Abstract
The Saccharomyces cerevisiae Ku complex, while important for nonhomologous DNA end joining, is also necessary for maintaining wild-type telomere length and a normal chromosomal DNA end structure. Yeast cells lacking Ku can grow at 23°C but are unable to do so at elevated temperatures due to an activation of DNA damage checkpoints. To gain insights into the mechanisms affected by temperature in such strains, we isolated and characterized a new allele of the YKU70 gene, yku70-30ts. By several criteria, the Yku70-30p protein is functional at 23°C and nonfunctional at 37°C. The analyses of telomeric repeat maintenance as well as the terminal DNA end structure in strains harboring this allele alone or in strains with a combination of other mutations affecting telomere maintenance show that the altered DNA end structure in yeast cells lacking Ku is not generated in a telomerase-dependent fashion. Moreover, the single-stranded G-rich DNA on such telomeres is not detected by DNA damage checkpoints to arrest cell growth, provided that there are sufficient double-stranded telomeric repeats present. The results also demonstrate that mutations in genes negatively affecting G-strand synthesis (e.g., RIF1) or C-strand synthesis (e.g., the DNA polymerase α gene) allow for the maintenance of longer telomeric repeat tracts in cells lacking Ku. Finally, extending telomeric repeat tracts in such cells at least temporarily suppresses checkpoint activation and growth defects at higher temperatures. Thus, we hypothesize that an aspect of the coordinated synthesis of double-stranded telomeric repeats is sensitive to elevated temperatures.
DNA double-strand breaks (DSBs) belong to the most disruptive forms of DNA damage. If left unrepaired, they lead to broken chromosomes, which can cause cell death. On the other hand, incorrectly repaired DSBs can give rise to chromosomal aberrations such as rearrangements, deletions, and chromosome fusions (reviewed in references 17, 34, and 61). In a cell with DSBs, surveillance mechanisms called DNA damage checkpoints are activated in order to prevent DNA replication and/or cell division before the damage is repaired (79, 85). DNA damage checkpoints are likely to be responsible for activating the repair mechanisms, and they are thought to provide cells with time to complete the repair process by slowing down cell cycle progression.
Telomeres are special functional complexes at the ends of eukaryotic chromosomes. Unlike DNA DSBs, the ends of eukaryotic chromosomes are stable and do not activate the DNA damage checkpoints. Thus, telomeres protect the chromosome ends from fusion and degradation and allow the cell to differentiate a chromosomal DSB from a natural end (48, 52, 57, 70). In the vast majority of eukaryotes, including yeast and mammals, telomeres consist of simple repeated DNA sequences and their associated proteins. The strand running 5′ to 3′ toward the end contains clusters of three or four guanines and is commonly referred to as the G-rich strand (81). For example, in the yeast Saccharomyces cerevisiae, telomeric DNA consists of ∼300 bp of C1-3A/TG1-3 sequences (76, 77, 84). In organisms where the DNA structure of the very end of the chromosome is known, the G-rich strand is extended to form a single-strand tail (38, 41, 51, 54). In budding yeast, long overhangs of the G-rich strand are only detected during S phase (21, 82) and the exact end structure outside of S phase is currently unknown. However, the assays used to analyze end structures in yeast exclude the possibility of G-strand or C-strand extensions of more than 20 bases (82).
In addition to the end protection function, telomeres are essential for ensuring the complete replication of the chromosomes (32, 78, 84). In most eukaryotes, the end replication problem is solved by telomerase. This ribonucleoprotein complex extends the G-rich strand of the telomere using telomeric repeats encoded within the RNA component of the enzyme as a template (33, 71). In yeast, the telomerase RNA component is encoded by the TLC1 gene (73), while the product of the EST2 gene provides the DNA polymerase activity (15, 45). Mutations in either of these genes lead to a gradual loss of telomeric DNA and cause a loss of cell viability during vegetative subculturing (15, 45, 73).
The Ku complex is present in most eukaryotic organisms and is composed of an ∼70-kDa subunit (Ku70) and an ∼80-kDa subunit (Ku80), forming a heterodimer (reviewed in references 23 and 25). In vitro, this heterodimer binds DNA ends in a sequence-independent manner, and Ku has been shown to be directly involved in nonhomologous end joining (NHEJ), a DNA DSB repair mechanism present in both yeast and mammals (reviewed in reference 25). NHEJ consists of a direct ligation of two DNA ends, and in this mechanism Ku is believed to bind to DNA ends to prevent DNA degradation, facilitate alignment of the broken ends, and recruit DNA ligases and other factors (29, 53, 60, 66, 74). Surprisingly, it has also been shown that Ku is physically associated with yeast and mammalian telomeric DNA (31, 39) and that this association is important for telomere homeostasis and genome stability (16, 69).
In budding yeast, inactivation of the Ku complex, referred to as Yku and consisting of the Yku70p and Yku80p proteins, has several consequences for telomere maintenance and cell growth. Telomeric DNA is dramatically shortened, and the terminal DNA structure maintains long extensions of the G-rich strand throughout the cell cycle (9, 31, 63, 64). Furthermore, telomeric silencing is diminished in Yku− cells (10, 31, 42, 58), and the subnuclear localization of telomeres is modified (42). Finally, Yku− cells are unable to grow at elevated temperatures (9, 26). Recent evidence suggests that this temperature sensitivity phenotype is related to a telomere defect rather than an inability to repair spontaneous DSBs generated in vivo. First, temperature-resistant subclones of Yku− mutant cells seem to maintain their telomeres by recombinational mechanisms (27). This result suggests that telomere maintenance by recombinational mechanisms allows growth of Yku− cells at elevated temperatures, whereas telomerase-based maintenance does not. Second, a deletion of the last 25 amino acids from Yku70p results in telomere maintenance defects and temperature sensitivity, whereas the cells are fully proficient for NHEJ (22). Finally, overexpression of telomerase subunits in Yku− cells restores viability at 37°C and suppresses the DNA damage checkpoint activation that occurs in these mutants (58, 75). In Yku− cells incubated at 37°C, checkpoint activation is dependent on the RAD9 gene product (5, 75), a factor that has emerged as a candidate DNA damage sensor (18, 28, 50). Once the damage is recognized, a set of protein kinases, including Mec1p and Rad53p, are activated (reviewed in references 47 and 79). These kinases transduce the signal and regulate cellular responses by phosphorylating appropriate target proteins involved in the control of cell cycle transitions, the transcriptional induction of repair genes, the activation of repair proteins, or the chromatin structure (reviewed in references 47 and 79). At the level of transcriptional activation, it has been shown that the Rad53p kinase regulates cellular deoxynucleoside triphosphate (dNTP) pools via the transcriptional induction of the genes encoding subunits of ribonucleotide reductase (RNR genes) (3, 40). The demonstration that Rad53p activation and transcriptional induction of the RNR genes occur in Yku− cells confirmed that the DNA damage checkpoint is activated in these cells (5, 75).
The precise role of Yku in telomere maintenance and the reasons for checkpoint activation and growth arrest of Yku− cells at elevated temperatures have remained enigmatic. To address these questions, we identified and analyzed a temperature-sensitive (TS) allele of the YKU70 gene. Having used this allele as well as other strategies to modify telomere lengths in Yku− cells, we report here that, despite the presence of constitutive G-rich overhangs, Yku− cells with long telomeres can grow normally at elevated temperatures. In all conditions tested, DNA damage checkpoint activation and absence of growth at 37°C qualitatively correlated with short telomeres. This new allele also allowed us to demonstrate that telomerase is not involved in the initial generation of the G-rich overhangs. Furthermore, a mutation in the gene encoding DNA polymerase α (Polα) that compromises C-strand synthesis suppresses the telomere length phenotype in Yku− cells. These results suggest that, in Yku− cells, the ability to coordinately generate a minimal amount of double-stranded telomeric repeats at 37°C is compromised. This defect apparently leads first to DNA damage checkpoint activation and ultimately to permanent cell cycle arrest and cell death.
MATERIALS AND METHODS
Yeast strains and plasmids.
