Abstract
In the budding yeast Saccharomyces cerevisiae, entry into meiosis and its successful completion depend on two positive regulators, Ime1 and Ime2. Ime1 is a transcriptional activator that is required for transcription of IME2, a serine/threonine protein kinase. We show that in vivo Ime2 associates with Ime1, that in vitro Ime2 phosphorylates Ime1, and that in living cells the stability of Ime1 depends on Ime2. Diploid cells with IME2 deleted show an increase in the level of Ime1, whereas haploid cells overexpressing IME2 show a decrease in the stability of Ime1. Furthermore, the level of Ime1 depends on the kinase activity of Ime2. Using a mutation in one of the ATPase subunits of the proteasome, RPT2, we demonstrate that Ime1, amino acids 270 to 360, is degraded by the 26S proteasome. We also show that Ime2 itself is an extremely unstable protein whose expression in vegetative cultures is toxic. We propose that a negative-feedback loop ensures that the activity of Ime1 will be restricted to a narrow window.
Successful progression and completion of the mitotic cell cycle depends on transcriptional and proteolytic regulation. These two processes determine the availability of cyclins and cyclin-dependent kinase (CDK) inhibitors that govern the sequential activation of CDKs (18, 28, 31, 35). Initiation and progression through the meiotic cycle should also be subjected to transcriptional and proteolytic regulation. Indeed, in budding yeast a transcriptional cascade governs initiation and progression through the meiotic cell cycle (4). Yet there is no direct evidence concerning proteolysis of either positive or negative meiotic regulators. This report focuses on the regulated degradation of one of the two positive regulators of meiosis in Saccharomyces cerevisiae, Ime1, by the other, Ime2.
IME1 encodes a transcriptional activator (30, 47) that is necessary for the transcription of meiosis-specific genes (48). Ime1 is tethered to promoters of early meiosis-specific genes, such as IME2, by a specific DNA-binding protein, Ume6 (39). Diploid cells with deletions of IME1 arrest at G1 prior to the initiation of premeiotic DNA replication (22). The transcription of IME1 is regulated by nutrients. In vegetative cultures with glucose as the sole carbon source, IME1 is silent, but in the presence of acetate, low levels of IME1 mRNA are observed (22). Under meiotic conditions, i.e., nitrogen depletion and the presence of a nonfermentable carbon source such as acetate, transcription of IME1 is induced transiently in MATa/MATα diploids (22). It is not known whether this transient transcription reflects transient availability of the Ime1 protein. In addition, the IME1 promoter is subject to positive autoregulation (40, 43, 44), as well as negative-feedback regulation by both Ime1 and Ime2 (43, 48, 49).
Another important regulator of meiosis and sporulation is the serine/threonine protein kinase Ime2 (12, 24, 34, 48, 49). Diploid cells with deletions of IME2 show a 5- to 12-h delay in the transcription of early meiosis-specific genes, a reduction in their level of expression, and extremely low RNA levels of middle and late genes (34, 49). These mutant cells show a delay in the initiation of premeiotic DNA synthesis and the commitment to meiotic recombination, as well as a delay in nuclear division (12). Overexpression of Ime2 bypasses the requirement of IME1 for transcription of meiosis-specific genes (34), supporting the idea that Ime2 functions as a positive regulator. That Ime2 is a negative regulator is evident from the observations that (i) in diploid cells with deletions of IME2, transcription of IME1 and early meiosis-specific genes is nontransient in comparison to that in wild-type cells (34, 43, 49) and (ii) levels of Pol1, Pol12, and Sic1 proteins are reduced in ime2 diploid cells (7, 12), and in the cases of Pol1 and Pol12 protein availability is also nontransient (12). Ime2 is also required to restrict DNA replication and nuclear division. In the absence of Ime2, a second round of premeiotic DNA synthesis and three rounds of nuclear division are observed (12).
Phosphorylation plays a central role in regulating proteolysis (5). For example, phosphorylation of the CDK inhibitor Sic1 by the Cln2/Cdc28 complex leads to its degradation by the ubiquitin/proteasome pathway (28, 31). The predicted amino acid sequence of Ime2 shows that Ime2 is related to human CDK2 (38.7% identity and 58.9% homology). This raises the possibility that, similarly to the function of Cdc28 in vegetative cultures, phosphorylation by Ime2 may regulate the stability of specific substrates in meiotic cultures. Indeed, the meiotic level of Sic1 is increased in cells with deletions of IME2 (7), suggesting that Ime2 is required for the degradation of Sic1 (7).
In this report we show that Ime2 interacts with and in vitro phosphorylates the C-terminal domain of Ime1, amino acids 270 to 360 [Ime1(270-360)]. We show that IME1 encodes an unstable protein, whose degradation by the proteasome depends on the kinase activity of Ime2.
MATERIALS AND METHODS
Plasmids.
Since in many cases multiple steps were involved in plasmid construction, here we describe only the structure of the plasmids. Details are available upon request. pGEX-4T-1 carries glutathione S-transferase (GST) expressed from the tac promoter (Promega). pGAD2F carries pADH1-gal4(768-881) on a pBR322 2μm LEU2 vector (3). pGBDU-C(1) carries pADH1-gal4(1-147) on a pBR322 2μm URA3 vector (21). pAS128 carries ime1-lacZ on a pBR322 2μm CEN6 URA3 vector (ime1 is from −4401 to +205 [43]). YEp53 carries IME1 from −621 to +2100 on a 2μm URA3 LEU2 vector. YEp1202 carries pADH1-gal(1-147)-IME1(6-360) on a pBR322 2μm HIS3 URA3 vector (30). YEp1406 carries pADH1-gal(768-881)-IME1(270-360) on a pBR322 2μm LEU2 vector (30). YIp1577 carries ime2::hisG-URA3-hisG on pGEM3Z (Promega). IME2 is disrupted at the codon encoding amino acid 369. YEp1906 carries the pGAL1-3HA chimera on a pBR322 2μm TRP1 vector. YEp1916 carries the pADH1-gal4(1-147)-HA-IME2(1-645) chimera on a BS 2μm TRP1 vector. YIp1930 carries ime2Δ::hisG-URA3-hisG on a pUC118 vector. ime2 has a deletion in the area encoding amino acids 36 to 439. YEp1931 carries pIME1(−1368-−31)-gal4(1-147)-ime1(270-360) on a pBR322 2μm TRP1 vector. YEp1952 carries pIME1(−1368-−31)-gal4(1-147) on a pBR322 2μm TRP1 vector. YEp1968 carries pIME1(−1368-−31)-gal4(1-147)-HA-IME2 on a pBR322 2μm LEU2 vector. YEp1969 carries pADH1-gal4(1-147)-ime1(270-360) on a pBR322 2μm LEU2 vector. YEp1974 carries pADH1-gal4(1-147)-ime1(270-360) on a pBR322 2μm URA3 vector. YEp1985 carries pGAL1-3HA-IME2(1-645) on a pBR322 2μm TRP1 vector. YIp2117 carries ime2(1-490) on a pGEM vector. YEp2126 carries pADH1-gal4(1-147)-HA-ime2(Δ478-499)-ADHt on a BS 2μm TRP1 CYH2 vector. YEp2142 carries pGAL1-3HA-ime2(Δ478-499) on a pBR322 2μm TRP1 vector. YEp2225 carries pADH1-gal4(1-147)-ime2K97A on a pBR322 2μm TRP1 vector. YEp2264 carries pGAL1-3HA-ime2K97A(1-645) on a pBR322 2μm TRP1 vector. YIp2353 carries pGAL1-3×HA-IME2 on a pBR322 LEU2 vector . P2677 carries ptac-GST -ime1(270-360).
