Abstract
In an effort to define the molecular basis for morphogenesis of major sperm tail structures, including outer dense fibers, we recently cloned the Spag5 gene by virtue of its strong and specific leucine-zipper-mediated interaction with Odf1, the 27-kDa major outer dense fiber protein. Spag5 is expressed during meiosis and in round spermatids and is similar, if not identical, to Deepest, a putative spindle pole protein. Here we report the disruption of the Spag5 gene by homologous recombination. Spag5-null mice lack Spag5 mRNA and protein. However, male mice are viable and fertile. Analysis of the process of spermatogenesis and sperm produced in Spag5-null mice did not reveal a major phenotype as a consequence of the knockout event. This result suggests that if Spag5 plays a role in spermatogenesis it is likely compensated for by unknown proteins.
Spermatogenesis is the developmental program that guides spermatogonia to differentiate into functional spermatozoa. It includes mitotic and meiotic divisions, genetic recombination, and spermiogenesis, the process during which round spermatids undergo extensive morphologic transformation to produce the distinctive structures of spermatozoa. These include the acrosome, condensed nucleus, and tail. A main component of the mammalian sperm tail are the structures surrounding the axoneme, called outer dense fibers (ODFs) (5, 10). Nine ODFs extend throughout the midpiece and principal piece and are surrounded by the fibrous sheath (FS). At the annulus, demarcating the border between the midpiece and the principle piece, two of nine ODFs are replaced by the two longitudinal columns of the FS (10). During spermiogenesis, ODFs are assembled in a proximal-to-distal direction in step 8 to 19 spermatids, whereas the FS is assembled in a distal-to-proximal direction in step 2 to 17 spermatids (11, 12).
Previous studies of protein components of rat ODFs have revealed multiple polypeptides, including several major (i.e., 32-, 27.5-, 20-, 14.4-, 84-, and 80-kDa) and many minor peptides (18, 19, 27). These polypeptides are phosphorylated at serine residues and are resistant to solubilization in ionic detergents due to a high content of disulfide bonds (4-6). Earlier biochemical and structural studies had suggested that ODFs probably function as passive elastic structures and provide elastic recoil for the sperm tail (10). It has been demonstrated recently that ODFs appear to protect sperm against shear forces (1). Two ODF cDNAs from rat, encoding the Odf1 and Odf2 proteins, have been cloned and characterized by different groups by using different strategies (2, 3, 16, 25, 26). Odf1 encodes the major 27-kDa ODF protein and harbors a leucine zipper motif in the N terminus and 16 Cys-Gly-Pro repeats in the C terminus (22). Using the Odf1 leucine zipper as a bait in a yeast two-hybrid protein interaction screen, we cloned several testicular genes, including Odf2 and Spag4 (21, 22, 26). Odf2 encodes the major 84-kDa ODF protein which, like Odf1, localizes to the ODF (22). Spag4 is a 49-kDa ODF-associated protein, which is present on spermatid microtubules in the manchette and the axoneme, suggesting that Spag4 functions as a molecular link between ODFs and the axoneme (21).
We recently cloned Spag5 as an Odf1-interacting protein by using the yeast two-hybrid system (26). Spag5 has a molecular mass of ca. 200 kDa. Spag5 mRNA is predominantly expressed in testis in pachytene spermatocytes and to a lesser extent in round spermatids. Spag5 protein is synthesized de novo in spermatocytes and round spermatids. It contains two leucine zipper motifs in the C-terminal region, the downstream one of which mediates the interaction with Odf1. The protein-protein interaction between Odf1 and Spag5 indicated that Spag5 might participate in sperm tail morphogenesis; however, we were not able to detect Spag5 in mature spermatozoa, suggesting that it is not a major structural protein. Interestingly, we discovered that rat Spag5 is highly related to human Deepest protein (73% identity), a putative mitotic spindle coiled-coil protein. Indeed, of several Deepest splice variants, some retain the Spag5 leucine zippers, whereas other variants do not. Together with Spag5 protein expression in spermatocytes, our data suggest that Spag5 may play a functional role in meiosis, as well as in spermatid morphogenesis (14, 26).