BY4705a (MATa ade2ΔhisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0) and isogenic derivatives were used for all experiments, except when stated otherwise. BY4705a was derived from BY4705 (12) by HO-mediated switching of the mating type (A. Wolf and D. Gottschling, personal communication). To generate strain BYyku70Δ, the entire coding region of the YKU70 gene (−19 to +1841 with respect to the initiation codon) was replaced by the LEU2 gene in BY4705a. BYyku70Δ/rif1Δ was created by replacing the RIF1 gene (−1 to +5752 with respect to the initiation codon) by URA3 in strain BYyku70Δ. BYyku70Δ/rad9Δ was generated by replacing the RAD9 gene (−10 to +3940 with respect to the initiation codon) by HIS3 in strain BYyku70Δ. These strains were transformed with either an empty pRS314 vector (72), plasmid pKu70 (see below), or pKu70-30ts (see below) by standard methods (30). Strain SGY51 (mata yku70Δ::LEU2 tlc1Δ::LEU2 his3 lys2 trp1Δ63 ura3 ADE2 MET15 or met15) harboring plasmids pKu70-30ts and pAZ1 (7) (see below) was generated by crossing strain BYyku70Δ, containing pKu70-30ts, with RWY10 (matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 tlc1Δ::LEU2 VR-ADE2-TEL), carrying pAZ1. tlc1Δ/yku70Δ spores containing both plasmids (pKu70-30ts and pAZ1) were selected, and strains lacking TLC1 were recovered by selecting against the presence of pAZ1 on fluoroorotic acid (FOA) media (8). Strain SGY53 (mata tlc1Δ::LEU2 yku70Δ::LEU2 rif1Δ::kanMX4 his3 lys2 trp1Δ63 ura3 ADE2 MET15 or met15), harboring plasmids pKu70-30ts and pAZ1, was created by replacing the RIF1 gene with the kanMX4 marker in strain SGY51. To generate strain SGY56 (MATa/α pol1-17/POL1 yku70Δ::URA3/YKU70), strain RWY121 (MATa ura3-52 trp1-289 ade2-101 gal2 can1 his pol1-17 bar1Δ::kanMX) (2) was crossed with BYyku70ΔURA (MATα ade2ΔhisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 yku70Δ::URA3). The SGY56 diploid was sporulated and microdissected to generate the mutant strains presented in Fig. 2B.
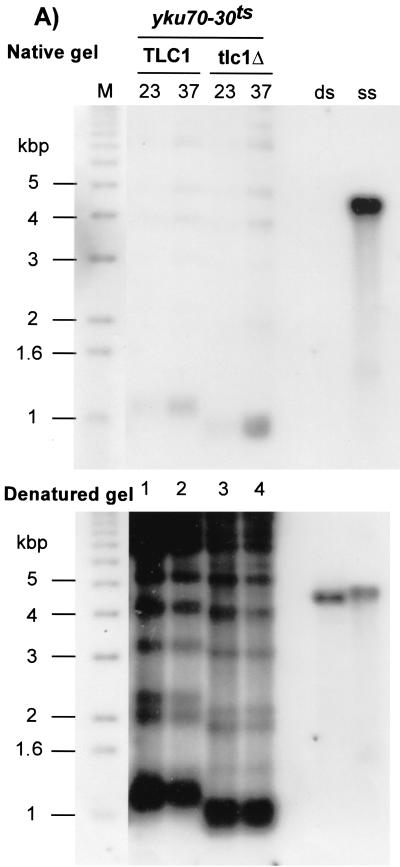
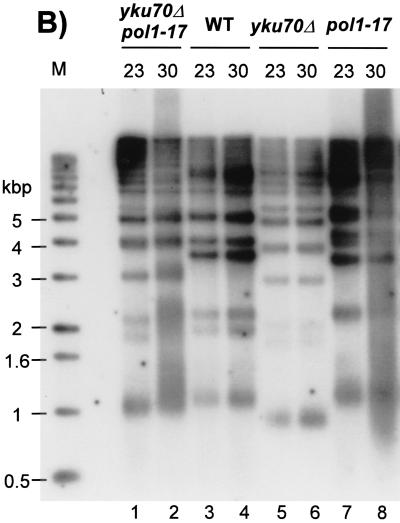
FIG. 2.
(A) Telomerase does not contribute significantly to the creation of G-rich overhangs in Yku− mutants. Strain SGY51 (yku70Δ tlc1Δ pKu70-30ts) and the same strain carrying in addition a plasmid-borne copy of TLC1(pAZ1) were grown at 23°C and then shifted for 8 h to 37°C. DNA was extracted, digested with XhoI, and analyzed by in-gel hybridization as described for Fig. 1B. Top, native gel; bottom, the same gel hybridized after denaturation. Lane M, end-labeled 1-kb ladder DNA serving as a size standard; lanes ss and ds, DNA with telomeric TG1-3 repeats in single-stranded and double-stranded forms, respectively. Note that in yku70-30ts cells telomere shortening occurs during the 30 generations of growth at 23°C that were necessary to produce the yku70-30ts tlc1Δ strain. (B) Telomere length analysis for pol1-17 yku70Δ double mutants. Strain SGY56 (MATa/α pol1-17/POL1 yku70Δ::URA3/YKU70) was sporulated and microdissected. Spores derived from a tetratype were grown, and total genomic DNA was extracted and digested with XhoI. The denatured gel from an in-gel analysis using the CA probe is shown. Lane M, end-labeled 1-kb ladder DNA serving as a size standard; lane 1, yku70Δ pol1-17 cells grown at 23°C; lane 2, the same strain as in lane 1 after a shift to 30°C for 16 h; lane 3, YKU70 POL1 wt cells grown at 23°C; lane 4, the same strain as in lane 3 after a shift to 30°C for 16 h; lane 5, yku70Δ POL1 cells grown at 23°C; lane 6, the same strain as in lane 5 after a shift to 30°C for 16 h; lane 7, YKU70 pol1-17 cells grown at 23°C; lane 8, the same strain as in lane 7 after a shift to 30°C for 16 h.
Telomeric silencing was tested with strain UCC3505 (MATa ura3-52 lys2-80 ade2-101 trp1Δ63 his3Δ200 leu2-Δ1 ppr1Δ::HIS3 adh4::URA3-TEL-VIIL VR-ADE2-TEL) (73), in which the YKU70 gene was replaced by the LEU2 marker; the resulting strain was referred to as UCC3505/yku70.
All deletions in strains were achieved by PCR-mediated gene disruption (6) and were verified by Southern blotting and determination of the corresponding phenotypes (data not shown).
pAZ1 is a pRS316(URA3 CEN3 ARSH4)-based plasmid (72) that contains a 5.5-kb genomic DNA fragment spanning the entire TLC1 locus (7). The pKu70 plasmid is pRS314(TRP1 CEN6 ARSH4) containing the wild-type (wt) YKU70 gene (nucleotides −386 to +1921 with respect to the initiation codon) under the control of its native promoter. The pKu70-GR plasmid was constructed in the following way. Nucleotides −385 to −3 with respect to the initiation codon of the YKU70 gene were amplified by PCR. XhoI and EcoRI recognition sites were appended to the upstream and downstream primers, respectively. The PCR product was cloned in pRS314 into the same sites to generate pKu70-G. One hundred twelve nucleotides downstream of the stop codon of the YKU70 gene were amplified by PCR, and EcoRI and BamHI recognition sites were appended to the upstream and downstream primers, respectively. This PCR product was cloned in pKu70-G at the same sites to generate pKu70-GR. The resulting plasmid was digested with EcoRI before transformation.
Yeast growth conditions.
Yeast strains were grown in standard media at various temperatures (67). For the determination of population doublings (numbers of generations), cell concentrations were measured by densitometry at 660 nm. Cells were diluted to 3 × 105 to 1 × 106 cells/ml in synthetic complete (SC)-Leu-Trp and regrown to 1 × 107 to 2 × 107 cells/ml.
Construction and selection of the yku70-30ts allele.
