Abstract
In metazoans, the 3′ end of histone mRNA is not polyadenylated but instead ends with a stem-loop structure that is required for cell cycle-regulated expression. The sequence of the stem-loop in the Drosophila melanogaster histone H2b, H3, and H4 genes is identical to the consensus sequence of other metazoan histone mRNAs, but the sequence of the stem-loop in the D. melanogaster histone H2a and H1 genes is novel. dSLBP binds to these novel stem-loop sequences as well as the canonical stem-loop with similar affinity. Eggs derived from females containing a viable, hypomorphic mutation in dSLBP store greatly reduced amounts of all five histone mRNAs in the egg, indicating that dSLBP is required in the maternal germ line for production of each histone mRNA. Embryos deficient in zygotic dSLBP function accumulate poly(A)+ versions of all five histone mRNAs as a result of usage of polyadenylation signals located 3′ of the stem-loop in each histone gene. Since the 3′ ends of adjacent histone genes are close together, these polyadenylation signals may ensure the termination of transcription in order to prevent read-through into the next gene, which could possibly disrupt transcription or produce antisense histone mRNA that might trigger RNA interference. During early wild-type embryogenesis, ubiquitous zygotic histone gene transcription is activated at the end of the syncytial nuclear cycles during S phase of cycle 14, silenced during the subsequent G2 phase, and then reactivated near the end of that G2 phase in the well-described mitotic domain pattern. There is little or no dSLBP protein provided maternally in wild-type embryos, and zygotic expression of dSLBP is immediately required to process newly made histone pre-mRNA.
Metazoan replication-dependent histone mRNAs are a unique class of RNA polymerase II transcripts that do not end in a polyadenylated tail. Instead, histone mRNAs end in a conserved stem-loop sequence (reviewed in references 9 and 33). In vertebrates and echinoderms, the 3′ end of mature histone mRNA is formed by an endonucleolytic cleavage of the nascent pre-mRNA in a reaction that requires both the stem-loop and a downstream element that binds to the U7 snRNP (21, 38, 47). The stem-loop is bound by a protein, the stem-loop binding protein (SLBP), that is required for histone pre-mRNA processing and is likely involved in all other steps in histone mRNA metabolism (24, 52). SLBP and U7 snRNP together recruit a protein complex capable of carrying out the pre-mRNA processing reaction (9).
In cycling mammalian cells, histone mRNAs accumulate only during S phase, since large amounts of histone protein synthesis are required only during DNA replication. Much of this cell cycle regulation is mediated by the stem loop at the 3′ end of histone mRNA (25, 31), which in addition to being essential for pre-mRNA processing controls histone mRNA stability during the cell cycle (39). SLBP is a cell-cycle regulated protein, and is also present in high levels only during S-phase (53). The cell-cycle regulation of SLBP is largely responsible for the cell cycle regulated accumulation of histone mRNA. Whether this same mechanism is used to couple histone mRNA production with the cell cycle in other organisms is not known.
An exception to the tight coupling between histone mRNA accumulation and S phase occurs during oogenesis and early embryogenesis in many animal species. In Xenopus and Drosophila, the same histone mRNAs that are cell cycle regulated in somatic cells are stored in the developing oocyte and are present continuously during the very rapid early embryonic cycles (55, 56). These cell cycles lack gap phases, and many rounds of chromosome duplication and mitosis occur in the absence of zygotic transcription. The histone proteins used to package newly synthesized DNA during S phase at this time must either be stored in the oocyte or translated from maternal mRNA. The timing and mechanism of the switch from continuous to cell cycle-regulated histone biosynthesis has not been well described.
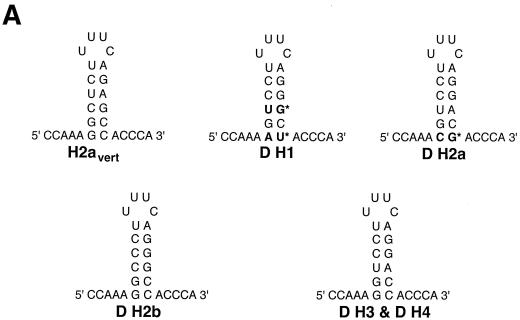
To address the question of how SLBP contributes to the developmental control of histone mRNA synthesis, we are characterizing SLBP function genetically in Drosophila. Drosophila has a single set of replication-dependent histone genes encoded in a 5-kb repeat present in approximately 100 tandem copies near the centromere on the left arm of chromosome 2. Each repeat unit contains one copy of each of the five histone genes (see Fig. 5A). Like the histone genes in other metazoans, these genes encode nonpolyadenylated mRNAs. The stem-loop sequences at the 3′ end of histone mRNA are well conserved in a wide variety of organisms, particularly the sequence of the stem, which is GGYYYU followed by a four-base loop, UYUN, and the complementary sequence ARRRCC (Fig. 1A). However, inspection of the sequences of the Drosophila histone genes revealed that while three of the histone mRNAs (H3, H4, and H2b) ended in a sequence identical to the consensus sequence, the histone H1 and H2a genes ended in a divergent sequence that could still form a stem-loop. The H2a gene has a CG as the first base pair of the stem, while the histone H1 gene has an AU as the first base pair of the stem and a UG as the third base pair of the stem (Fig. 1A). The same sequences are found in the genes for these same histone proteins in another Drosophila species, Drosophila hydei (30). None of the Drosophila histone genes have the most common stem-loop sequence present at the 3′ end of vertebrate histone mRNAs.
FIG. 5.
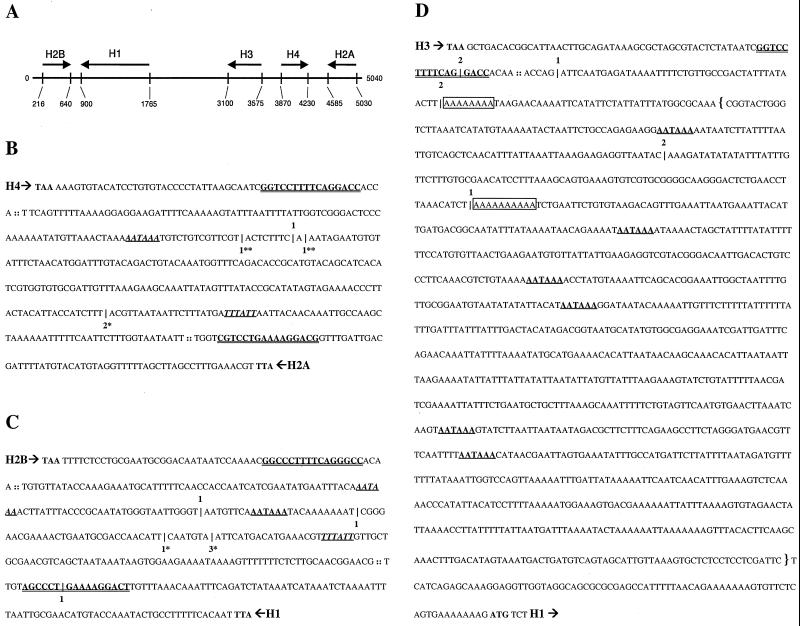
RT-PCR mapping of the location of cryptic polyadenylation sites in misprocessed histone messages from dSLBP mutant embryos. (A) Diagram of the 5-kb repeat unit of the Drosophila histone gene cluster. Arrows indicate the direction and length of each histone transcript, which do not contain introns. (B to D) Poly(A)+ mutant histone mRNA was amplified by RT-PCR using an oligo(dT) primer. Cloned products were randomly selected and sequenced to identify the location of polyadenylation. These sites of polyadenylation are indicated by a hatchmark (|) along with the number of times that the particular location was observed. RT-PCR products specific to the histone proceeding (5′ → 3′) top-to-bottom are shown with the number of products above the hatchmark. RT-PCR products specific to the histone proceeding (5′ → 3′) bottom-to-top are shown below the hatchmark. For each gene, the translation termination codon is shown in bold, the processing site is indicated (::), and the stem-loop sequence is depicted in bold and double underlined. Canonical polyadenylation signal sequences downstream of the processing sites are shown in bold with a single underline. Those signal sequences properly spaced with respect to polyadenylation sites determined by RT-PCR products are shown in italics. RT-PCR products corresponding in size to S1 products in Fig. 4 are shown with matching asterisks (∗, ∗∗). (B) H4/H2a intergenic sequence; (C) H2b/H1 intergenic sequence; (D) The downstream sequence of H3. RT-PCR products mapping to putative polyadenylation signal sequences are observed for histones H4, H2a, H2b, and H1. 3′-End mapping of misprocessed histone H3 mRNA was hampered by the presence of oligo(A) (boxed) sequences in the template mRNA. The DNA sequence included in the H3-ds probe used for detection of misprocessed histone H3 mRNA by embryonic in situ hybridization in Fig. 7 and 9 is indicated with brackets.
FIG. 1.
