Abstract
DNA mismatch repair (MMR) is a critical genome-stabilization system. However, the molecular mechanism of MMR in human cells remains obscure because many of the components have not yet been identified. Using a functional in vitro reconstitution system, this study identified three HeLa cell fractions essential for in vitro MMR. These fractions divide human MMR into two distinct stages: mismatch-provoked excision and repair synthesis. In vitro dissection of the MMR reaction and crucial intermediates elucidated biochemical functions of individual fractions in human MMR and identified hitherto unknown functions of human replication protein A (hRPA) in MMR. Thus, one fraction carries out nick-directed and mismatch-dependent excision; the second carries out DNA repair synthesis and DNA ligation; and the third provides hRPA, which plays multiple roles in human MMR by protecting the template DNA strand from degradation, enhancing repair excision, and facilitating repair synthesis. It is anticipated that further analysis of these fractions will identify additional MMR components and enable the complete reconstitution of the human MMR pathway with purified proteins.
DNA mismatch repair (MMR) is a mutation avoidance system that eliminates mispairs that accumulate in the genome during normal DNA metabolism. MMR corrects heteroduplex DNA that contains base-base mismatches and small insertion-deletion mispairs. These lesions are recognized by MMR proteins, the “wrong” base is excised from the newly synthesized strand of DNA, and a repair patch is synthesized by using the parental DNA strand as a template. In addition, MMR has been shown to maintain genomic stability by mediating DNA damage-induced apoptosis (for a review, see reference 32).
The Escherichia coli MMR pathway is well characterized. Eleven activities are required to carry out MMR in E. coli, including MutS, MutL, MutH, UvrD, ExoI, ExoVII, ExoX, RecJ, single-stranded DNA binding protein (SSB), polymerase III holoenzyme, and DNA ligase (8, 29, 63). These enzymes are necessary and sufficient to perform MMR in vitro. For human cells, less is known about the biochemistry and the components of the repair process, and the reaction is more complex. The human MutS (hMutS) and MutL (hMutL) homologs have been identified by studying mutant cells that demonstrate microsatellite instability and are deficient in MMR. The most commonly studied MMR-deficient cell lines are from patients who have hereditary nonpolyposis colorectal cancer (for reviews see references 7, 21, 22, 28, and 39). Like the E. coli MutS and MutL proteins, hMutS and hMutL are involved in the initiation phase of the repair reaction. However, unlike E. coli MutS and MutL, hMutS and hMutL are heterodimers (reviewed in reference 41). hMSH2 interacts with hMSH6 or hMSH3 to form heterodimeric hMutSα (12, 43) or hMutSβ (17, 44), respectively, and hMLH1 interacts with hPMS2, hPMS1, or hMLH3 to form three distinct hMutL heterodimers (16, 31, 33, 35, 50). Recently, DNA polymerase δ (37), proliferating cell nuclear antigen (PCNA) (3, 9, 10, 15, 18, 26, 61), human replication protein A (hRPA) (34), ExoI (2, 51, 54, 58, 59), and replication factor C (67) have been implicated in MMR. However, biochemical evidence for the involvement of many of these activities in human MMR is still lacking. In addition, in comparison with the E. coli pathway, many of the human components, e.g., a human MutH homolog(s) and a helicase(s), have not been identified.
Efforts have been made to identify novel components of MMR by characterizing human tumor cells that display microsatellite instability, but most of these tumor cell lines are defective in known hMSH2 or hMLH1. To identify novel proteins and to determine the involvement of known proteins in human MMR, it is necessary to establish a reconstituted in vitro assay system for human MMR by using fractionated extracts of wild-type cells. Similar approaches have been successful in studies of mammalian DNA replication (reviewed in reference 6), base excision repair (27), and nucleotide excision repair (1, 40). In this study, we have fractionated HeLa cell extracts and identified three essential fractions required for in vitro MMR. These three fractions identify two distinct stages in the human MMR reaction: nick-directed mismatch-provoked excision is the first stage, and repair DNA synthesis is the second stage. Purification of one fraction revealed the active component to be hRPA. hRPA is demonstrated to play multiple roles in the MMR pathway. The other two fractions comprise multiple activities required for MMR and require further fractionation.
MATERIALS AND METHODS
Fractionation of HeLa S3 nuclear extracts.
HeLa S3 cells were purchased from the National Cell Culture Center (Minneapolis, Minn.). Unless otherwise indicated, fractionation and chromatography were performed at 4°C.
(i) Ammonium sulfate precipitation.
Nuclear extracts prepared from HeLa S3 cells (20) were fractionated by using a two-step ammonium sulfate precipitation procedure. First, the nuclear extract was adjusted to 35% ammonium sulfate (0.21 g/ml) and the precipitate was collected by centrifugation. The supernatant was removed and adjusted to a final concentration of 65% ammonium sulfate by addition of 0.19 g of solid ammonium sulfate/ml. The precipitate was collected by centrifugation, and the supernatant was removed. The precipitates from both treatments were resuspended in and dialyzed against buffer A (25 mM HEPES [pH 7.5], 0.1 mM EDTA, 2 mM dithiothreitol [DTT], 0.1% phenylmethylsulfonyl fluoride [PMSF], 1 μg of leupeptin/ml) containing 0.1 M KCl. Samples were frozen in liquid nitrogen and stored at −80°C. The protein fractions that were insoluble in 35 or 65% ammonium sulfate were designated FI and FII, respectively.
(ii) Preparation of fractions SS1 and SS2.
FI was adjusted to a protein concentration of 5 mg/ml by using buffer B (25 mM Tris [pH 7.5], 10% glycerol, 0.01% NP-40, 0.1 mM EDTA, 2 mM DTT, 0.1% PMSF, 1 μg of leupeptin/ml) containing 0.5 M NaCl. The diluted sample was loaded onto a single-stranded DNA (ssDNA)-cellulose column (3 mg of DNA/g of cellulose; Sigma) as described elsewhere (25). The column was washed with buffer B containing 0.5 M NaCl until the flowthrough tested negative for protein by the Bradford assay (4). The bound proteins were eluted from the column with buffer B containing 2.0 M NaCl. The flowthrough and bound fractions, designated SS1 and SS2, respectively, were pooled, concentrated with 35% ammonium sulfate, dialyzed against buffer A containing 0.1 M KCl, and quickly frozen in liquid nitrogen. Samples were stored at −80°C.
(iii) Phosphocellulose chromatography of HeLa nuclear extracts.
Phosphocellulose chromatography was performed as described previously (33). Briefly, HeLa nuclear extracts (300 mg) were adjusted to a protein concentration of 5 mg/ml by dilution with buffer A and were applied to a phosphocellulose column (Whatman P-11, 6 cm by 10 cm2) equilibrated with buffer A containing 0.05 M KCl. The column was washed with 200 ml of the equilibration buffer and developed with a 0.8-liter linear gradient from 0.05 to 1.3 M KCl in buffer A at a flow rate of 1/2 column volume per h. Fractions were collected, dialyzed against buffer A containing 0.1 M KCl, and stored in aliquots at −80°C.
(iv) MonoQ chromatography of SS2.
Fraction SS2 (5 mg) was applied to a Pharmacia 5/5 MonoQ column equilibrated with buffer A containing 0.1 M KCl and was washed with 10 ml of the same buffer. The column was developed with a KCl gradient from 0.1 to 0.65 M. Fractions were collected and used in MMR assays.
Heteroduplex preparation and MMR assay.