Yeast strains.
The following yeast strains were used. Y153 is MATa URA3::pGAL1-lacZ LYS2::pGAL1-HIS3 his3-200 leu2-3,112 trp1-901 ade2-101 gal4Δ gal80Δ (17). Y742 is MATa ura3-52 leu2,3-112 trp1Δ his3Δ::hisG ade2-1 met GAL+. Y1009 is isogenic to Y742 but gal80::hisG. A one-step deletion protocol was used to replace the GAL80 allele in Y742 with a gal80::hisG-URA3-hisG fragment (from plasmid YIp1607 [39]). URA+ transformants were patched onto 5-fluoroorotic acid (5-FOA) plates to select for derivatives that had recombined out the URA3 gene. Y1071 is isogenic to Y1009 but ime2Δ36-439::hisG. A one-step deletion protocol was used to replace the IME2 allele in Y1009 with an ime2::hisG-URA3-hisG fragment (from plasmid YIp1930). URA+ transformants were patched onto 5-FOA plates to select for derivatives that had recombined out the URA3 gene. Y422 is MATa/MATα ura3-52/ura3-52 trp1Δ/trp1Δ leu2-3,112/leu2-3,112 ade2-1/ade2-R8 his4-519/HIS4 his6-1/HIS6 gal/GAL+ can1/CAN1. Y1039 is isogenic to Y422 but his3Δ::hisG/HIS3 HIS4/HIS4 gal80::hisG/gal80::hisG ime2::hisG-URA3-hisG/ime2::hisG-URA3-hisG. A one-step deletion protocol was used to replace the IME2 allele in the haploid parents with a DNA fragment (from plasmid YIp1577) that carries the ime2::hisG-URA3-hisG gene. Y1248, MATa ura3-52 leu2-3,112 trp1Δ::hisG his3Δ200 lys2-801, is a trp1Δ::hisG derivative of sub62 (38). Y1249 is isogenic to sub62 but rpt2Δ::HIS3 carrying rpt2RF on a CEN LEU2 plasmid. It is a trp1Δ::hisG derivative of DY62 (38). Y1255 is isogenic to Y742 but pGAL1::IME2(1-645). The pGAL1::IME2 chimera was integrated at the leu2-3,112 allele by transformation of Y742 with YIp2353 digested with PpuMI.
Media and genetic techniques.
PSP2 (minimal acetate medium) and SPM (sporulation medium), have been described previously (23). SD (synthetic dextrose) medium has been described previously (46). SG and SR are synthetic media with 2% galactose or raffinose, respectively. Meiosis in diploids was induced by growing cells in PSP2 supplemented with the required amino acids to 107 cells/ml, washing once with water, and resuspending in SPM. Meiosis in haploids was induced by growing cells in SD medium to stationary phase, washing once with water, and resuspending in SPM at a titer of 107 cells/ml. Yeast transformation with lithium acetate was carried out as described previously (13). Standard methods for DNA cloning and transformation were used (41). Site-directed mutagenesis was performed as described previously (26). Expression of genes from the inducible GAL1 promoter was accomplished by pregrowing cells with raffinose as the sole carbon source. For β-galactosidase assays, proteins were extracted from at least three independent transformants and assayed for β-galactosidase activity as described previously (32, 37). Results are given in Miller units.
Pulse-chase assay.
A total of 5 × 108 cells were concentrated in 400 μl of SD without methionine and cysteine, pulse-labeled for 4 to 7 min with EXPRE35S35S (NEN), pelleted, and chased in 3 to 4 ml of SD medium containing 200 mM cold methionine and cysteine. At various times of chasing, proteins were extracted, immunoprecipitated with antibodies directed against Gal4(1-147), and separated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels as described previously (25). Levels of radioactivity in bands were quantitated by using a Fujix Bas 1000 PhosphorImager. In addition, gels were exposed to X-ray film.
Antibodies.
Mouse monoclonal antibodies directed against Gal4(bd), amino acids 1 to 147, were purchased from Santa Cruz, and mouse monoclonal antibodies against the hemagglutinin (HA) epitope (12CA5) were purchased from Boehringer.
Preparation of yeast protein extracts and Western analysis.
Protein extracts were prepared from trichloroacetic acid-treated cells as described previously (10). The Western blotting procedure used was essentially that described previously (10, 11).
Purification of proteins from bacteria.
GST-Ime1(270-360) and Gst1 were expressed from the tac promoter by inducing bacterial BL21 cells carrying P2677 or pGEX-4T-1, respectively, with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Following 5 h of incubation at 30°C, proteins were isolated on glutathione-Sepharose beads (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Immunoprecipitation, immunoblot analysis, and kinase assay.
Immunoprecipitation, immunoblot analysis, and kinase assays were carried out essentially as described by Bowdish et al. (1). Cells grown in synthetic media with raffinose as the sole carbon source to a density of 107 cells/ml were harvested by centrifugation, resuspended in SG medium, and incubated at 30°C for 16 h. For immunoprecipitation, a 15-ml culture was pelleted, washed with ice-cold water, and frozen in a dry-ice-ethanol bath. Cell lysates were prepared by vortexing a cell suspension in an extraction buffer in the presence of glass beads (10 37-s bursts). Lysates were cleared by using two spins. All samples were normalized to contain the same amount of total protein (2). The lysate (500 μg) in immunoprecipitation buffer (IP buffer) was incubated with 2 μg of anti-HA antibodies for 2 h at 4°C with gentle shaking. Immune complexes were collected on protein A-Sepharose beads by gently shaking at 4°C for 1 h. Beads were pelleted by gentle centrifugation and washed twice with IP buffer, once with IP buffer minus ovalbumin, and once with kinase buffer. For the kinase assay, 2.5 μl of [γ-32P]ATP (6,000 Ci/mmol) (NEN) and 5 μg of an external substrate (if necessary) were added, and the reaction mixture was incubated at room temperature for 1 h. Reactions were terminated by addition of an equal volume of elution buffer (125 mM Tris [pH 6.8], 10% β-mercaptoethanol, 4% SDS, 20% glycerol, 25 mM EDTA, and 46% urea). Proteins were separated from the radioactive ATP on P-6 Micro-Bio-Spin chromatography columns (Bio-Rad). Samples were loaded onto two SDS-polyacrylamide gel electrophoresis (PAGE) gels following the addition of the sample buffer. One gel was dried and exposed to X-ray film. The second gel was processed for Western analysis by using anti-12CA5 antibodies (IP-Western).
RESULTS
Ime2 interacts with Ime1.
As described above, Ime1 activates the transcription of IME2, whereas Ime2 is required to shut off the transcription of IME1 and early meiosis-specific genes (34, 43, 48, 49). In addition, the transcription of IME1 is subject to positive autoregulation. These results suggest that Ime2 regulates the stability of the Ime1 protein. We used the two-hybrid method to determine the association between Ime1 and Ime2. We compared the levels of Gal1-LacZ in haploid cells expressing Gal4(bd)-Ime2 to those in cells also expressing Ime1 from a truncated promoter that allowed expression through a promoter present on the vector plasmid pBR322. Since Ime1 is a transcriptional activator (30, 47), there was no need to add a heterologous activation domain. Table 1 shows that Ime2, by itself, possesses a weak transcriptional activity that is increased fivefold under meiotic conditions (SPM medium). However, a substantial increase in expression (4- to 16-fold) was evident in the presence of Ime1, suggesting that Ime2 interacts with Ime1. In order to identify the domain in Ime1 that associates with Ime2, the level of expression of gal1-lacZ was determined in cells expressing both Gal4(bd)-Ime2 and Gal4(ad)-Ime1(270-360). Table 1 shows that the expression of gal1-lacZ was increased (6- to 12-fold) under all growth conditions. These results suggest that Ime2 interacts with Ime1 and that this association takes place through the interaction domain of Ime1, amino acids 270 to 360 [Ime1(id)].
