Abstract
Approximately 30 copies of the Ty1 retrotransposon are present in the genome of Saccharomyces cerevisiae. Previous studies gave insights into the global regulation of Ty1 transcription but provided no information on the behavior of individual genomic elements. This work shows that the expression of 31 individual Ty1 elements in S288C varies over a 50-fold range. Their transcription is repressed by chromatin structures, which are antagonized by the Swi/Snf and SAGA chromatin-modifying complexes in highly expressed Ty1 elements. These elements carry five potential Gcn4 binding sites in their promoter regions that are mostly absent in weakly expressed Ty1 copies. Consistent with this observation, Gcn4 activates the transcription of highly expressed Ty1 elements only. One of the potential Gcn4 binding sites acts as an upstream activating sequence in vivo and interacts with Gcn4 in vitro. Since Gcn4 has been shown to interact with Swi/Snf and SAGA, we predict that Gcn4 activates Ty1 transcription by targeting these complexes to specific Ty1 promoters.
The Ty retrotransposons of yeast are structurally and functionally related to retroviruses. They transpose via an RNA intermediate, encode a reverse transcriptase, and produce virus-like particles. The Ty1 copia-like retrotransposon is the most abundant in Saccharomyces cerevisiae.
Ty1 transcripts are initiated in the 5′ long terminal repeat (LTR; also called δ) and are terminated in the 3′ LTR. They account for nearly 10% of total mRNA in haploid cells. Two TATA boxes and an upstream activating sequence (UAS) element are located in the 5′ LTR (13, 17). Additional regulatory sequences are located downstream of the LTR in the internal portion of Ty1, within the first kilobase of the element (reviewed in reference 6). The Ste12 and Tec1 transcriptional activators of the Kss1 mitogen-activated protein kinase pathway recognize a sequence located within the TY1A open reading frame (4). In haploid cells, Ste12 and Tec1 are required for Ty1 transcription (30, 34). In diploid cells, they activate Ty1 transcription only under conditions that induce the Kss1 invasive-filamentous pathway (34). Ty1 transcription is also dependent on Gcr1, a transcriptional activator of glycolytic enzymes. Gcr1 recognizes a sequence located upstream of the TATA boxes in the 5′ LTR (13, 50). In addition, the Mcm1, Tea1, and Rap1 DNA-binding factors have been shown to interact with a region in Ty1 called the downstream enhancer-like region and to activate Ty1 transcription (15, 20, 21). Likewise, Mot3, a zinc finger DNA binding protein, binds the Ty1 LTR in vitro and represses Ty1 transcription modestly in vivo (32). Finally, an a1/α2 repressor complex binding site located in the TY1A sequence may contribute to the repression of Ty1 transcription in diploid cells (16). Given this complexity, the Ty1 promoter is closer to higher eukaryotic promoters, which contain many regulatory elements over a long distance, than to bona fide S. cerevisiae promoters, which are usually compact and have fewer elements located upstream of the TATA box.
Insertions of Ty1 elements into the promoter sequence of a gene can modify the transcriptional regulation of that gene, either increasing or decreasing its expression (reviewed in reference 6). Analysis of extragenic suppressors of inactivating Ty1 insertions that reestablish adjacent gene expression, called spt for suppressors of Ty, led to the identification of several genes required for Ty1 expression (reviewed in reference 53). In spt3, spt7, spt8, and spt20 mutants, Ty1 mRNA levels are strongly reduced. The Spt3, Spt7, Spt8, and Spt20 proteins are components of the SAGA (Spt-Ada-Gcn5 acetyltransferase) histone acetyltransferase complex (19). Independently, it was shown that Ada2, Ada3, and Gcn5 proteins, components of the SAGA complex, are also required for Ty1 transcription (42). SAGA combines histone acetylation, activator interactions, and TATA box binding protein interactions (46). Transcription of Ty1 also depends on the Snf2, Snf5, and Snf6 proteins of the Swi/Snf nucleosome-remodeling complex (9, 22). Swi/Snf perturbs nucleosome structure in a way that increases accessibility to nucleosomal DNA (reviewed in reference 52). Both SAGA and Swi/Snf complexes activate the transcription of a subset of genes including HO, SUC2, INO1, and Ty1 elements (42, 54). In contrast, the Isw1 and Isw2 chromatin-remodeling ATPases, which affect δ element chromatin, inhibit Ty1 transcription (27). The involvement of SAGA, Swi/Snf, Isw1, and Isw2 complexes known to regulate RNA polymerase II access to DNA strongly suggests that Ty1 transcription may be regulated by particular chromatin conformations. This hypothesis is strengthened by the fact that nucleosome depletion suppresses Ty1 transcriptional defects observed in snf2 mutants, which are unable to antagonize repressive chromatin structures (25).
Thirty-two copies of Ty1 are present in the genome of strain S288C (28). Previous measurements of steady-state Ty1 mRNA levels gave an overview of the regulation of resident Ty1 element expression. However, they provided no information on the individual behavior of these elements. A study on de novo Ty1 insertions showed that the position of integration influences Ty1 expression (7, 11). For example, transcription of Ty1 elements located in the RDN1 locus is silenced (7). To obtain insight into the expression of resident Ty1 elements, we constructed haploid strains, each expressing lacZ under the transcriptional control of a different Ty1 element at its original locus (34). The results presented below show that genomic Ty1 elements are expressed at very different levels and suggest explanations for this behavior.
MATERIALS AND METHODS
Yeast strains and media.
Yeast were grown in rich YPD, Hartwell’s synthetic complete (HC), and synthetic minimum (SD) media as described elsewhere (1). Unless otherwise stated, 2% glucose was used as the carbon source. Yeast transformations were performed by the lithium acetate procedure (1). Strains are derivatives of FYBL1-23D, matα flo8-1 ura3-Δ851 trp1Δ63 his3Δ200 (isogenic to S288C [34]) or Y328, MATα gcn4-2 ura3-52 leu2-3,112 (gcn4-2 is a partial deletion of GCN4 [2]).