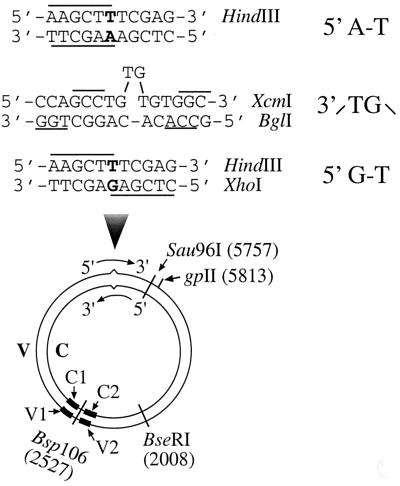
DNA heteroduplexes used in this study were 6.4-kb circular molecules (Fig. 1) containing either a G-T mismatch with a strand break 125 bp 5′ to the mismatch (5′ G-T substrate) or a TG dinucleotide insertion-deletion mispair with a strand break 181 bp 3′ to the mispair (3′ /TG\ substrate). The G-T substrate was constructed by hybridizing Sau96I-digested f1MR3 double-stranded DNA (dsDNA) with f1MR1 ssDNA as described previously (55). These two phages are identical in DNA sequence except at position 5632, so that a unique G-T mismatch forms when they hybridize to each other. The 3′ /TG\ substrate was prepared as described previously (46) by first hybridizing Sau96I-digested f1MR23 dsDNA with f1MR24 ssDNA. The nicked heteroduplex was ligated in the presence of ethidium bromide to introduce supercoiling into the substrate. DNA substrates were purified by CsCl density gradient banding, and the supercoiled substrate was incubated with gpII protein, an endonuclease that specifically cleaves supercoiled filamentous phage DNA at the origin of replication in the viral strand (38). This procedure generates a strand break in the viral strand (V) 181 bp 3′ to the heterology (see Fig. 1). A homoduplex (5′ A-T) was also constructed as described for the 5′ G-T heteroduplex, but using both single-stranded and double-stranded f1MR1 DNAs.
FIG. 1.
DNA substrates. The heteroduplexes were constructed from f1MR phage series (see Materials and Methods) to contain (i) a G-T mismatch and a strand break (at the Sau96I site) in the complementary strand 125 bp 5′ to the mismatch (5′ G-T) or (ii) a TG dinucleotide insertion-deletion mismatch with a strand break 181 bp 3′ to the mispair (3′ /TG\ substrate). The mismatches were within overlapping recognition sites for two restriction endonucleases so that the DNA is resistant to hydrolysis by both enzymes. Preferential repair on the nicked strand renders the repair products sensitive to HindIII (in the case of the 5′ G-T substrate) or BglI (in the case of the 3′ /TG\ substrate). A homoduplex (5′ A-T) was also constructed in a manner identical to the construction of the 5′ G-T substrate. V and C, viral strand and cDNA strand, respectively. C1, C2, V1, and V2 represent oligonucleotide probes (solid bars) complementary to the Bsp106-flanking sequences in the complementary (C1 and C2) and viral (V1 and V2) strands.
Unless otherwise specified, the MMR assay was performed as described previously (20) in a 15-μl reaction mixture containing 100 ng (24 fmol) of heteroduplex DNA, 50 to 60 μg of nuclear extract or fractionated protein, 10 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 1.5 mM ATP, and 0.1 mM each deoxynucleoside triphosphate. After incubation at 37°C for 15 min, DNA samples were recovered by phenol extraction and ethanol precipitation and were digested with the restriction enzymes Bsp106 and HindIII (5′ G-T substrate) or BglI (3′ /TG\ substrate). Reaction products were analyzed by 1% agarose gel electrophoresis and visualized by UV illumination in the presence of ethidium bromide.
Southern blot analyses.
MMR reactions were carried out as described above or as indicated. Reaction products were linearized with Bsp106, electrophoresed in an alkaline agarose gel (1.5%), and transferred onto a nylon membrane. Membranes were blotted with 32P-labeled oligonucleotide probes (purchased from Gibco) complementary to sequences that flank the Bsp106 restriction site. Oligonucleotides were specific for DNA fragments C1 (5′-ATGGTTTCATTGGTGACGTT-3′), C2 (5′-GATTCTGTCGCTACTGATTAC-3′), V1 (5′-AACGTCACCAATGAAACCAT-3′), or V2 (5′-CAGCACCGTAATCAGTAGCG-3′) (see Fig. 1 for details). Blots were exposed to X-ray film, and reaction products were visualized by autoradiography.
Antibodies and Western blot analyses.
Antibodies against hMSH2, hMLH1, and PCNA were purchased from Oncogene Sciences (Boston, Mass.), and antibodies against the 70- and 34-kDa subunits of hRPA either were a generous gift from Zhengxiang Pan (Mount Sinai School of Medicine, New York, N.Y.) or were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Protein samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes for Western blot analysis. Membranes were hybridized with antibodies against the 70- and 34-kDa subunits of hRPA. Bound antibody was detected by chemiluminescence using a secondary antibody conjugated with horseradish peroxidase (Amersham).
Expression and purification of recombinant hRPA.
The RPA expression vector was kindly provided by Marc Wold, and hRPA was purified as described elsewhere (19). Briefly, 2 liters of culture was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) when the optical density at 600 nm reached 0.6 to 0.8. After 2 h of incubation at 37°C, the cells were collected and resuspended in buffer HI (30 mM HEPES [pH 7.8], 0.01% NP-40, 0.25 mM EDTA, 1 mM DTT, and 0.25% inositol) containing the protease inhibitors PMSF, pepstatin A, and leupeptin as described elsewhere (47, 48). The resulting supernatant was loaded onto a 20-ml blue-Sepharose column equilibrated in HI buffer containing 0.1 M KCl. After a wash with the same buffer, the column was sequentially washed with HI buffer containing 0.8 M KCl and 0.5 M NaSCN. The hRPA protein was eluted with HI buffer containing 1.5 M NaSCN. Fractions containing hRPA were further purified using a 5-ml hydroxylapatite column and a 2-ml Q-Sepharose column as described previously (47, 48). hRPA was identified by its DNA binding activity to the 5′-[32P]dT30 DNA substrate under conditions of DNA excess. This was accomplished by the addition of unlabeled dT30 DNA and enabled accurate determination of activity and exclusion of contaminating activities. This procedure resulted in >95% protein purity. hRPA was dialyzed in HI buffer and stored at −80°C.
DNA ligase and DNA polymerase assay.
DNA ligase activity in FII was determined by its ability to convert a nicked circular substrate into supercoiled DNA. The 5′ G-T substrate was incubated with 30 μg of FII at 20°C for 2 h in the presence of ethidium bromide (0.29 nM/μg of DNA), 30 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 1 mM DTT, 26 μM NAD+, and 50 μg of bovine serum albumin/ml. As a positive control, the nicked DNA substrate was also incubated with E. coli DNA ligase (New England Biolabs) at a final concentration of 4 U/μg of DNA. DNA samples were recovered, fractionated in a 1% agarose gel, and visualized under UV illumination. DNA-dependent DNA polymerase activity was determined by measuring the incorporation of [32P]TMP using a poly(dA)/oligo(dT)16 substrate. Reactions were performed in 20 μl at 37°C as previously described (56). Reactions were terminated by addition of EDTA to a final concentration of 20 mM, and unincorporated nucleotides were removed by Sephadex G-50 spin column chromatography. Products were separated by electrophoresis on 8% polyacrylamide-7 M urea DNA sequencing gels. Products were detected by autoradiography.
RESULTS
Fractionation of HeLa nuclear extracts and reconstitution of MMR.