TABLE 1.
Two-hybrid assay between Ime1 and Ime2
Ime2 phosphorylates Ime1 in vitro.
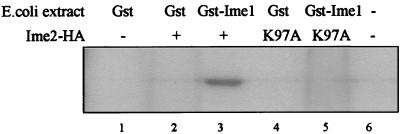
The interaction between Ime1 and Ime2 suggests that Ime2 might phosphorylate Ime1. In yeast cells, HA-tagged Ime1(id) is physically associated with another kinase, as deduced from the observation that immunoprecipitation and an in vitro kinase assay lead to Ime2-independent phosphorylation (data not shown). We think that this phosphorylation might depend on Rim11, a kinase that associates with and phosphorylates Ime1 in vitro (1, 39). In order to circumvent this problem, Ime1(270-360) tagged with GST was isolated from Escherichia coli. HA-tagged Ime2 expressed from the GAL1 promoter was immunoprecipitated from wild-type yeast cells (strain Y742) carrying 3HA-IME2, using antibodies directed against the HA epitope. The immunoprecipitated proteins were subjected to an in vitro kinase assay with the addition of either E. coli-expressed Gst-Ime1(id) or GST proteins. Reaction mixtures that contained both GST-Ime1(id) and HA-Ime2 gave rise to a distinct phosphorylated band with the apparent molecular weight of GST-Ime1 (Fig. 1, lane 3). This band was missing in reactions that contained only Gst1 (Fig. 1, lane 2). In kinase assays that followed immunoprecipitation of HA-tagged proteins from a strain carrying either a kinase-dead allele of IME2 (ime2K97A [a detailed, in vivo analysis of this allele is given below and in Fig. 4]) or the vector plasmid, Ime1 was not phosphorylated (Fig. 1, lanes 1, 4, 5, and 6). We conclude that Ime2 phosphorylates Ime1(270-360).
FIG. 1.
Ime2 phosphorylates Ime1 in vitro. Yeast cells (strain Y742) carrying either pGAL1-3HA-IME2(1-645) (YEp1985) (lanes 2 and 3), pGAL1-3HA-ime2K97A(1-645) (YEp2264) (lanes 4 and 5), or pGAL1-3HA (YEp1906) (lanes 1 and 6) grown in SR medium to 107 cells/ml were transferred to SG medium for 16 h. Proteins were extracted and immunoprecipitated with antibodies directed against 12CA5. GST-Ime1(270-360) (lanes 2 and 4) and Gst1 (lanes 1 and 3) isolated on glutathione-Sepharose beads were added to the immunoprecipitated extracts, subjected to a kinase reaction, and separated on an SDS-10% PAGE gel.
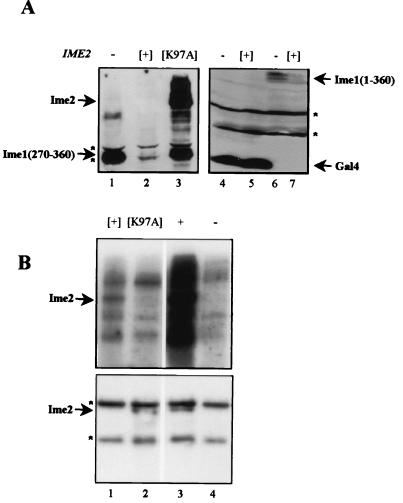
FIG. 4.
The stability of Ime1 depends on the kinase activity of Ime2. (A) Western blot analysis. Proteins were extracted from cells grown in SD medium to 107 cells/ml and loaded onto an SDS-PAGE gel, and immunoblot analysis was performed using antibodies directed against Gal4(bd). A MATa ime2Δ strain (Y1071) carrying either pADH1-gal4(1-147)-ime1(270-360) (YEp1969) (lanes 1 to 3), pADH1-gal4(1-147) [pGBDU-C(1)] (lanes 4 to 5), or pADH1-gal4(1-147)-ime1(1-360) (YEp1202) (lanes 6 to 7) was used. This strain also carried any one of the following plasmids: YEplac112 (14) (lanes 1, 4, and 6) (control), YEp1916 (lanes 2, 5, and 7) (pADH1-gal4(1-147)-HA-IME2), and P2225 (lane 3) (pADH1-gal4(1-147)-ime2K97A. (B) Cells grown in SR medium to 107/ml were transferred to SG medium for 16 h. Proteins were extracted, immunoprecipitated with antibodies directed against 12CA5, subjected to a kinase reaction, and separated on an SDS-10% PAGE gel. (Upper panel) kinase assay; (lower panel) IP-Western. Strains used were MATa LEU2::pGAL1-3HA-IME2(1-645) (Y1255) (lane 3) and its isogenic MATa strain (Y742) carrying pGAL1-3HA-IME2(1-645) (YEp1985) (lane 1), pGAL1-3HA-ime2K97A (YEp2264) (lane 2), and pGAL1-3HA (YEp1906) (lane 4). Brackets indicate that the gene was present on a multicopy plasmid. Asterisks, nonspecific bands.
Transient accumulation of Ime1 in meiotic cultures.
The above results raise the possibility that, as with other kinases, association and phosphorylation of Ime1(id) by Ime2 in living yeast cells might tag Ime1 for degradation and consequently generate a reduction in the transcription of IME1 as well as early meiosis-specific genes. In this study, we applied Western blotting and pulse-chase assays to examine this hypothesis. We used the truncated form of Ime1, Ime1(id), since this domain is sufficient for interaction with Ume6, Rim11 (39), and Ime2 (Table 1), and since this domain is phosphorylated in vitro by Ime2 (Fig. 1). Furthermore, as will be shown later (see Fig. 4), the steady-state level of a truncated Ime1 carrying only this domain mimics that of the complete Ime1. Ime1(id) fused to Gal4(bd) was detected by antibodies directed against Gal4(bd). This epitope did not affect the steady-state level of Ime1 (detailed analysis is given below), and similar results were obtained when the smaller HA epitope, was used (data not shown).
Figure 2A shows Western blot analysis of vegetative and meiotic cultures of MATa/MATα diploid cells expressing either Gal4(bd) or Gal4(bd)-Ime1(id) from the IME1 promoter. Two specific, closely migrating bands, corresponding to Gal4(bd)-Ime1(id), were observed in the immunoblot (Fig. 2), whereas Gal4(bd) appeared as a single band (Fig. 2). This is in contrast to the single band observed when Ime1(id) was expressed in E. coli (Fig. 1). These results suggest that in yeast cells, Ime1(id) is subject to posttranslational modification(s). This suggestion was confirmed by a pulse-chase experiment, revealing an increase in the molecular weight of Ime1 upon a metabolic pulse-chase (see Fig. 5).