The YKU70 gene (nucleotides −350 to + 1921 with respect to the initiation codon) was randomly mutagenized by PCR in a buffer containing 10 mM Tris (pH 9), 50 mM KCl, 0.2 mM dATP, 0.2 mM dGTP, 1 mM dCTP, 1 mM dTTP, 7 mM MgCl2, and 1.5 U of Taq DNA polymerase. The PCR product was gel purified and cotransformed with EcoRI-linearized pKu70-GR into strain UCC3505/yku70Δ, and transformants were selected on SC-Trp (gap repair method). Cell patches for 7,500 transformants were rereplica plated on SC-Trp or FOA-Trp media and incubated for 3 days at 23 or 37°C. Strains displaying the desired phenotypes (see Results) were used for plasmid rescue into Escherichia coli, and the inserts in the recovered plasmids were sequenced. The yku70-30ts allele encodes the following mutations: Thr134Ser, Leu170Phe, and Phe215Ile. The pRS314 vector containing this allele is referred to as pKu70-30ts. Currently, we do not know whether all three mutations are necessary to confer the TS character of this allele.
Nucleic acid methods.
Southern blotting and in-gel hybridization analyses were performed using the CA oligonucleotide probe (CA probe) described previously (21). To determine the lengths of terminal restriction fragments, genomic DNA was digested with XhoI and separated by electrophoresis on 1% agarose gel. The DNA was then blotted onto a nylon membrane and probed with a 600-bp KpnI fragment derived from the telomere-proximal side of the subtelomeric Y′ element (46) (Y′ probe). Blots were scanned with a Packard InstantImager, and the sizes of the terminal restriction fragments (TRFs) were determined with the Imager software. Broad peaks corresponding to the TRFs were visualized with the software, and the migration distances of the tops, middles, and bottoms of the peaks with respect to the wells were defined. The migration distances were then converted by the software to DNA sizes by using the parameters determined by the migration of the molecular size markers (100-bp ladder; AP Biotech). The XhoI restriction site situated in the Y′ subtelomeric element is 870 bp from the start of the telomeric TG1-3 sequences (46). Consequently, the apparent lengths of telomeric repeats were determined by subtracting 870 bp from the lengths determined for the TRFs.
For RNA analysis by Northern blotting, cells were grown to mid-log phase at 23 or 37°C for various numbers of generations and total RNA was isolated by glass bead-mediated cell disruption in LETS buffer (0.01 M Tris [pH 7.5], 0.1 M LiCl, 0.01 M EDTA, 0.2% sodium dodecyl sulfate) (67). Standard techniques were used for gel electrophoresis (68). RNA (25 μg/sample) was blotted to a Hybond N+ membrane (Amersham Pharmacia Biotech) and hybridized according to the manufacturer's instructions. RNA blots were probed sequentially with 32P-labeled DNA fragments specific for the RNR2, RNR3, and RNR4 genes, as described previously (14). A probe specific for the RNA of the yeast ACT1 gene was used as an RNA loading control. The blots were scanned with a Molecular Dynamics PhosphorImager, and the radioactive signals were quantified with ImageQuant software. The RNR transcript levels were arbitrarily set at 1 in the wt cells grown at 23°C, and RNR levels in Yku− mutants were normalized accordingly.
RESULTS
Identification of a TS allele of YKU70.
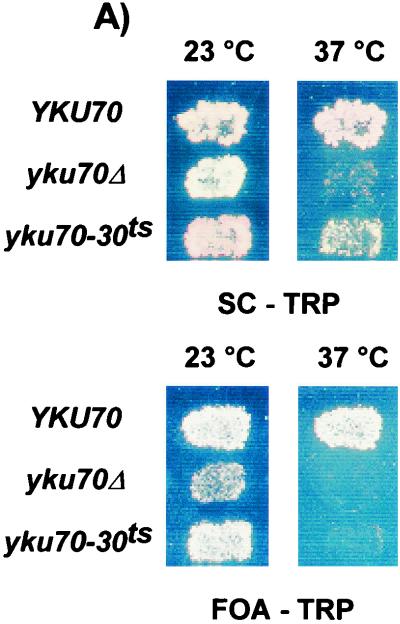
The ability to repress the transcription of telomere-proximal genes as well as viability at elevated temperature are drastically reduced in cells devoid of Yku (9, 10, 26, 31, 42, 58). We used these two properties to select for TS alleles of the YKU70 gene. The whole YKU70 gene was mutagenized by random PCR and expressed in a yku70Δ haploid strain (see Materials and Methods). We screened for mutants that could silence telomere-proximal reporter genes at 23°C and that showed reduced cell viability at 37°C. To assay telomere-proximal silencing, we monitored the expression of the ADE2 gene inserted near the telomeric repeats of chromosome V-R and the expression of the URA3 gene inserted near the telomeric repeats of chromosome VII-L (73). In cells with a wt allele of YKU70, the reporter genes were repressed and cells accumulated a red pigment (ADE2 assay; Fig. 1A, top). In addition, due to repression of the telomeric URA3 gene in wt cells, most cells were able to grow on medium containing 5-FOA, a drug toxic for cells expressing URA3 (URA3 assay; Fig. 1A, bottom). As previously reported, a strong derepression of these loci was observed in yku70Δ cells, leading to white patches in the ADE2 assay and significantly reduced growth in the URA3 assay, even at 23°C (Fig. 1A). Cells harboring the yku70-30ts allele, one of the alleles recovered in our screen, behaved very similarly to wt cells when grown at 23°C: they displayed accumulation of red pigment in the ADE2 assay and good viability in the URA3 assay (Fig. 1A). However, compared to YKU70 cells, pKu70-30ts-harboring cells displayed an important reduction in viability at 37°C (Fig. 1A).
FIG. 1.
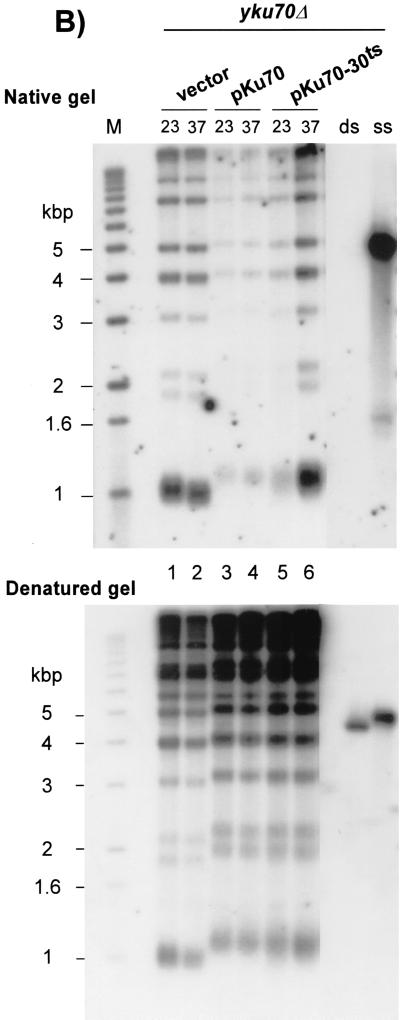
(A) Identification of a TS allele of YKU70. The entire YKU70 gene was randomly mutagenized, and the mutant alleles were expressed in a yku70Δ strain carrying the ADE2 and URA3 genes as reporter genes for telomere position effect (strain UCC3505/yku70Δ). Cells containing an empty vector (yku70Δ) or the vector containing the wt YKU70 gene (pKu70; YKU70) were used as controls for silencing status and growth at high temperature. Patches of cells were grown at 23°C on SC-Trp and then rereplica plated on the same medium at 37°C (ADE2 assay, top), or on FOA-Trp at 23 and 37°C (URA3 assay, bottom) and incubated for 3 days. (B) yku70-30ts cells grown at 37°C exhibit telomeric single-stranded G-rich DNA. Strain BYyku70Δ transformed with the indicated plasmids was grown in liquid culture at 23°C and then shifted to 37°C for 8 h. Genomic DNA was extracted and digested with XhoI, and telomeres were analyzed for G tails by nondenaturing in-gel hybridization with a CA oligonucleotide probe (top). To control for DNA loading, the DNA was denatured in the gel and rehybridized to the same probe (bottom). Lane M, end-labeled 1-kb ladder DNA serving as a size standard; lanes ss and ds, DNA with telomeric TG1-3 repeats in single-stranded and double-stranded forms, respectively, as described previously (21).