To investigate a possible role of Spag5 in mammalian spermatogenesis, we generated a null mutation in the Spag5 gene by homologous recombination in mouse embryonic stem (ES) cells. Mice homozygous for the Spag5 null mutation lacked detectable Spag5 protein, but were viable and fertile, and appeared normal. No gross morphologic defects were observed in spermatogenesis.
MATERIALS AND METHODS
Isolation of Spag5 genomic clones.
The Spag5 cDNA clone (26) was used as a probe to screen a 129/Sv genomic library constructed in the λ FIX II vector (Stratagene, La Jolla, Calif.). Since the Spag5 gene is closely linked in tandem to the aldolase C gene, positive clones were further separated into two groups based on Southern blot analysis by using the Spag5 cDNA probe and aldolase C cDNA probe, respectively. One 16.5-kb genomic clone, which reacted only with the 5′ probe fragment of the Spag5 cDNA clone, was chosen for the construction of a Spag5 targeting vector.
Construction of Spag5 gene targeting vector.
The Spag5 targeting vector was constructed by first subcloning a 9.5-kb PacI and NotI fragment of Spag5 genomic DNA into pBluescript II SK(+) (Stratagene). Next, deletions were introduced with the Erase-a-Base System (Promega, Madison, Wis.) from the PacI site and from the NotI site. Deletion clones were sequenced, and exons and introns were verified. Two of these were chosen to construct a Spag5 deletion mutant: a 3.2-kb DNA fragment which remained after deletion of 3′ sequences of the PacI-NotI fragment was ligated to a 5.6-kb DNA fragment which remained after deletion of 5′ sequences of the PacI-NotI fragment (Fig. 1A). Thus, the resulting mutant Spag5 genomic fragment lacks 0.7 kb of genomic sequences. A 3.5-kb ClaI pKT3LoxA neomycin resistance cassette, which contains a Cre-excisable neomycin resistance gene under the control of the polymerase II promoter (24), was inserted between the 3.2- and 5.6-kb Spag5 genomic fragments. Insertion of the neomycin resistance cassette introduced (i) extra HindIII and SacI sites that were exploited in Southern blot analysis for genotyping and (ii) stop codons. The Spag5 targeting construct was released from the pBluescript vector by NotI restriction enzyme digestion and subcloned into the λ 2TK phage vector containing thymidine kinase genes in both left and right polylinkers (24).
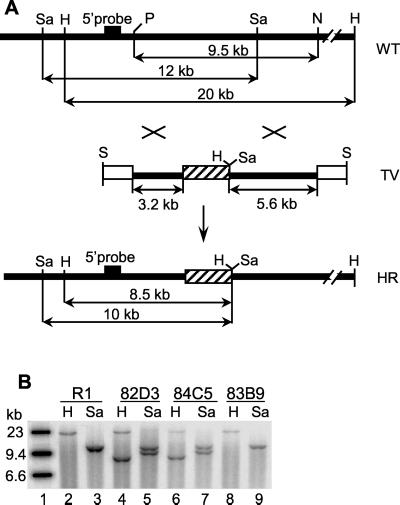
FIG. 1.
Inactivation of the Spag5 gene in mouse ES cells and mice. (A) Strategy used for homologous recombination of the Spag5 gene. A 0.7-kb genomic Spag5 fragment was replaced by the neomycin resistance cassette (cross-hatched box), and the indicated resulting fragment was introduced in a vector such that two thymidine kinase genes (open boxes) flank the recombinant genomic fragment. The result of homologous recombination is shown. Wild-type Spag5 genomic DNA generates 20- and 12-kb DNA fragments after restriction with HindIII and SacI, respectively. After homologous recombination HindIII and SacI generate 8.5- and 10-kb fragments, respectively. The probe used to detect such recombination events in Southern blotting analyses is located outside the genomic fragment employed (black box). Restriction enzyme sites: H, HindIII; N, NotI; P, PacI; S, SalI; Sa, SacI. (B) Southern blot genotyping of ES cells was performed by using HindIII or SacI digestions with the indicated 5′ probe. The 20-kb HindIII fragment of the wild-type Spag5 allele shifts to an 8.5-kb HindIII fragment of the recombined allele (indicated by H). The 12-kb SacI fragment of the wild-type Spag5 allele shifts to a 10-kb SacI fragment of the recombinant allele (indicated by Sa). R1, wild-type R1 ES cells; 82D3 and 84C5, two ES cell clones demonstrating one wild-type and one recombined allele; 83B9, an ES clone with only wild-type alleles.