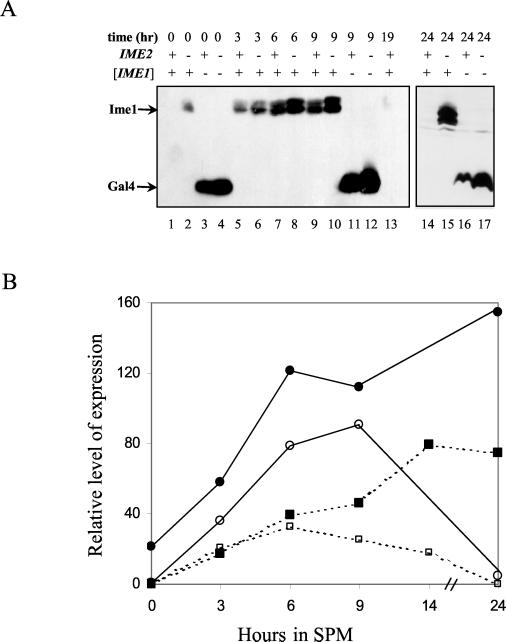
FIG. 2.
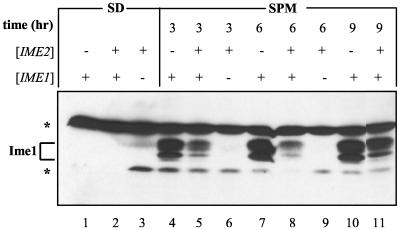
Ime2 modulates the level of Ime1 in diploid cells. Proteins were extracted from meiotic cultures at 0 (lanes 1 to 4), 3 (lanes 5 to 6), 6 (lanes 7 to 8), 9 (lanes 9 to 12), 19 (lane 13), and 24 (lanes 14 to 17) h of incubation in SPM. Strains used were MATa/MATα IME2/IME2 (Y422) (lanes 1, 3, 5, 7, 9, 11, 13, 14, and 16) and MATa/MATα ime2Δ/ime2Δ (Y1039) (lanes 2, 4, 6, 8, 10, 12, 15, and 17). (A) Cells carried the following genes on a 2μm plasmid: pIME1-gal4(1-147)-ime1(270-360) (P1931) (lanes 1, 2, 5 to 10, and 13 to 15), and pIME1-gal4(1-147) (P1952) (lanes 3, 4, 11, 12, 16, and 17). Immunoblot analysis was performed by using antibodies directed against Gal4(bd). Brackets indicate that the gene was present on a multicopy plasmid. (B) The relative levels of Ime1 (solid lines and circles) were quantitated from the results shown in panel A, and those of Ime1-LacZ (dashed lines and squares) were calculated from β-galactosidase assays on three transformants carrying pAS128. Solid symbols, strain Y422; open symbols, strain Y1039.
FIG. 5.
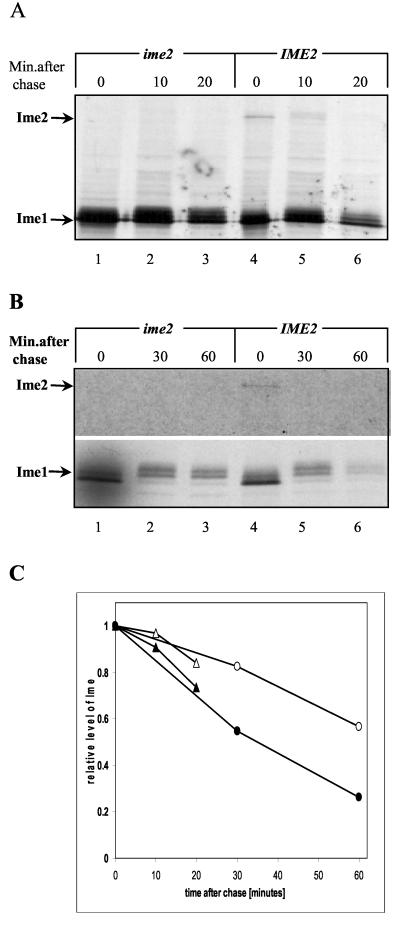
Ime2 regulates the half-life of Ime1. Shown are results of a pulse (7 min)-chase analysis of Ime1(id) and Ime2 tagged with Gal4(bd). (A) Proteins were extracted from cells grown to a density of 107/ml in SD medium. A MATa ime2Δ strain (Y1071) carrying pADH1-gal4(1-147)-ime1(270-360) on a 2μm plasmid (P1969) (lanes 1 to 6) or pADH1-gal4(1-147)-IME2 on a 2μm plasmid (P1916) (lanes 4 to 6) was used. (B) Proteins were extracted from cells grown in SD medium to stationary phase, washed in water, and incubated in SPM at a titer of 107 cells/ml for 4 h. A MATa ime2Δ strain (Y1071) carrying pIME1-gal4(1-147)-ime1(270-360) (P1931) (lanes 1 to 6) or pIME1-gal4(1-147)-IME2 (P1968) (lanes 4 to 6) was used. The film exposure was 15 times longer for the meiotic culture than for the mitotic one. This phenomenon is typical for meiotic cultures that become nonpermissive for the incorporation of compounds from the media (23). (C) The intensities of the bands corresponding to Gal4(bd)-Ime1(id) were quantitated by PhosphorImager analysis and are expressed as percentages of those present during initiation of the chase. Triangles, vegetative cultures; circles, meiotic cultures. Open symbols, cells with deletions of IME2; solid symbols, cells overexpressing Ime2. Brackets indicate that the gene was present on a multicopy plasmid.
In vegetative growth medium with acetate as the sole carbon source, Ime1 could not be detected (Fig. 2A, lane 1). Upon the switch to meiotic conditions (nitrogen depletion), there was a transient increase in the level of Ime1, reaching a peak at about 9 h (Fig. 2A, lanes 5, 7, and 9), after which Ime1 declined to undetectable levels at 19 and 24 h (Fig. 2A, lanes 13 and 14). Figure 2B shows the relative levels of Gal4(bd)-Ime1(id) and Ime1-LacZ. The level of Gal4(bd)-Ime1 was quantitated from the Western blotting data shown in Fig. 2A, by using nonspecific bands as an internal control. The level of Ime1-LacZ was determined by measuring the activity of β-galactosidase in wild-type diploid cells carrying ime1(−4401-+201)-lacZ (Fig. 2B). Our results demonstrate that the behavior of the Gal4(bd)-Ime1(id) fusion protein correlates well with the behavior of IME1 mRNA as determined by Northern analysis (22, 34) or by fusion of the IME1 5′ untranslated region to lacZ (Fig. 2B) (43). Moreover, the transient availability of the Ime1 protein suggests that it is unstable, since a stable protein is expected to show a constant level (see Discussion).
As expected, the level of Gal4(bd) was also transiently induced in meiosis (Fig. 2, lanes 3, 11, and 16), mimicking the transient activity of the IME1 promoter (43). However, at all times, the level of Gal4(bd) was higher than that of the Gal4(bd)-Ime1(id) fusion protein (Fig. 2A; compare lanes 1 and 3, 7 and 11, and 14 and 16). The most dramatic effect was observed in vegetative cultures, in which the level of Gal4(bd) is about 100 times higher than that of Gal4(bd)-Ime1(id). This result suggests that Ime1(id) is an unstable protein.
Ime2 regulates the level of Ime1.