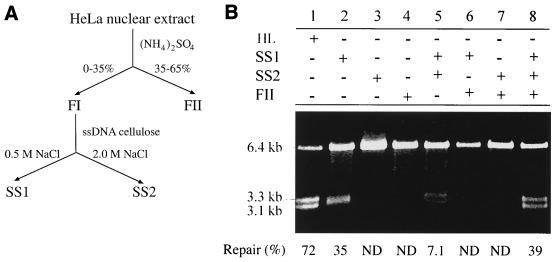
HeLa cell nuclear extracts were fractionated using ammonium sulfate precipitation followed by ssDNA-cellulose column chromatography as diagrammed in Fig. 2A. The three resulting fractions, SS1, SS2, and FII, were assayed for the ability to correct a heteroduplex containing a G-T mismatch and a strand break 125 nucleotides 5′ to the mismatch (5′ G-T substrate [see Fig. 1]). The product of the MMR reaction was detected by restriction enzyme cleavage with endonucleases HindIII (the scoring enzyme) and Bsp106, which yields two DNA fragments of 3.3 and 3.1 kb. Cleavage by HindIII occurs only when the mismatch is correctly repaired (i.e., by extracts of MMR-proficient cells such as HeLa cells [Fig. 2B, lane 1]). Assay results showed that no fraction or combination of two fractions was proficient in MMR (Fig. 2B, lanes 2 to 7). However, MMR activity was detected when SS1, SS2, and FII were all included in the reaction (Fig. 2B, lane 8), indicating that all three of these fractions are required for reconstitution of human MMR in vitro. Similar results were obtained in an MMR assay using an insertion-deletion heteroduplex substrate with a strand break 3′ to the heterology (3′ /TG\ substrate [Fig. 1]); SS1, SS2, and FII were all required for strand-specific repair of this substrate (data not shown). Therefore, the MMR reaction carried out in vitro using these three protein fractions appears to possess the major characteristics of human MMR (for a review, see reference 39): strand specificity (repair targeted on nicked strand), bidirectionality (mismatch removed from orientation of 5′ to 3′ or 3′ to 5′), and broad substrate capability (processing both base-base mismatches and insertion-deletion mismatches).
FIG. 2.
Reconstitution of MMR in vitro. (A) Fractionation of a HeLa nuclear extract into three components required for MMR. (B) Reconstitution of MMR requires SS1, SS2, and FII. The DNA substrate (100 ng of the 5′ G-T heteroduplex) was incubated for 15 min at 37°C in the reaction buffer with fractions as indicated. Amounts of protein used were 15 μg of SS1, 1.5 μg of SS2, or 30 μg of FII. DNA was extracted, treated with HindIII and Bsp106, electrophoresed on an agarose gel, and visualized by ethidium bromide staining under UV illumination. ND, not detectable.
Interestingly, although fraction SS1 did not repair a mismatched substrate to yield 3.3- and 3.1-kb fragments after restriction enzyme cleavage, it produced a novel 3.2-kb DNA species (Fig. 2B, lane 2). This species seems to disappear upon addition of SS2 to the SS1 reaction (Fig. 2B, lane 5), suggesting that SS2 either inhibits the formation of this species by SS1 or converts it to something else (see below for details). In addition, a small amount of the 3.3-kb fragment was observed in the products of reactions containing SS1 and SS2 (lane 5), indicating that limited repair may have occurred.
hRPA substitutes for SS2.
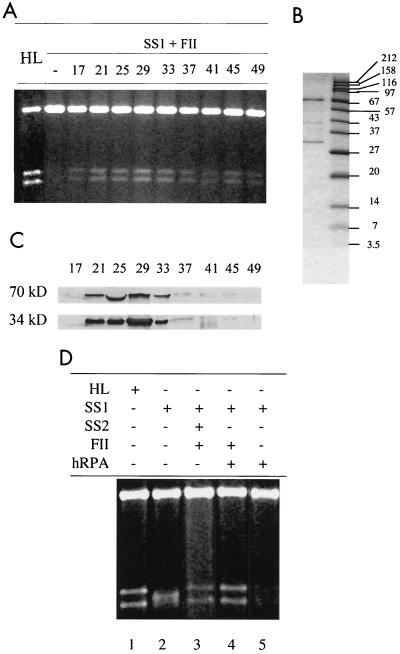
Fractions SS1, SS2, and FII are complex protein samples, and one or more components in each fraction could be required for the MMR reaction. To determine if any of these fractions contains a single MMR activity, in vitro reconstitution was performed by supplementing a combination of two fractions with individual factors derived from HeLa nuclear extracts that were chromatographed as discrete species on a P-11 phosphocellulose column (see Materials and Methods for details). No phosphocellulose column fraction could substitute for SS1 or FII (data not shown), but the requirement for SS2 was eliminated by a broad range of phosphocellulose fractions, with peak fractions at 21 to 33 (Fig. 3A). This result was confirmed by fractionating SS2 by fast- performance liquid chromatography on a MonoQ column and assaying the fractions for the ability to restore MMR proficiency to SS1-FII. The SS1-FII-complementing activity was detected in a high-salt fraction of the MonoQ column (data not shown). These complementation experiments strongly suggest that SS2 contains a single activity that is required in order to reconstitute MMR in vitro.
FIG. 3.
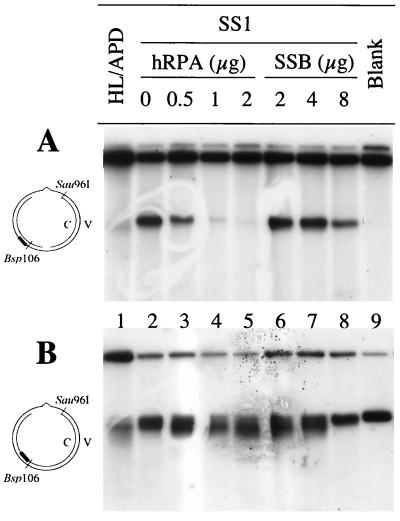
hRPA substitutes for SS2. (A) Reconstitution of MMR in SS1-FII by phosphocellulose P-11 fractions. MMR assays were performed on reaction mixtures containing 100 ng of G-T heteroduplex, 15 μg of SS1, 30 μg of FII, and 2 μl of a P-11 fraction as indicated. (B) SDS-PAGE of a MonoQ fraction that contains two major polypeptides with molecular sizes of 70 and 34 kDa and restores MMR to SS1-FII. Fraction SS2 was purified by Pharmacia HR 5/5 MonoQ column chromatography (see Materials and Methods), and the activity complementing SS1-FII was detected by an MMR assay (data not shown), electrophoresed on an SDS-12% polyacrylamide gel, and stained with Coomassie brilliant blue. (C) Western blot analyses of P-11 fractions. P-11 fractions (50 μl) were precipitated by an equal volume of 20% trichloroacetic acid and neutralized with Tris base prior to electrophoresis on an SDS-12% PAGE gel. Proteins were transferred to a nitrocellulose membrane and analyzed by Western blot analysis using antibodies against the 70- or the 34-kDa subunit of hRPA. (D) hRPA substitutes for SS2 in MMR. When present, hRPA was at 1.0 μg.
The SS1-FII-complementing MonoQ fraction was subjected to electrophoresis on a denaturing SDS-polyacrylamide gel. Several polypeptides were detected, including two major species with molecular sizes of 70 and 34 kDa (Fig. 3B). These polypeptides are similar in size to two of the three subunits of hRPA (a trimer of 70-, 34-, and 13-kDa subunits), suggesting that the protein in SS2 that is required for MMR may be hRPA. To explore this possibility, antibodies against the 70- and 34-kDa subunits of hRPA were tested for reactivity with the SS1-FII-complementing activity. Indeed, the 70- and 34-kDa polypeptides in the MonoQ fraction were recognized by the corresponding hRPA antibodies (data not shown). The same antibodies also reacted with the 70- and 34-kDa subunits in the complementing P-11 fraction. As shown in Fig. 3C, the peak of antibody reactivity corresponds to the peak of the SS1-FII-complementing activity in the fractions from the P-11 column. These results suggest that hRPA may be the component in fraction SS2 that is required for reconstitution of MMR in the presence of SS1-FII. This possibility was confirmed by substituting pure recombinant hRPA in the MMR assay. As shown in Fig. 3D, recombinant hRPA indeed substitutes for SS2, allowing the MMR reaction product to form (lane 4) and the 3.2-kb putative repair intermediate to disappear (lane 5).
SS1 nicks ssDNA.