Generation of Spag5-deficient mice.
R1 ES cells were obtained from J. Rossant and were maintained and cultured as described previously (17). The Spag5 targeting vector was separated from the λ phage arms by SalI restriction enzyme digestion. A total of 150 μg of SalI-digested Spag5 targeting vector was introduced into 5 × 106 to 7 × 106 R1 ES cells by electroporation at 240 V and 500 μF (Bio-Rad Gene Pulser). Electroporated cells were plated in 100-mm dishes and were cultured for 48 h. Positive and negative selections were carried out by using Geneticin (G418) at 300 U/ml and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU) at 2 × 10−7 M, respectively. Surviving clones were screened for morphology, and selected colonies were cloned on days 8, 9, and 10 of the selection. ES cell clones containing a modified Spag5 allele were identified by Southern blot analysis with a 5′ probe external to the Spag5 genomic DNA present in the vector. Southern blot transfer and hybridization were performed as described previously (7). Potential targets were rescreened by Southern blot analysis. The karyotype of targeted ES cell clones was determined to verify their diploid status by using a described method (13). Several Spag5 targeted ES cell clones demonstrating diploid karyotypes were injected into C57BL/6 (B6) blastocysts 3.5 days postcoitum, which were next transferred into 2.5-day postcoitum CD1 foster mothers. Male chimeras, ranging from 5 to 80% agouti pigmentation, were bred to C57BL/6 females, and agouti pups were genotyped to confirm germ line transmission of the targeted Spag5 allele. Spag5 heterozygotes were intercrossed to produce Spag5-null mice.
Analysis of Spag5−/− male mice. (i) Histology.
Testes from Spag5+/− and Spag5−/− mice were dissected and fixed in 4% paraformaldehyde at 4°C overnight, dehydrated through a graded series of ethanol, and embedded in paraffin. Sections (5 μm) were cut and stained with hematoxylin and eosin and examined by light microscopy.
(ii) Epididymal sperm counts.
Next, 3- to 4-month-old male Spag5+/− and Spag5−/− mice were sacrificed by cervical dislocation. Epididymides, along with the vas deferens, were dissected and cut into small pieces in a dish containing 5 ml of Dulbecco modified Eagle medium with 10% fetal calf serum. Sperm were allowed to release into the medium during incubation at 32°C in a 5% CO2 humidified incubator for 3 h. Total sperm counts were determined by using a hemocytometer.
(iii) Sperm motility assay.
For the sperm motility assay, 3- to 4-month-old male Spag5+/− and Spag5−/− mice were sacrificed by cervical dislocation. Caudal epididymides were cut into small pieces in human tubal fluid (HTF) (20) buffer saturated with CO2 and incubated at 37°C in a 5% CO2 humidified incubator for 30 min. Sperm in HTF buffer were diluted 10× in CO2-saturated HTF medium and incubated at 37°C in a 5% CO2 humidified incubator for 1 h. Sperm motility was examined under phase-contrast microscopy and registered on videotape.
(iv) RNA analysis.
mRNA from testes of 3- to 4-month-old Spag5+/− and Spag5−/− mice were isolated by using a QuickPrep Micro mRNA Purification Kit (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). Northern blot analyses were performed essentially as described previously (7) by using Hybond-N+, positively charged nylon membranes (Amersham Pharmacia Biotech). Hybridization probes were generated by using random hexamer primers (Boehringer Mannheim) and [α-32P]dCTP (Amersham Pharmacia Biotech).
(v) Spag5 protein analysis.
Total testicular germ cells were isolated from Spag5+/− and Spag5−/− mice and incubated with radiolabeled [35S]methionine in Dulbecco modified Eagle medium with 10% dialyzed fetal calf serum in a 5% CO2 humidified incubator for 3 h. Protein extracts were prepared, and immunoprecipitations were carried out by using two different anti-Spag5 polyclonal antibodies, the respective preimmune sera, and anti-Odf2 polyclonal antibodies as described previously. Immunoprecipitated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography as described previously (22).
RESULTS
Targeted disruption of Spag5 in ES cells.