To obtain 31 strains, each expressing lacZ from a different Ty1 element, FYBL1-23D was transformed with a ′ty1a′-lacZ-URA3-′ty1b′ DNA fragment carrying lacZ fused in-frame to TY1A (at coordinate 1571 of Ty1-H3 [5]). In this construct, the ′ty1a′ and ′ty1b′ sequences come from Ty1-H3 (coordinates 1144 to 1571 and 2171 to 3726, respectively) that is highly homologous to the genomic Ty1 elements. Ura+ recombinants were analyzed by PCR and Southern blotting to identify which Ty1 element carried the fusion (34). Because we failed to obtain lacZ fusions at Ty1(BL), Ty1(LR2), and Ty1(ML2) by this method, we targeted integration at these three loci by transforming FYBL1-23D with three ty1a-lacZ-URA3-downTy1 constructs. In these constructs, the downTy1 portion corresponds to a sequence of approximately 800 bp immediately downstream of either Ty1(BL), Ty1(LR2), or Ty1(ML2) that was amplified by PCR from genomic DNA. Ultimately, fusions to lacZ were obtained with all elements present in S288C, except Ty1(H). The TY1A-lacZ fusions were named according to the Ty1 sequence annotation by the Munich Information Center for Protein Synthesis (http://www.mip5.mpg.de/yeast/), e.g., TY1A(PR1)-lacZ corresponds to lacZ fused to the first Ty1 element of chromosome XVI to the right of the centromere. Their precise coordinates can be obtained upon request.
The strain carrying a TY1Ah(ML2)-lacZ fusion at position 815 of Ty1(ML2) was obtained upon transformation of FYBL1-23D, with a ′tyah′-lacZ-URA3-downTy1(ML2) construct. In this construct, the ′ty1ah′ sequences come from Ty1-H3 (coordinates 238 to 815) and downTy1(ML2) corresponds to 800 bp immediately downstream of Ty1(ML2). Null alleles of STE12 and TEC1 were obtained in the FYBL1-23D derivatives by one-step gene replacement using PCR fragments of the TRP1 gene, amplified with long primers containing 5′ and 3′ sequences of STE12 and TEC1, respectively (3). In the same way, the SNF2, SPT3, and SPT7 genes were replaced by the KanMX gene (33). To delete HTA1-HTB1, we first replaced LYS2 with the lys2-128δ allele, which confers lysine auxotrophy, by homologous recombination using the upLYS2-KanMX-lys2-128δ construct of pPL159. Then we replaced HTA1-HTB1 with HIS3 and selected His+ recombinants that were lysine prototrophs, as described previously (25). LV261 contains a DPS1-lacZ fusion integrated at the DPS1 locus and was obtained by transforming FYBL1-23D with an integrative URA3 plasmid containing a DPS1-lacZ fusion.
LV555 was constructed by replacing HIS3 with the KanMX gene in Y328, and LV568 was obtained by integrating a ty1ah-lacZ-URA3-downTy1(PR1) construct in LV555 at Ty1(PR1). In the ty1ah-lacZ-URA3-downTy1(PR1) construct, the ty1a sequences come from Ty1-H3 (coordinates 238 to 815) and the downTy1(PR1) sequences correspond to approximately 800 bp immediately downstream of Ty1(PR1) that was amplified by PCR from genomic DNA.
All gene replacements were confirmed by PCR analysis.
Plasmids.
The pPL159 plasmid containing the upLYS2-KanMX-lys2-128δ construct was obtained in two steps. First, we subcloned the 0.9-kb EcoRI-BglII fragment of pGS64 (a generous gift from F. Winston) containing the lys2-128δ allele in pMTL22 (8). Second, we introduced 5′ to lys2-128δ a BamHI-EcoRI PCR-amplified fragment containing the sequences upstream of LYS2 from coordinates −294 to −242 and the KanMX sequence. The upLYS2-KanMX-lys2-128δ construct was isolated from the resulting pPL159 plasmid as a 2.5-kb BamHI-BglII fragment.
Plasmid p164 (GCN4 URA3 CEN) and plasmid p238, which overproduces Gcn4 (GCN4c URA3 CEN), generous gifts from A. Hinnebusch, have been described previously (43). Plasmids pAM19 (GCN4 HIS3 CEN) and pAM25 (GCN4c HIS3 CEN) were obtained by introducing the 2.2-kb EcoRI-SalI fragment of p164 or the 2-kb SalI-PvuII fragment of p238, respectively, into pRS313 (44).
The lacZ gene is expressed from all the HIS4 regulatory sequences in pHIS4-lacZ (2μm URA3) (a generous gift from B. Daignan-Fornier). Plasmid pHIS4ΔUAS-lacZ was constructed by inserting a 192-bp PstI-HindIII PCR fragment, containing HIS4 sequences from coordinates −144 to +36, into Yep354 (2μm URA3) (36). The HIS4ΔUAS-lacZ fusion lacks the binding sites for all known activators of HIS4 transcription (i.e., Gcn4, Bas1, and Rap1) and retains only the TATA box (at position −123) and the transcription start site (at position −65) (2). Plasmids pHIS43H-lacZ and pHIS43L-lacZ were constructed in three steps. First, we generated double-stranded DNA molecules containing either the region of 12 nonconserved residues of the highly expressed Ty1(PR1) element (H molecules) or the region of 12 nonconserved residues of the poorly expressed Ty1(DR3) element (L molecules) by annealing the following oligonucleotides: for the H sequence, O-AM155 (5′ TCGAGGAATCCCAACAATTATCTCAACATTCACCCATTTCTCATGGTAGCGCCTGTGCTTCGGTCGACG 3′) and O-AM156 (5′ TCGACGTCGACCGAAGCACAGGCGCTACCATGAGAAATGGGTGAATGTTGAGATAATTGTTGGGATTCC 3′), and for the L sequence, O-AM153 (5′ TCGAGGAATCCCAACAATTATCTAATTACCCACATATATCTCATGGTAGCGCCTGTGCTTCGGTCGACG 3′) and O-AM154 (5′ TCGACGTCGACCGAAGCACAGGCGCTACCATGAGATATTATGTGGGTAATTAGATAATTGTTGGGATTCC 3′). Sequences corresponding to the 12-residue nonconserved region are underlined. H and L sequences correspond to sequences of Ty1(PR1) and Ty1(DR3) from coordinates 296 to 354, respectively. In a second step, the H and L DNA molecules were introduced at the XhoI site of pMTL22 (8), in which the SalI site was destroyed. Plasmids pAM29, containing three copies of H, and pAM31, containing three copies of L, were recovered and sequenced. Finally, the 3H and 3L sequences were inserted as 200-bp BglII-StuI fragments into pHIS4ΔUAS-lacZ digested with BamHI and SmaI to yield plasmids pHIS43H-lacZ and pHIS43L-lacZ, respectively.
In vitro DNA-protein binding assay.