Since Ime2 associates with and in vitro phosphorylates the C-terminal domain of Ime1, we compared the levels of Gal4(bd)-Ime1(id) in wild-type and ime2Δ diploid cells. In vegetative medium with acetate as the sole carbon source, Gal4(bd)-Ime1(id) was observed only in the absence of IME2 (Fig. 2A; compare lanes 1 and 2). Under meiotic conditions, the level of Gal4(bd)-Ime1(id) was elevated in ime2Δ cells in comparison to that in wild-type cells (Fig. 2A [compare lanes 5 and 6, 7 and 8, and 9 and 10] and B). At 24 h in SPM, Gal4(bd)-Ime1(id) was detected only in ime2Δ diploid cells (Fig. 2A; compare lanes 14 and 15). These results suggest that Ime2 causes the level of Ime1(id) to fall. The level of Gal4(bd) was also affected by Ime2, as determined by Western blot analysis (Fig. 2A; compare lanes 3 and 4, 11 and 12, and 16 and 17). However, at time zero, for example, deletion of IME2 led to a 21-fold increase in the level of Gal4(bd)-Ime1(id) and only a 1.2-fold increase in the level of Gal4(bd), suggesting that Ime2 affects the stability of Ime1. Moreover, although Ime2 is required to promote the decline in the transcription of IME1, it has no effect on the level of Ime1-LacZ at early meiotic times (Fig. 2B) (43), suggesting a direct effect of Ime2 on the stability of Ime1.
In order to strengthen the above conclusion, we determined the level of Ime1 in haploid cells with deletions of IME2 in comparison to that in haploid cells overexpressing IME2. Since neither Ime1 nor Ime2 is normally expressed in haploid cells (22, 48), both genes were expressed from a truncated version of the IME1 promoter in which the mating-type control was deleted. In vegetative growth medium with glucose as the sole carbon source, IME1 is not transcribed (22), and accordingly, Gal4(bd)-Ime1(id) was not detected (Fig. 3, lanes 1 and 2). Overexpression of Ime2 in meiotic cultures led to about a twofold decrease in the level of Gal4(bd)-Ime1(id) (Fig. 3; compare lanes 4 and 5, 7 and 8, and 10 and 11). Thus, the effect of Ime2 on the level of Ime1 could be studied in both haploid and diploid cells. Interestingly, in haploid cells Gal4(bd)-Ime1(id) appeared as four specific bands (Fig. 3). The nature of this modification will be described elsewhere (I. Rubin-Bejerano and Y. Kassir, unpublished data).
FIG. 3.
Ime2 modulates the level of Ime1 in haploid cells. Proteins were extracted either from cells grown in SD medium to 107 cells/ml (lanes 1 to 3) or from meiotic cultures at 3 (lanes 4 to 6), 6 (lanes 7 to 9), or 9 (lanes 10 and 11) h in SPM. A MATa ime2Δ strain (Y1071) carrying the following genes on a 2μm plasmid was used: pIME1-gal4(1-147)-HA-IME2 (P1968) (lanes 3, 6, and 9), pIME1-gal4(1-147)-ime1(270-360) (P1931) (lanes 1, 4, 7, and 10), or both P1968 and P1931 (lanes 2, 5, 8, and 11). Immunoblot analysis was performed using antibodies directed against Gal4(bd). Brackets indicate that the gene was present on a multicopy plasmid. Asterisks, nonspecific bands.
Ime2 regulates the stability of Ime1 in vegetative cultures.
The level of Gal4(bd) that is expressed from the IME1 promoter is increased in cells with deletions of IME2 (Fig. 2; compare lanes 4, 12, and 17 to lanes 3, 11, and 16, respectively). These findings can be explained by the effect of Ime2 on IME1 transcription. Since the transcription of IME1 is subject to positive autoregulation (40, 43), it is possible that the effect of Ime2 on the transcription of IME1 is indirect, via the effect on the stability of Ime1. That is, in the absence of Ime2, the level of endogenous Ime1 is increased, and consequently, transcription of the pIME1-gal4(1-147) chimera is elevated. In order to circumvent the contribution of transcriptional regulation to the level of Ime1, we expressed both Ime1 and Ime2 from the heterologous ADH1 promoter. Figure 4A shows that the level of Gal4(bd)-Ime1(id), as well as that of the complete Gal4(bd)-Ime1, is dramatically reduced in cells overexpressing Ime2 in a vegetative medium with glucose as the sole carbon source (compare lane 1 with lane 2 and lane 6 with lane 7). Quantitative analysis revealed 3.7- and 7.6-fold reductions in the levels of Ime1(id) and Ime1, respectively. On the other hand, overexpression of Ime2 had no effect on the level of Gal4(bd) (Fig. 4A; compare lanes 4 and 5) (0.83-fold). We conclude that Ime2 alters the steady-state level of Ime1. These results, together with the two-hybrid assay results (Table 1), demonstrate that the effect of Ime2 on the level of Ime1 is independent of meiotic conditions.
The kinase activity of Ime2 is required for destabilization of Ime1.
In order to determine whether the kinase activity of Ime2 is required to regulate the level of Ime1, we changed a conserved lysine at the putative ATP-binding site to alanine (ime2K97A). An in vitro kinase assay was used to verify that ime2K97A encodes a defective kinase. HA-tagged forms of Ime2 and Ime2K97A expressed from the GAL1 promoter were immunoprecipitated using antibodies directed against the HA epitope. Figure 4B shows that Ime2 is subject to autophosphorylation and could be detected in cells expressing the wild-type IME2 gene (lanes 1 and 3, upper panel). Autophosphorylation was absent in immunoprecipitated extracts isolated from either nontagged cells or cells expressing the mutant ime2K97A allele (Fig. 4B, lanes 2 and 4, upper panel). IP-Western analysis showed that the Ime2K97A protein was present in the immunoprecipitate (Fig. 4B, lower panel; compare lanes 2 and 4). We conclude that Ime2 is subject to autophosphorylation and that the ime2K97A allele leads to impaired kinase activity. Overexpression of the mutated kinase in vegetative cultures had no effect on the level of Gal4(bd)-Ime1(id) (Fig. 4A; compare lanes 2 and 3), suggesting that it is the kinase activity of Ime2 that modulates the stability of Ime1.
Ime2 affects the half-life of Ime1.
Pulse-chase experiments were performed to determine the effect of Ime2 on the half-life of Ime1. Gal4(bd)-Ime1(id) and Gal4(bd)-Ime2 were immunoprecipitated from a MATa ime2Δ strain using antibodies directed against Gal4(bd). Proteins were extracted from either vegetative (Fig. 5A) or meiotic (Fig. 5B) cultures. In the vegetative culture, Ime1 and Ime2 were expressed from the ADH1 promoter (Fig. 5A), and in the meiotic culture, these proteins were expressed from the IME1 promoter (Fig. 5B). Cells were labeled with [35S]methionine for 7 min and then chased for the indicated times. The results show that under both vegetative and meiotic conditions, the half-life of Gal4(bd)-Ime1(id) is about 30 min (Fig. 5C). In the absence of Ime2, Gal4(bd)-Ime1(id) was stabilized, and its half-life increased to more than 60 min (Fig. 5C).
Degradation of Ime1(270-360) is mediated by the 26S proteasome.