To generate ES cells with one disrupted Spag5 allele, we constructed a replacement-targeting vector TV (Fig. 1A). A neomycin resistance expression cassette replaced 0.7 kb of the Spag5 genomic DNA, which contains the first exon of the partial cDNA of Spag5 (Fig. 1A). ES cells that underwent homologous recombination were selected in the presence of G418 and FIAU. Figure 1A shows the position of the 5′ Spag5 genomic DNA probe used to identify Spag5 homologous recombinants by Southern blotting analysis, as well as the position of the HindIII and SacI restriction sites in the wild-type and recombined Spag5 genomic DNA. Two targeted ES cell clones, 82D3 and 84C5 (Fig. 1B), were identified by Southern blotting analysis, which showed a 20-kb HindIII fragment and a smaller 8.5-kb HindIII fragment generated by the mutated Spag5 allele in two ES clones (lanes 4 and 6). The original R1 ES cells and clone 83B9 only harbor the 20-kb HindIII fragment (lanes 2 and 8). The presence of the mutant Spag5 allele was confirmed by SacI analysis: a 12-kb SacI Spag5 fragment is present in R1 and 83B9 ES cells (lanes 3 and 9), but clones 82D3 and 84C5 contain, in addition to the wild-type allele, a 10-kb SacI Spag5 mutant fragment (lanes 5 and 7). The karyotypes of these clones were determined, and 84C5 was chosen for the generation of chimeric mice.
Generation of Spag5−/− mice.
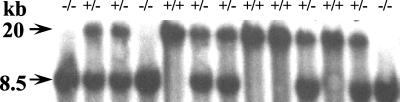
Cells from clone 84C5 were injected into C57BL/6 blastocysts to generate chimeric mice. Two male chimeras ranging in chimerism (5 and 80%) were bred with C57BL/6 females to produce agouti progeny. Spag5 heterozygous mice were identified by Southern blotting analysis. These Spag5+/− mice were interbred, and tail DNA from progeny was analyzed by Southern blotting by using HindIII and the probes described above. The results of this analysis are shown in Fig. 2. It is evident that Spag5 wild-type mice only harbor the HindIII 20-kb fragment, as expected, and that Spag5−/− mice only contain the 8.5-kb HindIII fragment. Heterozygous mice contain both fragments. The analysis of many breeding experiments revealed a Mendelian distribution of wild-type, heterozygous, and homozygous mice in offspring. These data, as well as the absence of any gross phenotypes, demonstrated that a Spag5−/− mouse is viable and apparently normal. Further breeding experiments showed that Spag5−/− mice are fertile (see below).
FIG. 2.
Transmission of the recombinant Spag5 allele. DNA was isolated from tail snips of F2 offspring resulting from breeding of Spag5+/− heterozygotic male and female mice. DNA was digested with HindIII and analyzed by Southern blotting with the probe indicated in Fig. 1A. The 20- and 8.5-kb HindIII fragments, representing the wild-type and recombinant alleles, respectively, are indicated. Note that Spag5−/− mice only harbor the 8.5-kb HindIII fragment.
RNA and protein analysis.
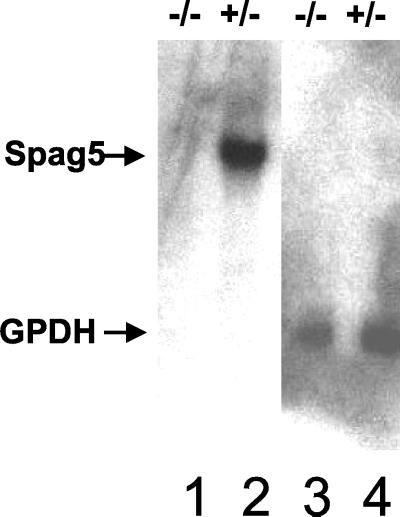
Previous studies showed that in the testes Spag5 mRNA was predominantly expressed in pachytene spermatocytes and round spermatids (23). Since the modified Spag5 allele has a deletion of the first exon of the partial Spag5 cDNA, it is expected that Spag5-null mice lack Spag5 mRNA and protein. To confirm this, mRNA from the testes of heterozygous and Spag5−/− mice were isolated. A Northern blot analysis (shown in Fig. 3) confirmed that the interruption of the Spag5 gene abolished production of full-length Spag5 mRNA in Spag5−/− mice compared to Spag5+/− mice (lanes 2 and 1, respectively).