The telomeric end structure is modified in yku70Δ cells, and, in contrast to what is found for wt cells, long overhangs of the G-rich strand are present throughout the cell cycle (31, 63). As a further test for the temperature sensitivity of the yku70-30ts allele, the telomeric DNA end structure present in cells harboring the yku70-30ts allele was analyzed for cells grown at 23°C or shifted to 37°C for 8 h (about two generations). G-rich overhangs were then probed by in-gel hybridization. As shown in Fig. 1B, the amount of single-stranded G-rich overhangs detected on telomeres derived from yku70-30ts cells grown at 23°C is comparable to that detected on telomeres derived from YKU70 cells. However, after two generations of growth at 37°C, a strong signal for G-rich extensions is detected. The intensity of the signal is comparable to that of the one detected in DNA derived from yku70Δ cells (Fig. 1B, compare lanes 2 and 6). To control for DNA loading, the DNA was denatured in gel and rehybridized to the same probe (Fig. 1B, bottom).
These results indicate that, when cells harboring the yku70-30ts allele are grown at 23°C, the repression of telomere-proximal genes is normal (Fig. 1A). In addition telomere lengths as well as the telomeric DNA end structure are indistinguishable from those found in wt cells (Fig. 1B). However, the function of Yku in maintaining a normal telomeric end structure is lost when yku70-30ts cells are incubated at 37°C (Fig. 1B), the cells lose the ability to form colonies, and the ability to repress telomere-proximal genes is reduced (Fig. 1A). Furthermore, in an HO endonuclease cleavage assay developed by Haber and coworkers to measure NHEJ efficiency (43, 56), cells harboring the yku70-30ts allele are at least 10-fold more sensitive than wt cells to HO expression when the assay is performed at 37°C, while displaying the same resistance as wt cells at 23°C (data not shown). Thus, for all the phenotypes analyzed, cells harboring the yku70-30ts allele behave as if the Yku complex is functional at 23°C but nonfunctional at 37°C.
Telomerase is not involved in the initial generation of G-rich overhangs after loss of Yku function.
Yku− cells possess overhangs of the G-rich strand of >30 bases throughout the cell cycle (31, 63). One obvious candidate for the creation of these single-stranded extensions was telomerase. However, because of the nearly synthetic-lethal interaction between Yku and telomerase (31, 58), it was impossible to test directly the importance of this enzyme in the creation of the G-rich overhangs. The yku70-30ts allele allowed us to address this question. A deletion of TLC1, the gene encoding the RNA component of telomerase, was introduced in a strain harboring the yku70-30ts allele as the sole source of Yku70p (see Materials and Methods). This strain survives due to the presence of TLC1 on a URA3/CEN plasmid and/or by growth at 23°C, the permissive temperature for the yku70-30ts allele. Cells lacking TLC1 were selected for by growth on FOA-Trp media at 23°C. The FOA-resistant cells lack functional telomerase by two criteria. First, the cells lost viability after subculturing for about 40 to 60 generations at 23°C (data not shown). Second, compared to telomeres derived from TLC1 yku70-30ts cells, the lengths of the telomeres derived from tlc1Δ yku70-30ts cells were reduced in cells grown for about 25 generations in the absence of TLC1 (Fig. 2A, bottom, compare lanes 1 and 3). The Yku complex was inactivated in these cells by shifting the culture to 37°C for 8 h, which is sufficient to inhibit Yku function in these cells (Fig. 1B). As controls, telomerase-positive cells were also shifted to 37°C for the same amount of time. Total genomic DNA was extracted, and the G-strand overhangs were analyzed by in-gel hybridization. The signal for single-stranded G-rich DNA detected on telomeres derived from tlc1Δ yku70-30ts cells is comparable to the one detected on telomeres from TLC1 yku70-30ts cells (Fig. 2A, top, compare lanes 2 and 4). This result indicates that at least part of the initial creation of the G-rich overhangs observed in Yku− mutants is independent of telomerase.
Cells harboring specific TS alleles of the gene encoding the catalytic subunit of Polα, such as pol1-17, also have an altered telomeric end structure (2). As in Yku− mutants, the G-strand overhangs in Polα mutants are initially formed in a telomerase-independent manner. However, in contrast to Yku− mutants, pol1-17 mutants display a telomerase-dependent telomere elongation during outgrowth at a semipermissive temperature rather than telomere shortening (1, 2). In light of these observations, we were interested to address the relationship between Yku and Polα with respect to telomere length maintenance. To test this, pol1-17 yku70Δ double mutants were created by sporulating diploid strain SGY56 (see Materials and Methods) and tetrad dissection. Telomere length in cell cultures derived from all four spores of a tetratype was determined (Fig. 2B). Compared to those in the yku70Δ mutants, the telomeres in the pol1-17 yku70Δ double mutant are elongated, even when the cells are grown at 23°C, a temperature at which the pol1-17 mutation by itself only causes a modest phenotype (Fig. 2B, compare lanes 1 and 5). Telomere lengths are even more increased when the double mutants are grown at 30°C, the semipermissive temperature for the pol1-17 mutants (Fig. 2B, compare lanes 1 and 2). Thus, the defect in pol1-17 cells causes telomere elongation even if combined with a Yku− mutation.
For Yku− cells, the lengths of telomeric repeats correlate with the capacity to grow at elevated temperature.
Despite the presence of G-rich overhangs, the number of the telomeric repeats, and hence telomere length, were not diminished significantly in yku70-30ts cells grown for two generations at 37°C (Fig. 1B). Furthermore, as shown in Fig. 1A (ADE2 assay), the viability of yku70-30ts cells at 37°C was not affected to the same extent as that of yku70Δ cells. However, in initial experiments we observed that yku70-30ts cells lose telomeric repeats and viability after more-prolonged exposures to high temperature (see below and data not shown). These observations suggested a possible correlation between the length of the telomeric repeats and the viability of Yku− cells at 37°C. To explore this possibility, we measured these two parameters during outgrowth of Yku− mutants. In these analyses, cells devoid of Yku (yku70Δ strains) and strains harboring the yku70-30ts allele were used. Furthermore, a deletion of the RIF1 gene was introduced into those strains to generate yku70Δ rif1Δ and yku70-30ts rif1Δ double mutants. The RIF1 gene encodes a protein that localizes to telomeres, where it acts as an inhibitor of telomere elongation (55). As a result, rif1Δ cells have elongated telomeres (36).
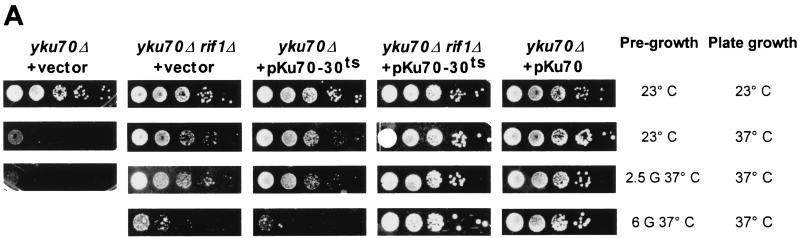
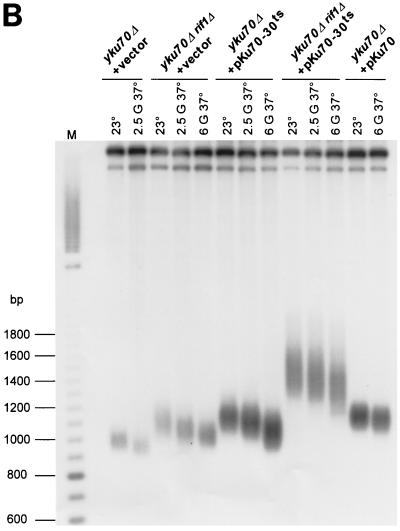
These different strains were grown in liquid medium at 23°C and then shifted to 37°C for 2.5 or 6 generations (or until loss of growth), and the viability at 37°C was assessed by plating 10-fold serial dilutions of the cultures at 37°C (Fig. 3A). Growth capacity was qualitatively assayed on plates based on sizes of colonies on the spots with diluted cultures and on which single-cell-derived colonies can be visualized. Cells were classified into three categories: (i) pregrowth and culture conditions allowed the formation of individual colonies similar to wt, (ii) individual colonies were smaller than wt (microcolonies) yet clearly visible, and (iii) individual microcolonies failed to form in spots of the diluted culture. We note that cells from some cultures incapable of forming microcolonies were able to form lawns at low dilutions. In parallel, aliquots of the same cultures used to determine viability were used to isolate genomic DNA and measure telomere repeat lengths at each time point by Southern blotting (Fig. 3B; all the data are summarized in Table 1). Thus, the DNA was digested with the XhoI restriction enzyme and the telomeric restriction fragments were detected with a telomere-proximal Y′ probe. Grown at 23°C, the different strains used here displayed telomeric repeat lengths of about 275 bp for wt, 130 bp for yku70Δ, 250 bp for yku70Δ rif1Δ, 265 bp for yku70-30ts, and 620 bp for yku70-30ts rif1Δ cells (Fig. 3B, lanes 23°; Table 1). A strain with a deletion of RIF1 alone (YKU70 rif1Δ) had telomere lengths very similar to those for the yku70-30ts rif1Δ strain grown at 23°C and displayed the growth characteristics of a wt strain in all conditions tested (data not shown). All strains form normal-size colonies when pregrown at 23°C and then incubated on plates at 23°C (Fig. 3A, top row).