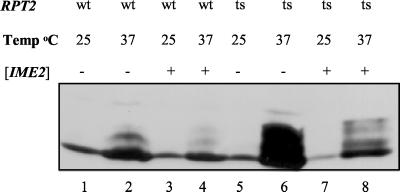
Regulated degradation of proteins takes place in the 26S proteasome (for a review, see reference 16). The ATPase Rpt2 is an integral component of the proteasome that is essential for viability (38). Cells carrying a specific point mutation in RPT2, rpt2RF, show a defect in degradation of short-lived proteins, including known substrates of the proteasome (38). rpt2RF cells are temperature sensitive, apparently reflecting the increased requirement for protein degradation at elevated temperatures (38; M. Glickman, personal communication). We used this mutation to determine the effect of the 26S proteasome on the stability of Ime1. The steady-state level of Ime1(270-360) was determined in wild-type and isogenic rpt2RF strains expressing Gal4(bd)-Ime1(id) and Gal4(bd)-Ime2 from the ADH1 promoter. Proteins were extracted for Western blot analysis from cells grown in vegetative glucose medium at the permissive temperature of 25°C and following 6 h of incubation at 37°C (Fig. 6). The level of Gal4(bd)-Ime1(id) increased in wild-type strains incubated at 37°C in comparison to 25°C (Fig. 6; compare lanes 1 and 2). Nevertheless, a dramatic increase (sixfold) in the level of Gal4(bd)-Ime1(id) was observed in the rpt2RF cells incubated at 37°C (Fig. 6; compare lane 2 with lane 6 and lane 4 with lane 8). We conclude that Ime1(270-360) is most probably degraded by the 26S proteasome. Overexpression of Ime2 led to a decrease (threefold) in the level of Ime1, even in the rpt2RF mutant (Fig. 6; compare lane 2 with lane 4 and lane 6 with lane 8). The effect of Ime2 could also be observed at the permissive temperature (data not shown). Since the rpt2RF mutation causes a defect in activity rather than complete inactivation of the proteasome (a lethal event), the effect of Ime2 was still observed.
FIG. 6.
Ime1 is degraded by the proteasome. Proteins were extracted from cells that were grown in SD medium at 25°C (lanes 1, 3, 5, and 7) and incubated for 6 h at 37°C (lanes 2, 4, 6, and 8). Strains used were Y1248 (lanes 1 to 4) and its isogenic rpt2RF strain (Y1249) (lanes 5 to 8) carrying YEp1974 [pADH1-gal4(1-147)-ime1(270-360)] and YEpLac112 (14) (control) (lanes 1, 2, 5, and 6) or YEp1974 and YEp1916 [pADH1-gal4(1-147)-HA-IME2] (lanes 3, 4, 7, and 8). Immunoblot analysis was performed using antibodies directed against Gal4(bd). Brackets indicate that the gene was present on a multicopy plasmid.
IME2 encodes an extremely unstable protein.
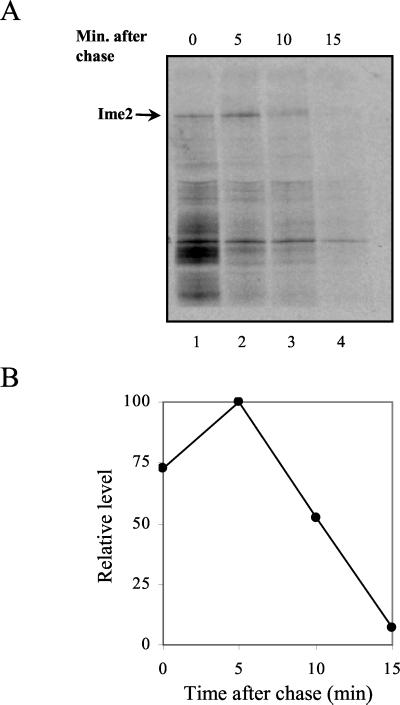
We also studied the stability of Ime2 using a pulse-chase assay. Figure 5 shows that Gal4(bd)-Ime2 could not be detected when pulsed for 7 min and chased for longer than 20 min. In order to determine the half-life of Ime2, we shortened the time of labeling and the length of the chase. Gal4(bd)-Ime2 expressed in meiotic cultures from the IME1 promoter was labeled with [35S]methionine for 4 min and immunoprecipitated after a chase of 5, 10, or 15 min. Figure 7 shows that Gal4(bd)-Ime2 is an extremely unstable protein with a half-life of about 5 min (Fig. 7B). A change in the molecular weight of Gal4(bd)-Ime2 (Fig. 5A, lane 5) was observed upon the chase, suggesting that in vivo, Ime2 is subject to posttranslational modifications.
FIG. 7.
IME2 encodes an unstable protein. Pulse (4 min)-chase analysis of Ime2 tagged with Gal4(bd). (A) Proteins were extracted from meiotic cultures at 4 h in SPM. A MATa strain (sub62) (38) carrying pIME1-IME2 on a 2μm plasmid (YEp1968) was used. (B) Quantitative analysis of the level of Ime2.
Expression of Ime2 in vegetative cultures is toxic.
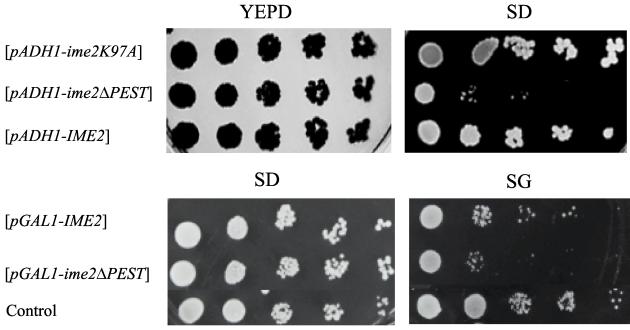
Usually, the detection of an unstable protein such as Ime2 requires overexpression. Overexpression of Ime2 was accomplished by expressing it from the strong ADH1 or GAL1 promoter, as well as by increasing the number of IME2 copies by inserting it on a 2 μm plasmid. Nevertheless, under these conditions Ime2 was not detected by either Western blot or IP-Western analysis (Fig. 4A, lane 1, and Fig. 4B, lower panel, lane 1). Ime2 could be detected only by using more-sensitive assays, such as a metabolic labeling (Fig. 5 and 7) or kinase activity (Fig. 4B, upper panel, lane 1) assay. Surprisingly, Ime2 could be detected by Western blot analysis when the gene was integrated in the genome (Fig. 4B, lower panel, lane 3). These results, coupled with the short half-life of Ime2, suggest that expression of this protein in vegetative cultures might be toxic, thus selecting against cells overexpressing Ime2. Serial dilution analysis confirmed this suggestion (Fig. 8). The efficiency of plating of cells carrying IME2 on a 2μm plasmid was slightly reduced under selection conditions in minimal SD medium in comparison to that in the rich yeast extract-peptone-dextrose (YEPD) medium (Fig. 8, third row from top). Moreover, the kinase-defective Ime2K97A protein, which was easily detected in Western blot and IP-Western analyses (Fig. 4A, lane 3, and Fig. 4B, lower panel, lane 2), did not reduce the efficiency of plating (Fig. 8, top row). When Ime2 was expressed from the inducible GAL1 promoter and the efficiencies of plating on glucose (SD) and galactose (SG) media were compared, toxicity was observed only in the presence of galactose (Fig. 8). The amino acid sequence of IME2 reveals putative PEST sequences. Such sequences are involved in facilitating the degradation of proteins by the proteasome (5), and could be responsible for the low stability of Ime2. Expression of an Ime2 derivative with the PEST sequence (amino acids 478 to 499) deleted led to a more-pronounced decrease in the efficiency of plating (Fig. 8), suggesting that deletion of this sequence stabilized Ime2. Microscopic observation of vegetative cells expressing Ime2 showed an accumulation of large and unbudded cells (data not shown), suggesting that these cells are inefficient in entering the cell cycle. Alternatively, the reduction in viability might reflect entry of haploid cells into the meiotic pathway, as previously suggested (33).
