome rearrangements. Many transposases exhibit particular target preferences. One of the best-studied examples is the bacterial transposon Tn7, which employs two transposition pathways. In the first, the TnsD protein binds a specific DNA sequence, attTn7, and stimulates transposition by distorting the target DNA (15). In the absence of TnsD, Tn7 prefers to integrate near regions of triplex DNA, which are thought to mimic the distortion induced by TnsD binding (32, 33). In the alternate pathway, another accessory protein, TnsE, targets transposition events to altered DNA structures associated with replication (27). Tn10, another transposase that shares many functional similarities with the RAG proteins, also induces DNA distortion near the integration site (28).
Although early studies showed that the RAG transposase exhibits a modest preference for target sequences with high G+C content (9), no specific preferred target sequences were identified in plasmid substrates (1, 9). Given that other transposases recognize targets bearing particular DNA distortions and that the RAG proteins both distort DNA during hairpin formation (5, 12, 31) and recognize distorted DNA structures such as DNA hairpins and single-strand-double-strand DNA junctions (3, 36, 38), we hypothesized that the RAG transposase may prefer targets bearing particular structural features. We therefore tested a variety of DNA target structures to ascertain their effects on transposition. We found that transposition is markedly stimulated by—and specifically targeted to—DNA hairpins. Transposition is also targeted to other distorted DNA structures, such as single-strand-double-strand DNA junctions. These data indicate that distorted DNA plays an important role in target site selection by the RAG transposase and suggest the intriguing possibility that transposition may be targeted to particular genomic locations in vivo.
MATERIALS AND METHODS
Proteins.
All experiments were performed with recombinant truncated “core” RAG-1 and RAG-2 glutathione S-transferase fusions copurified from Chinese hamster ovary (RMP41) cells as previously described (37). Recombinant human HMG-1 was purified from Escherichia coli as previously described (12).
DNA substrates.
Uncleaved 12 and 23 RSS donor molecules were generated by annealing appropriate pairs of oligonucleotides (DAR39/40 and DG61/62, respectively) (19). Precleaved 12 and 23 RSS oligonucleotides were created by annealing DG10 and DG4 to their complements (19). The hairpin oligonucleotide used as a target for transposition was HY9 (5′-TAGCTCGAGACCTATAGGTCTCGA-3′), and the double-stranded control was FM116/117 (19). The hairpin oligonucleotide used for detection of target capture complexes was HY9. MM30t/b (25) was used as the nonhairpin double-stranded control. All oligonucleotides were purchased from GIBCO Life Technologies and gel purified before use. The cruciform-containing plasmid was pUC8F14C (designated hereafter F14C) (26). Plasmids containing various other cruciform-forming inverted repeats (26) were also used as targets; pUC8 was used as a non-cruciform-containing control. In competition experiments, pcDNA1/AMP was used as the competing long plasmid target. The pJH299 plasmid substrate was used for hybrid joint assays as previously described (37).
Creation of topoisomers.
The use of eukaryotic topoisomerase extracts for this purpose has been described previously (14). F14C plasmid was incubated overnight in a volume of 40 μl with topoisomerase alone or with 0.05, 0.1, 0.5, or 0.9 ng of ethidium bromide per μl. Reaction mixtures were then incubated with 40 μl of stop buffer, which included proteinase K to a final concentration of 175 ng/μl and sodium dodecyl sulfate (SDS) to a final concentration of 0.1%. Products were phenol extracted twice, ethanol precipitated, and resuspended, and aliquots were analyzed by agarose gel electrophoresis. Poststaining of the gel with ethidium bromide revealed that, as expected, topoisomerase relaxed the plasmid and that the degree of supercoiling increased as a function of the concentration of ethidium bromide in the incubation. Aliquots of DNA purified from the topoisomerase reactions were used as targets for transposition.
Plasmid target transposition reactions.
Transposition was performed as described previously (25). Briefly, radiolabeled 12 RSS and unlabeled 23 RSS donor molecules (0.4 pmol each) were incubated with 200 ng of each of the RAG proteins and 30 ng of HMG-1 in 5 mM Ca2+ at 37°C for 20 min. Target was added as indicated for each experiment, and reaction mixtures were incubated for 20 min at 37°C. Divalent metal ion (Mg2+ or Mn2+) was added to a final concentration of 3 mM (in a final volume of 30 μl), and the mixtures were incubated at 37°C for 30 min. After transposition, mixtures were treated for 30 min at 37°C with 30 μl of stop buffer, which included proteinase and SDS as described above. Competition reaction products were then resolved on gels and visualized. Targeting reaction products were phenol-chloroform extracted, digested for 1.5 h with ScaI, treated with proteinase-SDS, phenol-chloroform extracted again, and then separated on gels. All plasmid transposition products were resolved on 1% alkaline agarose gels, dried, and visualized by autoradiography. Products were quantitated with ImageQuant software, with intensities normalized to free donor as a control for potential losses due to extraction.
Oligonucleotide target transposition reactions.
Transposition into oligonucleotide targets was performed as described above, using 0.2 pmol of each precleaved donor molecule (the 12 RSS was radiolabeled), and 0.5 pmol of target. MgCl2 (3 mM) was used as the divalent metal ion. Reaction mixtures were treated with proteinase K and SDS, resolved on denaturing 6% acrylamide gels as described previously (37), and visualized by autoradiography.
Sequencing of integration events.