FIG. 3.
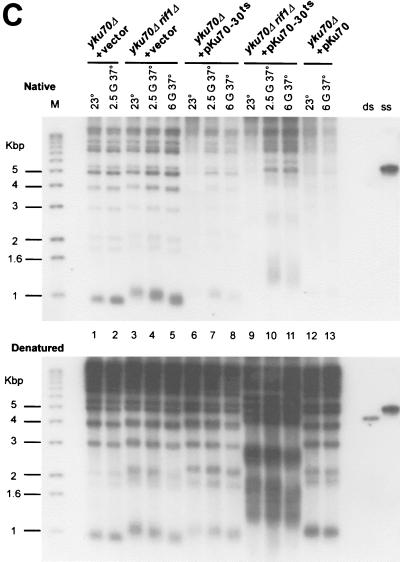
(A) Effects of deleting the RIF1 gene on growth of Yku− mutants at elevated temperatures. Cells with the indicated genotypes were grown at 23°C to mid-log phase and then diluted into prewarmed medium and regrown for 2.5 or 6 generations at 37°C. Following the growth period in liquid medium, 10-fold serial dilutions of the cells were spotted on SC-Trp and the plates were incubated for 3 days at 37°C. As controls, 10-fold serial dilutions of the cultures prior to the shift to 37°C were also spotted onto SC-Trp and incubated at 23 (top row) or 37°C (second row from top). (B) Telomere lengths in the strains analyzed in panel A. Genomic DNAs from the cultures tested in panel A were digested with XhoI, and the TRFs were detected by standard Southern blotting using a probe that is specific for telomere-proximal Y′ sequences. Lane M, end-labeled 100-bp ladder DNA serving as a size standard. G, generations. (C) In Yku− cells, deletion of the RIF1 gene does not restore a normal terminal DNA structure on telomeres. Aliquots of the genomic DNAs tested in Fig. 3B were analyzed by in-gel hybridization as for Fig. 1B. G-rich overhangs were detected with a CA probe (top). The rehybridization of the gel after denaturation (bottom) indicates that similar amounts of DNA were loaded in each lane.
TABLE 1.
Telomere lengths and growth phenotypes of Yku mutants
| Strain genotype | Telomere lengtha and growthb under conditions of:
|
|||||
|---|---|---|---|---|---|---|
| Pregrowth at 23°C, plate growth at 23°C
|
Pregrowth for 2.5 generations at 37°C, plate growth at 37°C
|
Pregrowth for 6 generations at 37°C, plate growth at 37°C
|
||||
| Telomere length (bp) | Growth | Telomere length (bp) | Growth | Telomere length (bp) | Growth | |
| yku70Δ | 130 (55-230) | + | 95 (40-180) | − | N.A.d | N.A. |
| yku70Δ rif1Δ | 250 (100-380) | + | 205 (80-330) | +/− | 155 (75-280) | − |
| yku70-30ts | 265 (160-395) | + | 220 (110-360) | +/− | 180 (76-330) | − |
| yku70-30ts rif1Δ | 620 (385-880) | + | 565 (360-875) | + | 465 (280-720) | + |
| YKU70 | 275 (185-370) | + | N.D.c | + | 265 (145-360) | + |
Telomere length after the pregrowth period was calculated from Southern analysis (Fig. 3B) as described in Materials and Methods. The means and ranges are indicated.
The growth phenotype was evaluated from Fig 3A. +, formation of colonies similar to wt; +/−, formation of microcolonies; −, microcolonies not detectable in spots of the diluted culture.
N.D., not determined.
N.A., not applicable due to cell mortality.
Yku70Δ cells grown for 2.5 generations in liquid medium at 37°C do not grow further when incubated at the same temperature on plates (Fig. 3A, first column from left) or in liquid culture (data not shown). Interestingly, yku70Δ cells that have elongated telomeres conferred by the RIF1 deletion show an important growth recovery at the same time point, without however being able to form wt size colonies (Fig. 3A, second column). The Southern analysis revealed that culturing at 37°C resulted in a gradual telomere shortening. From 250 bp, the length at 23°C, the lengths of telomeric repeats were decreased to reach a mean of 205 bp after 2.5 generations and 155 bp after 6 generations at 37°C (Fig. 3B). After six generations of pregrowth in liquid medium at 37°C, the cells were not able to form colonies on the plates. The profile of viability and telomere loss for cells harboring the yku70-30ts allele was very comparable the one for yku70Δ rif1Δ cells. A shift to 37°C produced a rapid telomere shortening accompanied by a marked decrease in viability (Fig. 3). The yku70-30ts rif1Δ cells, which have the most-elongated telomeres of all the Yku− mutants tested, showed an interesting phenotype. Even after six generations of growth at 37°C, these yku70-30ts rif1Δ cells showed no growth defect and formed wt size colonies when plated at 37°C (Fig. 3A, fourth column). At this time point, these cells possessed a mean telomere length of 465 bp, more than twice the lengths of telomeres in yku70-30ts and yku70Δ rif1Δ cells at the same time point (Fig. 3B).
These results were in agreement with a model in which the viability of Yku− cells at 37°C is dictated by the overall lengths of telomeric repeats. However it was possible that the gain of viability caused by the deletion of the RIF1 gene was due to a correction of the aberrant terminal structure present in Yku− cells. Thus, we analyzed the G-strand overhangs in yku70Δ rif1Δ and yku70-30ts rif1Δ cells (Fig. 3C) at each of the time points where viability was tested. Comparable signals for G-strand overhangs were detected on telomeres derived from yku70Δ cells and yku70Δ rif1Δ cells (Fig. 3C, top, compare lanes 1 and 2 and 3 and 4). Also, G-strand overhangs are clearly detectable on telomeres derived from yku70-30ts rif1Δ cells shifted to 37°C for 2.5 or 6 generations (Fig. 3C, lanes 10 and 11). These results demonstrate that a deletion of the RIF1 gene does not restore a normal terminal DNA structure to telomeres in Yku− cells and show that the aberrant end structure present on telomeres of Yku− cells per se does not interfere with growth at high temperatures.
The DNA damage checkpoint is activated in Yku− mutants with short telomeres.