His-tagged Gcn4 protein was overproduced and purified from a strain carrying a pET28b derivative, containing almost the entire GCN4 open reading frame (kindly provided by M. Proft [40]). The “3H” and “3L” DNA probes were purified from pAM29 and pAM31, respectively, as XhoI-BglII fragments and were labeled by filling their protruding ends with Klenow polymerase. His-tagged Gcn4 protein (5 to 160 ng) was incubated for 20 min at room temperature with 0.2 to 0.3 ng of a 32P-radiolabeled probe in 20 μl of binding buffer (10 mM Tris-HCl [pH 7.9], 1 mM EDTA, 500 mM Nacl, 15% glycerol, 3 μg of bovine serum albumin). Reaction products were separated by electrophoresis at 4°C on a 5.5% polyacrylamide gel (80:1) in low-ionic-strength buffer (6.7 mM Tris-HCl [pH 7.9], 3.3 mM sodium acetate, 1 mM EDTA).
Northern blot analysis and β-galactosidase assays.
Cultures and procedures were performed as described previously (34). β-Galactosidase units are expressed in nanomoles of 2-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein. Values are averages of at least three independent measurements. Standard deviations are <10%.
RESULTS
Ty1 copies are expressed at different levels from relatively weak promoters.
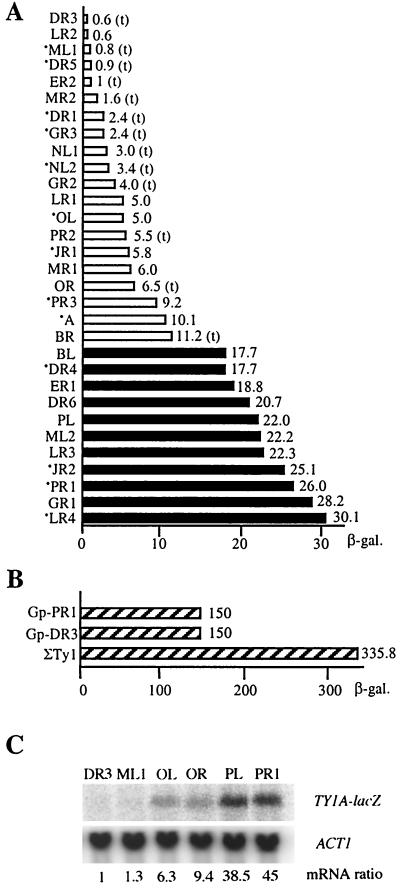
To determine the relative transcription level of each of the individual Ty1 elements present in S. cerevisiae S288C, we constructed 31 strains, each expressing a lacZ chromosomal fusion from the transcriptional signals of a different Ty1 element. These fusions were named according to the Ty1 sequence annotation by MIPS (see Materials and Methods) (34). Expression of the 31 TY1A-lacZ fusions was determined by measuring the β-galactosidase activity of each strain. Our results indicate that the relative level of expression of the Ty1 elements varies over a 50-fold range [0.6 U of β-galactosidase for the least-expressed TY1A(DR3)-lacZ fusion versus 30.1 U for the best-expressed TY1A(LR4)-lacZ fusion (Fig. 1A)]. Total RNA was extracted from six representative strains producing different levels of β-galactosidase activity. Northern blot analysis using a lacZ probe showed a clear correlation between β-galactosidase activity and steady-state levels of TY1A-lacZ transcripts in these strains (Fig. 1C) (34). Our results also indicated that 75% of total β-galactosidase activity comes from 11 elements only. We arbitrarily named these elements “highly expressed elements” in this study. In contrast, the 20 elements expressed at lower levels are referred to as “weakly expressed elements.”
FIG. 1.
Expression of native Ty1 elements in haploid cells. (A) β-Galactosidase activities of the 31 TY1A-lacZ fusions expressed in FYBL1-23D derivatives. Open bars, Ty1 elements defined in the text as weakly expressed elements; solid bars, Ty1 elements defined in the text as highly expressed elements; t, single-nucleotide change in the Tec1 binding site; *, β-galactosidase obtained with several strains containing an independently obtained fusion at the same Ty1 locus. Strains were grown in HC medium lacking uracil. β-Galactosidase activities were assayed as described in Materials and Methods and are shown at the right of each bar. (B) Comparison of the sum of the β-galactosidase activities obtained with the 31 TY1A-lacZ fusions (ΣTy1) with the β-galactosidase activities in the presence of galactose of Gp-PR1 and Gp-DR3, two TY1A-lacZ fusions in which the U3 region of the 5′LTR has been replaced by the GAL1 promoter at Ty1(PR1) and Ty1(DR3), respectively (34). In the presence of glucose, these two fusions gave approximately 1 U of β-galactosidase activity. (C) Northern blot analysis of total RNA in wild-type cells. For each strain, 10 μg of RNA was loaded onto the gel. Steady-state levels of TY1A-lacZ mRNA were normalized to ACT1 mRNA levels. The ratios given are relative to the TY1A(DR3)-lacZ/ACT1 ratio, arbitrarily set at 1.
To determine the strength of Ty1 promoters, we next compared the sum of the β-galactosidase activities of the 31 TY1A-lacZ fusions with the activity of TY1A-lacZ fusions expressed from the inducible GAL1 promoter. The total β-galactosidase activity of the 31 TY1A-lacZ fusions was estimated to be 335.8 U (Fig. 1B). The expression of 31 fusions from the Ty1 promoter sequences was only twofold higher than the activity obtained with the GAL1p-TY1A(DR3)-lacZ or GAL1p TY1A(PR1)-lacZ fusion under conditions of galactose induction (Fig. 1B, Gp-DR3 and Gp-PR1). In these two fusions, the genuine promoter of Ty1(DR3) or Ty1(PR1) is replaced by the inducible GAL1 promoter at the original location (34). Previous Northern blot analyses estimated that steady-state levels of induced GAL1 mRNA were 0.2 to 1% of total mRNA and that steady-state levels of Ty1 mRNA were more than 10% of total mRNA (12, 14, 47). Therefore, we expected approximately 10- to 50-fold-higher levels of β-galactosidase activity from the 31 TY1A-lacZ fusions than from a single fusion expressed from the GAL1 promoter. This prediction is far from the observed twofold difference (335.8 versus 150 U [Fig. 1B]). Since the TY1A-lacZ mRNA molecules expressed from the Ty1 endogenous promoter and from the GAL1 promoter are identical (5, 34), the twofold variation in β-galactosidase levels reveals a difference in promoter activity rather than in mRNA stability or translatability. Therefore, the discrepancy in β-galactosidase activity between the 31 TY1A-lacZ fusions and a single fusion expressed from the GAL1 promoter can only be attributed to a difference in promoter strength and suggests that the Ty1 promoters, even those of highly expressed elements, are relatively weak.