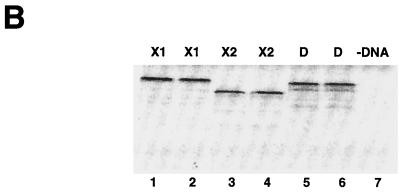
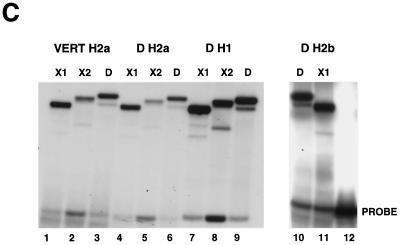
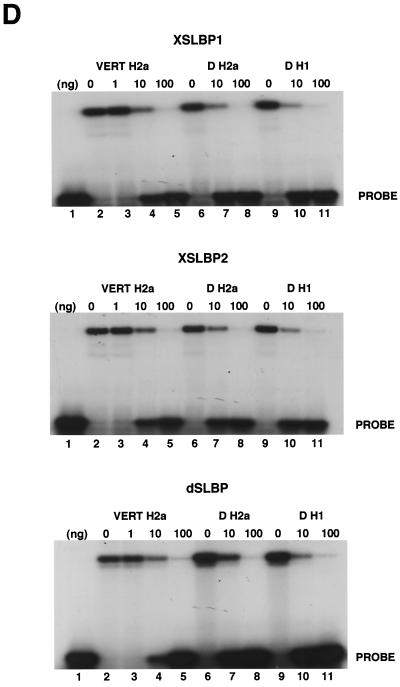
dSLBP binds to all five histone mRNAs. (A) The sequence of the 3′ end of the most common stem-loop sequence in vertebrate histone mRNAs (H2avert) and the sequence of the 3′ end of the five Drosophila histone mRNAs is shown. ∗, nucleotide pairs that are different than the canonical sequence and that have not yet been observed in any other stem-loop sequence. (B) DNAs encoding dSLBP (lanes 1 and 2), xSLBP1 (lanes 3 and 4), and xSLBP2 (lanes 5 and 6) were incubated in a reticulocyte lysate-coupled transcription-translation system in duplicate in the presence of [35S]methionine. The products were analyzed on an SDS-10% polyacrylamide gel and detected by autoradiography. Lane 7, lysate incubated without added DNA. Parallel reaction mixtures incubated in the absence of [35S]methionine were used for the mobility shift assays (C and D). (C) The indicated stem-loop sequences (lanes 1 to 3, H2avert; lanes 4 to 6, Drosophila H2a; lanes 7 to 9, Drosophila H1; lanes 10 and 11, Drosophila H2b) labeled with 32PO4 were tested for their ability to bind to Xenopus SLBP1 (X1 [lanes 1, 4, 7, and 11]), Xenopus SLBP2 (X2 [lanes 2, 5, and 8]), or Drosophila SLBP (D [lanes 3, 6, and 9]). Lane 12, probe incubated in buffer. (D) xSLBP1, xSLBP2, and dSLBP (indicated above each panel) were incubated with the H2avert probe (lanes 2 to 5), dH2a probe (lanes 6 to 8), or dH1 probe (lanes 9 to 11), and the complexes were detected by mobility shift assay. Increasing amounts of unlabeled H2avert RNA (indicated above each lane) were mixed with the probe prior to addition of the SLBPs. Lane 1, probe incubated in buffer. The complexes were analyzed by electrophoresis on an 8% polyacrylamide gel.
We previously isolated mutations of the single Drosophila dSLBP gene and used these to show that dSLBP function is required for both maternal and zygotic production of mature histone H3 and H4 mRNAs (48). However, whether the other three histone mRNAs required dSLBP for processing was not determined. Here we used a variety of molecular and cytological assays to conclusively demonstrate that all five Drosophila histone mRNAs bind dSLBP and that dSLBP function is required for the production of each of the five histone mRNAs in vivo. Moreover, our previous data suggested that, in the absence of dSLBP, the H3 and H4 mRNAs are converted to poly(A)+ forms, possibly by using cryptic polyadenylation signals located downstream of the normal processing site (48). Now we show that all five Drosophila histone genes contain polyadenylation sites that function in the absence of dSLBP. These polyadenylation signals may provide a failsafe mechanism to ensure that transcription of each of the five highly expressed and closely spaced histone genes do not interfere with one another. We also use the dSLBP mutant phenotype to characterize the developmental transition from continuous to regulated histone expression during embryogenesis. In dSLBP mutant embryos, aberrant, polyadenylated forms of the histone mRNAs accumulate as soon as zygotic transcription of the histone genes begins at the embryonic blastoderm stage, indicating an early requirement for zygotic dSLBP function. At this stage of development, the rapid cell cycles lack a G1 phase. Nevertheless, we show that histone gene transcription occurs in late G2 phase in a temporally programmed manner coincident with the mitotic domain pattern. Thus, even in rapid embryonic cell cycles that lack a G1 phase histone mRNA accumulation is cell cycle dependent, and efficient histone mRNA accumulation requires dSLBP-dependent 3′ end processing.
MATERIALS AND METHODS
Gel mobility shift assay.
The probes used in the mobility shift assay were synthesized from DNA templates that contain a stem-loop sequence cloned behind a T7 RNA polymerase promoter. DNA fragments (∼1 kb) were produced by PCR using a primer internal in the pGEM3 plasmid and a primer that ended at the stem-loop. These fragments were used to synthesize RNA labeled at the 5′ end using [γ-32P]ATP (7). Unlabeled RNAs for use as competitors were synthesized from the same templates. Drosophila SLBP and Xenopus SLBP1 and SLBP2 cDNAs were cloned into pXRFM (50), transcribed into synthetic mRNAs, and translated in a rabbit reticulocyte lysate (29) for use in mobility shifts assays (7, 52). To ensure that these proteins were present in similar concentrations, [35S]methionine was included in a parallel translation assay that was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Northern analysis.
All dSLBP alleles employed have been described previously (48). Df(3R)3450 removes the entire dSLBP locus; dSLBP15 is null or nearly null and produces no detectable dSLBP protein; and dSLBP10 is hypomorphic and produces reduced amounts of dSLBP protein. Flies containing this mutation are viable but female-sterile. Zero- to 2-h-old maternally mutant embryos were collected from dSLBP10/Df(3R)3450 females and stored at −80°C. Live, zygotically mutant embryos were obtained using dSLBP mutant stocks containing a TM3 Ser P[act-GFP] balancer chromosome (43). Three-hour-old embryo collections were aged 13 h at room temperature, dechorionated, and then hand sorted into green fluorescent protein (GFP−) and GFP+ (wild-type) populations by visualization using a Zeiss Stemi SV11 Apo stereomicroscope equipped with epifluorescence. Our ability to correctly select dSLBP mutant embryos was tested by separating viable dSLBP10 homozygous (GFP-negative) embryos and allowing them to mature to adult flies, which can then be scored phenotypically as mutant. Based on this test, we could hand sort hundreds of mutant embryos with 100% efficiency. Batches of hand-sorted embryos were frozen at −80°C and then combined for RNA preparation. Total RNA was prepared from frozen embryos using Trizol reagent (Gibco). Poly(A)+ RNA was recovered from total RNA using the PolyATract isolation system (Promega). Then, 0.5 μg of total RNA/lane or 0.4 μg of poly(A)+ RNA/lane was transferred to a nylon membrane and probed with 32P-labeled, random primed (Roche) PCR products corresponding to most of the coding regions of H2a, H2b, H3, and H4 (GenBank accession no. X14215).
Embryo in situ hybridization.
Embryos collected from dSLBP mutant stocks were aged at room temperature until the desired stage, dechorionated, and fixed with 37% formaldehyde for 5 min. Histone mRNA was detected by in situ hybridization using digoxigenin-labeled riboprobes complementary to the coding region of H2a, H2b, and H3, or the 3′ flanking region of H3 (see Fig. 5D) as previously described (48).
Western analysis.
Appropriately aged embryos were dechorionated and homogenized into SDS-containing sample buffer at a concentration of 1 embryo/μl. Homozygous dSLBP mutant embryos were hand sorted as described above. The protein equivalent of 20 embryos was loaded per gel lane. For analysis of dSLBP phosphorylation, 50 embryos were homogenized in lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% NP-40, 50 mM NaF, and protease inhibitor cocktail from Sigma) and incubated either with or without 20 U of CIP (New England Biolabs) at 37°C for 30 min. dSLBP protein was detected by Western blotting using affinity-purified rabbit polyclonal antibodies (48).
RT-PCR.
The polyadenylation sites of mis-processed histone mRNAs were determined using the 3′ RLM-RACE kit (Ambion). A single reverse transcriptase (RT) reaction was followed by two nested PCRs. The RT reaction mixture contained the oligo(dT) 3′ RACE adapter primer (5′-GCGAGCACAGAATTAATACGACTCACTATAGGT12) and 100 ng of poly(A)+ RNA isolated from 13- to 16-h-old embryo collections obtained from dSLBP15/TM3 flies. The first PCR contained 10% of the RT reaction mixture either with or without RT (as a negative control), the 3′ RACE outer primer (5′-GCGAGCACAGAATTAATACGACT), and a gene-specific outer primer (Table 1). The second, nested PCR contained 4% the volume of the first PCR, the 3′ RACE inner primer (5′-CGCGGATCCGAATTAATACGACTCACTATAGG), and a gene-specific inner primer (Table 1). Negative-control reaction mixtures lacked RT and generated no PCR product. RT-PCR products were cloned at random using the TOPO TA cloning kit (Invitrogen) and were sequenced.
S1 nuclease protection assay.
An H2a full-length PCR product was generated from the Drosophila histone gene cluster clone encompassing the coding region and 3′ untranslated region (UTR) of H2a using a forward primer (5′-GTGGCAAAGTGAAGGGAAAGGCAA) and a reverse primer (5′-TAATTTTATTGGTCGGGACTCCCAA). This product was cloned into a TA vector (Invitrogen), digested with BspEI, and 3′ end labeled with Klenow enzyme (New England Biolabs) in the presence of [α-32P]dCTP. An H1-specific probe was made in a similar way. Forward (5′-CCCACCAGCGACAGTTGAGAAGAA) and reverse primers (5′-CAACCACCAATCGAATATGAATT) were used to generate a PCR product encompassing the coding region and 3′ UTR of H1. The cloned PCR product was digested with HindIII, and 3′ end labeled with Klenow in the presence of [α-32P]dCTP. A positive-control RNA corresponding to the sense strand of H2a was synthesized using T7 enzyme, and the full-length H2a clone was digested with StuI. A sense strand H1 RNA positive control was generated using SP6 and the full-length H1 clone digested with StyI. Approximately 20 ng of 32P-labeled probe was hybridized with 1.5 μg of total embryonic RNA or 2 μg of control yeast tRNA and digested with S1 nuclease (22). The protected fragments were resolved on a 6% polyacrylamide-7 M urea gel and detected by autoradiography.
RESULTS
dSLBP binds each of the five histone stem-loops with similar affinity.