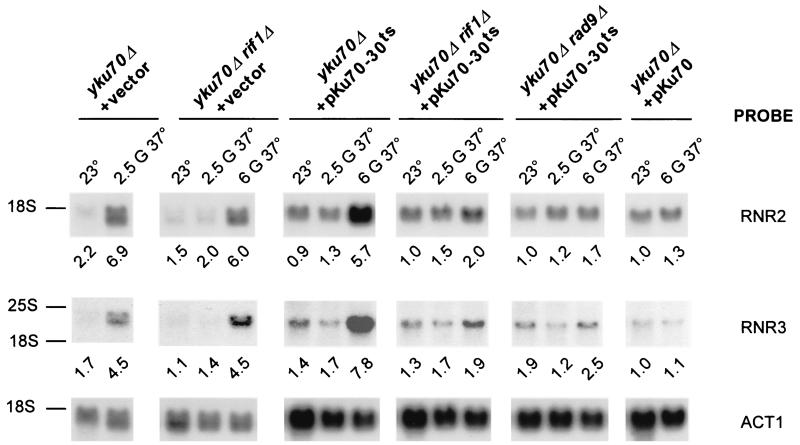
When incubated at 37°C, Yku− cells arrest in the G2/M phase of the cell cycle (5, 26). This arrest is dependent on the Rad9p protein, a component of the DNA damage checkpoint (5, 75), suggesting that DNA damage is detected in Yku− mutants grown at high temperature. Because Yku− mutants with elongated telomeres gain viability at elevated temperature (Fig. 3A), we wondered whether activation of the DNA damage checkpoint was correlated with overall telomere length, rather than DNA end structure (see above). The DNA damage checkpoint response is initiated by recognition and/or processing of the damage, and it has been proposed that several proteins are involved in these processes. These include the Rad17p, Rad24p, Mec3p, Ddc1p, Rad9p, and Lcd1/Ddc2p proteins, all of them being essential for efficient checkpoint functions in both the G1 and G2/M phases of the cell cycle (47, 59, 79). Once the damage is recognized, the Mec1p and Rad53p kinases are activated and phosphorylate appropriate substrates. One of the downstream effects in response to DNA damage is the induction of ribonucleotide reductase gene (RNR) transcription (3, 24). Since this transcriptional induction is under the control of the DNA damage checkpoint signaling pathway, a measurement of the RNR transcript levels reflects the checkpoint activation status. We used this assay to monitor checkpoint activation in cells derived from the same cultures used to assay cellular viability as well as telomere length and structure (Fig. 3). Total RNA was prepared, and the levels of transcripts specific for the RNR2, RNR3, and RNR4 genes were detected and quantified by Northern analysis (Fig. 4 and data not shown). The RNR transcript levels in the wild-type cells grown at 23°C were arbitrarily set at 1, and levels in Yku− mutants were normalized accordingly. A robust transcriptional activation of the RNR genes is detected in yku70Δ cells grown for 2.5 generations at 37°C (Fig. 4, first column from left). Interestingly, the checkpoint activation is delayed in yku70Δ rif1Δ and yku70-30ts mutants. In these strains, the highest transcriptional activation observed required six generations of growth at 37°C (Fig. 4, second and third columns). In yku70-30ts rif1Δ cells, which have extremely elongated telomeres and no observable growth defect after six generations at 37°C, only a marginal activation of RNR transcription is detectable (Fig. 4, fourth column). Furthermore, the transcriptional activation of RNR genes in yku70-30ts cells is almost completely abolished by deletion of the RAD9 gene (Fig. 4, fifth column), indicating that the detected transcriptional induction of the RNR genes is due to RAD9-dependent DNA damage checkpoint activation. Deletion of RAD9 does not, however, rescue the growth defect of yku70-30ts cells at 37°C (data not shown). This result suggests that the growth arrest at 37°C in these cells is not solely due to a prolonged cell cycle arrest.
FIG. 4.
DNA damage checkpoint activation in Yku− cells correlates with short telomeres. Total RNA was extracted from the cells tested in Fig. 3A and transferred to nylon membranes. The membranes were successively probed to detect RNR2, RNR3, and actin transcripts. To correct for RNA loading, the signals specific for the RNR transcripts were normalized to the actin signal. The RNR transcript levels were arbitrarily set at 1 in the wt cells grown at 23°C, and RNR transcript levels in Yku− mutants were determined accordingly. The migration positions of the 25S rRNA (3,392 nucleotides) and 18S rRNA (1,798 nucleotides) are indicated as molecular weight references. G, generations.
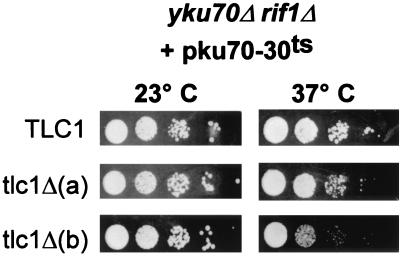
It has been shown that overexpression of telomerase subunit Est2p or TLC1 suppresses checkpoint activation and restores viability at high temperature in Yku− cells without apparently modifying telomere length (75). To explain these observations, it has been proposed that, in the absence of Yku, components of yeast telomerase could play a telomere-capping function important for viability at high temperatures (75). To explore this possibility, we asked if inactivation of telomerase in Yku− mutants with elongated telomeres would also cause a major capping defect that would result in a dramatic and immediate decrease in viability such as is observed for telomerase− Yku− cells. We used strain SGY53 (tlc1Δ yku70Δ rif1Δ) containing both pKu70-30ts and pAZ1(TLC1) in these assays. In this strain, telomeres are elongated due to the rif1 deletion (Fig. 3). By a passage on FOA-Trp media at 23°C, we selected an SGY53 strain that contained pKu70-30ts as the sole source of Yku. This strain (tlc1Δ yku70Δ rif1Δ pKu70-30ts) and the strain still containing the wt TLC1 gene (tlc1Δ yku70Δ rif1Δ pKu70-30ts pAZ1[TLC1]) were then grown in liquid medium at 23°C, and viability of the Yku− mutant strains was determined by plating 10-fold serial dilutions of the cultures at 37°C. As shown in Fig. 5, the combined absence of telomerase and Yku in this strain with elongated telomeres reduced viability at 37°C only modestly, in contrast to the lethality observed with yku70Δ tlc1Δ strains. This result suggests that telomerase inactivation does not modify the capping status of telomeres in Yku− mutants, provided that there is a significant number of telomeric repeats present.
FIG. 5.
A combined absence of telomerase and Yku does not lead to immediate growth arrest in cells with elongated telomeric repeat tracts. Strain SGY53 (yku70Δ tlc1Δ rif1Δ pKu70-30ts) and the same strain carrying in addition a plasmid-borne copy of TLC1(pAZ1) were grown at 23°C in liquid medium. To assess viability of these strains, 10-fold serial dilutions of the cells were spotted on SC-Trp plates and incubated at 23 or 37°C. tlc1Δ(a) and -(b), two independent clones of the strain without pAZ1.
DISCUSSION
Yeast cells devoid of Yku exhibit excess of single-stranded G-rich DNA at their telomeres (31, 63). One obvious candidate for the creation of these overhangs was telomerase. The yku70-30 TS allele of the YKU70 gene described in this study allowed us to determine that, upon loss of Yku function, telomerase per se is not an essential factor for the creation of the overhangs (Fig. 2A). These data are consistent with our earlier findings, which indicated that the cell cycle-dependent occurrence of G-rich overhangs at yeast telomeres can also be detected in cells lacking telomerase (21, 80). It is therefore possible that the overhangs observed in Yku− mutants are generated via the same mechanisms as the S-phase-specific overhangs in wt cells but would become permanent in Yku− mutants.