FIG. 3.
Spag5−/− mice lack Spag5 mRNA. mRNA was isolated from the testes of Spag5−/− mice (lane 1) and Spag5+/− heterozygotic mice (lane 2), and 5 μg of mRNA from each was analyzed by Northern blotting hybridization with a 5′ Spag5 cDNA fragment as probe. After autoradiography, filters were stripped of the probe and rehybridized to a GAPD (glyceraldehyde-3-phosphate dehydrogenase) cDNA probe to confirm the quantity and quality of mRNA for both preparations (lanes 3 and 4).
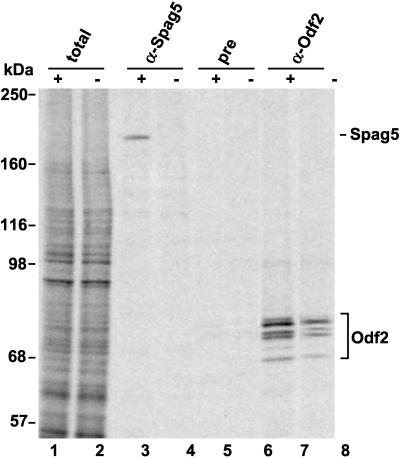
To analyze the production of Spag5 protein in homozygous mutant animals, testicular germ cells were isolated from Spag5+/− and Spag5−/− mice and metabolically labeled with [35S]methionine. Two different anti-Spag5 polyclonal antibodies were used to immunoprecipitate radiolabeled Spag5. Immunoprecipitation of Odf2 was included as control, as were relevant preimmune antisera. Figure 4 shows the results. The overall pattern of radiolabeled proteins in cells from these mice is indistinguishable (lanes 1 and 2). The 200-kDa Spag5 protein was detected in wild-type germ cells (lane 3) and, importantly, was absent in testicular germ cells from Spag5−/− mice, as predicted (lane 4). The 200-kDa Spag5 protein was not detected by preimmune antisera as shown previously (lanes 5 and 6). Odf2 was synthesized in both Spag5+/− and Spag5−/− mouse strains: the slight decrease in de novo Odf2 synthesis observed in Spag5−/− cells is reproducible, and the reason for this decrease is unknown (compare lanes 5 and 8; see Discussion). These results show that Spag5−/− mice suffer a loss of the 200-kDa Spag5 protein, but apparently this does not interfere with their normal development.
FIG. 4.
The Spag5 null mutation abolishes testicular Spag5 protein synthesis. The synthesis of Spag5 protein in testicular germ cells was analyzed and compared in Spag5+/− heterozygotic mice (lanes 1, 3, 5, and 7 [indicated by “+”) and Spag5−/− mice (lanes 2, 4, 6, and 8 [indicated by “−”). Isolated germ cells were incubated in the presence of [35S]methionine, and radiolabeled de novo-synthesized proteins were characterized directly (lanes 1 and 2) or after immunoprecipitation with anti-Spag5 antibodies (lanes 3 and 4), preimmune antiserum (lanes 5 and 6), or anti-Odf2 antibodies (lanes 7 and 8). Immunoprecipitated proteins and whole lysates were analyzed by SDS-PAGE and autoradiography. Lanes 1 and 2 show the total labeled protein pattern. Spag5 is indicated, as are the Odf2 peptides. Note the absence of Spag5 protein in the Spag5−/− mice.
Physiological consequences of Spag5 gene inactivation.