Products of transposition into F14C were amplified with a primer specific to either the 12 RSS donor (PCR 12 RSS, 5′-CAGGGTTTTTGTTCCAGTCTGTAGCAC-3′) or the 23 RSS donor (PCR 23 RSS, 5′-GCCAGACAGTGGAGTACTACCAC-3′) and one specific to the F14C backbone (either CruciformFor [5′-CGTATCACGAGGCCCTTTCGTC-3′] or CruciformRev [5′-CGCCACCTCTGACTTGAGCGTC-3′]). These PCR products were cloned with the TOPO TA cloning kit (Invitrogen) and sequenced from the PCR cloning vector (PCR2.1-TOPO) with the included primers.
Physical analysis of target capture complexes.
Target capture complexes were detected as described previously (25). Uncleaved donor 12 and 23 RSS (0.13 pmol of each) were incubated with 200 ng of each of the RAG proteins, 20 ng of HMG-1, and 5.4 mM Ca2+ for 10 min at 37°C. Reaction mixtures were then supplemented with MnCl2 (5 mM, final concentration) in a final volume of 10 μl and 0.5 pmol of radiolabeled target. Reaction mixtures were incubated for 15 min at 37°C and resolved on a 4-to-20% gradient native acrylamide gel. Gels were then dried and visualized by autoradiography.
Detection of hybrid joints.
The hybrid joint assay was described previously (21, 30). Briefly, 100 ng of plasmid pJH299 was incubated with RAG proteins and HMG-1 for 20 min at 37°C in 4 mM Ca2+. Mg2+ was then added to a final concentration of 4 mM along with either Tris-EDTA or plasmid competitor (100, 300, or 500 ng of F14C or pUC8) in a final volume of 10 μl. Reaction mixtures were then incubated at 37°C for 70 min. One microliter of the reaction mixture was analyzed by PCR with DR55 and ML68 for 23 cycles. Products were run on a 6% native acrylamide gel and visualized by Vistra Green staining.
RESULTS
Experimental design.
To determine whether DNA secondary structure affects target site selection by the RAG transposase, we tested a series of target plasmids containing sequences that form various altered DNA structures, all cloned at the same site in the pUC8 backbone. Previous work using these plasmids has shown that the inserts form cruciforms (from an inverted repeat sequence), Z-DNA, triplex DNA, and unwound DNA under the conditions employed in our experiments (39, 42). These targets were incubated with the RAG proteins under standard conditions that yield both single- and double-ended transposition events. Although single-ended insertions simply nick the target, double-ended insertions attack both strands, characteristically separated by a 5-bp stagger (1, 9), producing a double-stranded break in the target DNA. Under the conditions used in our experiments, most transposition events are double ended and occur with an efficiency of approximately 1 to 2% (1, 9, 25).
Transposition reactions with the various target plasmids were performed as previously described (25), and the products were digested with a restriction enzyme (ScaI) that cuts the plasmid only once. Nontargeted transposition into random sites on these plasmids should not produce distinct fragments after ScaI digestion. If, however, transposition occurred preferentially at a single site, digestion with ScaI should yield two distinct fragments of characteristic sizes (Fig. 1A). To simplify analysis of the data, products were analyzed by denaturing agarose gel electrophoresis, so that both single-ended and double-ended transposition events at a given site would yield fragments of the same size on a gel. (All experiments were also analyzed by native gel electrophoresis. In all cases, both single- and double-ended transposition events were observed [data not shown].)
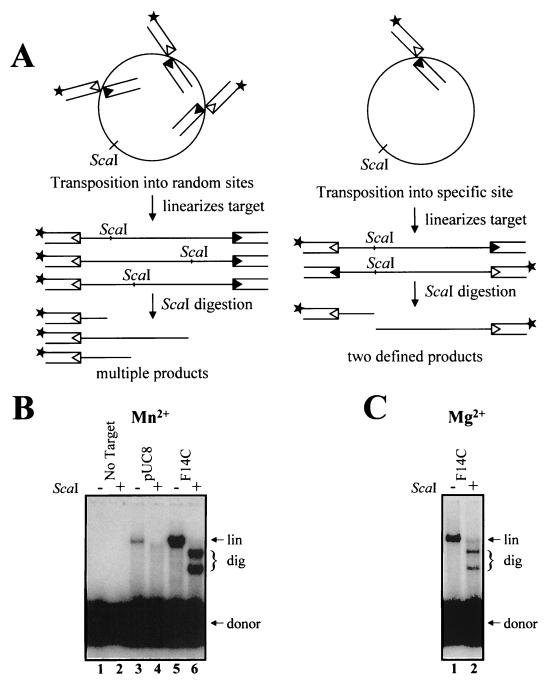
FIG. 1.
Preferential transposition into an inverted repeat. (A) Schematic of transposition into plasmid targets. Oligonucleotide donors contained either a 12 RSS (open triangles) or a 23 RSS (filled triangles). The 12 RSS-containing oligonucleotide was 32P end labeled (asterisk). Transposition into random sites followed by ScaI digestion generates products of many different sizes. Transposition into a single hot spot, however, followed by ScaI digestion yields two distinct products. (B) Transposition reactions were carried out as described in the text, with either no target or 0.11 pmol of F14C or pUC8 plasmid target. MnCl2 (3 mM) was used as the divalent metal ion. (lin, linearized target DNA; dig, products resulting from ScaI digestion). (C) Transposition reactions were carried out as for panel B, with 3 mM MgCl2.
Inverted repeats are preferred sites for RAG-mediated transposition.