Only a single SLBP was identified in the Drosophila genome by both using a screen for proteins that bind the histone stem-loop and database searching via sequence similarity to the vertebrate and Caenorhabditis elegans SLBP (48). Since there were variant stem-loops in the H1 and H2a histone mRNAs, we tested whether the single dSLBP was capable of binding the 3′ end of all five Drosophila histone mRNAs with similar affinity (Fig. 1). We also determined whether vertebrate SLBPs recognized these unusual Drosophila stem-loops, since the variant H1 and H2a stem-loop sequences could be structurally equivalent to the canonical sequence (Fig. 1A). Two different SLBPs in Xenopus (xSLBP1 and xSLBP2) have been previously identified. xSLBP1 is required for histone pre-mRNA processing and is the functional homologue of the single mammalian SLBP, and xSLBP2 is an oocyte-specific SLBP that does not function in processing (50). The RNA binding domain (RBD) of xSLBP2 differs in 14 of the 73 positions in the RBD of xSLBP1, similar to the differences between xSLBP1 and dSLBP. We tested the ability of dSLBP, xSLBP1, and xSLBP2 to bind the various Drosophila stem-loops. dSLBP, xSLBP1, and xSLBP2 were synthesized in vitro in a rabbit reticulocyte lysate (Fig. 1B). Mobility shift assays were used to determine whether each of these SLBPs bound the major vertebrate stem-loop sequence (H2avert) as well as the Drosophila H2a, H1, and H2b stem-loop sequences. An RNA-protein complex between each of the four stem-loops and each SLBP protein were readily detected (Fig. 1C). To determine whether the proteins bound the different stem-loops with similar affinity, increasing amounts of wild-type competitor stem-loop RNA were added to each binding reaction mixture. The binding of dSLBP, xSLBP1, and xSLBP2 were all competed to similar extents by the competitor RNA, indicating that each SLBP bound the three different probes with similar affinity (Fig. 1D). Therefore, despite the divergent sequences in some Drosophila stem-loops, the single Drosophila dSLBP and the two types of Xenopus SLBP bind the 3′ end of all the Drosophila histone mRNAs with similar affinity.
Production of all five replication-associated histone mRNAs requires dSLBP.
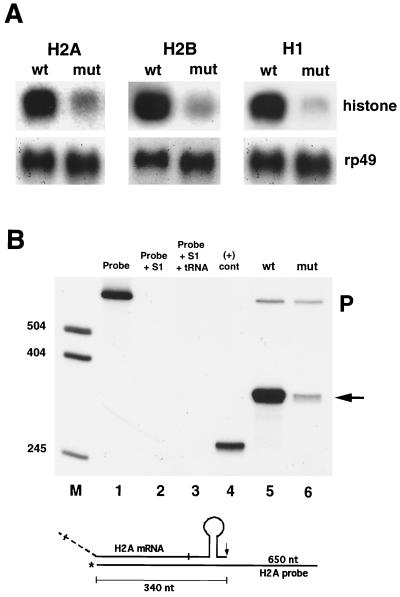
The binding data predict that dSLBP should be required for the accumulation of all five replication-associated histone pre-mRNAs in vivo. Moreover, the similarity in binding affinity between dSLBP and the various stem-loop sequences suggests that each mRNA might be affected equally after reduction of dSLBP function. To test these hypotheses, we analyzed histone mRNA isolated from dSLBP mutant embryos by Northern and S1 nuclease protection assays. dSLBP10 is a viable, hypomorphic allele that causes maternal effect lethality. dSLBP10/Df(3R)3450 hemizygous females lay normal numbers of eggs that do not develop because of chromosome segregation defects that occur during the pre-blastoderm syncytial nuclear cycles (48). In our previous study, we demonstrated that histone H3 and H4 maternal mRNA were depleted in eggs laid by dSLBP10 mutant females (48). Here, we found that histone H2a, H2b, and H1 mRNA is also severely reduced relative to the wild type in total RNA samples prepared from 0- to 2-h-old eggs laid by dSLBP10/Df(3R)3450 mutant females (Fig. 2A). An S1 nuclease protection assay was developed to map the 3′ end of H2a mRNA in order to measure both the relative abundance and processing of this message (see Materials and Methods). In wild-type 0- to 2-h-old embryos, a single 340-nucleotide (nt) protection product was observed (Fig. 2B, lane 5) which correlates precisely with the expected pre-mRNA processing site in the H2a gene (see Fig. 5B). Consistent with the Northern analysis, the level of this S1 protection product was significantly reduced in 0- to 2-h-old embryos collected from dSLBP10 hemizygous females (Fig. 2B, lane 6), and there was no evidence of H2a mRNAs with altered 3′ ends. Thus, dSLBP function is required for maternal production of all five replication-associated histone mRNAs during oogenesis. Moreover, these data suggest that the mitotic defects seen in the nuclear division cycles in eggs laid by dSLBP10 mutant females (48) are likely a result of inadequate biosynthesis of all five histone proteins.
FIG. 2.
The dSLBP10 maternal effect lethal allele causes reduced deposition of maternal histone mRNA. (A) Total RNA extracted from 0- to 2-h-old embryos laid by wild-type or dSLBP10/Df(3R)3450 mutant mothers were subjected to Northern analysis using radiolabeled probes for H2a, H2b, and H1. The blots were stripped and reprobed with rp49 as a loading control. (B) Detection of H2a mRNA by S1 nuclease protection. A 3′ end-labeled 650-nt H2a probe (lane 1) was incubated with S1-containing buffer (lane 2), nonspecific yeast tRNA (lane 3), a synthetic partial histone H2a mRNA that should protect a 265-nt fragment (lane 4), or total RNA from 0- to 2-h-old wild-type (lane 5) or dSLBP10/Df(3R)3450 embryos (lane 6). The S1 nuclease-resistant fragments were resolved by gel electrophoresis. In both samples, a fragment of 340 nt is observed which corresponds to histone H2a mRNA ending at the normal processing site (arrow). The upper 650-nt fragment is undigested probe (P). A diagram of the H2a S1 assay is shown on the bottom.
A different embryonic phenotype is observed when maternal dSLBP function is normal and zygotic dSLBP function is compromised. We previously reported that in this situation, longer forms of both histone H3 and H4 mRNA are observed in Northern blot analysis of a mixed population of embryos derived from dSLBP/+ heterozygous parents (48). These longer, aberrant mRNAs were recovered in the poly(A)+ fraction, suggesting that they are polyadenylated. These data were interpreted as an indication that histone H3 and H4 pre-mRNAs are not processed properly in dSLBP mutants but instead are expressed primarily as poly(A)+ mRNAs via usage of downstream cryptic polyadenylation signals.
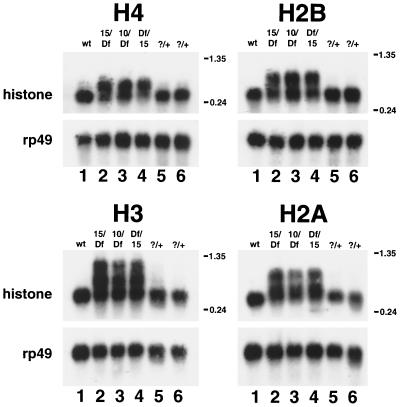
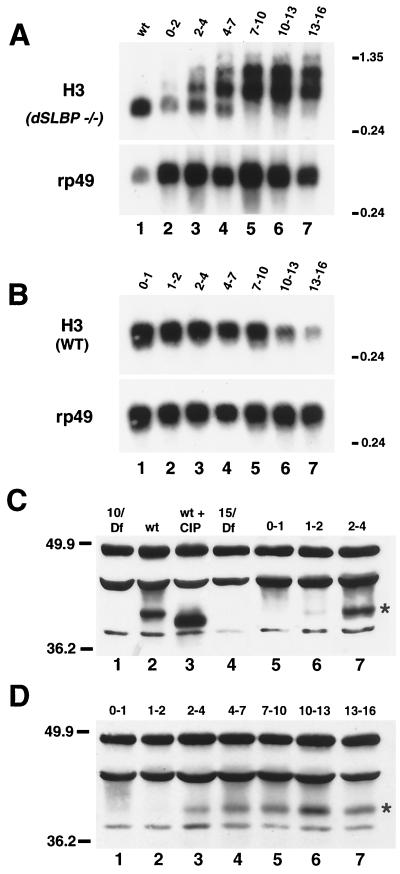
Since a homogeneous population of dSLBP mutant embryos was not analyzed in these initial experiments, the relative proportion of wild-type to mutant mRNA after loss of dSLBP function could not be determined. Moreover, we did not test whether the other histone mRNAs (H2a, H2b, and H1) were misprocessed and converted to polyadenylated mRNAs. To address these issues, dSLBP mutant embryos were recovered by hand sorting a population of dechorionated eggs collected from stocks carrying a balancer chromosome that expresses GFP. In this situation, 25% of the embryos are GFP− and represent dSLBP mutants. Total RNA was extracted from homogeneous mutant and age-matched wild-type control embryos and were analyzed by Northern blotting using a coding region probe for histones H2a, H2b, H3, and H4 (Fig. 3). For each gene, a single, processed mRNA was detected in RNA from wild-type embryos (Fig. 3, lanes 1) as well as GFP+ siblings of the mutant embryos (Fig. 3, lanes 5 and 6). (A small amount of misprocessed H3 mRNA was detected in the GFP+ sample [lane 5], which contains two of three dSLBP/+ heterozygous embryos, suggesting a slight haploinsufficiency for H3 processing; see Fig. 7B and D below.) Longer, misprocessed mRNAs were readily observed by Northern blotting for all four core histone mRNAs in each mutant RNA sample (Fig. 3, lanes 2 to 4). For each histone, there was a greater proportion of misprocessed mRNA in embryos hemizygous for the dSLBP15 allele (Fig. 3, lanes 2 and 4) than in embryos hemizygous for the dSLBP10 allele (Fig. 3, lane 3). This is most readily apparent for the histone H3 and H2a mRNAs and is consistent with our previous observation that dSLBP10 is hypomorphic and expresses a reduced amount of dSLBP protein relative to the wild type, while dSLBP15 produces no detectable protein and is probably null (48). Similar phenotypes were observed in reciprocal crosses involving the dSLBP15 and Df(3R)3450 alleles (Fig. 3, compare lanes 2 and 4). This result indicates either that maternal dSLBP function does not persist until these late embryonic stages or that dSLBP15 is a null allele comparable in strength to the deficiency.
FIG. 3.
Altered histone mRNA processing in dSLBP mutant embryos. Total RNA isolated from 13- to 16-h-old wild-type or hand-selected GFP-negative mutant embryos was subjected to Northern analysis using a radiolabeled H4, H3, H2b, or H2a probe. Lane 1, yw67 wild-type; lane 2, dSLBP15/TM3 [actin-GFP] males mated to Df(3R)3450/TM3 [actin-GFP] females; lane 3, dSLBP10/TM3 [actin-GFP] males mated to Df(3R)3450/TM3 [actin-GFP] females; lane 4, Df(3R)3450/TM3 [actin-GFP] males mated to dSLBP15/TM3 [actin-GFP] females (the reciprocal cross to lane 2); lanes 5 and 6, GFP-positive embryos (two of three of which are dSLBP/+, and one of three are +/+) selected from the same collections used for lane 2 and lane 4, respectively. Each blot was stripped and reprobed with a radiolabeled rp49 probe as a loading control.