Spag5 is transcribed and translated in pachytene spermatocytes and spermatids, respectively (23). Spag5−/− mice develop normally. To assess a possible physiological role of Spag5 in spermatogenesis, the fertility of Spag5−/− mice was tested. Male Spag5−/− mice were bred with wild-type, Spag5+/− mice and Spag5−/− female mice. Mating results from these three breeding experiments showed that both Spag5 null males and Spag5 null females are fertile. Both Spag5−/− males and females had normal mating and breeding behavior. The litter size from these matings was indistinguishable from those of wild-type matings, and the progeny all appeared normal. Previous studies demonstrated that male mice are classified as fertile even if they produce <10% of the normal number of mature sperm (8). To verify whether the Spag5−/− gene knockout resulted in abnormal sperm counts, without a consequence in fertility, mature sperm counts of wild-type and Spag5−/− mice were determined and male reproduction organs were examined for gross abnormalities. No significant sperm count differences were detected between wild-type and Spag5−/− mice (Table 1). The weight of the testes and epididymides from wild-type and Spag5−/− mice also showed no statistically significant differences (Table 1). Spag5 was originally cloned by a functional interaction assay with Odf1, a 27-kDa major ODF protein, in a yeast two-hybrid system (26), which suggested that Spag5 may fulfill a function during spermatid development. To determine whether loss of Spag5 affects sperm morphology and motility, these parameters were examined. Spermatozoa were isolated from the caudal epididymis and capacitated in the HTF medium (20). Capacitated spermatozoa were examined by phase-contrast microscopy for motility and morphology. Hyperactive spermatozoa from both wild-type and Spag5−/− mice had indistinguishable flagella that traced similar figures and showed similar forward progression. To further investigate possible effects of loss of Spag5 on the process of spermatogenesis, the histology of testes and epididymis from Spag5−/− and wild-type mice was evaluated. The interstitial and seminiferous tubule compartments appeared normal in Spag5−/− mice and were not distinguishable from those of wild-type mice. Spermatogonia, spermatocytes, and round spermatids were found at successively higher levels within the epithelium, and mature sperm cells were released into the tubular lumen. Furthermore, the thickness of the seminiferous epithelium, the diameter of the seminiferous tubules, and the size of the tubular lumen appeared normal in Spag5−/− mice.
TABLE 1.
Body weight, organ weight, and sperm counts of wild-type and Spag5 mutant micea
| Genotype | Mean ± SD
|
|||
|---|---|---|---|---|
| Body wt (g) | Testis wt (mg) | Epididymis wt (mg) | Sperm countb (107) | |
| +/+ | 25.0 ± 0.9 | 66.3 ± 4.5 | 37.7 ± 4.6 | 1.8 ± 0.3 |
| −/− | 30.6 ± 4.0 | 71.8 ± 12.0 | 35.9 ± 4.3 | 1.9 ± 0.3 |
Values are means ± standard deviations of individual determinations made for three +/+ and seven −/− 3-month-old mice.
Sperm counts are the numbers of sperm obtained from two epididymides.
DISCUSSION
A difficulty in the study of mammalian male germ cells is that spermatogenesis cannot yet be recapitulated in cell culture. However, the use of transgenic and knockout mice allows us to ask questions related to gene function during spermatogenesis. The present work was undertaken in an effort to define the physiological role of the Deepest/Spag5 gene in mammalian spermatogenesis. We generated a null mutation in the Spag5 gene by homologous recombination in mouse ES cells, which were used to produce heterozygous and homozygous Spag5 knockout mice. Mice homozygous for the mutation lacked Spag5 mRNA and protein in germ cells. Males deficient in Spag5 protein did not exhibit a detectable abnormality in spermatogenesis. They were fertile and had normal testis weights, mature sperm count and motility, and normal testis histology.
The absence of any detectable abnormality in Spag5-deficient testes is unexpected in view of the expression pattern of Spag5 mRNA and protein, which is predominantly in spermatocytes and round spermatids (23). Furthermore, analysis of sperm tail protein interactions in the yeast two-hybrid system suggested that cooperation among several sperm tail proteins, including Odf1, Odf2, Spag4, and Spag5, may be essential during morphogenesis of ODF and axoneme (22-24). In the yeast system, the interaction of Spag5 and Odf1 is mediated by leucine zipper motifs (23). Deletion of either the leucine zipper of Spag5 or the one of Odf1 completely abolished this interaction. The presence of another, putative, leucine zipper motif in Spag5, which does not interact with Odf1, also suggested that Spag5 might interact with other proteins (23). In addition, the high degree of similarity of Spag5 to Deepest, a putative mitotic spindle protein, pointed to the possibility for another role of Spag5 during meiosis. Together, these data had suggested that the loss of one of the sperm tail-interacting proteins likely alters the profile of sperm tail morphogenesis with changes in fertility and sperm structure as a consequence.