Before digestion, transposition of labeled donor RSS into each plasmid target produced a linear product, as expected (Fig. 1B, lanes 3 and 5). Transposition into the control target (pUC8) was observed in undigested samples (Fig. 1B, lane 3), but no discrete bands were observed after digestion with ScaI (lane 4). Transposition into target plasmids containing Z-DNA, triplex, and unwound structures occurred with an efficiency similar to that observed with the pUC8 control, but digestion of the products with ScaI failed to produce distinct bands (data not shown). In contrast, digestion of the products of transposition into the F14C plasmid (which contains a 106-nucleotide inverted repeat capable of forming a cruciform with a 53-bp stem) produced two prominent fragments of sizes consistent with transposition into the inverted repeat (Fig. 1B, lane 6). Although Mn2+ supported more efficient transposition overall, targeted transposition was observed both in Mn2+ (Fig. 1B) and in Mg2+ (Fig. 1C). Quantitation of three experiments revealed that 85 to 100% of transposition events were targeted to the inverted repeat in Mn2+ and in Mg2+. This was determined by comparing the intensity of the sum of the two ScaI fragments (Fig. 1B, lane 6, and 1C, lane 2) to the intensity of the undigested linear products, which represent all transposition events (Fig. 1B, lane 5, and 1C, lane 1) (see Materials and Methods for details). In addition to targeting transposition to a specific site, the presence of the inverted repeat substantially stimulates the reaction (Fig. 1B, compare lanes 3 and 5). Quantitation of three independent experiments revealed an average stimulation of 10-fold. These results demonstrate that transposition shows a strong preference for the inverted repeat-containing target.
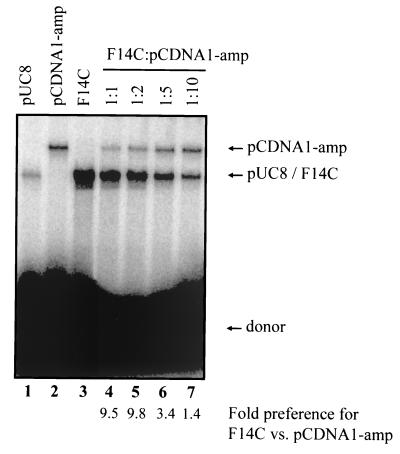
To further investigate target preferences, we performed transposition reactions using mixtures of F14C (2.8 kb) and various amounts of a larger competitor plasmid, pCDNA1/amp (4.8 kb). Transposition into these two targets can be readily distinguished (Fig. 2, lanes 2 and 3). Because pUC8 (the backbone of F14C) and pCDNA1/amp are used with similar efficiency (within a factor of two) as targets for transposition (Fig. 2, lanes 1 and 2), any observed differences between F14C and pCDNA1/amp should be attributable to the presence of the inverted repeat sequence. When the two plasmids were present in a 1:1 mass ratio, transposition into F14C was preferred by a factor of 10 (Fig. 2, lane 4; see quantitation below gel lanes). When the competitor plasmid was present at a two- and fivefold excess, transposition into F14C was still preferred (Fig. 2, lanes 5 and 6); only a 10-fold excess of competitor was able to abolish the preference for F14C (lane 7). These data are in agreement with the roughly 10-fold preference observed for F14C over pUC8 shown above and demonstrate that even in the presence of excess competitor target molecules, the inverted repeat stimulates RAG-mediated transposition.
FIG. 2.
Inverted repeats stimulate transposition. Transposition reactions were carried out as for Fig. 1B, without ScaI digestion. The target consisted of either a single plasmid or a mixture of F14C and pCDNA1-amp in the indicated mass ratios. For mixtures containing a mixture of targets, the preference for F14C was calculated as the amount of linearized F14C divided by the amount of linearized pCDNA1-amp. All lanes are from the same gel.
Cruciform structures are efficient targets for RAG-mediated transposition.
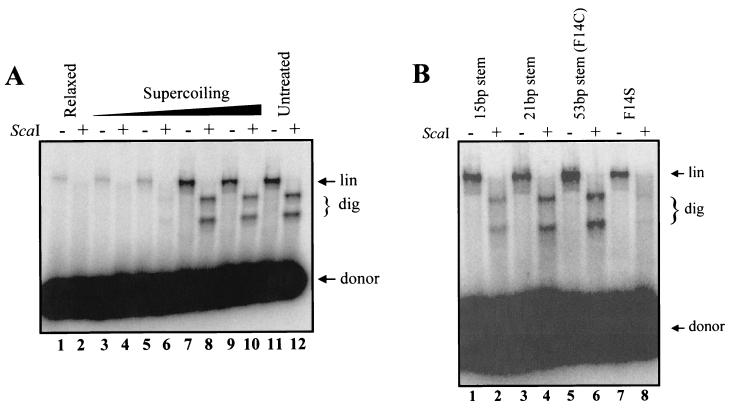
Because the inverted repeat used in the F14C plasmid forms a cruciform structure that would be stable under our experimental conditions (39), we hypothesized that this structure might serve as a hot spot for RAG-mediated transposition. Alternatively, some other feature of the inverted repeat sequence could serve as a target for the RAG proteins. To determine whether the cruciform structure itself stimulates transposition, we took advantage of the fact that cruciform formation is highly dependent upon the presence of supercoiling (17, 22, 39, 42). We generated a panel of topoisomers of F14C by treating the plasmid with topoisomerase in the presence of various concentrations of ethidium bromide (14). In the absence of ethidium, topoisomerase completely relaxes the plasmid, preventing cruciform formation. Gel electrophoresis revealed that the products of the topoisomerase reactions ranged from relaxed to highly supercoiled, depending on the concentration of ethidium bromide in the incubation (data not shown). Although the relaxed plasmid was a substrate for transposition, no targeted events were detected (Fig. 3A, lanes 1 and 2). Efficient targeting of transposition to the inverted repeat was observed once a certain concentration of ethidium was reached (corresponding to increased superhelical density, facilitating cruciform formation) (Fig. 3A, lanes 7 to 12). These data demonstrate that targeted transposition is a function of structural features that are influenced by supercoiling, such as formation of a cruciform, and not of the sequence itself. As an additional control, we tested another inverted repeat target substrate, F14S, in which the central 14 bp of the insert contain mirrored, rather than inverted, repeat symmetry and consequently do not readily form cruciforms at natural superhelical densities (42). Targeted transposition was not observed with the pUC8F14S (F14S) plasmid (Fig. 3B, lanes 7 and 8). These results indicate that transposition is indeed targeted to cruciform structures. This conclusion is further supported by our observation that the hairpin ends of self-complementary oligonucleotides serve as hot spots for RAG-mediated transposition (see below).