FIG. 7.
In situ hybridization with a downstream probe complementary to the 3′ untranscribed region of histone H3 mRNA to detect misprocessed H3 mRNA in vivo. The sequence used to generate the antisense H3-ds probe for use in detecting only misprocessed forms of histone H3 is shown by brackets in Fig. 5D. Each panel shows a whole embryo oriented as in Fig. 6 that was hybridized with the H3-ds probe (A to D, F, and H) or a coding region H3 probe. (E and G). The embryos stained with the H3-ds probe are siblings collected from heterozygous dSLBP15/+ parents and provide examples of the three different phenotypic classes observed. (A) Stage 12 +/+. Most cells are beginning to enter cell cycle 17 at this time. (B) Stage 12 dSLBP15/+; (C) stage 13 +/+; (D) stage 13 dSLBP15/+; (E) stage 12 yw67 wild type; (F) stage 12 dSLBP15/dSLBP15; (G) stage 14 yw67 wild type; (H) stage 14 dSLBP15/dSLBP15. Persistence of misprocessed histone H3 message is observed in nonreplicating cells of the pharanx, anterior midgut, hindgut, and anal pads (indicated from left to right, respectively, by arrows in H and G).
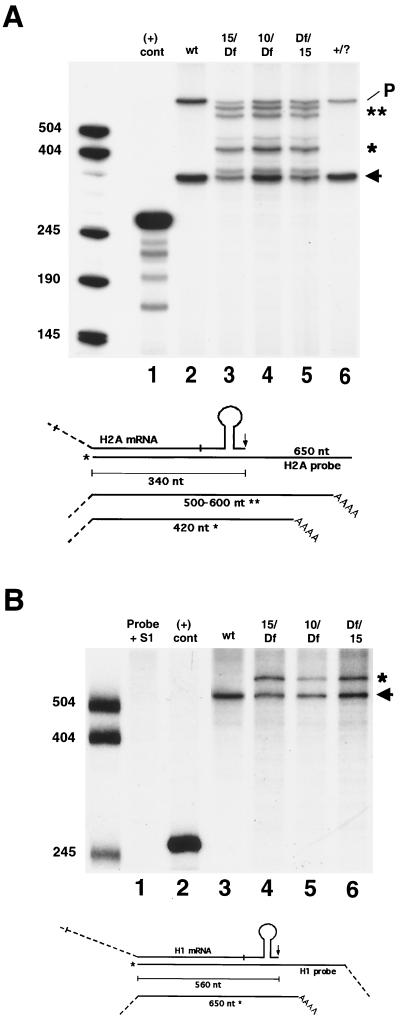
In addition to the misprocessed forms of histone mRNAs, dSLBP mutant embryos contain an RNA species that comigrates with the processed form of each histone mRNA (Fig. 3, lanes 2 to 4) on Northern blots. This was somewhat surprising and suggested that some pre-mRNA processing may still occur in the mutant embryos. To better determine the different forms of histone H2a mRNA present in dSLBP mutants, we used the S1 nuclease protection assay with RNA extracted from GFP− selected embryos. With wild-type embryos, only a 340-nt product representing the processed form of histone H2a mRNA was observed (Fig. 4A, lane 2). Mutation of dSLBP results in the appearance of several longer products that represent mutant histone H2a mRNAs (Fig. 4A, lanes 3 to 5). One major product of 420 nt and several products in the range of 500 to 600 nt could be detected, corresponding to polyadenylated transcripts detected by RT-PCR (see below). There was also clearly a protected product that comigrated with the processed form of H2a mRNA in each of the dSLBP mutant RNA samples (Fig. 4A, lanes 3 to 5). Consistent with the Northern blot results, the laser densitometry of the autoradiographs indicates that the ratio of mutant to processed forms of histone H2a mRNA was greater in the dSLBP15 samples (1.26 and 0.96; Fig. 4A, lanes 3 and 5, respectively) than in the dSLBP10 sample (0.65; Fig. 4A, lane 4). In the dSLBP10 embryos, there was an approximately equal amount of processed and mutant mRNA (Fig. 4A, lane 4). Only processed H2a mRNA was detected in samples prepared from GFP+ sibling embryos (Fig. 4A, lane 6).
FIG. 4.
S1 nuclease protection analysis of histone mRNA in dSLBP mutant embryos. (A) Total RNA isolated from 13- to 16-h-old wild-type or hand-selected GFP-negative mutant embryos was subjected to S1 nuclease protection analysis using a 32P end-labeled histone H2aprobe as described for Fig. 2B. Lane 1, in vitro-transcribed H2a RNA as a positive control, designed to yield a 265-nt protection product; lane 2, yw67 wild type. The arrow indicates the 340-nt H2a-protected fragment corresponding to the normally processed form, and the upper band is undigested probe (P) (Fig. 2). Lane 3, dSLBP15/TM3 [actin-GFP] males mated to Df(3R)3450/TM3 [actin-GFP] females; lane 4, dSLBP10/TM3 [actin-GFP] males mated to Df(3R)3450/TM3 [actin-GFP] females; lane 5, Df(3R)3450/TM3 [actin-GFP] males mated to dSLBP15/TM3 [actin-GFP] females (the reciprocal cross to lane 3); lane 6, GFP-positive embryos (two of three of which are dSLBP/+ and one of three of which are +/+) selected from the same collections used for lane 2. Multiple protected fragments corresponding to misprocessed forms of H2a mRNA are observed. The major misprocessed forms are indicated (∗ and ∗∗). A diagram of the S1 nuclease assay is shown below the gel. (B) S1 analysis using a 32P end-labeled probe designed to detect histone H1 mRNA. Lane 1, H1 probe plus S1 nuclease; lane 2, H1 probe plus a positive control synthetic H1 RNA that generates a predicted fragment of 260 nt; lane 3, yw67 wild-type RNA; lane 4, dSLBP15/TM3 [actin-GFP] males mated to Df(3R)3450/TM3 [actin-GFP] females; lane 5, dSLBP 10/TM3 [actin-GFP] males mated to Df(3R)3450/TM3 [actin-GFP] females; lane 6, Df(3R)3450/TM3 [actin-GFP] males mated to dSLBP15/TM3 [actin-GFP] females. A diagram of the S1 nuclease assay is shown below the gel. Only the normally processed form of H1 mRNA is observed in the wild-type sample at the expected size of 560 nt (arrow). Mutant extracts contain an additional fragment corresponding to misprocessed H1 mRNA at 650 nt (asterisk).
Unlike the four core histones, it was unclear from Northern blot experiments whether mutation of dSLBP caused a failure of histone H1 mRNA processing, since only a slight increase in the size of histone H1 mRNA was seen (not shown). An S1 nuclease protection assay was developed for histone H1 mRNA to resolve this issue. In wild-type RNA preparations, only a single protected fragment of 565 nt is detected using a probe starting near the 5′ end of the coding region (Fig. 4B, lane 3). This is the expected size for a histone H1 mRNA molecule cleaved at the typical processing site immediately following the stem-loop (Fig. 5C). Mutation of dSLBP results in the appearance of a single, longer protected fragment of approximately 650 nt in length (Fig. 4B, lanes 4 to 6). The predicted similarity in size of this unique mutant H1 mRNA to wild-type H1 mRNA [∼1,000 nt versus 900 nt without the poly(A) tail] explains our failure to readily resolve the two species via Northern blot analysis. As observed for histone H2a mRNA, processed histone H1 mRNA is also present in dSLBP mutants (Fig. 4B, lanes 4 to 6), and the ratio of mutant to processed H1 mRNA in the dSLBP15 mutant (1.08) is approximately twice that of the dSLBP10 mutant (0.55), as determined by laser densitometry. The twofold difference in the proportion of correctly processed mRNA is similar to that observed with the H2a mRNA by the S1 nucelase assay and the other mRNAs by Northern blotting (Fig. 3). Taken together, the Northern blot and S1 nuclease protection data indicate that dSLBP is involved in processing each of the five replication associated histone pre-mRNAs.
In dSLBP mutant embryos, histone mRNAs are polyadenylated via downstream polyadenylation signals.
In our previous study, we showed that improperly processed histone H3 and H4 mRNA in dSLBP mutants binds to oligo(dT) cellulose, suggesting that these RNAs are polyadenylated like other polymerase II mRNAs. This could occur if transcription continues past the normal processing site until a polyadenylation signal is encountered and the polyadenylation machinery is recruited. To generate additional evidence for this hypothesis, the 3′ end of the mutant histone mRNAs was determined by RLM-RACE RT-PCR. Poly(A)+ RNA made from whole-embryo populations obtained from dSLBP15 heterozygous parents was primed for RT by using an oligo(dT) primer. RT products were amplified by two nested PCRs, cloned, and sequenced at random to determine sites of polyadenylation. A summary of the data is presented in Fig. 5, which shows both the location and the number of times a particular polyadenylation site was identified for each histone gene (Fig. 5B to D).
The Drosophila histone genes are arranged in a 100-fold-repeated cluster containing one copy of each of the five histone genes (Fig. 5A). There is a short intergenic region (<400 nt) shared by the 3′ end of the H4 and H2a genes, and another is shared by the 3′ ends of the H2b and H1 genes. The 3′ end of histone H3 is ∼1.3 kb upstream of the 5′ end of the H1 gene. At least one canonical (AAUAAA) poly(A) signal sequence is present in the region 3′ of all five histone genes. For every gene except H3, at least one polyadenylation site was mapped 20 to 30 nt downstream of a consensus poly(A) signal sequence (Fig. 5B and C). Priming from endogenous oligo(A) stretches in the 3′ flanking region of the H3 gene likely prevented the recovery of RT-PCR products representing full-length, polyadenylated H3 mRNAs (Fig. 5D). Nevertheless, these data are consistent with the presence of long misprocessed H3 mRNA detected by Northern blot analysis of dSLBP mutant RNA (Fig. 3). The H2a RT-PCR products revealed that several poly(A) addition sites occurred substantially downstream of the first canonical poly(A) signal sequence located 53 nt after the normal processing site. Multiple misprocessed H2a mRNAs were also detected by the S1 nuclease protection assay. These data suggest that utilization of the first cryptic polyadenylation site is not 100% efficient. Curiously, clones with the poly(A) sequence starting in the stem-loop of both histone H1 and H3 mRNAs were also recovered at low frequency from the RT-PCR. The origin of these RT-PCR products is unclear, but they have also been observed by others (2).