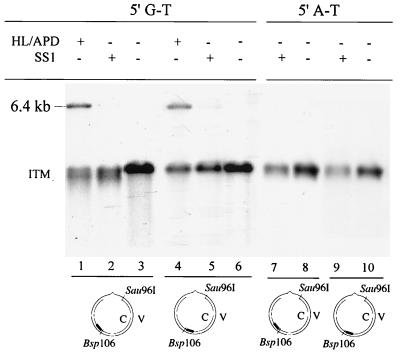
A novel 3.2-kb species was observed in reactions with fraction SS1, but the level of this species was dramatically reduced when SS2 was added to the reaction mixtures (Fig. 2B, lane 5). The identity of this species is unknown; however, the size of the fragment generated suggests that the plasmid (6.4 kb) is digested into two DNA fragments of equal size, 3.2 kb each. As all reaction products were digested with Bsp106 prior to electrophoresis, a double strand break must be generated 180° from the Bsp106 site. This position corresponds with the Sau96I site that was used to generate the nick in the complementary strand; therefore, to generate a double strand break, SSI must nick the viral strand at a position close to the nick on the complementary strand.
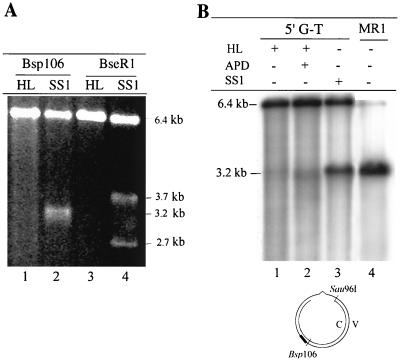
If this is the case, digestion of the 6.4-kb G-T circular DNA substrate with Bsp106 or BseRI will produce two fragments; otherwise, a single species will be observed. The 5′ G-T substrate was incubated with SS1 and digested with the restriction enzyme Bsp106. The product of this reaction was a ∼3.2-kb DNA fragment (Fig. 4A, lane 2), with an electrophoretic mobility identical to that of the product of Bsp106-HindIII double digestion of the MMR reaction product (Fig. 2B). In contrast, digestion with BseRI created a 3.7- and a 2.7-kb fragment (Fig. 4A, lane 4). These results suggest that a strand break is made on the viral strand at or near the Sau96I site when the DNA substrate is incubated in the presence of SS1.
FIG. 4.
SS1 carries out ssDNA incision. Repair reactions were performed as described in the legend to Fig. 2B. DNA products were recovered and digested with the indicated restriction enzyme. (A) Analysis of the repair product on a native agarose gel. DNA was cleaved with Bsp106 (lanes 1 and 2) or BseRI (lanes 3 and 4). (B) Analysis of the repair product on an alkaline agarose gel. DNA was digested with Bsp106 and analyzed by Southern blotting. The membrane was hybridized with the 32P-labeled oligonucleotide 5′-AACGTCACCAATGAAACCAT-3′ (probe V1 [solid bar]), which is complementary to the 3′ flanking sequence of Bsp106 on the viral strand. Lane 4 (MR1) contained double-stranded f1MR1 DNA digested with Bsp106 and Sau96I, which served as a marker to locate the single-stranded nick of the viral strand by SS1. APD, aphidicolin; HL, HeLa nuclear extract; MR1, flMR1 dsDNA.
Southern hybridization was performed to map SS1-induced nicks in the viral strand of the 5′ G-T substrate. After digestion with Bsp106, a full-length (6.4-kb) viral strand was detected in HeLa nuclear extracts without (Fig. 4B, lane 1) or with (Fig. 4B, lane 2) aphidicolin, a DNA polymerase inhibitor. However, after incubation with SS1 (Fig. 4B, lane 3), a defined fragment was detected that has the same length as the fragment derived from Sau96I-Bsp106 double digestion of f1MR1 dsDNA (compare lane 3 with lane 4), suggesting that the viral strand was nicked at or near the Sau96I site.
Fraction SS1 carries out nick-directed and mismatch-dependent excision.
A possible mechanism to create the nick of the viral strand is excision of the complementary strand to generate a nuclease-susceptible ssDNA region around the preexisting strand break. The excision activity must also then be present in SS1. To test this hypothesis, we performed experiments to determine if excision occurs in the SS1 reaction. In HeLa nuclear extracts, MMR excision intermediates contain a single-stranded region spanning the shorter patch between the nick and the mismatch in a circular substrate. The extent of the ssDNA region can be mapped relative to a restriction endonuclease cleavage site by Southern blot analysis (14, 18). Figure 5 shows such an analysis with the 5′ G-T substrate. As expected, hybridization with the complementary fragment C1 (from the Bsp106 site to the mismatch to the strand break [Fig. 1]) placed the untreated substrates at the Sau96I site (Fig. 5, lane 3, 3.2 kb). After incubation with HeLa nuclear extracts in the presence of aphidicolin, a group of smeared molecules approximately 200 bp shorter than the untreated substrate were detected (Fig. 5; compare lanes 1 and 3). These molecules are excision intermediates that accumulate when DNA synthesis is blocked (14, 18). When the DNA substrate was incubated with SS1, similar reaction products were observed (Fig. 5, lane 2), suggesting that SS1 can carry out excision along the short path between the strand break and the mismatch, thereby leading to a ssDNA region in the viral strand. Southern hybridization was also carried out on the same blot with complementary fragment C2 (from Bsp106 to BseRI to the strand break [Fig. 1]). There was no evidence of excision in this region of the substrate, which is the longer path from the nick to the mismatch (Fig. 5, lanes 4, 5, and 6). These results suggest that SS1 may be capable of nick-directed mismatch-provoked excision.
FIG. 5.
Mismatch-provoked and nick-directed excision by SS1. Reactions were performed by incubating a DNA heteroduplex (5′ G-T) or homoduplex (5′ A-T) with SS1 (30 μg) or HeLa nuclear extracts (HL, 50 μg) containing 100 nM aphidicolin (APD), as indicated. Products were cleaved with Bsp106, electrophoresed on alkaline agarose gels (1.5%), and transferred to nylon membranes as described in Materials and Methods. The membrane was hybridized with probe C1 (5′-32P-ATGGTTTCATTGGTGACGTT-3′) (lanes 1, 2, 3, 7, and 8) or probe C2 (5′-32P-GATTCTGTCGCTACTGATTAC-3′) (lanes 4, 5, 6, 9, and 10), which are complementary to the 5′ and 3′ flanking sequences of the Bsp106 site in the complementary strand, respectively. HL, HeLa nuclear extract; ITM, excision intermediates; solid bars, 32P-labeled probes.
To further determine if excision in SS1 is dependent on the mismatch, a Sau96I-nicked homoduplex (5′ A-T) was subjected to Southern hybridization analysis. As shown in Fig. 5, lanes 7 and 9, little (if any) excision was detected on either side of the strand break. Therefore, we conclude that SS1-mediated excision is conducted in a manner dependent on both a preexisting strand break and a mismatch.
hRPA protects the template strand from incision by nucleases.
While the excision reaction is required for MMR, the viral strand nicking activity is detrimental to the process, and it is likely that one component of the MMR pathway inhibits this reaction. As mentioned above, addition of SS2 to SS1 blocks the production of the 3.2-kb DNA fragment by SS1, suggesting that SS2 may possess this inhibition function. We therefore assessed hRPA for the ability to prevent SS1 from nicking DNA substrates. As shown in Fig. 6A, the amount of nicked viral strand decreased as the concentration of hRPA increased. One microgram of hRPA is sufficient to prevent SS1 (30 μg) from making strand breaks in the ssDNA region during the MMR reaction (Fig. 6A, lane 4). Interestingly, E. coli SSB binds ssDNA and is functionally similar to hRPA, but SSB does not substitute for hRPA in the reaction with human MMR proteins: as much as 8 μg of SSB failed to protect the continuous strand of the substrate from nicking in the presence of SS1 (Fig. 6A, lane 8). These results suggest that hRPA interacts specifically with human MMR proteins.
FIG. 6.
Role of hRPA in MMR. The DNA substrate was incubated with SS1 in the presence or absence of hRPA or SSB, as indicated, and products were analyzed by Southern blot hybridization. The membrane was probed with 32P-labeled V1 (5′-AACGTCACCAATGAAACCAT-3′) (A) or 32P-labeled C1 (5′-ATGGTTTCATTGGTGACGTT-3′) (B). APD, aphidicolin; HL, HeLa nuclear extract; Blank, reaction containing no protein; solid bars, 32P-labeled probes.
hRPA enhances mismatch-provoked excision.