Ty1 transcription is inhibited by repressive chromatin structures that are differentially antagonized by the Swi/Snf chromatin-remodeling complex.
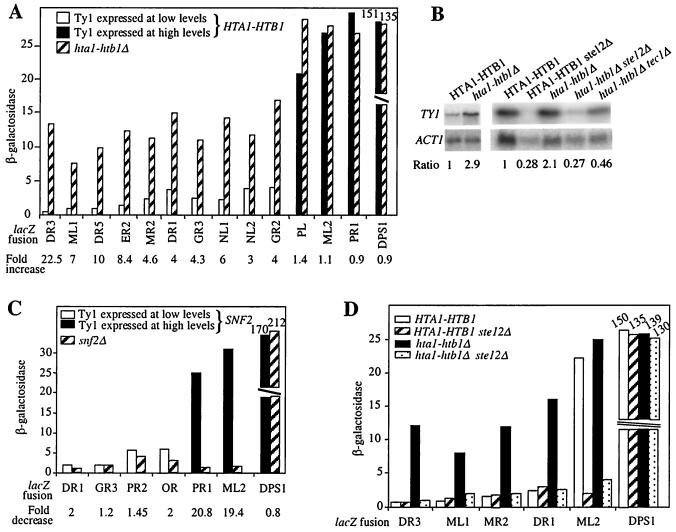
Deletion of the HTA1-HTB1 gene pair, one of the two sets of genes encoding histones H2A and H2B, leads to an unbalanced production of histones. The consequent alteration in chromatin structure correlates with an enhancement of Ty1 mRNA levels (25). This observation suggests that Ty1 transcription might be inhibited by nucleosomes. To determine whether repressive chromatin could be responsible for the low transcriptional levels of some Ty1 elements, we compared the β-galactosidase activities of several TY1A-lacZ fusions in wild-type and hta1-htb1Δ cells. The results show that the β-galactosidase activities of poorly expressed TY1A-lacZ fusions were 3- to 22-fold higher in hta1-htb1Δ mutants than in the wild type. In contrast, the expression of highly expressed fusions remained constant (Fig. 2A). This result suggests that repressive chromatin is partially responsible for the inefficient transcription of those Ty1 elements expressed at lower levels. In parallel, we compared steady-state levels of total Ty1 mRNA in wild-type and hta1-htb1Δ strains by Northern blot analysis. A 2.1- to 2.9-fold increase in total Ty1 mRNA was observed upon histone depletion, in agreement with previously published results (Fig. 2B) (25).
FIG. 2.
Ty1 transcription is repressed by chromatin. (A) β-Galactosidase activities determined in HTA1-HTB1 and hta1-htb1Δ cells. Ty1 elements expressed at low levels and Ty1 elements expressed at high levels are defined in the text. (B) Northern blot analysis of total RNA extracted from wild-type (HTA1-HTB1), hta1-htb1Δ, ste12Δ, hta1-htb1Δ ste12Δ, and hta1-htb1Δ tec1Δ cells. For each strain, 10 μg of RNA was loaded onto the gel. Ty1 mRNA levels were normalized to ACT1 mRNA levels. Ratios given are relative to the ratio measured in the HTA1-HTB1 strain, arbitrarily set at 1. (C) β-Galactosidase activities determined in SNF2 and snf2Δ cells. (D) β-Galactosidase activities determined in wild type (HTA1-HTB1), ste12Δ, hta1-htb1Δ, and hta1-htb1Δ ste12Δ cells. A DPS1-lacZ fusion integrated at the DPS1 gene in LV261, which encodes an aspartyl-tRNA synthetase and whose expression is not regulated by chromatin, served as a control in our experiments. All strains are FYBL1-23D derivatives and were grown in HC medium lacking uracil. β-Galactosidase activities were assayed as described in Materials and Methods.
Previous studies reported that Ty1 mRNA levels are severely reduced in mutants deficient in Swi/Snf chromatin-remodeling and SAGA histone acetyltransferase activities (9, 22, 42). Thus, we expected that inactivation of Swi/Snf would affect the transcription of those Ty1 copies requiring chromatin remodeling for increased expression. Since snf2Δ mutants lack the ATPase function essential for remodeling activity (31), we deleted SNF2 in several strains carrying representative TY1A-lacZ fusions. Transcription of the TY1A-lacZ fusions expressed at higher levels was dramatically diminished in the absence of SNF2 (19.4- and 20.8-fold reductions [Fig. 2C]), whereas the β-galactosidase activities of TY1A-lacZ fusions expressed at lower levels were not significantly affected by the snf2Δ mutation. We chose TY1A-lacZ fusions producing 2 to 6.5 U of β-galactosidase activity in a wild-type background to represent the poorly expressed elements, so that a decrease in activity could be easily detected.
To determine whether SAGA also contributed to the expression of highly transcribed Ty1 elements, we compared the expression of the highly transcribed Ty1(ML2) element in wild-type cells and spt3Δ or spt7Δ mutants. Deletion of SPT3 causes a subtle structural alteration of SAGA, whereas deletion of SPT7 completely disrupts SAGA (46). Previous results showed that in spt3 mutants, Ty1 transcription from the original start site decreases in favor of a new transcript initiated at approximately coordinate +1000 (55). To avoid any β-galactosidase expression from that transcript, we constructed a TY1A(ML2)h-lacZ fusion, in which lacZ is fused at position 815 of Ty1(ML2). In wild-type cells, this fusion yielded 87 U of β-galactosidase activity. In spt3Δ and spt7Δ mutants, β-galactosidase activity was decreased to 19 and 5 U, respectively. A dramatic decrease in activity was also observed in snf2Δ mutants (to approximately 1 U).
The reduction of Ty1(ML2) and Ty1(PR1) expression in the presence of mutations altering either SAGA or Swi/Snf activity indicates that these elements require chromatin remodeling to be expressed. These data, in addition to those obtained with hta1-htb1Δ cells, suggest that Ty1 elements are contained in repressive chromatin and that the Swi/Snf and SAGA complexes are able to antagonize this repressive chromatin at certain Ty1 loci, i.e., those elements that are highly expressed.
In hta1-htb1Δ mutants, transcription of weakly expressed Ty1 elements requires Ste12 and Tec1.