The locations of polyadenylation sites mapped by RT-PCR correlate well with the S1 nuclease protection results for histones H2a and H1 mRNAs. The most prevalent site detected by S1 nuclease mapping of histone H2a mRNA corresponds to a major site recovered from the RT-PCRs, about 80 nt from the normal 3′ end (Fig. 4A). This corresponds precisely to the poly(A) addition site identified in the shortest H2a RT-PCR product (Fig. 5B). In addition, the other major S1 protected fragments of 500 to 600 nt in length (150 to 250 nt greater than the processed form; Fig. 4A) may correspond to the two longest RT-PCR products recovered (Fig. 5B). For histone H1, the S1 assay produced a protected fragment of about 650 nt (Fig. 4B), which is approximately 90 nt greater in size than the processed form of histone H1 mRNA. Again, this fragment corresponds exactly in size to the only detected RT-PCR product generated from misprocessed histone H1 mRNA (Fig. 5C). These data confirm that in the absence of embryonic dSLBP function, all the replication-associated histone mRNAs become polyadenylated at downstream polyadenylation sites. These cryptic polyadenylation sites may serve to terminate transcription before it proceeds into the next gene and also may prevent generation of anti-sense RNA from the adjacent histone gene.
Polyadenylated histone mRNAs are not cell cycle regulated.
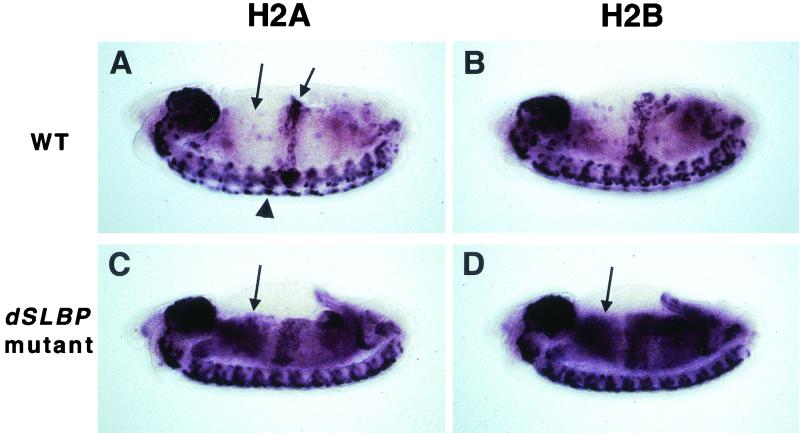
During development, there are no cycles with a G1 phase until the 17th embryonic cell cycle, at about 8 h of development at 25°C (14, 15). At this time, histone mRNAs are expressed in a stereotypic spatiotemporal pattern that coincides with cells in S phase of the cell cycle (26, 48). This is manifested as a diagnostic pattern of expression in wild-type germ-band-retracted embryos, where histone mRNA accumulates in proliferating cells of the central nervous system (CNS) and in endocycling cells in the central midgut (shown for histones H2a and H2b in Fig. 6A and B, respectively). We previously reported that at this stage, dSLBP mutant embryos accumulate histone H3 and H4 mRNA in the cytoplasm of nonreplicating cells, causing an aberrant in situ hybridization pattern (48). A similar result was observed with coding region probes for histones H2a and H2b (Fig. 6C and D, respectively). The pattern is consistent with persistence of histone mRNA in cells that have already exited S phase (e.g. the anterior midgut; Fig. 6A, left arrow), suggesting an increase in half life of the polyadenylated mRNAs relative to wild-type mRNAs that end in a stem-loop.
FIG. 6.
Altered histone mRNA expression in dSLBP mutant embryos. Each panel shows a whole mount in situ hybridization of a stage 14 embryo (anterior at left, dorsal at top) using a coding region histone probe. (A) Wild-type H2a expression is restricted to replicating cells, including those in the proliferating CNS (arrowhead) and endoreduplicating midgut (right arrow). The cells of the anterior midgut (left arrow) have exited S phase and do not express histone H2a mRNA. (B) Wild-type histone H2b expression is identical to H2a. (C) In dSLBP15/dSLBP15 mutant embryos, histone H2a mRNA persists in cells that have finished replicating (e.g. the anterior midgut; arrow), generating a diagnostic aberrant pattern. (D) H2b mRNA also accumulates in nonreplicating dSLBP15/dSLBP15 mutants cells (arrow) in a manner similar to H2a mRNA.
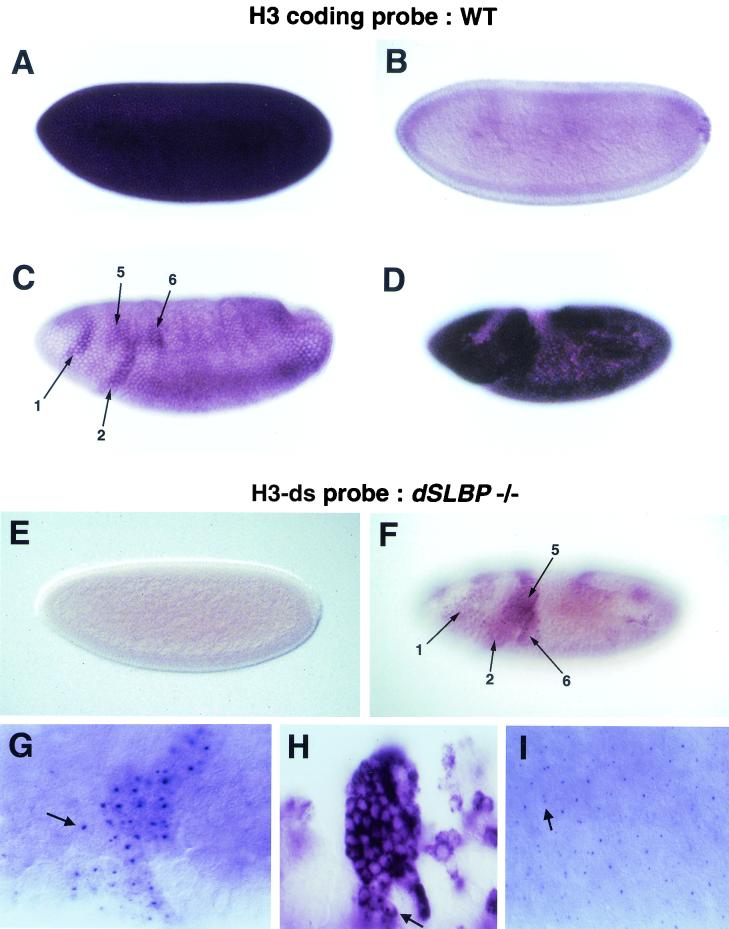
Because some histone mRNAs still accumulate in dSLBP mutant embryos (Fig. 3 and 4), in situ probes derived from the coding region of histone genes cannot distinguish between wild-type processed and mutant polyadenylated histone mRNAs. We therefore developed a method to specifically detect the polyadenylated forms of histone H3 in vivo. An antisense RNA probe complementary to the sequences downstream of the normal processing site of the H3 mRNA was synthesized (Fig. 5D, brackets). This probe (designated H3-ds) did not generate any hybridization signal in wild-type embryos at any stage (Fig. 7A and C and Fig. 10E). In contrast, three phenotypic classes of embryos were observed in collections from a dSLBP15/+ stock. Of 552 embryos scored (cycle 17 and older), 25% were unstained (not shown), 51% displayed staining in small numbers of cells in the CNS (Fig. 7B and D), and 24% displayed intense staining (Fig. 7F) in a pattern similar to that obtained for wild-type embryos with an H3 coding region probe (Fig. 7E). The 1:2:1 ratio of the three phenotypic classes strongly suggests that they represent the +/+; dSLBP15/+; dSLBP15/dSLBP15 genotypic classes. These data indicate that we can specifically detect misprocessed, polyadenylated histone H3 mRNA in dSLBP mutant embryos using the H3-ds probe. With this technique, several conclusions can be drawn. First, dSLBP is partially haploinsufficient for histone H3 processing, since a small amount of signal was detected with the H3-ds probe in heterozoygous embryos (Fig. 7B and D). Second, all tissues (e.g., CNS, peripheral nervous system [PNS], and midgut) containing replicating cells that normally accumulate histone H3 mRNA in the wild type also accumulate polyadenylated H3 mRNA in dSLBP homozygous mutants (Fig. 7F and H). Third, polyadenylated histone H3 mRNA is more stable than wild-type H3 mRNA, as evidenced by the accumulation of mRNA in the nonreplicating G phase in cells of endocycling tissues (compare arrows in Fig. 7G and H) (48).
FIG. 10.
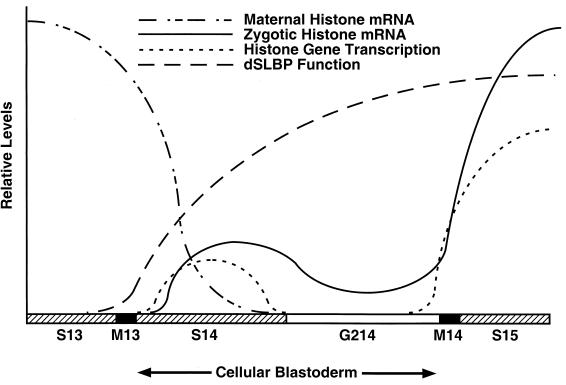
Model of histone gene expression in the early Drosophila embryo. Maternal histone mRNA is rapidly destroyed at interphase 14. Zygotic histone transcription occurs during S phase of cycle 14 as determined by transcription dots visible with the H3-ds 3′ flanking probe and terminates by the start of G2. Transcription reinitiates in late G2 of cycle 14 in each mitotic domain in anticipation of S phase of cycle 15. A low level of histone mRNA can be detected in all cells during G2 of cycle 14, probably because both the maternal mRNA and mRNA synthesized during S phase 14 are not quantitatively destroyed at the end of S phase 14. dSLBP protein is not provided maternally, and the timing of its accumulation zygotically matches the onset of histone transcription in S phase 14. When zygotic dSLBP is not provided, misprocessed histone messages begin to accumulate immediately in S phase 14 and subsequently in G2 phase 14.
dSLBP is required as soon as zygotic histone transcription begins.