The impact of hRPA on excision and repair of the nicked mismatch-containing DNA strand was assessed by using the Southern blots described above and probe C1. As shown in Fig. 6B, lanes 3 to 5, probe C1 detected increasingly shorter DNA fragments in reactions carried out with SS1 and increasing concentrations of hRPA; this suggests that hRPA may stimulate MMR-associated excision in the mismatch-containing DNA strand. The increase in the shorter DNA fragments was also observed when a lower concentration of E. coli SSB was incubated with SS1 (Fig. 6B, lanes 6 and 7), indicating that SSB at a low concentration can also stimulate MMR-associated excision. However, this stimulation was inhibited in the presence of a high concentration of SSB, as evidenced by the fact that intermediates in reactions containing 8 μg of SSB are almost identical to those in the original substrate (Fig. 6B; compare lanes 8 and 9). Inhibition of MMR-associated excision was not detected when 8 μg of hRPA was added to SS1 (data not shown). These results again suggest that a specific interaction between hRPA and other MMR proteins is required for the human MMR reaction.
FII carries out gap filling and ligation.
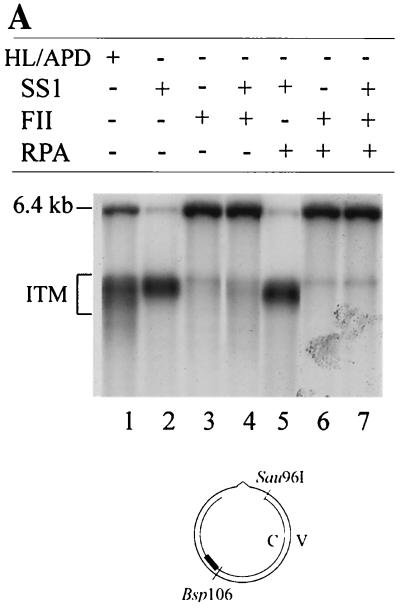
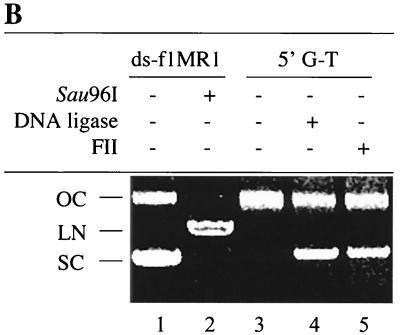
The above results indicate that SS1 carries out mismatch-provoked excision on the nick-containing DNA strand of heteroduplex DNA and that fraction SS2 (or hRPA) protects the template DNA strand from degradation. When the MMR reaction is carried out in the presence of SS1 and SS2 and in the absence of FII, the reaction is blocked at the DNA synthesis step (Fig. 4). Therefore, it seems possible that FII might perform gap-filling DNA synthesis and/or DNA ligation. To test this hypothesis, repair intermediates were characterized in reactions with or without FII. As shown in Fig. 7A, the nicked complementary strand was a 6.4-kb species in reactions containing FII (lane 3), SS1-FII (lane 4), hRPA-FII (lane 6), and SS1-hRPA-FII (lane 7). However, reaction products in all reactions, except in that containing SS1, hRPA, and FII, still contained a mismatch, indicating that repair had not occurred (see Fig. 2B). Therefore, the 6.4-kb species is likely to result from sealing of the strand break by a DNA ligase in FII. This is consistent with the observation that SS1 is very active in repair excision (Fig. 5, lane 2, and Fig. 7A, lane 2), but FII inhibits this excision activity (Fig. 7A, lane 4), possibly because it removes the DNA substrate by ligating strand breaks that are required for in vitro MMR (20, 57). These results strongly suggest that fraction FII contains a ligase activity required for in vitro MMR. Our ligase activity assay confirmed that this was indeed the case. Like E. coli DNA ligase, FII could convert a nicked circular DNA substrate into a supercoiled molecule in the presence of ethidium bromide (Fig. 7B).
FIG. 7.
FII contains gap-filling and ligase activities required for MMR. (A) FII carries out gap-filling and ligase reactions. The DNA substrate was incubated with SS1, hRPA, and FII as indicated, and products were analyzed by Southern hybridization using probe C1 (5′-32P-ATGGTTTCATTGGTGACGTT-3′). (B) Conversion of nicked circular DNA into supercoiled DNA by FII. The 5′ G-T substrate was incubated with E. coli DNA ligase (lane 4) at a final concentration of 4 U/μg of DNA or with 30 μg of FII (lane 5) at 20°C for 2 h in the presence of ethidium bromide (0.29 nM/μg of DNA). Double-stranded f1MR1 replicative-form DNA with (lane 2) or without (lane 1) Sau96I digestion was used as a reference for linear (LN), supercoiled (SC), and open circular (OC) DNA, as indicated. (C) DNA polymerase activity in FII. FII was assayed for DNA polymerase activity as described in Materials and Methods. Reaction mixtures contained 7.5 (lanes 1 and 4), 15 (lanes 2 and 5), or 30 (lanes 3 and 6) μg of protein and were incubated for 15 min at 37°C. To test for inhibition by monoclonal antibody SJK132-20 (MoAb), reaction mixtures were incubated with 1 μg of antibody for 15 min on ice prior to initiation of the polymerase reactions (lanes 4 to 6). (D) The gap-filling activity of FII requires hRPA. The DNA heteroduplex was incubated with SS1-hRPA, and the repair intermediates were purified by phenol extraction and ethanol precipitation. Intermediates were incubated with FII in the presence or absence of hRPA or SS1. Products were either analyzed by Southern blotting using the 32P-labeled probe C1 (5′-ATGGTTTCATTGGTGACGTT-3′) after cleavage by Bsp106 (lanes 1 to 5) or digested by Bsp106 and HindIII, followed by agarose gel electrophoresis as described in the legend to Fig. 2B to determine if the mispaired base is removed during the gap-filling reaction (lanes 6 to 8). APD, aphidicolin; ITM, excision intermediates; solid bar, 32P-labeled probe.
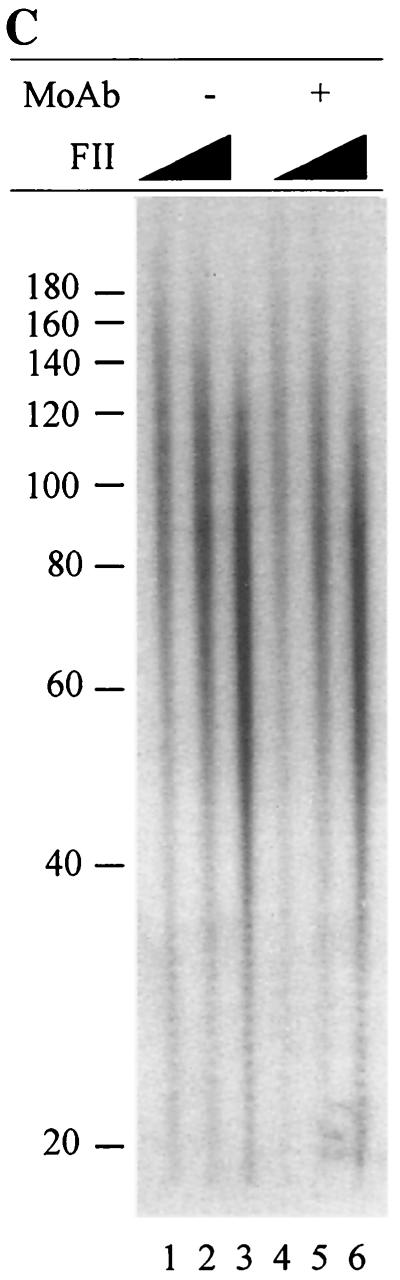
FII was assayed for DNA-dependent DNA polymerase activity by using a primer extension assay. The reactions monitored the extension of an oligo(dT)16 primer annealed to a poly(dA)1,500 template using an [α-32P]TTP substrate. Increasing concentrations of FII were incubated with the DNA substrate, and incorporation of [α-32P]TMP into high-molecular-weight products was determined. Reaction products were separated by denaturing PAGE and detected by autoradiography. Results are presented in Fig. 7C; the products observed were 50 to 100 bases in length and increased in intensity with increasing concentrations of FII. These results demonstrate that FII contains a DNA-dependent DNA polymerase activity. Preincubation of FII with the DNA polymerase α-specific inhibitory monoclonal antibody (Fig. 7C, lanes 4 to 6) SJK 132-20 revealed minimal inhibition, suggesting that the polymerase responsible for the extension observed is either DNA polymerase δ or DNA polymerase ɛ. However, it should be noted that the conditions employed for the assay were optimized for DNA polymerase δ (56) and that the FII fraction also likely contains DNA polymerase α.