The transcriptional activators Ste12 and Tec1 bind Ty1 DNA downstream of the TATA boxes and activate Ty1 transcription in wild-type cells (4, 30, 34). The role of these factors in the activation of Ty1 transcription is confirmed, in the case of Ste12, by our Northern blot analysis (Fig. 2B; compare HTA1-HTB1 with HTA1-HTB1 ste12Δ). The same analysis shows that in an hta1-htb1Δ mutant also, Ty1 transcription requires Ste12 and Tec1 (Fig. 2B; compare hta1-htb1Δ with hta1-htb1Δ ste12Δ and hta1-htb1Δ tec1Δ).
We also investigated the role of Ste12 in the transcriptional activation of several weakly expressed TY1A-lacZ fusions in hta1-htb1Δ strains. β-Galactosidase activity was diminished 4- to 12-fold in the absence of STE12 [Fig. 2D, fusions to Ty1(DR3), Ty1(ML1), Ty1(MR2), and Ty1(DR1)]. A similar decrease in β-galactosidase activity was observed in the absence of TEC1 in these strains (data not shown). Taken together, these results suggest that chromatin inhibits transcription of Ty1 elements expressed at low levels by preventing the binding of Ste12 and Tec1. Our results do not exclude the possibility that the recruitment of other transcriptional activators is also impaired by the presence of nucleosomes at Ty1.
Interestingly, the absence of STE12 in HTA1-HTB1 cells caused a decrease in β-galactosidase activity produced by the highly expressed TY1A(ML2)-lacZ fusion similar to that observed for poorly expressed TY1A-lacZ fusions in hta1-htb1Δ cells. Since TY1A(ML2)-lacZ requires chromatin remodeling by Swi/Snf and SAGA for high-level expression, this result suggests that chromatin remodeling at Ty1(ML2) also allows subsequent transcriptional activation by Ste12.
Identification of five potential Gcn4 binding sites in the highly expressed Ty1 elements.
Recent studies implicate several DNA-binding transcriptional activators in the targeting of Swi/Snf and SAGA complexes to specific promoters (reviewed in reference 41). We asked whether already known transcriptional regulators of Ty1 might be involved in recruiting Swi/Snf or SAGA to Ty1 promoter regions. We first searched for differences in the binding sites of Ty1 transcriptional factors that might account for differences in expression. Alignments of the Gcr1, Ste12, Mcm1, Tea1, and Rap1 binding sites of each of the Ty1 elements did not reveal any correlation between a deviation from the consensus sequence and expression of the TY1A-lacZ fusions (data not shown). The Tec1 binding site, on the other hand, was not well conserved among Ty1 elements; several weakly expressed Ty1 elements had the same single-nucleotide change (underlined) in the Tec1 consensus binding site (GAATA instead of GAATG) (Fig. 1A). To determine whether this change was responsible for the low expression level of some Ty1 elements, we restored a GAATA Tec1 binding site to the consensus sequence in a TY1A(DR3)-lacZ fusion in which lacZ was fused to TY1A at coordinate +425. Expression of the TY1A(DR3)-lacZ fusion showed the same Tec1 dependency with the GAATA and GAATG binding sites (data not shown). This finding suggests that the single-nucleotide difference in the Tec1 binding site found in some poorly expressed Ty1 elements is not responsible for the difference in expression.
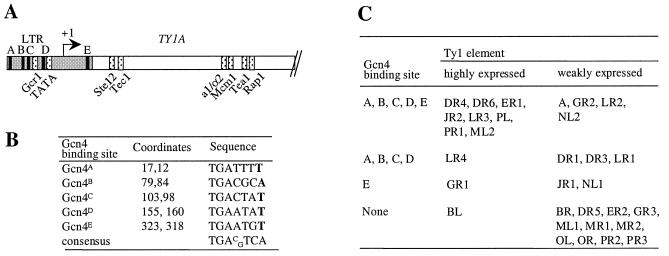
We next looked for differences outside of the motifs recognized by known regulators of Ty1 expression in the first kilobase of Ty1, which contains all the sequences regulating Ty1 transcription. We identified a region of 12 nonconserved residues in the LTR region downstream of the transcription start site, at positions 315 to 326. Using the Saccharomyces cerevisiae Promoter Database (SCPD; http://cgsigma.cshl.org/jian/), we identified a potential Gcn4 binding site (TGAATG) in this region at positions 323 to 318 (Fig. 3A and B, Gcn4E site). Four additional potential Gcn4 binding sites were pinpointed in the LTR regions of several elements as well (Fig. 3A and B, Gcn4A to Gcn4D sites). Interestingly, 8 out of 11 highly expressed elements possessed all five potential Gcn4 binding sites (Gcn4A to Gcn4E), while the full complement of binding sites was present in only 4 of the 20 poorly expressed elements (Fig. 3C). Gcn4 is a transcriptional activator of amino acid biosynthetic genes that binds to multiple sites upstream of the genes it regulates (24). The identification of five potential Gcn4 binding sites in several of the highly expressed elements suggested that Gcn4 might specifically activate their transcription.
FIG. 3.
Potential Gcn4 binding sites in the 5′ LTRs of Ty1 elements. (A) Locations of the five potential Gcn4 binding sites in the 5′ LTR. A, B, C, D, and E, Gcn4A, Gcn4B, Gcn4C, Gcn4D, and Gcn4E sites, respectively; stippled boxes, motifs recognized by the transcriptional regulators indicated below the boxes. (B) Coordinates and sequences of the Gcn4A, Gcn4B, Gcn4C, Gcn4D, and Gcn4E sites. Coordinates are given relative to the numbering of Ty1-H3 (5). For each binding site, the order of the coordinates represents its orientation in the Ty1 sequence. Although Gcn4 recognizes a 7-bp core sequence, the SCPD database identified sequences of 6 bp as potential Gcn4 binding sites. The base ignored by SCPD is given in boldface for each Gcn4 binding site. (C) Inventory of the five potential Gcn4 binding sites in Ty1 elements.
Gcn4 overproduction activates Ty1 transcription.