FIG. 3.
Cruciform structures are hot spots for transposition. (A) F14C was treated with topoisomerase as described in the text. Topoisomers were used as targets for transposition as described for Fig. 1B. Products were resolved on a 1% alkaline agarose gel, dried, and visualized by autoradiography. (B) Plasmids containing various cruciform-forming inverted repeats were used as targets and also show targeting of transposition.
F14C contains an inverted repeat sequence of 106 bases, which forms cruciform arms approximately 53 bp in length (42). To test the ability of the RAG transposase to target transposition to cruciforms of differing length and sequence, we employed plasmid targets containing inverted repeat sequences capable of forming cruciform arms of approximately 21 and 15 bp (26). In addition to F14C, plasmids capable of forming 21- and 15-bp stems contained preferred target sites (Fig. 3B, lanes 2, 4, and 6) (80 to 100%; quantitated as described above for Fig. 1). This reinforces the observation that targeting is not sequence specific but instead depends on target structure.
Transposition is targeted specifically to hairpin ends.
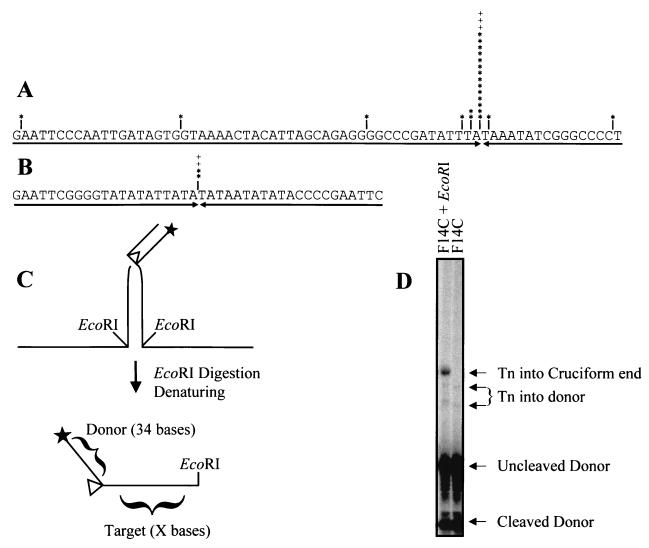
In order to determine the precise sites of integration into the cruciform arms, we cloned and sequenced transposition products. As summarized in Fig. 4A, 21 of 25 transposition events into F14C (84%) occurred within two nucleotides of the tip of the predicted hairpin end of the cruciform arm; 17 of 25 events (68%) occurred precisely at the hairpin tip. To verify that targeting to the hairpin end is not a sequence-specific phenomenon, we sequenced several products of transposition into the plasmid capable of forming the cruciform with a 21-bp stem, which has a different inverted repeat sequence than F14C. As shown in Fig. 4B, 100% of the integrations occurred precisely at the hairpin tip.
FIG. 4.
Transposition is targeted to the hairpin ends of cruciform arms. (A) Products of transposition into F14C were PCR cloned and sequenced. Sites of integration are marked (∗, clones sequenced from the 12 RSS; +; clones sequenced from the 23 RSS). (B) Products of transposition into the 21-bp stem used in Fig. 3B were sequenced as for panel A. (C) Schematic of integration into a cruciform end, followed by digestion with EcoRI. For clarity, the 23 RSS donor and the complementary strand are not shown. (D) Products of transposition into F14C were digested with EcoRI and run on a denaturing 6% acrylamide gel. A single product is formed, indicating transposition into a single site. The top of the gel was removed for layout purposes; all bands present on the entire gel are visible in this figure.
To confirm that most transposition events occur at or very near the hairpin tip of the cruciform arm, we mapped the sites of integration into F14C by EcoRI digestion of the bulk population of products, followed by electrophoresis through a sequencing gel. Since the inverted repeat sequence was cloned into pUC8 by using an EcoRI site, digestion at that site after transposition yields products that are the length of the cleaved donor plus the amount of target DNA between the integration site and the EcoRI site (schematized in Fig. 4C). The site of integration relative to the end of the inverted repeat was determined by subtracting the length of the labeled, cleaved donor 12 RSS (34 bases) from the size of the fragments. As shown in Fig. 4D, the integration sites in most transposition products map to the hairpin tip. Minor products resulting from transposition into the donor RSS were also observed; these products account for approximately 11% of detectable transposition events (see below for further discussion).