FIG. 8.
Overexpression of Ime2 in vegetative cultures is toxic. MATa strains (Y742) carrying various IME2 plasmids grown on SD plates were plated on YEPD (rich glucose medium), SD (minimal glucose medium), and SG (minimal galactose medium) plates following serial 10-fold dilutions. Plasmids used were YEp1916 (pADH1-IME2), YEp2126 (pADH1-ime2ΔPEST), YEp2225 (pADH1-ime2K97A), YEp1985 (pGAL1-IME2), and YEp2142 (pGAL1-ime2ΔPEST). As a control, Y742 carried the vector plasmid, YEpLac112 (14). Brackets indicate that the gene was present on a multicopy plasmid.
DISCUSSION
Transient availability of Ime1 in meiosis.
The transcription of all meiosis-specific genes, including that of the gene encoding their main transcriptional activator, Ime1, is transient (22, 27, 48). However, transient transcription does not necessarily reflect a transient availability of proteins, which requires that the encoded proteins are unstable. For instance, even though transcription of POL1 and CLB5 is periodic in the mitotic cell cycle, and identically regulated (29, 42), the steady-state level of Pol1 is constant (10), whereas the level of Clb5 is periodic (20). The periodicity in the level of Clb5 is accomplished by regulated degradation (20). In this report we show that the level of Ime1 parallels its transcription pattern, increasing under meiotic conditions, reaching a peak, and then declining (Fig. 2). These results suggest that Ime1 is an unstable protein. In vegetative cultures with acetate as the sole carbon source, Ime1 was undetectable (Fig. 2), although the presence of a low but substantial level of IME1 mRNA was reported (22). This phenomenon is due to translational regulation, which leads to inefficient translation of IME1 mRNA in the presence of a nitrogen source (45). We also show that Ime1 is subject to extensive degradation that is regulated by Ime2 (Fig. 2). Apparently, these two mechanisms combine to ensure the absence of Ime1 from vegetative, mitotic cultures.
The stability of Ime1 depends on the kinase activity of Ime2.
Several approaches were used to show that Ime2 reduces the stability of Ime1. First, the level of Ime1 was lower in the wild-type diploid strain than in the ime2Δ isogenic strain under vegetative conditions, as well as throughout the meiotic process (Fig. 2). Second, we compared the level of Ime1 in haploid cells with deletions of IME2 to that in haploid cells overexpressing Ime2. The results showed that as in diploid cells, the level of Ime1 increased upon a shift to meiotic conditions. However, lower levels were observed in the cells overexpressing Ime2 (Fig. 3), pointing again to the role of Ime2 in destabilizing Ime1. Third, the effect of Ime2 was observed when both Ime1 and Ime2 were expressed from the heterologous ADH1 promoter (Fig. 4). This approach enabled us to override the previously reported effect of both Ime1 and Ime2 on the transcription of IME1 (40, 43, 48, 49). Finally, using pulse-chase analysis, we showed that under both vegetative and meiotic conditions, overexpression of Ime2 shortened the half-life of Ime1 (Fig. 5).
The effect of Ime2 on the half-life of Ime1 (Fig. 5) seems minor in comparison to the effect of Ime2 on the steady-state level of Ime1 as studied by Western blot analysis (Fig. 2, 3, and 6). Two properties of Ime2 may explain this disparity. First, Ime2 is an extremely unstable protein with a half-life at least 5 to 6 times shorter than that of Ime1 (Fig. 5 and 7). Second, overexpression of Ime2 is toxic. Thus, at any given time, the Ime2 protein is absent in many cells. Therefore, in the Western blot analysis, which measures the accumulative levels of Ime1, the effect is more significant, and it better reflects the normal physiological conditions of the cells.
Phosphorylation is a key mechanism in regulating the stability of proteins. This event facilitates the covalent linkage of the 8-kDa ubiquitin to these substrates and thus tags them for degradation by the 26S proteasome (for a review, see reference 5). Since Ime2 is a serine/threonine protein kinase (49), it was tempting to postulate that phosphorylation of Ime1 by Ime2 tagged it for degradation by the 26S proteasome. Several experimental approaches supported this hypothesis. (i) Using the two-hybrid assay, we showed an association between Ime1 and Ime2 (Table 1). (ii) By an in vitro kinase assay, we showed that Ime2 phosphorylated the C-terminal domain of Ime1 (Fig. 1). (iii) A point mutation in IME2, K97A, that abolished its in vitro kinase activity was unable to promote destabilization of Ime1 (Fig. 4). (iv) Using a mutation in RPT2, an essential ATPase subunit in the regulatory particle of the proteasome (15, 38), we showed that in these rpt2RF cells the stability of Ime1 was temperature sensitive (Fig. 6), suggesting that Ime1 is degraded by the 26S proteasome. (v) In rpt2RF cells overexpressing Ime2, a distinct, less mobile form of Ime1 was observed (Fig. 6; compare lanes 6 and 8). Since this form was undetected in cells that do not express Ime2, we postulate that it might represent an Ime2-dependent, phosphorylated form of Ime1. We further suggest that in wild-type cells this form is rapidly degraded and thus undetectable.
The importance of transient availability of Ime1.
Ime1 is the master regulator required to initiate meiosis in S. cerevisiae. Ime1 is subject to multiple levels of regulation, ensuring that entry into meiosis is an alternative to the mitotic cycle and will take place only in the absence of nitrogen and glucose sources. How do cells regulate the availability and activity of Ime1? First, IME1 is not transcribed in vegetative growth medium with glucose as the sole carbon source, while low levels are observed when glucose is replaced with acetate (22). In addition, in the presence of a nitrogen source, the translation of IME1 mRNA is inefficient (43). In this report we showed an additional level of regulation in which Ime1 is subject to regulated degradation. Finally, the activity of Ime1 is also subject to nutrient regulation, as Ime1 is excluded from the nucleus in vegetative cells expressing the G1 cyclins (6), and the association of Ime1 with Ume6 or the ability of this complex to activate transcription depends on the absence of both glucose and nitrogen (39).
The above discussion explains why Ime1 is not available and active in vegetative cultures. It should also be emphasized that the transient expression of Ime1 is required for efficient meiosis. Mild overexpression of Ime1 in meiotic cultures leads to a reduction in the percentage of sporulation, an increase in the level of dyads, and an increase in the level of nondisjunction (43). Extensive overexpression of Ime1 is accomplished in cells with deletions of IME2 (Fig. 2). It is possible that certain phenotypes previously observed in these cells, specifically, the second round of DNA replication, and the third round of nuclear division observed at late meiotic times (12), are due to accumulation of Ime1 rather than to the absence of Ime2.
A feedback loop controlling meiosis.