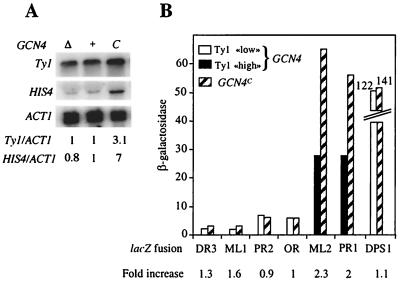
To establish whether Gcn4 could activate Ty1 expression, we first compared Ty1 mRNA levels in isogenic strains bearing wild-type GCN4, a gcn4Δ allele, or a GCN4c constitutive allele that overproduces Gcn4. We noticed a threefold increase in Ty1 mRNA levels when Gcn4 was overproduced (Fig. 4A; compare lanes + and C). This result establishes that Gcn4 activates Ty1 transcription. However, no decrease in Ty1 mRNA levels was observed in gcn4Δ cells compared to levels in wild-type cells (Fig. 4A; compare lanes Δ and +). These observations suggest that Gcn4 is not produced at sufficient levels in wild-type cells to activate Ty1 expression. Gcn4 may also compete with some factor(s) to stimulate Ty1 transcription. In the absence of Gcn4, this factor(s) would ensure the activation of Ty1 transcription. Gcn4 is a basic leucine zipper DNA binding protein of the AP-1 family of transcriptional activators (reviewed in reference 24). In a preliminary search for activators competing with Gcn4 in a rich environment, we investigated the effects on Ty1 transcription of Bas1 and Yap1, two other AP-1-like proteins (35, 48). When gcn4Δ bas1Δ or gcn4Δ yap1Δ cells were grown in rich medium, Ty1 transcription did not significantly decrease, suggesting that neither Bas1 nor Yap1 activates Ty1 transcription under these conditions (data not shown).
FIG. 4.
Gcn4 overproduction activates Ty1 transcription. (A) Northern blot analysis of total RNA with strains expressing different GCN4 alleles. Lanes: Δ, Y328 transformed with YCp50; +, Y328 transformed with p164 (GCN4 URA3 CEN); C, Y328 transformed with p238 (GCN4c URA3 CEN), which overproduces Gcn4. Strains were grown in synthetic complete medium (HC) lacking uracil. For each sample, 10 μg of total RNA was loaded onto the gel. The HIS4 gene, which is activated by Gcn4, served as a positive control. Ty1 and HIS4 mRNA levels were normalized to those of ACT1. The mRNA ratios given are relative to the ratio obtained for the Y328/p164 strain, arbitrarily set at 1. (B) β-Galactosidase activities of TY1A-lacZ fusions in wild-type cells (GCN4) and in wild-type cells overexpressing GCN4 (GCN4c). Ty1 “low,” weakly expressed Ty1 elements; Ty1 “high,” highly expressed Ty1 elements; GCN4, strains transformed with pRS313 (HIS3 CEN); GCN4c, strains transformed with pAM25 (GCN4c HIS3 CEN). Strains are FYBL1-23D derivatives. Cells were grown in HC medium lacking uracil and histidine.
The identification of five Gcn4 binding sites in the 5′ LTR of many Ty1 elements expressed at higher levels suggested that Gcn4 specifically stimulates transcription of these elements. We compared the expression of several TY1A-lacZ fusions in wild-type cells in the absence and in the presence of the constitutive GCN4c allele. As predicted, overproduction of Gcn4 increased the β-galactosidase activities of the highly expressed TY1A(ML2)-lacZ and TY1A(PR1)-lacZ fusions 2.3- and 2-fold, respectively (Fig. 4B). Both fusions contain five Gcn4 binding sites. In contrast, the GCN4c allele did not significantly affect the transcription of lacZ fusions to Ty1(DR3), Ty1(ML1), Ty1(OR), or Ty1(PR2), none of which have the five Gcn4 binding sites.
Gcn4 activates Ty1 transcription under conditions of amino acid limitation.
Ttranslation of GCN4 is activated in response to amino acid limitation (24). As a consequence, transcription of genes regulated by Gcn4 increases. To determine whether Ty1 transcription is also stimulated by starvation, we compared Ty1 mRNA levels in cells grown in rich medium with those in cells grown under conditions of amino acid limitation.
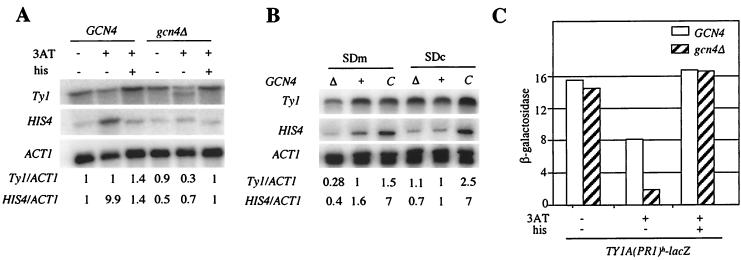
In the presence of 3-aminotriazole (3AT), an inhibitor of the HIS3 gene product that induces histidine starvation, we observed a 3.3-fold reduction in Ty1 mRNA levels in gcn4Δ cells compared to wild-type cells (Fig. 5A; compare GCN4+3AT −his with gcn4Δ +3AT −his). The difference in Ty1 mRNA levels was specific to histidine starvation, as no difference in transcript levels was detected in the presence of both 3AT and histidine (Fig. 5A; compare GCN4 +3AT +his with gcn4Δ +3AT +his). Likewise, Ty1 mRNA levels were threefold lower in gcn4Δ cells than in wild-type cells in minimal SD medium containing only arginine and leucine, required by the strain's auxotrophy (Fig. 5B; compare SDm Δ with SDm +). These results imply that Ty1 transcription depends partially on Gcn4 under amino acid limitation. Interestingly, the decrease in Ty1 transcript levels observed in gcn4Δ cells with amino acid starvation was always accompanied by the appearance of a new Ty1 mRNA species of smaller molecular size (Fig. 5A; also slightly detectable in Fig. 5B). Primer extension analyses suggest that this new mRNA is similar to a species of smaller molecular size already described for spt3 mutants and corresponds to an initiation approximately 800 bp downstream of the main transcription initiation (reference 55 and our unpublished data).
FIG. 5.