Transposition into hairpin and 3′-flap-containing oligonucleotides.
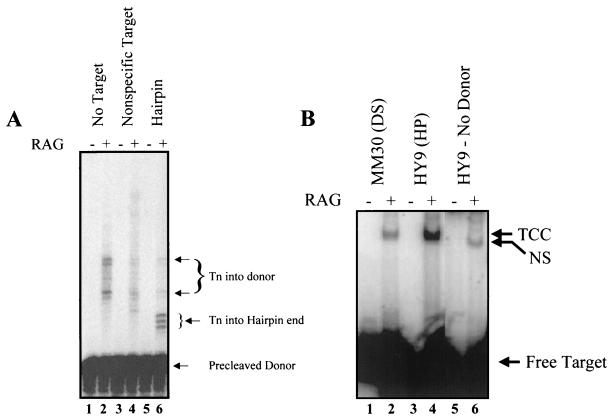
Several structural features of the cruciform might be important for stimulating transposition, such as the branched structure at the cruciform base or the hairpin-like character of the cruciform ends. To further elucidate the structural requirements for targeting transposition, we employed HY9, an oligonucleotide target that forms a hairpin, thereby mimicking one hairpin arm of a cruciform. This hairpin target produced three distinct transposition products (Fig. 5A, lane 6), differing in size by 1 nucleotide, that correspond to transposition at or very near the hairpin tip. In contrast, a control double-stranded oligonucleotide target elicited no targeted transposition (Fig. 5A, lane 4). Transposition is thus directed to a hairpin tip and does not require the presence of a cruciform structure.
FIG. 5.
Transposition is targeted to hairpins. (A) Transposition was carried out as described in the text. “Precleaved donor” refers to an oligonucleotide donor that corresponds to the RSS after cleavage, i.e., the donor terminates at the site of cleavage (19). The double-stranded control oligonucleotide was FM116/117, and the hairpin oligonucleotide was HY9. Some products of transposition into free donor molecules are visible in the absence of target. The nonspecific target shows no unique bands, while the hairpin target shows several products which correspond to transposition into the hairpin end. (B) The formation of target capture complexes (TCC) was assayed with double-stranded (MM30) and hairpin (HY9) oligonucleotides. HY9 forms substantially more target capture complexes than the MM30 control. The nonspecific shift of target in the absence of donor is indicated (NS). All lanes are from the same gel.
The presence of some RAG-dependent products in the absence of target (Fig. 5A, lanes 1 and 2) suggests that the donor RSS oligonucleotides themselves can serve as targets for transposition. This interpretation is supported by additional data (G. S. Lee and D. B. Roth, unpublished observations). In the presence of a nonhairpin target, transposition into the donor RSS was somewhat diminished (Fig. 5A, lane 4), perhaps because the excess target serves as a competitor. Transposition into the donor RSS was further diminished by addition of a hairpin oligonucleotide (Fig. 5A, lane 6), indicating that the hairpin specifically and efficiently outcompetes the donor RSS for use as a target for transposition. Thus, the presence of a hairpin end is sufficient to direct transposition by the RAG proteins.
Because hairpin tips typically contain localized DNA distortions (reviewed in reference 11), we wondered whether other distortions recognized by the RAG proteins might target transposition. One such structure is the junction between single- and double-stranded DNA present at ends containing 3′ extensions (3′ flaps), which are cleaved specifically by an endonucleolytic activity of the RAG proteins (36). Indeed, using such an oligonucleotide as a target, we found that transposition was specifically directed to the single-strand-double-strand junction (data not shown). Thus, like hairpins, single-strand-double-strand junctions are preferred targets for RAG-mediated transposition.
To investigate the mechanism responsible for directing transposition to hairpin ends, we measured the association of RAG-RSS complexes with hairpin targets using a physical assay. We had previously used a short double-stranded oligonucleotide target, MM30, to detect stable RAG-RSS-target complexes by electrophoretic mobility shift analysis (25). RAG-RSS-target complexes with HY9, the hairpin oligonucleotide, are much more abundant than complexes formed with a non-hairpin-containing double-stranded oligonucleotide of similar length, MM30 (sevenfold by phosphorimager quantitation) (Fig. 5B, compare lanes 2 and 4). Without donor RSS, the hairpin oligonucleotide is bound only weakly by the RAG proteins, producing a nonspecific complex of somewhat higher mobility (Fig. 5B, lane 6). Such nonspecific binding was also observed for MM30 (25) (data not shown). These data demonstrate that the presence of a hairpin in the target substantially enhances formation of stable target capture complexes.
Hairpin-containing competitor targets inhibit hybrid joint formation.
The use of hairpin ends as preferred targets for RAG-mediated transposition is reminiscent of a previously described reaction in which the RAG proteins use the 3′ OH of a signal end to attack a hairpin coding end, forming products termed open-and-shut joints (if the signal end reattaches itself to the original coding end) or hybrid joints (if the signal end attacks the other coding end) (21). In vivo, hybrid joints are formed from plasmid substrates (16) and from endogenous antigen receptor loci (4, 24). The fact that high levels of hybrid joints form in the absence of the double-strand break repair factors normally required for formation of coding and signal joints led us to hypothesize that hybrid joints can be formed in vivo directly by the RAG proteins (4, 7). The existence of hybrid joints provided the strongest evidence that the signal and coding ends actually reside in the same DNA-protein complex (a four-ended postcleavage complex) in vivo (4, 7), because it was assumed that the coding and signal ends must coexist in the same complex to facilitate their joining. Our data suggest an alternate, transposition model for the formation of hybrid joints, in which signal end-RSS complexes attack free hairpin coding end targets, bypassing the requirement for a four-ended complex.