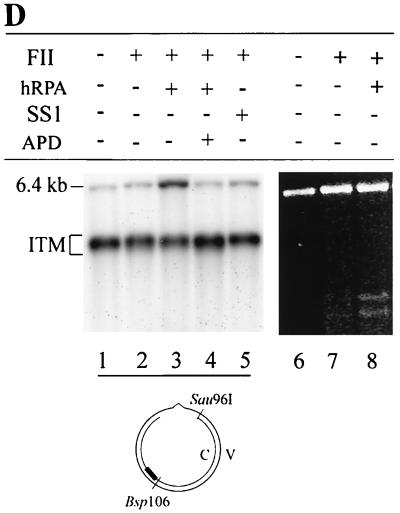
The gap-filling activity of FII is dependent on hRPA.
It is not clear whether the gap-filling step of the in vitro MMR reaction is carried out by FII alone or by FII with another component(s) in SS1 and/or SS2 (hRPA). Previous studies indicate that ssDNA binding activity is essential for DNA repair synthesis during mammalian nucleotide excision repair (53). Thus, it seems possible that FII-mediated gap filling requires hRPA. To address this issue, the excision and repair synthesis steps of the human MMR reaction were carried out sequentially. The 5′ G-T substrate was incubated with SS1-hRPA, and the repair intermediates from this reaction were purified and used as a substrate for a gap-filling reaction in the presence or absence of FII and SS1 or hRPA. The reaction products were analyzed by Southern blotting. As shown in Fig. 7D, reactions containing FII alone (lane 2) or FII with SS1 (lane 5) had about the same amount of full-length dsDNA (6.4 kb) as the control reaction (lane 1), suggesting that gapped intermediates recovered from the SS1-hRPA reaction are not converted into full-length dsDNA in these reactions. However, when the reaction was carried out in the presence of FII and hRPA (lane 3), more than 30% of the gapped intermediates were converted to full-length molecules, indicating that gap filling and ligation had occurred. In the presence of aphidicolin, no full-length ligated products were formed (Fig. 7D, lane 4), suggesting that DNA synthesis is required to form this product. Interestingly, when E. coli SSB was substituted for hRPA in the gap-filling reaction, approximately 30% of the gapped substrates were converted to the 6.4-kb product in the presence of FII and E. coli SSB (data not shown). This result suggests that hRPA may bind and protect the ssDNA region during repair synthesis associated with MMR. To determine if the removal of the mispaired base is associated with hRPA-FII-mediated gap filling, DNA products were subjected to digestion by restriction enzymes HindIII (scoring enzyme) and Bsp106 (see Fig. 1 for assay details). As shown in Fig. 7D, the expected repair products (3.1- and 3.3-kb fragments) were observed in reactions containing FII and hRPA (lane 8), but minimal levels (if any) were observed in reactions containing FII only (lane 7). It was noted that the repair rate (16%) in the partial reaction (gapped substrate, FII, and hRPA) was lower than that (39%) in a normal reaction (nicked substrate, SS1, SS2, and FII [see Fig. 2B, lane 8]). On the other hand, a similar lower rate (22%) was also observed when whole HeLa cell nuclear extracts were tested for processing of the gapped DNA substrate (data not shown) (compare the repair rate of 22% in this gap-filling reaction with that of 72% in a normal reaction [Fig. 2B, lane 1]). Therefore, the DNA substrate used in the gap-filling reaction seems to be a limiting factor. Nevertheless, these observations clearly indicate that the hRPA-dependent gap filling by FII employed the continuous strand as a template for repair resynthesis and that the mispaired base in the nicked strand was removed during the process.
DISCUSSION
Identifying components of the human MMR reaction.
Human MMR is a complex process that involves many proteins and protein complexes. hMutS and hMutL are essential MMR components which have been identified and characterized in human cells, but other components have not been well characterized. In this study, a HeLa nuclear extract was separated into three fractions, SS1, SS2 and FII, each of which is required in order to reconstitute MMR in vitro (see Fig. 2B). When MMR reactions were carried out in the presence of one or two of these three fractions, it was evident that the human MMR reaction proceeds in two stages: mismatch-provoked excision and DNA repair synthesis. This marks the first time that the human MMR reaction has been dissected and reconstituted in vitro.
SS1 is capable of initiating strand-specific MMR in a manner dependent on a mismatch and a strand break, as evidenced by the fact that the repair excision intermediates generated by SS1 are almost identical to those produced in HeLa nuclear extracts containing aphidicolin (Fig. 6A, lanes 1 and 2). In addition, SS1-mediated excision was not detected when a homoduplex was used in the reaction (Fig. 5, lanes 7 and 9). Previous studies have shown that defects in human MutS and MutL homologs and PCNA abolish mismatch-provoked excision (9, 18, 23, 46, 61). In E. coli, a helicase and at least four exonucleases are involved in MMR-associated excision (8, 29, 63), and human MMR may be mechanistically similar. Therefore, it is conceivable that SS1 from HeLa cells includes hMutS, hMutL, PCNA, helicase, and nuclease activities. Western blot analysis indeed revealed that MSH2, MLH1, and PCNA are present in significant amounts in SS1. Although a helicase(s) and an exonucleases(s) that participate in human MMR have not yet been identified or characterized, ExoI, which possesses a 5′-to-3′ exonuclease activity, has been implicated in MMR (2, 51, 54, 58, 59). It may be possible to purify an MMR-associated helicase(s) and exonucleases, including ExoI if it is required for MMR, by additional fractionation of SS1. Biochemical analyses of these protein fractions will improve understanding of the mechanism of MMR in human cells.
Although SS1 carries out nick-directed mismatch-provoked excision along the shorter path between the strand break and the mismatch with the 5′ G-T substrate, SS1 does not carry out this reaction with the 3′ /TG\ substrate (data not shown). This is presumably because SS1 cannot carry out excision in the 3′-to-5′ direction from the nick to the mismatch. Previous studies indicate that HeLa cells possess a bidirectional mechanism to remove a mismatch (14). Depending on the location of a strand break relative to the mismatch, MMR-associated excision occurs in a 5′-to-3′ or a 3′-to-5′ direction. Because MMR reconstituted in vitro in the presence of SS1-hRPA-FII corrects the 3′/TG substrate in a strand-specific manner (data not shown), it is likely that 3′-to-5′ excision activity is present in SS2 and/or FII. Further studies are required in order to determine which of these two fractions contains 3′-to-5′ excision activity.