Amino acid limitation affects Ty1 transcription in the absence of Gcn4. (A) Northern blot analysis of cells grown under histidine starvation. GCN4, strain Y328 transformed with p164 (GCN4 URA3 CEN); gcn4Δ, Y328 transformed with YCp50. 3AT (10 mM) and histidine (120 μM) were added (+) or not (−) to cultures grown at 22°C in minimal medium supplemented with arginine, isoleucine, tryptophan, leucine, and valine (SDc), as described previously (43). For each sample, 10 μg of total RNA was loaded onto the gel. Ty1 and HIS4 mRNA levels were normalized to those of ACT1. The mRNA ratios given are relative to that for the GCN4 strain grown in SDc lacking 3AT and histidine. The HIS4 gene, which is activated by Gcn4, served as a positive control. (B) Northern blot analysis with strains expressing different GCN4 alleles. Δ, Y328 transformed with YCp50; +, Y328 transformed with p164; C, Y328 transformed with p238 (GCN4c URA3 CEN), which overproduces Gcn4. SDm, minimal medium supplemented with arginine and leucine. SDc is explained in the legend to panel A. In SDc, GCN4 expression is not activated, whereas GCN4 expression is activated in SDm due to amino acid limitation. Precultures and cultures were grown as described previously (43), except that cultures were grown to mid-log phase at 22°C. Ty1 and HIS4 mRNA levels were normalized to those of ACT1. The mRNA ratios given are relative to the ratio under the nonactivating conditions in SDc medium with strain Y328/p164. (C) β-Galactosidase activities of the TY1A(PR1)h-lacZ fusion under conditions of histidine starvation. Growth conditions are described in the legend to panel A. gcn4Δ, strain LV568 transformed with pRS313 (HIS3 CEN) (44). GCN4, strain LV568 transformed with pAM19 (GCN4 HIS3 CEN). Strains are Y328 derivatives.
Although amino acid limitation induces the synthesis of Gcn4 (24), it did not increase Ty1 mRNA levels in wild-type cells (Fig. 5A [compare GCN4 −3AT −his with GCN4 +3AT −his] and B [compare SDm + with SDc +). Likewise, overexpressing GCN4 in wild-type cells grown under amino acid limitation conditions did not activate Ty1 transcription as it did in cells grown in rich medium (Fig. 5B; compare SDm + with SDm C). Taken together, these results suggest that amino acid limitation restricts the effect of increased expression of GCN4 on Ty1 transcription (see Discussion).
To determine whether the expression of a Ty1 element containing five potential Gcn4 binding sites is regulated by histidine starvation, we compared the β-galactosidase activities of a TY1A(PR1)h-lacZ fusion in wild-type and gcn4Δ cells grown in the presence or absence of 3AT. We fused lacZ at position 815 of Ty1(PR1) in order to avoid β-galactosidase synthesis from the smaller-molecular-size Ty1 mRNA species detected in gcn4Δ cells with histidine starvation (Fig. 5A). Expression of TY1A(PR1)h-lacZ was 4.4-fold lower in gcn4Δ cells than in wild-type cells in the presence of 3AT (Fig. 5C). The reduction in TY1A(PR1)h-lacZ expression was the consequence of histidine starvation, since β-galactosidase activities were similar in wild-type and gcn4Δ cells when histidine and 3AT were simultaneously added to the cultures. Therefore, Gcn4 is required for the expression of Ty1(PR1) under conditions of histidine starvation.
In wild-type cells, addition of 3AT induced a twofold reduction in β-galactosidase activity, although it had no effect on total Ty1 mRNA levels (compare Fig. 5A, GCN4 +3AT −his, with Fig. 5C, GCN4 +3AT −his). We explained this difference, which had already been observed with the HIS4 gene (43), by a general translation defect caused by histidine starvation in the presence of 3AT.
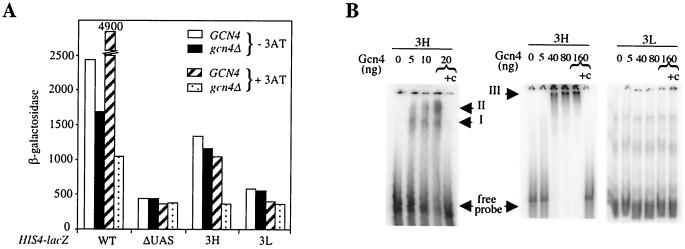
The 12-residue nonconserved region of the 5′ LTR which contains the Gcn4E binding site functions as a Gcn4-dependent UAS in vivo and interacts with Gcn4 in vitro.
The Gcn4E site present in the 12-residue nonconserved region of the highly expressed elements is relatively far from the consensus sequence recognized by Gcn4 (23) (Fig. 3). To determine whether this sequence plays a role in the activation of these elements by Gcn4, we inserted, upstream of a HIS4ΔUAS-lacZ reporter gene, either three copies of the Gcn4E-containing region or three copies of the equivalent region lacking Gcn4E (see Materials and Methods). The different constructs, called 3H and 3L, respectively, were carried on a 2μm plasmid. We compared the expression of the different constructs in wild-type and gcn4Δ cells grown in the presence or absence of 3AT. Under histidine starvation conditions (with 3AT), Gcn4 activated the transcription of HIS4ΔUAS-lacZ threefold in the presence of the 3H sequence but not at all in the presence of the 3L sequence (Fig. 6A). This result indicates that the 12-residue nonconserved region of the highly expressed elements confers Gcn4-dependent UAS activity under histidine starvation conditions. In the absence of 3AT, insertion of the 3H construct, but not of the 3L construct, induced a 2.5- to 3-fold increase in β-galactosidase activity in both wild-type and gcn4Δ cells. These data suggest that factors other than Gcn4 can also activate transcription through the nonconserved region of the highly expressed Ty1 elements when histidine is not limiting.
FIG. 6.
The Gcn4E binding site contributes to the activation of Ty1 transcription by Gcn4. (A) β-Galactosidase activities of the different HIS4-lacZ constructs in strains grown under normal conditions or under conditions of histidine starvation. 3AT was added to the cultures as described in the legend to Fig. 5A. GCN4, strain LV555 transformed with pAM19 (GCN4 HIS3 CEN); gcn4Δ, strain LV555 transformed with pRS313; WT, HIS4-lacZ fusion; ΔUAS, HIS4ΔUAS-lacZ fusion; 3H, HIS43H-lacZ fusion; 3L, HIS43L-lacZ fusion. The different HIS4-lacZ constructs are carried on 2μm URA3 plasmids as described in Materials and Methods. LV555 is a Y328 derivative. (B) Gel retardation assays. 3H, DNA probe containing 3 copies of the Gcn4E motif; 3L, DNA probe without the Gcn4E motif. Gcn4, purified His-tagged Gcn4 protein; +c, cold competitor (250 ng of pHIS43H-lacZ); I, II, and III, His-tagged Gcn4/DNA complexes.