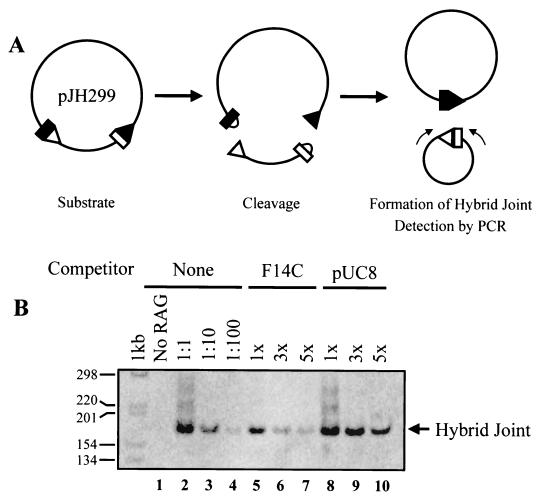
We designed competition experiments to test these rival models. The previously accepted model predicts that the postcleavage complexes should be quite stable. Thus, addition of free target molecules should not inhibit hybrid joint formation. In contrast, the transposition model posits a dynamic postcleavage complex that does not stably retain hairpin coding ends. Thus, according to this model, hybrid joint formation should be quite sensitive to the presence of hairpin-containing competitor targets. The results of such a competition experiment are shown in Fig. 6. The RAG proteins were incubated with a plasmid substrate (pJH299) in the presence of Ca2+ (to allow binding, but not catalysis). Competitor DNA molecules with or without cruciforms (F14C and pUC8, respectively) were then added along with Mg2+ to initiate catalysis. Hybrid joints were detected by PCR as diagrammed in Fig. 6A. Incubation with various levels of pUC8 competitor had little or no effect on the efficiency of hybrid joint formation, as expected (Fig. 6B, lanes 8 to 10). Also as expected, addition of F14C did not affect the efficiency of cleavage (data not shown). F14C did, however, significantly inhibit hybrid joint formation: even a threefold molar excess of F14C resulted in a 10- to 100-fold decrease in the level of hybrid joints. (In several experiments, a fivefold excess of F14C inhibited hybrid joint formation 100-fold, on average.) These data support a model in which hybrid joint formation occurs via a transposition mechanism, capturing hairpin ends from solution, rather than in the context of a stable postcleavage complex.
FIG. 6.
A cruciform-containing plasmid specifically inhibits hybrid joint formation. (A) Schematic of the hybrid joining assay. A plasmid substrate, pJH299, is incubated with RAG proteins. Hybrid joints are detected by PCR with primers at the indicated sites. (B) The presence of F14C inhibits formation of hybrid joints in this assay, while the presence of pUC8 control has little or no effect.
DISCUSSION
The terminal two to four nucleotides of fully self-complementary DNA hairpins generally adopt distorted forms (e.g., unpaired bases and buckled base pairs) that serve as targets for structure-specific nucleases (reviewed in reference 11). We have demonstrated that the presence of cruciform or hairpin structures stimulates transposition, with the vast majority of integration events occurring within two nucleotides of the hairpin tip. Furthermore, these structures are greatly preferred over competitor targets. Distortion at the hairpin tip thus appears to provide a strong impetus for transposition. We have also shown that another type of DNA distortion, a single-strand-double-strand DNA junction, can serve as a preferred target for RAG-mediated transposition. Such structures are generated during recombination and DNA repair. As discussed below, protein binding can also generate DNA distortions. Thus, there are many potential distorted DNA structures that might serve as hot spots for RAG-mediated transposition in vivo.
Implications for the transposition mechanism.
As noted above, the Tn7 and Tn10 transposases strongly prefer targets containing distorted DNA. The human immunodeficiency virus and avian sarcoma virus integrases also display a preference for targets located in the arms of cruciform structures (13). A preference for certain distorted DNA structures provides another point of similarity between the transposase-integrase superfamily and the V(D)J recombinase, supporting the notion that the RAG proteins, and by extension adaptive immunity as a whole, may have evolved from an ancient transposon.
The RAG transposase may, like Tn7, preferentially bind to distorted DNA (33). Our observation that the presence of hairpins in the target stimulates formation of stable transposase-target complexes supports this possibility. Generating a distorted DNA intermediate could be a rate-limiting step in RAG-mediated transposition. If this is the case, the presence of an already-distorted molecule could accelerate catalysis. This second possibility is supported by recent work showing that strand transfer is markedly slower than target capture (25). In fact, the presence of distorted DNA could work at both levels, augmenting stable binding and facilitating catalysis. Regardless of the mechanism, targeting of transposition to distorted DNA may explain the preference for G+C rich target sequences observed in an earlier study (9), because these sequences may form distorted DNA structures.
The region(s) of the RAG proteins responsible for capturing the target and delivering it to the active site have yet to be established. We have shown that active-site amino acids in RAG-1 are important for stable target binding (25). The present study suggests that the domain(s) that bind the hairpin coding ends after cleavage (in the postcleavage complex) may also participate in target selection.
Has transposition already been observed in vivo?
RAG-mediated transposition of a segment of DNA to an unrelated target molecule, though well established in vitro, has not yet been demonstrated in vivo. This has prompted the suggestion that transposition in vivo may be carefully regulated (1, 6). Our discovery that transposition is targeted efficiently and specifically to sequences bearing certain structural features provides a powerful new tool with which to search for RAG-mediated transposition in vivo.