Such single-stranded DNA overhangs are important determinants for recognition as DNA damage (43). Once such DNA sensed, the cell cycle is arrested and the damage is repaired. Curiously, the constitutive telomeric overhangs observed in Yku− cells appear not to cause a cell cycle arrest, at least when the cells are incubated at 23°C (75). However, Yku− cells are unable to grow at 37°C, and it was possible that, at this temperature, the single-stranded overhangs were recognized as damage and the DNA damage checkpoints were activated. This argument is complicated by the fact that, in Yku− cells, the double-stranded tracts of telomeric repeats are extremely short (9, 64), and any failure of mechanisms to maintain that double-stranded portion may have the same effect, independently of the presence of the overhangs. To address the question as to whether the overhangs are responsible for temperature sensitivity of Yku− cells at 37°C, we created situations in which the double-stranded tracts were elongated to at least wt levels but in which the terminal structure still comprised the overhangs. We used the fact that a deletion of the gene encoding the Rif1p protein, which is part of the telomeric chromatin complex, induces a lengthening of the telomeric repeat tracts (11, 36, 55, 83). Thus, yku70Δ rif1Δ cells incubated at 23°C harbor telomeric tracts that are comparable in length to those found in wt cells or yku70-30ts cells (Table 1). The telomeric DNA end structure in these cells did, however, comprise single-stranded overhangs, indicative of an absence of Yku function (Fig. 3C), and these cells were able to form colonies at 37°C when there was no preexposure to 37°C (Fig. 3A). Similarly, yku70-30ts rif1Δ cells were able to form wt size colonies at 37°C on plates, even after a pregrowth of six generations at 37°C in liquid media, after which G-strand extensions are detectable (Fig. 3). Thus, clearly the inability to from colonies at 37°C did not correlate with the presence of the G-strand overhangs. In these experiments, we also assessed the activation of the RAD9-dependent DNA damage checkpoint by analyzing RNR transcript levels on Northern blots after an incubation of the various cells at 37°C (Fig. 4). For all situations tested, we observed that, if the preincubation of the cells in liquid media had no effect on RNR transcript levels, the cells were able to form colonies on plates incubated at 37°C and, conversely, if the RNR transcript levels were induced, no more colony formation on plates was discernible. For example, yku70Δ rif1Δ cells or yku70-30ts cells incubated in liquid media at 37°C for 2.5 generations displayed G-strand overhangs and virtually no RNR gene induction and were able to form colonies on the plates afterward (Fig. 3A and 4). In these same cells, after six generations of growth at 37°C, the detection of single-stranded overhangs is unchanged, yet RNR transcript levels are induced and the cells are not able to form colonies anymore. We conclude that the altered terminal DNA structure found in Yku− cells alone is not sufficient to induce DNA damage checkpoint activation and is not the cause for the growth arrest of such cells at 37°C. Consistent with these results, the altered DNA end structure in Yku− cells remains unchanged when single telomerase subunits are expressed, yet such cells are also able to grow at 37°C (75).
However, our experiments demonstrate that the apparent lengths of the TRFs correlated with suppression of checkpoint activation and the capacity to grow at 37°C (Fig. 3 and 4 and Table 1). For example, at 23°C, yku70Δ rif1Δ and yku70-30ts cells maintain telomeres of about wt lengths. Without preincubation at 37°C or after a pregrowth for only two generations at 37°C the telomeres are longer than those found in yku70Δ cells and these cells are still able to form colonies on plates incubated at 37°C. However, during growth at 37°C, telomeric repeats are being lost and, after about six generations of growth, the lengths reach levels that are quite comparable to those in yku70Δ cells grown at 23°C (Table 1). Importantly, after these six generations of preincubation at 37°C, these same cells are not able to form colonies anymore at 37°C. Most significantly, yku70-30ts rif1Δ cells harbor elongated telomeres comparable in length to those of rif1Δ cells at 23°C (Fig. 3B, Table 1, and data not shown). Such yku70-30ts rif1Δ cells are able to form normal colonies, even after pregrowth for six generations at 37°C (Fig. 3A). Again, telomeric repeats are being lost during growth at 37°C, but the average length of the telomeres after six generations of growth still was significantly larger than that in yku70Δ cells (465 versus 130 bp; Table 1). Thus, in all cases investigated here, the apparent lengths of telomeric tracts, and in particular the sizes of the shortest telomeres, correlated with RNR transcript induction and the ability to form colonies at 37°C.
Recent evidence suggests that Yku itself may be important for recruitment or activation of telomerase (62). In all of our experiments, we observed progressive losses of telomeric repeats when Yku− cells were shifted to 37°C. Using gels such as that shown in Fig. 3B, we estimate that the loss rates after abolition of Yku function are about 10 to 15 bp/generation, even in the presence of telomerase. These loss rates are about two- to threefold higher than those occurring due to an absence of telomerase (44, 49, 73). Moreover, yku70Δ rif1Δ strains harbor telomeres of almost normal length, and yku70Δ cells are able to maintain very short telomeric repeats, when these strains are grown at 23°C. These data lend support to the idea that Yku is involved in protecting yeast chromosome ends from sequence losses other than or in addition to those occurring due to a lack in G-strand synthesis alone.
Thus, our results are consistent with the hypothesis that, in Yku− cells, some aspect of the mechanisms involved in maintaining a minimal tract of double-stranded telomeric repeats is TS. Recently, it has been postulated that factors involved in lagging-strand synthesis are important for telomere length regulation and new-telomere synthesis (2, 19, 65). For example, a mutation in the gene encoding Polα (pol1-17) will lead to extensive, but transient, G-rich overhangs at telomeres and an overall lengthening in telomeric repeats (2, 13). This same mutation can at least partially restore the telomere-shortening phenotype of Yku− cells (Fig. 2B). While it is unclear how the pol1-17 mutation eventually leads to telomeric repeat expansion, our results do suggest that, in the absence of Yku, C-strand synthesis is a crucial determinant for maintaining a functional repeat tract. Therefore, we favor the idea that Yku has a function in telomere replication. In the absence of Yku, the telomeric C-rich strand may be incompletely replicated and the naturally transient extensions would become permanent. In this situation, in order to compensate for incomplete C-strand replication and the resulting increased rates of loss of telomeric sequences, a more active G-strand elongation by telomerase would be required to generate stable telomeres. When telomeric repeat tracts are still relatively long, the Rif-mediated inhibition of telomere extension could prevent sufficient Yku-independent elongation, leading to a rapid overall telomere shortening. However, at least at 23°C, once the telomeres reach very short lengths, this cis inhibition would drop to a level that allows the Yku-independent telomere-lengthening process to counteract further losses.
In this scenario, Yku could play a stabilizing role in the setup of the complete terminal complex required for the elongation of both telomeric strands. This idea would explain why both mutations relieving inhibition of G-strand synthesis (rif1Δ) and mutations affecting actual C-strand synthesis (pol1-17) can suppress the telomere length deficiency in cells lacking Yku. This stabilizing activity of Yku would explain why Yku-independent telomere maintenance, while in principle possible, is inefficient at 23°C, leading to very short telomeres and incomplete synthesis of the C-rich strand, and is unable to maintain a minimal double-stranded telomeric repeat tract at 37°C.
Formally, we cannot rule out the possibility that very short telomeres combined with an altered DNA end structure trigger a reorganization of the global telomere architecture toward a capped state that is TS. Upon shift to higher temperatures, the capping function would be lost and the ends would be recognized as DSBs. However, we were unable to uncover such an unstable capping state in cells with elongated telomeres (Fig. 5). Thus, in both scenarios, ultimately it is the ability to maintain a critical minimal amount of double-stranded telomeric repeats which is the critical determinant for the growth properties of Yku− mutants at high temperature.
Cells suffering damage to their DNA activate DNA damage checkpoints in order to slow down cell cycle progression and turn on repair mechanisms. Surprisingly, Yku− cells preincubated at 37°C in which the RAD9-dependent checkpoint is activated are unable to form colonies, even when plates are incubated at 23°C (data not shown). Thus, despite activation of the DNA damage checkpoints, massive cell death or permanent cell cycle arrest occurs when Yku− cells are incubated for prolonged periods at 37°C. This suggests that the incurred damages are too important to be repaired or that, because the chromosome ends are now recognized as DSBs, they are repaired as such and telomere-telomere fusions and other aberrant chromosome rearrangement events occur. Eventually, such events could lead to unrecoverable chromosomal aberrations, which would be responsible for cell death.