We were interested in learning when de novo zygotic transcription and processing of histone mRNA begins during embryogenesis and whether dSLBP function is required during the first 16 cycles, which lack the G1 phase. To investigate the developmental time at which misprocessed histone messages are produced in dSLBP mutants, we prepared poly(A)+ RNA extracted from staged embryos collected from heterozygous dSLBP15/+ flies and performed Northern blot analysis with a histone H3-coding region probe (Fig. 8A). As noted previously (48) and shown here (Fig. 3), little or no polyadenylated maternal histone mRNA was detected in 0- to 2-h-old embryos (Fig. 8A, lane 2). However, as soon as zygotic gene expression begins at ∼2 h after egg deposition (AED) polyadenylated H3 mRNA is detected (Fig. 8A, lane 3). Polyadenylated H3 mRNA continued to accumulate throughout embryogenesis (Fig. 8A, lanes 4 to 7) and remained at a high level even in late-stage mutant embryos (Fig. 8A, lane 7). In contrast, the level of wild-type H3 mRNA was high early in embryogenesis when cells were replicating (Fig. 8B, lanes 1 to 5) and then declined as most cells exit the cell cycle and become quiescent after stage 11 (Fig. 8B, lanes 6 to 7). This is consistent with our interpretation that polyadenylated H3 mRNA persists in unreplicating cells because it has a longer half-life than wild-type H3 mRNA.
FIG. 8.
dSLBP function is required as soon as zygotic transcription begins. Appearance of misprocessed histone mRNA in dSLBP mutant embryos is coincident with the normal onset of dSLBP protein expression. (A) Total RNA from 13- to 16-h-old wild-type embryos (lane 1) and the poly(A)+ RNA fraction from appropriately aged embryos collected from dSLBP15/+ parents corresponding to 0 to 2 h (lane 2), 2 to 4 h (lane 3), 4 to 7 h (lane 4), 7 to 10 h (lane 5), 10 to 13 h (lane 6), and 13 to 16 h (lane 7) AED was subjected to Northern analysis using a radiolabeled H3 coding region probe. Polyadenylated H3 mRNA begins to accumulate at ∼2 h of embryogenesis, which is the time when most zygotic gene expression begins. The blot was stripped and reprobed for rp49 as a loading control. (B) Total RNA isolated from wild-type embryos aged 0 to 1 h (lane 1), 1 to 2 h (lane 2), 2 to 4 h (lane 3), 4 to 7 h (lane 4), 7 to 10 h (lane 5), 10 to 13 h (lane 6), and 13 to 16 h (lane 7) AED was probed with the histone H3 coding region. (C and D) Western analysis of embryonic extracts using affinity-purified anti-dSLBP rabbit antibodies. dSLBP protein is indicated with an asterisk. All other bands represent nonspecific cross-reactingproteins (48). (C) Lane 1, 0- to 2-h-old embryos collected from dSLBP10/Df(3R)3450 females; lane 2, wild-type 13- to 16-h-old embryos; lane 3, wild-type 13- to 16-h-old embryo extract treated with CIP. Note that dSLBP migrates faster after phosphatase treatment. Lane 4, 13- to 16-h-old dSLBP15/Df(3R)3450 embryos. Extracts of wild-type yw67 embryos collected 0 to 1 h (lane 5), 1 to 2 h (lane 6), 2 to 4 h (lane 7) AED. (D) Extracts of wild-type yw67 embryos collected 0 to 1 h (lane 1), 1 to 2 h (lane 2), 2 to 4 h (lane 3), 4 to 7 h (lane 4), 7 to 10 h (lane 5), 10 to 13 h (lane 6), and 13 to 16 h (lane 7) AED. These are the same embryo collections used to prepare the RNA for panel B.
The normal profile of dSLBP protein accumulation during embryogenesis was determined by Western blot analysis using an antibody directed against full-length dSLBP (Fig. 8C to D). As observed previously (48), dSLBP was detected as a single species in extracts derived from 13- to 16-h-old wild-type embryos (Fig. 8C, lane 2) but not in 13- to 16-h-old dSLBP15 mutant embryos (Fig. 8C, lane 4). Phosphatase treatment of the embryonic extract quantitatively increased the mobility of dSLBP, indicating that the vast majority of dSLBP was phosphorylated in vivo (Fig. 8C, lane 3). A similar level of phosphorylation of dSLBP was observed in S2 tissue culture cells (not shown). Surprisingly, dSLBP was not detected in extracts prepared from 0- to 1-h-old embryos (Fig. 8C, lane 5 and 8D, lane 1) even though these embryos contained large amounts of histone mRNA whose accumulation requires dSLBP (Fig. 2). dSLBP rapidly accumulated in 2- to 4-h-old embryos and was maintained at a relatively constant level throughout the first 16 h of embryogenesis (Fig. 8C, lane 7, and D, lanes 3 to 7). Given the sensitivity of our antibody, we would have readily detected dSLBP if it were present in the 0- to 1-h-old embryos in as little as 10% of the amount present in 2- to 4-h-old embryos. These results suggest that dSLBP is not provided maternally in significant amounts, consistent with the early zygotic requirement for dSLBP function demonstrated genetically in Fig. 8A. It is likely that dSLBP protein is present during oogenesis and may be destroyed prior to the deposition of RNAs and proteins into the egg. All the dSLBP was phosphorylated at all stages examined, suggesting that the active form of dSLBP is phosphorylated. Taken together, these data clearly demonstrate that dSLBP function is required for histone pre-mRNA processing as soon as zygotic histone transcription begins in the early embryo.
Histone transcription occurs in early embryonic mitotic domain pattern.
To determine more precisely the onset of polyadenylated histone H3 expression in dSLBP mutant embryos, we detected histone gene transcription by in situ hybridization of dSLBP mutant embryos with the H3-ds probe. Single fixed embryos can be staged very accurately with respect to developmental time, allowing us to precisely match the pattern of histone mRNA expression to the stage of the cell cycle. We first established the early pattern of wild-type H3 mRNA accumulation in individual embryos. The first 13 cycles of embryogenesis are nuclear S-M cycles that lack gap phases, occur in a syncytium, and are controlled exclusively by maternally derived gene products. Histone H3 mRNA is present at high levels throughout this time (Fig. 9A). Cellularization occurs during interphase 14, which also marks the appearance of the first G2 phase and the switch from maternal to zygotic control of development (18). Histone H3 expression was high during S phase of cycle 14 (not shown), but by late cellular blastoderm, as cells entered G2 phase, most, but not all, H3 mRNA disappeared (Fig. 9B). During gastrulation, different groups of cells entered mitosis 14 at different times, giving rise to the well-described mitotic domain pattern of the early embryo (17, 18). These post-blastoderm cell divisions (i.e., cycles 14 to 16) lack a G1 phase and are regulated at the G2-M transition via developmentally controlled transcription of stringcdc25, which activates cdc2 (13, 14). Interestingly, H3 mRNA begins to accumulate in the mitotic domain pattern in wild-type embryos, although the pattern is somewhat masked by a low level of ubiquitous histone mRNA (Fig. 9C). The ubiquitous mRNA is derived from perdurance of a small amount of maternal mRNA and/or zygotic RNA that accumulated during S14 (see below). Both by careful staging of the embryos and by double staining with the DNA binding dye DAPI, we determined that this accumulation begins immediately before mitosis 14 while the cells are still in G2. Thus, the histone genes are likely responding to the developmental signals that control the G2-M transition at this stage of development (see Discussion). By germ-band-extended stages, all cells that had passed into S phase 15 accumulated histone H3 mRNA (Fig. 9D).
FIG. 9.
Zygotic histone transcription occurs in the mitotic domain pattern in the early embryo. (A to D) In situ hybridization of yw67 wild-type embryos using an H3 coding region probe. (A) A high level of maternal histone H3 mRNA is deposited into the egg. (B) Much lower levels are observed in cellular blastoderm embryos. The detectable H3 message may result from a failure to destroy all maternal H3 mRNA and/or from the initiation of zygotic histone transcription during S phase 14. (C) As embryos begin to gastrulate in cycle 14, zygotic H3 mRNA begins to accumulate in the mitotic domain pattern. The arrows indicate mitotic domains 1, 2, 5, and 6. The mitotic domain pattern is somewhat masked by the presence of ubiquitous H3 mRNA. (D) H3 mRNA accumulation becomes widespread in germ band extended embryos, although the mitotic domain pattern is still evident. (E to I) In situ hybridization of embryos collected from dSLBP15/+ parents using the histone H3-ds probe. (E) No misprocessed maternal H3 mRNA is detected in either pre-blastoderm or blastoderm embryos regardless of genotype. (F) During gastrulation de novo synthesis of misprocessed histone H3 mRNA in dSLBP15/dSLBP15 embryos begins to appear in the mitotic domain pattern, with domains 1, 2, 5, and 6 indicated by arrows. This embryo is slightly older than the embryo shown in panel C. (G) Higher magnification of misprocessed histone H3expression reveals nascent transcription dots (arrow) in the nucleus of cells from dSLBP15/dSLBP15 mutant embryos. Mitotic domain 2 is shown. (H) Stage 11 (cycle 17) dSLBP15/dSLBP15 embryo showing that misprocessed H3 mRNA eventually accumulates in the cytoplasm. Nascent transcription dots can also be seen at this stage (arrow). (I) A cellularizing blastoderm embryo where nascent transcription dots are observed during S14. A single transcription dot is visible in some nuclei, while others have two (arrow). One versus two likely depends on the degree of pairing between homologs and/or the orientation of the nucleus with repect to the focal plane (28).
Using the histone H3-ds probe, we determined that early embryos had no detectable polyadenylated maternal histone message regardless of genotype (Fig. 9E), consistent with the Northern blot and S1 nuclease protection assays. However, as gastrulation began in dSLBP15 mutant embryos, polyadenylated histone H3 message accumulates in regions of the embryo corresponding to early mitotic domains (Fig. 9F). The mitotic domain pattern of histone expression can be seen with much greater clarity than with the coding region probe in wild-type embryos because only de novo transcription of misprocessed histone H3 RNA is being detected, and the maternal histone H3 mRNA does not contain the probe sequences which are all from the 3′ flanking region. There was a lag in the accumulation of polyadenylated histone H3 mRNAs in dSLBP15 mutant embryos (Fig. 9F) when compared to the expression of processed histone H3 in wild-type embryos at the same stage (Fig. 9D). This suggests that production of mature, polyadenylated histone mRNA is inefficient at this time.