Our results also suggest that authentic transposition events may have already been detected in living cells. Previous work has shown that the RAG proteins can use the 3′ OH of a signal end to attack the hairpin coding end and form a product known as a hybrid joint (21). Hybrid joints attributed to RAG-mediated joining have been detected in vivo in mice (4) and in cultured cells (7). Formation of these products had been considered a very specialized reaction, necessitating the retention of both coding and signal ends by the RAG proteins in a postcleavage complex. According to the established model, the close proximity of the signal ends to the coding ends in this complex facilitates hybrid joint formation by a reaction that can be considered a special case of transposition involving a target (the hairpin end) that was generated in the same DNA-protein complex (4, 21). We have shown, however, that the RAG transposase can efficiently capture hairpin targets de novo. In fact, exogenous hairpins can effectively inhibit formation of hybrid joints, which suggests that the postcleavage complex is not static but is actually much more dynamic than was previously thought. Based on these results, we suggest that formation of hybrid joints in vivo may proceed by a transposition reaction involving capture of free coding ends by a RAG-RSS complex. If this is the case, hybrid joints suddenly acquire biological significance. Rather than yielding immunologically irrelevant (and wasteful) by-products, hybrid joint formation may actually protect the organism by channeling dangerous transposition intermediates into harmless products.
Targeted transposition and oncogenic chromosome translocations.
The discovery that the RAG proteins can mediate transposition in vitro brought with it the realization that such a reaction could produce genomic instability in lymphocytes (9). Our data imply that particular genomic locations, specifically those containing distorted DNA structures, might be hot spots for transposition in vivo. Particular sequence elements clearly favor the formation of distorted DNA. For example, triplet repeat sequences form hairpin structures in vitro and are associated with genomic instability in yeasts, bacteria, and mammals (20). Recent evidence indicates that triplet repeats and palindromes form hairpins in vivo in yeasts, bacteria, and mice (23, 35). Protein binding can also create altered DNA structures; for example, the Tn7-encoded TnsD protein targets transposition by binding to and distorting DNA (32), and retroviral integrases target integration events to DNA distorted by nucleosome binding (29).
Lymphoid malignancies are commonly associated with chromosome translocations, many of which are thought to be mediated by the V(D)J recombinase (reviewed in reference 34). In these translocations, RSS are found at the breakpoint in the T-cell receptor or immunoglobulin genes, and RSS-like sequences have been identified at approximately 50% of the breakpoints in the partner chromosomes (18). In such cases, it is easy to imagine that the RAG proteins mediate cleavage of both chromosomes. It has been less obvious, however, what process(es) might be responsible for inducing rearrangement at the remaining breakpoints, which do not contain RSS. Indeed, breakpoints often cluster in regions that appear devoid of RSS-like sequences (10, 18). What could account for the tendency of the chromosome to suffer cleavage in these so-called breakpoint cluster regions? It has been suggested that some (unknown) nuclease might recognize some feature of these regions and promote cleavage (10, 18). We propose a new, more unifying model in which the RAG proteins themselves are responsible for targeting translocations to breakpoint cluster regions. Indeed, analysis of two different breakpoint cluster regions implicated in common human lymphomas revealed oligopurine-oligopyrimidine sequences that showed hypersensitivity to S1 nuclease (18), a nuclease that recognizes alternative DNA structures, including cruciforms and hairpins. We hypothesize that single-ended transposition events join antigen receptor loci to partner sites that lack RSS elements but bear particular structural features that target transposition. The RAG proteins may thus be far more responsible for generating oncogenic translocations than has been previously suspected.
Acknowledgments
We thank Vicky Brandt for editorial assistance; M. Gellert, Ilana Goldhaber-Gordon, L. Hanakahi, J. Qiu, L. E. Huye, J.-Y. Masson, A. N. Miller, and H. Yarnall-Schultz for valuable comments on the manuscript; and L. E. Huye for HMG1 protein. D. Guzman provided secretarial assistance.
D.B.R. is an Assistant Investigator of the Howard Hughes Medical Institute. This work was supported by a grant from the National Institutes of Health (AI-36420).