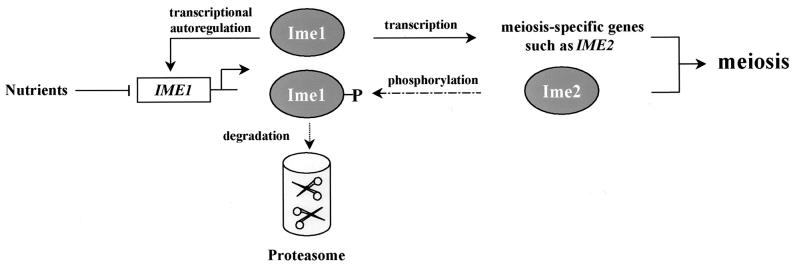
Entry into, and progression through, meiosis depends on a transcriptional cascade that is set off by the transcriptional activator Ime1 and then by the serine/threonine protein kinase Ime2 (for a review, see reference 27). IME2 is an early meiosis-specific gene whose expression is regulated by Ime1 (48, 49). In this report we showed that in vitro Ime2 phosphorylates Ime1, and in vivo its kinase activity is required for reducing the steady-state level of Ime1. We suggest, therefore, that phosphorylation of Ime1 by Ime2 marks Ime1 for degradation by the 26S proteasome. This decrease in the level of Ime1 is intensified by the fact that Ime1 exhibits positive autoregulation (40, 43). This autoregulation is required to relieve the repression mediated by Sok2 (44). Thus, the decline in the level of Ime1 is due to a decline in its transcription and an increase in its degradation. This feedback loop is schematically illustrated in Fig. 9. We suggest that the effect of Ime2 on the stability of Ime1 is responsible for its role in negatively regulating the transcription of IME1. In the absence of Ime2, transcriptional autoregulation by the nondegraded Ime1 continues, leading to accumulation of IME1 mRNA. This feedback loop ensures that Ime1 will be active for only a limited time and that following activation of the meiotic pathway, its transcription will be shut off. A similar feedback loop was reported for the transcription factor p53 and its regulated gene, Mdm2 (36). Mdm2 is an E3 ubiquitin ligase (8, 19), and degradation of p53 by the proteasome depends on the formation of the Mdm2/p53 complex (36).
FIG. 9.
A feedback loop controlling meiosis. Shown is a schematic illustration of the transcriptional cascade and feedback loops that control entry into meiosis.
Ime1 and Ime2 maintain antagonistic relationships, so it is important to delay the function of Ime2 in order to ensure that Ime1 will be accessible for a sufficient time to promote transcription of early meiosis-specific genes. This lag between the functions of the two proteins is accomplished by two mechanisms. Transcription of IME2 is delayed in relation to transcription of IME1 and, as shown in this report, IME2 itself encodes an unstable protein whose half-life is shorter than that of Ime1.
Acknowledgments
We thank D. Cassel and M. Glickman for critical reading of the manuscript. We thank S. Elledge, S. Fields, D. Finley, B. Futcher, M. Johnston, N. Kleckner, M. Ptashne, A. Sugino, and I. Yamashita for kindly providing plasmids and yeast strains.
This work was supported by a grant from the Israel Academy of Sciences.
REFERENCES
- 1.Bowdish, K. S., H. E. Yuan, and A. P. Mitchell. 1994. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol. Cell. Biol. 14:7909-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. (Erratum, 282:1421.) [DOI] [PubMed]
- 5.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colomina, N., E. Gari, C. Gallego, E. Herrero, and M. Aldea. 1999. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 18:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 8.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 9.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 10.Foiani, M., G. Liberi, G. Lucchini, and P. Plevani. 1995. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol. Cell. Biol. 15:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foiani, M., F. Marini, D. Gamba, G. Lucchini, and P. Plevani. 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foiani, M., E. Nadjar-Boger, R. Capone, S. Sagee, T. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278-288. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 15.Glickman, M. H., D. M. Rubin, V. A. Fried, and D. Finley. 1998. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18:3149-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glickman, M. H., D. M. Rubin, H. Fu, C. N. Larsen, O. Coux, I. Wefes, G. Pfeifer, Z. Cjeka, R. Vierstra, W. Baumeister, V. Fried, and D. Finley. 1999. Functional analysis of the proteasome regulatory particle. Mol. Biol. Rep. 26:21-28. [DOI] [PubMed] [Google Scholar]
- 17.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 18.Hershko, A. 1997. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr. Opin. Cell Biol. 9:788-799. [DOI] [PubMed] [Google Scholar]
- 19.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 20.Irniger, S., and K. Nasmyth. 1997. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110:1523-1531. [DOI] [PubMed] [Google Scholar]
- 21.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassir, Y., D. Granot, and G. Simchen. 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853-862. [DOI] [PubMed] [Google Scholar]
- 23.Kassir, Y., and G. Simchen. 1991. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 194:94-110. [DOI] [PubMed] [Google Scholar]
- 24.Kominami, K., Y. Sakata, M. Sakai, and I. Yamashita. 1993. Protein kinase activity associated with the IME2 gene product, a meiotic inducer in the yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 57:1731-1735. [DOI] [PubMed] [Google Scholar]
- 25.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 27.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Lew, D. J., T. Weinert, and J. R. Pringle. 1997. Cell cycle control in Saccharomyces cerevisiae, p. 607-696. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Lowndes, N. F., A. L. Johnson, and L. H. Johnston. 1991. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans factor. Nature 350:247-250. [DOI] [PubMed] [Google Scholar]
- 30.Mandel, S., K. Robzyk, and Y. Kassir. 1994. IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev. Genet. 15:139-147. [DOI] [PubMed] [Google Scholar]
- 31.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Mitchell, A. P., and K. S. Bowdish. 1992. Selection for early meiotic mutants in yeast. Genetics 131:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagano, M. 1997. Cell cycle regulation by the ubiquitin pathway. FASEB J. 11:1067-1075. [DOI] [PubMed] [Google Scholar]
- 36.Piette, J., H. Neel, and V. Marechal. 1997. Mdm2: keeping p53 under control. Oncogene 15:1001-1010. [DOI] [PubMed] [Google Scholar]
- 37.Rose, M., and D. Botstein. 1983. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101:167-180. [DOI] [PubMed] [Google Scholar]
- 38.Rubin, D. M., M. H. Glickman, C. N. Larsen, S. Dhruvakumar, and D. Finley. 1998. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17:4909-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubin-Bejerano, I., S. Mandel, K. Robzyk, and Y. Kassir. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16:2518-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagee, S., A. Sherman, G. Shenhar, K. Robzyk, N. Ben-Doy, G. Simchen, and Y. Kassir. 1998. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1985-1995. (Erratum, 18:3645.) [DOI] [PMC free article] [PubMed]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schwob, E., and K. Nasmyth. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7:1160-1175. [DOI] [PubMed] [Google Scholar]
- 43.Shefer-Vaida, M., A. Sherman, T. Ashkenazi, K. Robzyk, and Y. Kassir. 1995. Positive and negative feedback loops affect the transcription of IME1, a positive regulator of meiosis in Saccharomyces cerevisiae. Dev. Genet. 16:219-228. [DOI] [PubMed] [Google Scholar]
- 44.Shenhar, G., and Y. Kassir. 2001. A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:1603-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherman, A., M. Shefer, S. Sagee, and Y. Kassir. 1993. Post-transcriptional regulation of IME1 determines initiation of meiosis in Saccharomyces cerevisiae. Mol. Gen. Genet. 237:375-384. [DOI] [PubMed] [Google Scholar]
- 46.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 47.Smith, H. E., S. E. Driscoll, R. A. Sia, H. E. Yuan, and A. P. Mitchell. 1993. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics 133:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, H. E., and A. P. Mitchell. 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, M., H. Kawaguchi, Y. Sakata, K. Kominami, M. Hirano, H. Shima, R. Akada, and I. Yamashita. 1990. Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221:176-186. [DOI] [PubMed] [Google Scholar]