The second stage of human MMR in this reconstituted assay system is DNA repair synthesis. During this stage of the reaction, SS2 and FII convert gapped intermediates into closed dsDNA molecules. We have shown that hRPA can substitute for SS2. The components of FII that play a role in MMR are not yet clear, but they apparently include DNA polymerase and ligase activities required for MMR. FII does not carry out strand-specific MMR (Fig. 2), but it does generate a 6.4-kb full-length product that is resistant to restriction enzyme cleavage. This product is probably generated by a very active ligase in FII that ligates the nick in the DNA substrate. This possibility is consistent with the fact that FII inhibits SS1-mediated nick-directed excision on heteroduplex DNA, presumably by ligating the nick before excision takes place. Indeed, our in vitro ligase assay shows that FII is capable of sealing single-stranded breaks and converting a nicked circular DNA substrate into supercoiled DNA (Fig. 7B). We also show that FII contains a DNA polymerase activity. For example, FII and hRPA convert gapped MMR reaction intermediates generated by SS1-hRPA into full-length dsDNA molecules (Fig. 7D) (see discussion below), and this conversion is inhibited by aphidicolin, indicating involvement of DNA polymerase α, δ, or ɛ in the gap-filling reaction. DNA polymerase activity is indeed identified in FII by using a traditional in vitro DNA replication assay system (Fig. 7C). Even though our data cannot rule out the involvement of polymerase α in MMR, the gap-filling activity detected in FII is not conducted by polymerase α, as evidenced by the fact that an antibody specific for DNA polymerase α has essentially no effect on the DNA polymerization reaction in FII (Fig. 7C). Thus, it is highly likely that DNA polymerase δ (37) and/or ɛ is responsible for the gap-filling activity in FII. Western blot analyses revealed that FII also contains abundant MSH2, MLH1, and PCNA. However, unlike SS1, FII is unable to conduct repair excision, suggesting that components required for excision, e.g., exonucleases, are not present in FII.
Role of hRPA in MMR.
hRPA is a human ssDNA binding protein that is essential for cellular DNA metabolism (62) involving ssDNA intermediates including DNA replication (13, 64, 65), homologous recombination (36), base excision repair (45), and nucleotide excision repair (11). Recently, Lin et al. have demonstrated that incubation of an anti-hRPA antibody with HeLa extracts inhibits strand-specific MMR in vitro, suggesting that hRPA is also involved in MMR (34). However, the actual role of hRPA in MMR is not known. Using an in vitro reconstitution system, we demonstrate here that recombinant hRPA can substitute for one of the required HeLa nuclear fractions in MMR. Analysis of critical intermediates reveals that hRPA plays multiple roles in human MMR in vitro.
First, hRPA protects single-stranded regions of the template DNA strand from nuclease degradation. While SS1 is capable of mismatch-provoked excision in the nicked strand, it also makes a single-stranded break in the continuous strand that serves as a template for repair synthesis. Since such a strand break in the template strand has never been observed in the whole-cell nuclear extract, a DNA binding activity required for MMR must be responsible for protecting the single-stranded region in the template strand. We found that this activity is hRPA, as evidenced by the fact that addition of hRPA to the SS1 reaction mixture blocks the single-stranded nicking in the continuous DNA strand, suggesting that hRPA may bind to the single-stranded region, protecting it from attack by nucleases and ensuring that the template DNA strand is suitable for repair DNA synthesis.
Second, hRPA may also directly contribute to mismatch-provoked excision, because hRPA enhances repair excision by SS1 (Fig. 6B). How might hRPA participate in repair excision? It is known that in E. coli, MMR-associated excision requires DNA helicase II to unwind the DNA helix (29), and a similar activity should also be required for the human reaction. Possibly, hRPA facilitates DNA unwinding by a DNA helicase activity that participates in MMR. Recently, it was shown that yeast RPA stimulates MER3 helicase during meiotic crossing over (42). hRPA plays a similar role in simian virus 40 (SV40) DNA replication, during which hRPA and T antigen interact physically and unwind the SV40 origin of replication (5, 24, 66). A DNA helicase activity that is stimulated by hRPA has been purified from human cells (52), and it is possible that this helicase could play a role in human MMR.
Finally, hRPA plays a crucial role in repair DNA synthesis during human MMR. FII is required to convert gapped intermediates into complete circular DNA, and this conversion also requires hRPA (Fig. 7D). However, the mechanism by which hRPA stimulates repair synthesis during human MMR is not exactly clear. Previous studies have demonstrated that hRPA stimulates the activity of DNA polymerases α, δ, and ɛ (24, 25, 30, 60), all of which are sensitive to aphidicolin and may participate in MMR-associated DNA repair synthesis. In fact, evidence suggests that DNA polymerase δ is involved in human MMR (37). Therefore, it is possible that hRPA may stimulate repair synthesis during MMR in addition to protecting the DNA template from degradation by nucleases. Recently, Ranalli et al. (49) have demonstrated that hRPA stimulates a ligase activity of DNA ligase I, which is probably required for MMR.
In summary, we have established an in vitro system that enables one to dissect the complex human MMR reaction into multiple stages. Analysis of individual stages of the reaction has revealed crucial repair intermediates, which help to identify the requirement for hRPA in MMR and define the roles of hRPA in the pathway. We believe that further analysis of the three fractions described here, especially fractions SS1 and FII, will identify additional components required for human MMR and will eventually elucidate the molecular mechanisms of the human pathway.
Acknowledgments
We thank Peggy Hsieh (National Institute of Diabetes and Digestive and Kidney Diseases) for comments on the manuscript, Zhengxiang Pan (Mount Sinai School of Medicine) for hRPA antibodies, Jianxin Wu and Scott McCulloch for help in phosphocellulose chromatography, and Lu Qiu for nuclear extract preparations.
This work was supported in part by grants CA82604 and CA85377 (to G.-M.L.) and CA82741 (to J.J.T.) from the National Cancer Institute.
REFERENCES
- 1.Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin, V. N. Podust, M. Protic, U. Hubscher, J. M. Egly, and R. D. Wood. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859-868. [DOI] [PubMed] [Google Scholar]
- 2.Amin, N. S., M. N. Nguyen, S. Oh, and R. D. Kolodner. 2001. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21:5142-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowers, J., P. T. Tran, A. Joshi, R. M. Liskay, and E. Alani. 2001. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol. 306:957-968. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brill, S. J., and B. Stillman. 1989. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature 342:92-95. [DOI] [PubMed] [Google Scholar]
- 6.Brush, G., and T. Kelly. 1996. Mechanisms for replicating DNA, p. 1-43. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Buermeyer, A. B., S. M. Deschenes, S. M. Baker, and R. M. Liskay. 1999. Mammalian DNA mismatch repair. Annu. Rev. Genet. 33:533-564. [DOI] [PubMed] [Google Scholar]
- 8.Burdett, V., C. Baitinger, M. Viswanathan, S. T. Lovett, and P. Modrich. 2001. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. USA 98:6765-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C., B. J. Merrill, P. J. Lau, C. Holm, and R. D. Kolodner. 1999. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 19:7801-7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, A. B., F. Valle, K. Drotschmann, R. K. Gary, and T. A. Kunkel. 2000. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 275:36498-36501. [DOI] [PubMed] [Google Scholar]
- 11.Coverley, D., M. K. Kenny, M. Munn, W. D. Rupp, D. P. Lane, and R. D. Wood. 1991. Requirement for the replication protein SSB in human DNA excision repair. Nature 349:538-541. [DOI] [PubMed] [Google Scholar]
- 12.Drummond, J. T., G. M. Li, M. J. Longley, and P. Modrich. 1995. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268:1909-1912. [DOI] [PubMed] [Google Scholar]
- 13.Fairman, M. P., and B. Stillman. 1988. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 7:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang, W. H., and P. Modrich. 1993. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J. Biol. Chem. 268:11838-11844. [PubMed] [Google Scholar]
- 15.Flores-Rozas, H., D. Clark, and R. D. Kolodner. 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 26:375-378. [DOI] [PubMed] [Google Scholar]