Several explanations could account for the lack of a clear phenotype in Spag5 knockout mice. First, the function of Spag5 may not be absolutely required for spermatogenesis. There is precedent for this possibility from studies of H1t, a unique histone variant specifically expressed in late spermatocytes and in round spermatids: H1t knockout mice have no discernible phenotype (9). Interestingly, that study demonstrated that the lack of H1t was partially compensated for by somatic H1s. Accordingly, an unidentified protein may exist that could compensate for the loss of the function of Spag5. Second, the defect in Spag5−/− mice may be too small to be detected by the techniques employed here. This would warrant a detailed ultrastructural examination of Spag5−/− spermatozoa, as well as studies that challenge spermatogenesis in Spag5−/− mice that could reveal a defect. Third, since spermatogenesis is a very complex process, involving many genes, inactivation of one gene may not have detectable phenotypic change. This notion has been observed previously in double- and triple-gene inactivation experiments. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Mice lacking any single receptor, or any combination of two receptors, are viable and fertile, but male mice that lack all three receptors produce no mature sperm, owing to the progressive death of differentiating germ cells (15). Hence, the generation of homozygous mutant mice with ablations in Spag5 and one or more of the other sperm tail genes may shed light on the genetic interrelationship of sperm tail genes in the morphogenesis of sperm tail.
It should also be noted that the predicted structure of the Deepest gene is complex, with 24 exons spread out over 21 kb of genomic DNA, and that several different Deepest cDNAs have been reported. Therefore, the possibility remains that Deepest mRNA splice variants exist that are not affected by the recombination employed here, which involved the replacement of two of the Spag5 exons by the neomycin resistance cassette. We have observed, in large-scale immunoprecipitations with anti-Spag5 antibodies, the presence of two other peptides of ca. 125 and 155 kDa. These two peptides were also detectable in Spag5-null mice and therefore are not Spag5-associated coimmunoprecipitated proteins (results not shown). Work is currently in progress to further identify the nature of these peptides as possibly Deepest protein variants.
Acknowledgments
We acknowledge the expert technical assistance of E. Rattner, S. Y. Liu, B. Carson, and J. Turnbull in the Embryonic Stem Cell Facility at the University of Calgary.
This work was supported by grants from the Canadian Institutes of Health Research to F.A.V.D.H. and D.E.R. and from the Alberta Cancer Board in support of the Embryonic Stem Cell Facility (D.E.R.).
REFERENCES
- 1.Baltz, J. M., P. O. Williams, and R. A. Cone. 1990. Dense fibers protect mammalian sperm against damage. Biol. Reprod. 43:485-491. [DOI] [PubMed] [Google Scholar]
- 2.Brohmann, H., S. Pinnecke, and S. Hoyer-Fender. 1997. Identification and characterization of new cDNAs encoding outer dense fiber proteins of rat sperm. J. Biol. Chem. 272:10327-10332. [DOI] [PubMed] [Google Scholar]
- 3.Burfeind, P., and S. Hoyer-Fender. 1991. Sequence and developmental expression of a mRNA encoding a putative protein of rat sperm outer dense fibers. Dev. Biol. 148:195-204. [DOI] [PubMed] [Google Scholar]
- 4.Calvin, H. I. 1979. Electrophoretic evidence for the identity of the major zinc-binding polypeptides in the rat sperm tail. Biol. Reprod. 21:873-882. [DOI] [PubMed] [Google Scholar]
- 5.Calvin, H. I., and J. M. Bedford. 1971. Formation of disulphide bonds in the nucleus and accessory structures of mammalian spermatozoa during maturation in the epididymis. J. Reprod. Fertil. 13(Suppl. 13):65-75. [PubMed]