At higher magnification of the dSLBP15 mutant embryos stained with the H3-ds probe, nascent transcription dots were clearly visible in cells in the mitotic domains (Fig. 9G, arrow). Large, nascent transcription dots have previously been observed using synthetic transgene constructs that lack transcription termination signals (54). Moreover, improper 3′ end processing blocks nuclear export and causes the accumulation of nascent transcripts in yeast (27). Thus, the large dots we see in early embryos likely represent H3 transcripts that have proceeded into the intergenic region. Nascent transcription is not detected in wild type embryos with a coding region probe, suggesting that the newly transcribed RNA in the mutant embryos is processed more slowly (and less efficiently) than the wild-type mRNA and remains associated with the chromatin. This would occur because normal processing of histone mRNA is blocked and is consistent with the usage of the cryptic polyadenylation signals being inefficient at this time. Later in embryogenesis, misprocessed polyadenylated histone H3 generated in dSLBP15 mutants accumulates in the cytoplasm (Fig. 7F and G and 9H) where it is likely translated, although nascent transcripts are still apparent (Fig. 9H, arrow).
In dSLBP mutants, nascent, misprocessed histone H3 mRNA could be detected with the H3-ds probe as early as the cellular blastoderm stage during S phase of cycle 14 (Fig. 9I). Nascent transcripts were visible in cells during S phase of cycle 14 but not in cells of the late blastoderm that had entered G2 of cycle 14 (not shown). The nascent transcripts became visible again in the late G2 phase of cycle 14 (e.g., Fig. 9G). This suggests that zygotic transcription of histone mRNA and dSLBP-dependent processing normally occurs during S phase of cycle 14. Thus, we can quite accurately measure the very onset of histone transcription as well as concomitant dSLBP-dependent processing. Given the low levels of dSLBP protein provided by the mother, zygotic dSLBP synthesis is immediately required for the normal production of histone mRNA in the early embryo.
DISCUSSION
In this report, we demonstrate that dSLBP is required for the production of all five replication-associated histone mRNAs in vivo. The consequences of dSLBP mutation for histone mRNA synthesis depends on developmental stage, and our studies provide a picture of how dSLBP function is integrated with the normal expression of histone mRNA during Drosophila embryogenesis.
Polyadenylation of histone mRNA occurs in the absence of dSLBP.
Each of the five Drosophila histone genes contains at least one cryptic polyadenlyation signal that is utilized in the absence of zygotic dSLBP expression. The efficiency of utilization of these signals likely depends on the gene, cell type, and stage of embryonic development. Poly(A)+ histone messages accumulate slowly in young dSLBP mutant embryos, despite high levels of histone gene expression early in wild-type embryogenesis. At late embryonic stages, the abundance of poly(A)+ histone mRNA is greater than the wild type, suggesting the possibility that the efficiency of polyadenylation increases as embryos age. The bulk of the poly(A)+ histone mRNA is present in the cytoplasm (reference 48 and Fig. 9H) and is likely translated. The poly(A)+ histone mRNAs probably accumulate because these mRNAs are not degraded at the end of S phase as are the normal histone mRNAs (25, 53). The consequence of the increase in stability is that accumulation of histone mRNA, and as a result most likely the synthesis of histone protein, is uncoupled from the cell cycle (48). The increased stability of polyadenylated histone mRNA was obvious only in continuously cycling cells (e.g., endocycling midgut cells) and not in cells that exited the cell cycle. For instance, there was no obvious persistence of polyadenylated histone mRNA in epidermal cells upon entering G0 for the first time during cell cycle 17. Therefore, the turnover of polyadenylated histone mRNA appears to vary depending on cell type or on differences between cycling (i.e., proliferating or endoreduplicating) and quiescent cells.
Why are consensus polyadenylation signals found downstream of each Drosophila histone gene? One possibility is that these signals are not actually cryptic but have a normal function. Termination of transcription requires the presence of a 3′ processing site, either a polyadenylation signal (3, 42) or a histone 3′ end (4). Since the histone genes are actively transcribed and the 3′ ends of adjacent genes are quite close together, these signals may have been selected to ensure termination and prevent either read-through into the next gene, possibly disrupting transcription or producing antisense histone mRNA that might trigger RNA interference (1).
In contrast to the embryonic phenotype, polyadenylated histone mRNAs do not accumulate during oogenesis in dSLBP10 hypomorphic mothers and the levels of maternal mRNAs for each of the five histones are reduced at least 10-fold. This suggests that mutation of dSLBP causes a dramatic defect in histone mRNA biosynthesis in the female germ line. In mammalian cells, mutation of the stem-loop prevents accumulation of histone mRNA and accumulation is restored only by addition of an efficient polyadenylation signal (40). Therefore, one possibility is that the cryptic polyadenylation signals are not utilized in the female germ line in the absence of dSLBP and the misprocessed histone transcripts are rapidly degraded and hence are not detectable in steady-state RNA (40). Other examples of tissue-specific differences in polyadenylation have been reported. For instance, usage of a cryptic polyadenylation site in the murine H2a-614 gene occurs during spermatogenesis (35, 36), although this site is not utilized in somatic cells even in the absence of the stem-loop (4). The molecular basis for differences in polyadenylation efficiency in different developmental contexts (e.g., germ line versus embryo) is not known.
Another possibility for the failure to accumulate maternal histone mRNA after reduction of dSLBP function is that polyadenylated histone mRNAs, if indeed synthesized, may not be efficiently transported from the nurse cells to the oocyte. In mammalian cells, the stem-loop has been implicated in the transport of mature histone mRNA from the nucleus to the cytoplasm, where SLBP is found as part of the mature mRNP complex (10, 24). Perhaps dSLBP is required for histone mRNA movement during nurse cell cytoplasmic transfer to the oocyte.
The role of SLBP in histone pre-mRNA processing.
Although Drosophila U7 snRNP has not yet been identified, it is likely that the overall mechanism of histone pre-mRNA processing is similar in Drosophila, vertebrates, and echinoderms. U7 snRNA is the smallest of the snRNAs (55 to 70 nt) in vertebrates (5, 38, 44) and echinoderms (47), and the sequence has not been well-conserved even among vertebrates. The 5′ end of the U7 snRNA is complementary to a short region in the histone downstream element, which itself is not well-conserved among the multiple histone genes in any species (51). There is a novel Sm site in U7 snRNA (23) which probably binds to the novel U7-specific Sm protein, Lsm10 (41), followed by a stem-loop whose sequence has not been conserved. Thus, it has not been possible to identify the U7 snRNA by searching the genomic sequences of Drosophila or C. elegans. Two mammalian U7-specific proteins have been identified: Lsm10 and a large zinc finger protein, hZFP100, (8, 41). Searches of the Drosophila and C. elegans genomes did not identify obvious homologues of the Lsm10 protein (D. Schumperli, personal communication). Because of the large number of zinc finger proteins in metazoan genomes, it is not possible to identify the orthologue of hZFP100 in Drosophila or C. elegans. Drosophila apparently does not contain the U11 snRNA or U11 specific proteins, although it contains ATAC introns that are spliced, and it does contain U12 snRNA (37). Thus, it is formally possible that the mechanism of histone mRNA formation is mechanistically different in Drosophila than in echinoderms and vertebrates.
Nevertheless, the molecular data demonstrate that dSLBP contributes in an important way to the processing of endogenous histone pre-mRNAs in vivo, and it is essential for the processing of all five Drosophila histone mRNAs in vitro (Z. Dominski and W. F. Marzluff, unpublished data). Unexpectedly, some histone mRNA ending at the normal 3′ end was present in the dSLBP15 mutants. Although dSLBP15 acts genetically as a null and produces no dSLBP protein detectable by Western blotting, this allele retains the entire dSLBP coding region and contains a P element sequence inserted in the 5′ UTR. Therefore, it is formally possible that very small amounts of dSLBP are expressed from dSLBP15. Very low levels of dSLBP may be able to act catalytically to allow substantial processing of histone mRNA. An alternative possibility is that dSLBP is not absolutely required for the processing reaction but rather acts to stimulate assembly of the 3′ end processing machinery. For instance, vertebrate SLBP bound to the stem-loop stabilizes the interaction of U7 snRNP with histone pre-mRNA (11). In vitro, processing can occur in the absence of SLBP if there is extensive base pairing between U7 snRNA and the histone pre-mRNA (7, 10, 11, 45). Whether all the histone mRNAs ending at the normal 3′ end in the dSLBP mutant embryos are produced by the normal processing pathway or an alternative pathway is not known. In vertebrates, SLBP participates in the processing reaction and remains associated with the mature histone mRNA (10, 24). Even if SLBP normally remains bound to the mRNA when transported to the cytoplasm, perhaps processed histone mRNA lacking bound dSLBP can still accumulate to measurable levels in the cell.
Developmental control of histone mRNA production.
The first 14 replication cycles of Drosophila embryogenesis occur normally in the absence of zygotic transcription (34), indicating that a supply of maternal histone protein and or mRNA is sufficient to support chromosome duplication and segregation until the blastoderm stage. As with other mRNAs encoding components required for DNA replication (12), maternal histone mRNA is depleted from the embryo at cycle 14 but is rapidly replaced by zygotic expression. A probe derived from sequences downstream of the normal histone H3 processing site allowed us to discern the de novo transcription pattern of histone H3 in dSLBP mutant embryos, since this probe does not detect accumulation of the processed, wild-type mRNA (Fig. 10). The earliest we could detect transcription, as indicated by nascent, misprocessed pre-mRNA, was during the synchronous 14th S phase that occurs during cellularization. By the beginning of G2 of cycle 14, which is the first G2 phase of development occurring just before the onset of gastrulation, histone H3 transcription is reduced as determined by the failure to detect transcription dots.