In summary, we found that, in the absence of the Yku complex, yeast telomeres acquire extensions of the G-rich strand independently of a functional telomerase and that the DNA damage checkpoint machinery is not activated by these extensions if a minimal tract of double-stranded telomeric repeats is maintained. However, upon a shift to 37°C, Yku− cells suffer increased overall telomere attrition, which is detrimental to cell viability. While some telomeric repeats can still be detected on chromosomal DNA derived from Yku− cells grown at 37°C, all the data are also consistent with the hypothesis that some chromosome ends have lost the repeats altogether. Recent results derived from the analysis of DNA isolated from telomerase− cells undergoing senescence support a similar conclusion (35, 37). Finally, our results further support the notion that the regulation and coordination of the synthesis of the G-rich strand by telomerase with that of the C-rich strand are important for maintaining a functional telomeric repeat tract. The Ku complex can also be detected on telomeres of mammalian cells (4, 39). In this context, Ku has been postulated to suppress genomic instability, including telomere fusions, and oncogenesis in mice (20). Moreover, recently it was reported that mammalian cells lacking Ku have shortened telomeres as well (16). Although there are differences in the terminal DNA structures between yeast and mammalian cells, a role for Ku in the efficient replication of both strands of telomeric repeats is consistent with the findings for both systems. Since the maintenance of functional telomeres is an essential process for cell viability and since deficiencies in this process increase susceptibility to cell transformation, the results presented here may be of importance for the understanding of complete telomere replication in mammalian cells as well.
Acknowledgments
We thank D. Gottschling, S. Elledge, J. Haber, S.-E. Lee, S. Labbé, and V. Lundblad for providing yeast strains and plasmids. We also thank Stéphanie Larose for her help with RNA preparations. Members of the Wellinger lab are thanked for their continued intellectual input, and K. Runge is thanked for his insightful comments on the manuscript.
This work was supported by CCS research grant 010049 from the National Cancer Institute of Canada (NCIC) and a core facility group grant by the Canadian Institutes of Health Research (MGC-48372). R.J.W. is a Chercheur-National of the FRSQ.
REFERENCES
- 1.Adams, A. K., and C. Holm. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams-Martin, A., I. Dionne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li, B. E. Lehnert, and E. H. Goodwin. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96:14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, G., and D. Rio. 1997. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:867-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beeler, T., K. Gable, C. Zhao, and T. Dunn. 1994. A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. J. Biol. Chem. 269:7279-7284. [PubMed] [Google Scholar]
- 8.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 9.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourns, B. D., M. K. Alexander, A. M. Smith, and V. A. Zakian. 1998. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18:5600-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 13.Carson, M. J., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 14.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Counter, C. M., M. Meyerson, E. N. Eaton, and R. A. Weinberg. 1997. The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. USA 94:9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d'Adda di Fagagna, F., M. P. Hande, W. Tong, D. Roth, P. M. Lansdorp, Z. Wang, and S. P. Jackson. 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11:1192-1196. [DOI] [PubMed] [Google Scholar]
- 17.Dasika, G. K., S. C. Lin, S. Zhao, P. Sung, A. Tomkinson, and E. Y. Lee. 1999. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 18:7883-7899. [DOI] [PubMed] [Google Scholar]
- 18.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 20.Difilippantonio, M. J., J. Zhu, H. T. Chen, E. Meffre, M. C. Nussenzweig, E. E. Max, T. Ried, and A. Nussenzweig. 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driller, L., R. J. Wellinger, M. Larrivee, E. Kremmer, S. Jaklin, and H. M. Feldmann. 2000. A short C-terminal domain of Yku70p is essential for telomere maintenance. J. Biol. Chem. 275:24921-24927. [DOI] [PubMed] [Google Scholar]
- 23.Dynan, W. S., and S. Yoo. 1998. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26:1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elledge, S. J., and R. W. Davis. 1987. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell. Biol. 7:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Featherstone, C., and S. P. Jackson. 1999. Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 434:3-15. [DOI] [PubMed] [Google Scholar]
- 26.Feldmann, H., and E. L. Winnacker. 1993. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 268:12895-12900. [PubMed] [Google Scholar]
- 27.Fellerhoff, B., F. Eckardt-Schupp, and A. A. Friedl. 2000. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics 154:1039-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Getts, R. C., and T. D. Stamato. 1994. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J. Biol. Chem. 269:15981-15984. [PubMed] [Google Scholar]
- 30.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravel, S., M. Larrivee, P. Labrecque, and R. J. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280:741-744. [DOI] [PubMed] [Google Scholar]
- 32.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337-365. [DOI] [PubMed] [Google Scholar]
- 33.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 34.Haber, J. E. 2000. Partners and pathways repairing a double-strand break. Trends Genet. 16:259-264. [DOI] [PubMed] [Google Scholar]
- 35.Hackett, J. A., D. M. Feldser, and C. W. Greider. 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106:275-286. [DOI] [PubMed] [Google Scholar]
- 36.Hardy, C. F., L. Sussel, and D. Shore. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6:801-814. [DOI] [PubMed] [Google Scholar]
- 37.Hemann, M. T., M. A. Strong, L.-Y. Hao, and C. W. Greider. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77. [DOI] [PubMed] [Google Scholar]
- 38.Henderson, E. R., and E. H. Blackburn. 1989. An overhanging 3′ terminus is a conserved feature of telomeres. Mol. Cell. Biol. 9:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu, H. L., D. Gilley, E. H. Blackburn, and D. J. Chen. 1999. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 96:12454-12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang, M., and S. J. Elledge. 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6105-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klobutcher, L. A., M. T. Swanton, P. Donini, and D. M. Prescott. 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl. Acad. Sci. USA 78:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde, E. J. Louis, and S. M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653-656. [DOI] [PubMed] [Google Scholar]
- 43.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 44.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 46.Louis, E. J., and J. E. Haber. 1990. The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics 124:533-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 48.Lundblad, V. 2000. DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res. 451:227-240. [DOI] [PubMed] [Google Scholar]
- 49.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 50.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 51.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 52.McClintock, B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26:234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McElhinny, N. S. A., C. M. Snowden, J. McCarville, and D. A. Ramsden. 2000. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 16:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra, K., and D. Shore. 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol. 9:1123-1126. [DOI] [PubMed] [Google Scholar]
- 56.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller, H. J. 1938. The remaking of chromosomes. Collecting Net 13:181-195, 198.
- 58.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 59.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 60.Pang, D., S. Yoo, W. S. Dynan, M. Jung, and A. Dritschilo. 1997. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 57:1412-1415. [PubMed] [Google Scholar]
- 61.Pastink, A., and P. H. Lohman. 1999. Repair and consequences of double-strand breaks in DNA. Mutat. Res. 428:141-156. [DOI] [PubMed] [Google Scholar]
- 62.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger, C. O. Johnson, M. Tzoneva, and D. E. Gottschling. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27:64-67. [DOI] [PubMed] [Google Scholar]
- 63.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 64.Porter, S. E., P. W. Greenwell, K. B. Ritchie, and T. D. Petes. 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price, C. M. 1997. Synthesis of the telomeric C-strand. A review. Biochemistry 62:1216-1223. [PubMed] [Google Scholar]
- 66.Ramsden, D. A., and M. Gellert. 1998. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 17:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 68.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul, and M. A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 71.Shippen-Lentz, D., and E. H. Blackburn. 1990. Functional evidence for an RNA template in telomerase. Science 247:546-552. [DOI] [PubMed] [Google Scholar]
- 72.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 74.Teo, S. H., and S. P. Jackson. 2000. Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr. Biol. 10:165-168. [DOI] [PubMed] [Google Scholar]
- 75.Teo, S. H., and S. P. Jackson. 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walmsley, R. W., C. S. Chan, B. K. Tye, and T. D. Petes. 1984. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310:157-160. [DOI] [PubMed] [Google Scholar]
- 77.Wang, S. S., A. F. Pluta, and V. A. Zakian. 1989. DNA sequence analysis of newly formed telomeres in yeast. Prog. Clin. Biol. Res. 318:81-89. [PubMed] [Google Scholar]
- 78.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239:197-201. [DOI] [PubMed] [Google Scholar]
- 79.Weinert, T. 1998. DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev. 8:185-193. [DOI] [PubMed] [Google Scholar]
- 80.Wellinger, R. J., K. Ethier, P. Labrecque, and V. A. Zakian. 1996. Evidence for a new step in telomere maintenance. Cell 85:423-433. [DOI] [PubMed] [Google Scholar]
- 81.Wellinger, R. J., and D. Sen. 1997. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 33:735-749. [DOI] [PubMed] [Google Scholar]
- 82.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 83.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
- 84.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]
- 85.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]