- 6.Calvin, H. I., and G. Bleau. 1974. Zinc-thiol complexes in keratin-like structures of rat spermatozoa. Exp. Cell Res. 86:280-284. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134-139. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, P. E., O. Chisholm, R. J. Arceci, E. R. Stanley, and J. W. Pollard. 1996. Absence of colony-stimulating factor-1 in osteopetrotic (csfmop/csfmop) mice results in male fertility defects. Biol. Reprod. 55:310-317. [DOI] [PubMed] [Google Scholar]
- 9.Fantz, D. A., W. R. Hatfield, G. Horvath, M. K. Kistler, and W. S. Kistler. 2001. Mice with a targeted disruption of the H1t gene are fertile and undergo normal changes in structural chromosomal proteins during spermiogenesis. Biol. Reprod. 64:425-431. [DOI] [PubMed] [Google Scholar]
- 10.Fawcett, D. W. 1975. The mammalian spermatozoon. Dev. Biol. 44:394-436. [DOI] [PubMed] [Google Scholar]
- 11.Irons, M. J., and Y. Clermont. 1982. Formation of the outer dense fibers during spermiogenesis in the rat. Anat. Rec. 202:463-471. [DOI] [PubMed] [Google Scholar]
- 12.Irons, M. J., and Y. Clermont. 1982. Kinetics of fibrous sheath formation in the rat spermatid. Am. J. Anat. 165:121-130. [DOI] [PubMed] [Google Scholar]
- 13.Kato, H., and K. Moriwaki. 1972. Factors involved in the production of banded structures in mammalian chromosomes. Chromosoma 38:105-120. [DOI] [PubMed] [Google Scholar]
- 14.Kierszenbaum, A. L. 2001. Spermatid manchette: plugging proteins to zero into the sperm tail. Mol. Reprod. Dev. 59:347-349. [DOI] [PubMed] [Google Scholar]
- 15.Lu, Q., M. Gore, Q. Zhang, T. Camenisch, S. Boast, F. Casagranda, C. Lai, M. K. Skinner, R. Klein, G. K. Matsushima, H. S. Earp, S. P. Goff, and G. Lemke. 1999. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398:723-728. [DOI] [PubMed] [Google Scholar]
- 16.Morales, C. R., R. Oko, and Y. Clermont. 1994. Molecular cloning and developmental expression of an mRNA encoding the 27-kDa outer dense fiber protein of rat spermatozoa. Mol. Reprod. Dev. 37:229-240. [DOI] [PubMed] [Google Scholar]
- 17.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oko, R., and Y. Clermont. 1988. Isolation, structure and protein composition of the perforatorium of rat spermatozoa. Biol. Reprod. 39:673-687. [DOI] [PubMed] [Google Scholar]
- 19.Olson, G. E., and D. W. Sammons. 1980. Structural chemistry of outer dense fibers of rat sperm. Biol. Reprod. 22:319-332. [DOI] [PubMed] [Google Scholar]
- 20.Quinn, P., J. F. Kerin, and G. M. Warnes. 1985. Improved pregnancy rate in human in vitro fertilization with the use of a medium based on the composition of human tubal fluid. Fertil. Steril. 44:493-498. [DOI] [PubMed] [Google Scholar]
- 21.Shao, X., H. A. Tarnasky, J. P. Lee, R. Oko, and F. A. van der Hoorn. 1999. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 211:109-123. [DOI] [PubMed] [Google Scholar]
- 22.Shao, X., H. A. Tarnasky, U. Schalles, R. Oko, and F. A. van der Hoorn. 1997. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. Leucine zippers mediate associations of Odf84 and Odf27. J. Biol. Chem. 272:6105-6113. [DOI] [PubMed] [Google Scholar]
- 23.Shao, X., J. Xue, and F. A. van der Hoorn. 2001. Testicular protein Spag5 has similarity to mitotic spindle protein Deepest and binds outer dense fiber protein Odf1. Mol. Reprod. Dev. 59:410-416. [DOI] [PubMed] [Google Scholar]
- 24.Tsuzuki, T., and D. E. Rancourt. 1998. Embryonic stem cell gene targeting using bacteriophage lambda vectors generated by phage-plasmid recombination. Nucleic Acids Res. 26:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner, K. J., R. M. Sharpe, J. Gaughan, M. R. Millar, P. M. Foster, and P. T. Saunders. 1997. Expression cloning of a rat testicular transcript abundant in germ cells, which contains two leucine zipper motifs. Biol. Reprod. 57:1223-1232. [DOI] [PubMed] [Google Scholar]
- 26.van der Hoorn, F. A., H. A. Tarnasky, and S. K. Nordeen. 1990. A new rat gene RT7 is specifically expressed during spermatogenesis. Dev. Biol. 142:147-154. [DOI] [PubMed] [Google Scholar]
- 27.Vera, J. C., M. Brito, T. Zuvic, and L. O. Burzio. 1984. Polypeptide composition of rat sperm outer dense fibers. A simple procedure to isolate the fibrillar complex. J. Biol. Chem. 259:5970-5977. [PubMed] [Google Scholar]