REFERENCES
- 1.Agrawal, A., Q. M. Eastman, and D. G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744-751. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. [DOI] [PubMed] [Google Scholar]
- 3.Besmer, E., J. Mansilia-Soto, S. Cassard, D. J. Sawchuk, G. Brown, M. Sadofsky, S. M. Lewis, M. C. Nussenzweig, and P. Cortes. 1998. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell 2:817-828. [DOI] [PubMed] [Google Scholar]
- 4.Bogue, M. A., C. Wang, C. Zhu, and D. B. Roth. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7:37-47. [DOI] [PubMed] [Google Scholar]
- 5.Cuomo, C. A., C. L. Mundy, and M. A. Oettinger. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 7.Han, J.-O., S. B. Steen, and D. B. Roth. 1997. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol. Cell. Biol. 17:2226-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 9.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463-470. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger, U., B. Purtscher, G. D. Karth, S. Knapp, C. Mannhalter, and K. Lechner. 1993. Mechanism of the chromosomal translocation t(14;18) in lymphoma: detection of a 45-Kd breakpoint binding protein. Blood 81:1833-1840. [PubMed] [Google Scholar]
- 11.Kabotyanski, E. B., C. Zhu, D. A. Kallick, and D. B. Roth. 1995. Hairpin opening by single-strand specific nucleases. Nucleic Acids Res. 23:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kale, S. B., M. A. Landree, and D. B. Roth. 2001. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol. Cell. Biol. 21:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz, R. A., K. Gravuer, and A. M. Skalka. 1998. A preferred target DNA structure for retroviral integrase in vitro. J. Biol. Chem. 273:24190-24195. [DOI] [PubMed] [Google Scholar]
- 14.Kochel, T. J., and R. R. Sinden. 1988. Analysis of trimethylpsoralen photoreactivity to Z-DNA provides a general in vivo assay for Z-DNA: analysis of the hypersensitivity of (GT)n B-Z junctions. BioTechniques 6:532-543. [PubMed] [Google Scholar]
- 15.Kuduvalli, P. N., J. E. Rao, and N. L. Craig. 2001. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 20:924-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis, S. M., J. E. Hesse, K. Mizuuchi, and M. Gellert. 1988. Novel strand exchanges in V(D)J recombination. Cell 55:1099-1107. [DOI] [PubMed] [Google Scholar]
- 17.Lilley, D. M. 1983. Dynamic, sequence-dependent DNA structure as exemplified by cruciform extrusion from inverted repeats in negatively supercoiled DNA. Cold Spring Harbor Symp. Quant. Biol. 47:101-112. [DOI] [PubMed] [Google Scholar]
- 18.Lu, M., N. Zhang, S. Raimondi, and A. D. Ho. 1992. S1 nuclease hypersensitive sites in an oligopurine/oligopyrimidine DNA from the t(10;14) breakpoint cluster region. Nucleic Acids Res. 20:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 20.McMurray, C. T. 1999. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. USA 96:1823-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melek, M., M. Gellert, and D. C. van Gent. 1998. Rejoining of DNA by the RAG1 and RAG2 proteins. Science 280:301-303. [DOI] [PubMed] [Google Scholar]
- 22.Mizuuchi, K., M. Mizuuchi, and M. Gellert. 1982. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J. Mol. Biol. 156:229-243. [DOI] [PubMed] [Google Scholar]
- 23.Moore, H., P. W. Greenwell, C. P. Liu, N. Arnheim, and T. D. Petes. 1999. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA 96:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morzycka-Wroblewska, E., F. Lee, and S. Desiderio. 1988. Unusual immunoglobulin gene rearrangement leads to replacement of recombination signal sequences. Science 242:261-263. [DOI] [PubMed] [Google Scholar]
- 25.Neiditch, M. B., G. S. Lee, M. A. Landree, and D. B. Roth. 2001. The RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol. Cell. Biol. 21:4302-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oussatcheva, E. A., L. S. Shlyakhtenko, R. Glass, R. R. Sinden, Y. L. Lyubchenko, and V. N. Potaman. 1999. Structure of branched DNA molecules: gel retardation and atomic force microscopy studies. J. Mol. Biol. 292:75-86. [DOI] [PubMed] [Google Scholar]
- 27.Peters, J. E., and N. L. Craig. 2000. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell 6:573-582. [DOI] [PubMed] [Google Scholar]
- 28.Pribil, P. A., and D. B. Haniford. 2000. Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. J. Mol. Biol. 303:145-159. [DOI] [PubMed] [Google Scholar]
- 29.Pruss, D., F. D. Bushman, and A. P. Wolffe. 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 91:5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu, J., S. B. Kale, H. Y. Schultz, and D. B. Roth. 2001. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell 7:77-87. [DOI] [PubMed] [Google Scholar]
- 31.Ramsden, D. A., J. F. McBlane, D. C. van Gent, and M. Gellert. 1996. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15:3197-3206. [PMC free article] [PubMed] [Google Scholar]
- 32.Rao, J. E., and N. L. Craig. 2001. Selective recognition of pyrimidine motif triplexes by a protein encoded by the bacterial transposon Tn7. J. Mol. Biol. 307:1161-1170. [DOI] [PubMed] [Google Scholar]
- 33.Rao, J. E., P. S. Miller, and N. L. Craig. 2000. Recognition of triple-helical DNA structures by transposon Tn7. Proc. Natl. Acad. Sci. USA 97:3936-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth, D. B., and N. L. Craig. 1998. VDJ recombination: a transposase goes to work. Cell 94:411-414. [DOI] [PubMed] [Google Scholar]
- 35.Samadashwily, G. M., G. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17:298-304. [DOI] [PubMed] [Google Scholar]
- 36.Santagata, S., E. Besmer, A. Villa, F. Bozzi, J. S. Allingham, C. Sobacchi, D. B. Haniford, P. Vezzoni, M. C. Nussenzweig, Z. Q. Pan, and P. Cortes. 1999. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell 4:935-947. [DOI] [PubMed] [Google Scholar]
- 37.Schultz, H. Y., M. A. Landree, J. Qiu, S. B. Kale, and D. B. Roth. 2001. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell 7:65-75. [DOI] [PubMed] [Google Scholar]
- 38.Shockett, P. E., and D. G. Schatz. 1999. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol. Cell. Biol. 19:4159-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinden, R. R. 1994. DNA structure and function. Academic Press, San Diego, Calif.
- 40.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature 302:575-581. [DOI] [PubMed] [Google Scholar]
- 41.van Gent, D. C., K. Hiom, T. T. Paull, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng, G. X., T. Kochel, R. W. Hoepfner, S. E. Timmons, and R. R. Sinden. 1991. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J. Mol. Biol. 221:107-122. [DOI] [PubMed] [Google Scholar]