- 16.Flores-Rozas, H., and R. D. Kolodner. 1998. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 95:12404-12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genschel, J., S. J. Littman, J. T. Drummond, and P. Modrich. 1998. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 273:19895-19901. [DOI] [PubMed] [Google Scholar]
- 18.Gu, L., Y. Hong, S. McCulloch, H. Watanabe, and G.-M. Li. 1998. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 26:1173-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 20.Holmes, J., S. Clark, and P. Modrich. 1990. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl. Acad. Sci. USA 87:5837-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh, P. 2001. Molecular mechanisms of DNA mismatch repair. Mutat. Res. 486:71-87. [DOI] [PubMed] [Google Scholar]
- 22.Jiricny, J. 1998. Replication errors: cha(lle)nging the genome. EMBO J. 17:6427-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kat, A., W. G. Thilly, W. H. Fang, M. J. Longley, G. M. Li, and P. Modrich. 1993. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. USA 90:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny, M. K., S. H. Lee, and J. Hurwitz. 1989. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc. Natl. Acad. Sci. USA 86:9757-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenny, M. K., U. Schlegel, H. Furneaux, and J. Hurwitz. 1990. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 265:7693-7700. [PubMed] [Google Scholar]
- 26.Kleczkowska, H. E., G. Marra, T. Lettieri, and J. Jiricny. 2001. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15:724-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klungland, A., and T. Lindahl. 1997. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16:3341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 29.Lahue, R. S., K. G. Au, and P. Modrich. 1989. DNA mismatch correction in a defined system. Science 245:160-164. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. H., Z. Q. Pan, A. D. Kwong, P. M. Burgers, and J. Hurwitz. 1991. Synthesis of DNA by DNA polymerase epsilon in vitro. J. Biol. Chem. 266:22707-22717. [PubMed] [Google Scholar]
- 31.Leung, W. K., J. J. Kim, L. Wu, J. L. Sepulveda, and A. R. Sepulveda. 2000. Identification of a second MutL DNA mismatch repair complex (hPMS1 and hMLH1) in human epithelial cells. J. Biol. Chem. 275:15728-15732. [DOI] [PubMed] [Google Scholar]
- 32.Li, G.-M. 1999. The role of mismatch repair in DNA damage-induced apoptosis. Oncol. Res. 11:393-400. [PubMed] [Google Scholar]
- 33.Li, G. M., and P. Modrich. 1995. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl. Acad. Sci. USA 92:1950-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Y. L., M. K. Shivji, C. Chen, R. Kolodner, R. D. Wood, and A. Dutta. 1998. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J. Biol. Chem. 273:1453-1461. [DOI] [PubMed] [Google Scholar]
- 35.Lipkin, S. M., V. Wang, R. Jacoby, S. Banerjee-Basu, A. D. Baxevanis, H. T. Lynch, R. M. Elliott, and F. S. Collins. 2000. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat. Genet. 24:27-35. [DOI] [PubMed] [Google Scholar]
- 36.Longhese, M. P., P. Plevani, and G. Lucchini. 1994. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol. Cell. Biol. 14:7884-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longley, M. J., A. J. Pierce, and P. Modrich. 1997. DNA polymerase δ is required for human mismatch repair in vitro. J. Biol. Chem. 272:10917-10921. [DOI] [PubMed] [Google Scholar]
- 38.Meyer, T. F., K. Geider, C. Kurz, and H. Schaller. 1979. Cleavage site of bacterophage fd gene II-protein in the origin of viral strand replication. Nature 278:365-367. [DOI] [PubMed] [Google Scholar]
- 39.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 40.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa, T., A. Datta, and R. D. Kolodner. 1999. Multiple functions of MutS- and MutL-related heterocomplexes. Proc. Natl. Acad. Sci. USA 96:14186-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa, T., H. Flores-Rozas, and R. D. Kolodner. 2001. The MER3 helicase involved in meiotic crossing over is stimulated by single-stranded DNA-binding proteins and unwinds DNA in the 3′ to 5′ direction. J. Biol. Chem. 276:31487-31493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palombo, F., P. Gallinari, I. Iaccarino, T. Lettieri, M. Hughes, A. D'Arrigo, O. Truong, J. J. Hsuan, and J. Jiricny. 1995. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science 268:1912-1914. [DOI] [PubMed] [Google Scholar]
- 44.Palombo, F., I. Iaccarino, E. Nakajima, M. Ikejima, T. Shimada, and J. Jiricny. 1996. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6:1181-1184. [DOI] [PubMed] [Google Scholar]
- 45.Parker, A., Y. Gu, W. Mahoney, S. H. Lee, K. K. Singh, and A. L. Lu. 2001. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J. Biol. Chem. 276:5547-5555. [DOI] [PubMed] [Google Scholar]
- 46.Parsons, R., G. M. Li, M. J. Longley, W. H. Fang, N. Papadopoulos, J. Jen, A. de la Chapelle, K. W. Kinzler, B. Vogelstein, and P. Modrich. 1993. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75:1227-1236. [DOI] [PubMed] [Google Scholar]
- 47.Patrick, S. M., and J. J. Turchi. 1998. Human replication protein A preferentially binds cisplatin-damaged duplex DNA in vitro. Biochemistry 37:8808-8815. [DOI] [PubMed] [Google Scholar]
- 48.Patrick, S. M., and J. J. Turchi. 1999. Replication protein A (RPA) binding to duplex cisplatin-damaged DNA is mediated through the generation of single-stranded DNA. J. Biol. Chem. 274:14972-14978. [DOI] [PubMed] [Google Scholar]
- 49.Ranalli, T. A., M. S. DeMott, and R. A. Bambara. 2002. Mechanism underlying replication protein A stimulation of DNA ligase I. J. Biol. Chem. 277:1719-1727. [DOI] [PubMed] [Google Scholar]
- 50.Raschle, M., G. Marra, M. Nystrom-Lahti, P. Schar, and J. Jiricny. 1999. Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J. Biol. Chem. 274:32368-32375. [DOI] [PubMed] [Google Scholar]
- 51.Schmutte, C., M. M. Sadoff, K. S. Shim, S. Acharya, and R. Fishel. 2001. The interaction of DNA mismatch repair proteins with human exonuclease I. J. Biol. Chem. 276:33011-33018. [DOI] [PubMed] [Google Scholar]
- 52.Seo, Y. S., S. H. Lee, and J. Hurwitz. 1991. Isolation of a DNA helicase from HeLa cells requiring the multisubunit human single-stranded DNA-binding protein for activity. J. Biol. Chem. 266:13161-13170. [PubMed] [Google Scholar]
- 53.Shivji, M. K., V. N. Podust, U. Hubscher, and R. D. Wood. 1995. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 34:5011-5017. [DOI] [PubMed] [Google Scholar]
- 54.Sokolsky, T., and E. Alani. 2000. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics 155:589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su, S.-S., R. S. Lahue, K. G. Au, and P. Modrich. 1988. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J. Biol. Chem. 263:6829-6835. [PubMed] [Google Scholar]
- 56.Tan, C. K., C. Castillo, A. G. So, and K. M. Downey. 1986. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J. Biol. Chem. 261:12310-12316. [PubMed] [Google Scholar]
- 57.Thomas, D. C., J. D. Roberts, and T. A. Kunkel. 1991. Heteroduplex repair in extracts of human HeLa cells. J. Biol. Chem. 266:3744-3751. [PubMed] [Google Scholar]
- 58.Tishkoff, D. X., N. S. Amin, C. S. Viars, K. C. Arden, and R. D. Kolodner. 1998. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 58:5027-5031. [PubMed] [Google Scholar]
- 59.Tran, P. T., J. A. Simon, and R. M. Liskay. 2001. Interactions of Exo1p with components of MutLα in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:9760-9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsurimoto, T., and B. Stillman. 1989. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 8:3883-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umar, A., A. B. Buermeyer, J. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65-73. [DOI] [PubMed] [Google Scholar]
- 62.Umezu, K., N. Sugawara, C. Chen, J. E. Haber, and R. D. Kolodner. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148:989-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viswanathan, M., V. Burdett, C. Baitinger, P. Modrich, and S. T. Lovett. 2001. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 276:31053-31058. [DOI] [PubMed] [Google Scholar]
- 64.Wobbe, C. R., L. Weissbach, J. A. Borowiec, F. B. Dean, Y. Murakami, P. Bullock, and J. Hurwitz. 1987. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl. Acad. Sci. USA 84:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wold, M. S., and T. Kelly. 1988. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 85:2523-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wold, M. S., J. J. Li, and T. J. Kelly. 1987. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc. Natl. Acad. Sci. USA 84:3643-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie, Y., C. Counter, and E. Alani. 1999. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn 3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics 151:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]