Interestingly, histone H3 transcription resumes in the mitotic domain pattern. Accurate staging of individual embryos by morphological criteria (18) and detection of chromosome behavior with DNA binding dyes allowed us to precisely determine the cell cycle phase at which histone transcription resumed. mRNA accumulation begins in late G2 of cycle 14 in anticipation of S phase 15, which begins immediately after mitosis, and continues throughout S phase 15. During cycle 15, the accumulation of histone mRNA was not strictly restricted to S phase. Either these G2-regulated cycles occur so rapidly and/or histone mRNA half-life is sufficiently long that some histone mRNA persists during G2 (i.e., it never falls to undetectable levels) before reaccumulation to high levels in S phase 16. Very tight coupling between histone mRNA accumulation and S phase occurred in later embryos (e.g., in germ band-retracted embryos >10 h), suggesting that the mechanism that destroys histone mRNA at the end of S phase becomes more efficient as development proceeds.
string is the only gene currently known to be transcribed coincident with all of the mitotic domains (13). string encodes a cdc25 phosphatase that activates cdc2 and induces the G2-M transition (20). In most cells, string transcription is causal for mitotic entry (15), and the control region of string serves as an integrator of developmental patterning information that connects the morphological events of embryogenesis with cell cycle progression (13). The string control region is very large and complex, being composed of modular enhancer elements that direct tissue and stage specific expression. As the control region of the histone genes is simple by comparison, the correlation of histone transcription with the mitotic domain pattern suggest that histone transcription may depend on string. String could either activate histone gene transcription directly by modifying the activity of a transcription factor (e.g., YY1 [16] or NPAT [32, 57]) or indirectly as a result of a regulatory cascade of events associated with cell cycle progression. That is, some G1-S events like histone expression might be triggered by String late in G2 at this stage of development because these cycles lack a bona fide G1 phase. In mammalian cells, histone gene transcription occurs in G1 and even serum-starved G0 cells at a reduced rate (20 to 30% of the S phase rate) (6), and the major regulation of accumulation occurs at the level of histone mRNA processing (46). A similar situation could occur in Drosophila embryos, such that once transcription is activated via the action of String late in G2 of cycle 14 it persists through the entire next cell cycle.
Processing is also stimulated at the G1-S transition in mammalian cells (25, 53), and zygotic dSLBP function is required as soon as zygotic histone transcription begins to process a newly made message. Little to no maternal dSLBP protein is present, providing an explanation for the very early zygotic requirement of dSLBP function. We were somewhat surprised by the lack of maternal dSLBP protein. Vertebrate SLBP accompanies histone mRNA to the cytoplasm, where it is thought to contribute to the stability of the histone mRNA as well as to stimulate histone mRNA translation (19, 49). Therefore, either dSLBP is not required for translation in the early embryo or translation of maternal histone mRNAs does not occur or is inefficient. Consequently, the histone protein needed during the early syncytial cycles must also be deposited into the egg. Our preliminary data indicate that dSLBP function is required in the germ line for production of this maternal histone protein (data not shown). dSLBP mutant embryos proceed normally through embryogenesis and hatch into viable larvae. Thus, the maternal histone protein and/or the production of histone protein from the low level of cytoplasmic histone poly(A)+ mRNA beginning at gastrulation is sufficient to support all the syncytial cycles as well as S phase 15 and 16. At subsequent stages, higher levels of poly(A)+ histone mRNA are present, and presumably provide enough histone protein required for the completion of embryogenesis.
Acknowledgments
We thank Zbig Dominski and Tin Tin Su for helpful discussions and Mark Peifer, Steve Crews, and Jeff Sekelsky for critical reading of the manuscript.
This work was supported by NIH grants GM58921 to W.F.M. and an NSF Career Award to R.J.D.
REFERENCES
- 1.Akhmanova, A., H. Kremer, K. Miedema, and W. Hennig. 1997. Naturally occurring testis-specific histone H3 antisense transcripts in Drosophila. Mol. Reprod. Dev. 48:413-420. [DOI] [PubMed] [Google Scholar]
- 2.Akhmanova, A., K. Miedema, H. Kremer, and W. Hennig. 1997. Two types of polyadenated mRNAs are synthesized from Drosophila replication-dependent histone genes. Eur J. Biochem. 244:294-300. [DOI] [PubMed] [Google Scholar]
- 3.Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 4.Chodchoy, N., N. B. Pandey, and W. F. Marzluff. 1991. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol. Cell. Biol. 11:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotten, M., O. Gick, A. Vasserot, G. Schaffner, and M. L. Birnstiel. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. EMBO J. 7:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLisle, A. J., R. A. Graves, W. F. Marzluff, and L. F. Johnson. 1983. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol. Cell. Biol. 3:1920-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominski, Z., J. A. Erkmann, J. A. Greenland, and W. F. Marzluff. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. 21:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski, Z., J. A. Erkmann, X. Yang, R. Sanchez, and W. F. Marzluff. 2002. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 16:58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 10.Dominski, Z., J. Sumerel, R. J. Hanson, and W. F. Marzluff. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1:915-923. [PMC free article] [PubMed] [Google Scholar]
- 11.Dominski, Z., L. X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duronio, R. J., and P. H. O'Farrell. 1994. Developmental control of a G1-S transcriptional program in Drosophila. Development 120:1503-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar, B. A., D. A. Lehman, and P. H. O'Farrell. 1994. Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development 120:3131-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar, B. A., and P. H. O'Farrell. 1989. Genetic control of cell division patterns in the Drosophila embryo. Cell 57:177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar, B. A., and P. H. O'Farrell. 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 62:469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliassen, K. A., A. Baldwin, E. M. Sikorski, and M. M. Hurt. 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18:7106-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foe, V. E. 1989. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development 107:1-22. [PubMed] [Google Scholar]
- 18.Foe, V. E., G. M. Odell, and B. A. Edgar. 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint, p. 149-300. In M. Bate and A. Martinez Arias (ed.), The development of Drosophila melanogaster, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 19.Gallie, D. R., N. J. Lewis, and W. F. Marzluff. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24:1954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier, J., M. J. Solomon, R. N. Booher, J. F. Bazan, and M. W. Kirschner. 1991. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell 67:197-211. [DOI] [PubMed] [Google Scholar]
- 21.Gick, O., A. Kramer, W. Keller, and M. L. Birnstiel. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graves, R. A., S. E. Wellman, I. M. Chiu, and W. F. Marzluff. 1985. Differential expression of two clusters of mouse histone genes. J. Mol. Biol. 183:179-194. [DOI] [PubMed] [Google Scholar]
- 23.Grimm, C., B. Stefanovic, and D. Schumperli. 1993. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 12:1229-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson, R. J., J. Sun, D. G. Willis, and W. F. Marzluff. 1996. Efficient extraction and partial purification of the polyribosome- associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35:2146-2156. [DOI] [PubMed] [Google Scholar]
- 25.Harris, M. E., R. Bohni, M. H. Schneiderman, L. Ramamurthy, D. Schumperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan, B., and H. Vaessin. 1997. Daughterless is required for the expression of cell cycle genes in peripheral nervous system precursors of Drosophila embryos. Dev. Genet. 21:117-122. [DOI] [PubMed] [Google Scholar]
- 27.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 28.Hiraoka, Y., A. F. Dernburg, S. J. Parmelee, M. C. Rykowski, D. A. Agard, and J. W. Sedat. 1993. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J. Cell Biol. 120:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingledue, T. C., 3rd, Z. Dominski, R. Sanchez, J. A. Erkmann, and W. F. Marzluff. 2000. Dual role for the RNA-binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA 6:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kremer, H., and W. Hennig. 1990. Isolation and characterization of a Drosophila hydei histone DNA repeat unit. Nucleic Acids Res. 18:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luscher, B., and D. Schumperli. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expr. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 34.Merrill, P. T., D. Sweeton, and E. Wieschaus. 1988. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development 104:495-509. [DOI] [PubMed] [Google Scholar]
- 35.Moss, S. B., P. B. Challoner, and M. Groudine. 1989. Expression of a novel histone 2B during mouse spermiogenesis. Dev. Biol. 133:83-92. [DOI] [PubMed] [Google Scholar]
- 36.Moss, S. B., R. A. Ferry, and M. Groudine. 1994. An alternative pathway of histone mRNA 3′ end formation in mouse round spermatids. Nucleic Acids Res. 22:3160-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mount, S. M., and H. K. Salz. 2000. Pre-messenger RNA processing factors in the Drosophila genome. J. Cell Biol. 150:F37-F44. [DOI] [PMC free article] [PubMed]
- 38.Mowry, K. L., and J. A. Steitz. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238:1682-1687. [DOI] [PubMed] [Google Scholar]
- 39.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey, N. B., A. S. Williams, J. H. Sun, V. D. Brown, U. Bond, and W. F. Marzluff. 1994. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol. Cell. Biol. 14:1709-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pillai, R. S., C. L. Will, R. Luhrmann, D. Schumperli, and B. Muller. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20:5470-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proudfoot, N. J. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14:105-110. [DOI] [PubMed] [Google Scholar]
- 43.Reichhart, J. M., and D. Ferrandon. 1998. Green Balancers. Dros. Inf. Serv. 81:201-202. [Google Scholar]
- 44.Soldati, D., and D. Schumperli. 1988. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 8:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spycher, C., A. Streit, B. Stefanovic, D. Albrecht, T. H. Koning, and D. Schumperli. 1994. 3′ End processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 22:4023-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stauber, C., and D. Schumperli. 1988. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 16:9399-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strub, K., G. Galli, M. Busslinger, and M. L. Birnstiel. 1984. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 3:2801-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, J., D. R. Pilch, and W. F. Marzluff. 1992. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 20:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Z. F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Z. F., T. Krasikov, M. R. Frey, J. Wang, A. G. Matera, and W. F. Marzluff. 1996. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1Mb. Genome Res. 6:688-701. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z. F., M. L. Whitfield, T. C. Ingledue, 3rd, Z. Dominski, and W. F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 53.Whitfield, M. L., L. X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkie, G. S., A. W. Shermoen, P. H. O'Farrell, and I. Davis. 1999. Transcribed genes are localized according to chromosomal position within polarized Drosophila embryonic nuclei. Curr. Biol. 9:1263-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodland, H. R. 1980. Histone synthesis during the development of Xenopus. FEBS Lett. 121:1-10. [DOI] [PubMed] [Google Scholar]
- 56.Woodland, H. R., and E. D. Adamson. 1977. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev. Biol. 57:118-135. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]