We also performed a gel mobility shift assay to determine whether Gcn4 binds the 3H and 3L constructs differentially in vitro. Three His-tagged Gcn4/DNA complexes were detected using the 3H DNA probe (Fig. 6B). Two low-molecular-weight complexes were present with an approximately 100- to 400-fold excess of His-tagged Gcn4 over the 3H DNA probe (Fig. 6B, 5 to 20 ng of protein, complexes I and II). With a larger excess of protein over DNA probe (an 800- to 3,200-fold excess), a complex of lower mobility was formed (Fig. 6B, 40 to 160 ng of protein, complex III). We presume that the three complexes detected reflect occupation of one, two, or all three Gcn4E sites of the 3H probe by Gcn4. Binding of His-tagged Gcn4 to the 3H probe was competed by an unlabeled plasmid containing the 3H sequences. In contrast, the 3L probe did not form a complex with His-tagged Gcn4 protein, even with a 3,200-fold excess of protein over the 3L DNA probe. In conclusion, the in vivo and in vitro experiments imply that the Gcn4E binding site directly contributes to the activation of Ty1 transcription by Gcn4.
DISCUSSION
Ty1 transcription is inhibited by repressive chromatin structures.
The first striking result of this work is that the relative transcriptional level of 31 Ty1 elements present in the genome of strain S288C varies over a 50-fold range. This difference in expression occurs despite a 94% nucleotide identity within the region required for Ty1 transcription. It is also noteworthy that only 11 elements are responsible for 75% of the β-galactosidase activity produced by all the elements. These elements could be responsible for most of the Ty1 retrotransposition activity observed in S288C. Moreover, the comparison of Ty1 and GAL1 promoter activities shows that Ty1 promoters are relatively weak despite Ty1 mRNA abundance. Thus, the high steady-state Ty1 mRNA levels observed in haploid cells (12, 14) might be the consequence of its accumulation rather than of elevated synthesis. This hypothesis is relevant since Ty1 mRNA is exceptionally stable in the cell (18).
It was previously shown that Ty1 transcription depends on Swi/Snf and SAGA complexes. This dependence on chromatin-modifying activities suggests that repressive chromatin inhibits the accessibility of transcriptional activators to Ty1 promoter sequences. Consistent with this idea, Ty1 mRNA levels increase in hta1-htb1Δ mutants, in which genes normally inhibited by nucleosomes are activated (25). We showed that in hta1-htb1Δ mutants, expression of Ty1 elements expressed at lower levels is significantly increased, while expression of Ty1 elements expressed at higher levels remains constant. By contrast, in snf2Δ mutants, which are deficient in Swi/Snf chromatin-remodeling activity, the expression of Ty1 elements normally expressed at higher levels is strongly decreased, whereas the expression of copies expressed at lower levels is unchanged. The lack of an increase in expression of the highly expressed copies in hta1-htb1Δ mutants could merely reflect our inability to increase a level of expression that is already maximal. Likewise, the absence of a decrease in expression of the poorly transcribed elements in snf2Δ mutants could result from our inability to decrease a level of expression that is already minimal. However, these hypotheses are unlikely for two reasons. First, our data indicate that the transcription of two highly expressed elements is augmented two- to threefold when Gcn4 is overexpressed, establishing that their expression is not maximal in hta1-htb1Δ mutants (Fig. 4B). Second, we previously showed that the transcription of poorly expressed Ty1 elements can be reduced in diploid cells, indicating that their expression is not minimal in an snf2Δ mutant (34). Therefore, we propose to sort the genomic Ty1 elements into two basic classes, depending on their responses to mutations affecting chromatin structures. The first class contains Ty1 elements which are weakly expressed because nucleosome-mediated repression is not antagonized by chromatin-modifying complexes. Elements of this class are expressed when the amounts of histones H2A and H2B are limiting, as is the case in hta1-htb1Δ cells. The second class contains Ty1 elements expressed at higher levels because the Swi/Snf complex antagonizes chromatin-mediated repression at these loci. Their expression might also depend on the SAGA complex, since transcription of the TY1A(ML2)h-lacZ fusion, which is highly expressed, is severely reduced in spt3Δ cells and in spt7Δ cells The fact that transcription of Ty1 elements expressed at higher levels does not increase in hta1-htb1Δ mutants suggests that Swi/Snf and SAGA activities are sufficient to completely remove repressive chromatin at these Ty1 loci. We cannot exclude the possibility that some elements are expressed at low or high levels for other reasons. For example, the specific location of Ty1(LR4), which overlaps the HAP1 open reading frame, may be responsible for its high expression (34). Likewise, the positions of some poorly expressed elements may affect their expression (see below). Further studies will be necessary to test more directly the hypothesis of different chromatin organizations at the promoters of several highly and poorly expressed Ty1 elements.
There is a correlation between the transcriptional levels of endogenous Ty1 elements and the presence of potential Gcn4 binding sites in their 5′ LTRs.
We found that 8 out of 11 highly expressed elements possess five potential Gcn4 binding sites (Gcn4A to Gcn4E) in their 5′ LTRs. The three Ty1 elements expressed at high levels that do not contain the five Gcn4 motifs were Ty1(BL), Ty1(GR1), and Ty1(LR4). Ty1(BL) belongs to a subtype of Ty1 elements whose sequence differs from the other Ty1 sequences (28), suggesting that expression of these elements is regulated differently. Ty1(GR1) contains only the Gcn4E motif, which may be sufficient for its high expression. As noted above, Ty1(LR4) is not regulated like the other Ty1 elements, probably because the HAP1 open reading frame overlaps its 5′ LTR (34). We also noticed that 4 out of the 20 elements expressed at lower levels contain the five potential Gcn4 binding sites (Fig. 3C). Among them, Ty1(LR2) is located adjacent to the ribosomal DNA region on chromosome XII. Since this region has a specific chromatin structure known to down-regulate RNA polymerase II-dependent promoters (7, 45), Ty1(LR2) expression is probably silenced. Ty1(NL2) overlaps the adjacent tD(GUC)N gene, encoding a tRNAAsp, by 6 nucleotides. Since the two sequences are in opposite orientations, the transcription of the tRNA gene by RNA polymerase III probably inhibits the transcription of Ty1(NL2), as has been previously shown with other adjacent RNA polymerase III- and RNA polymerase II-dependent genes (26). For the time being, we have no explanation for the low levels of expression of the two remaining elements, Ty1(A) and Ty1(GR2), despite the presence of five potential Gcn4 binding sites in their sequences.
Recently, Dudley et al. identified a region (called #12) in the 912δ element which is bound by an unknown factor and which activates transcription (13). This region overlaps the Gcn4C motif. Mutations in sequences now identified as Gcn4C decrease the UAS activity of this region (13). Our results indicate that the Gcn4E motif also acts as an UAS in vivo and interacts with Gcn4 in vitro. Taken together, these observations suggest that these two sites may contribute to the high expression of some Ty1 elements. In contrast, mutations in the Gcn4D sequence have a minor effect on the UAS activity of the 912δ element (13). It would be interesting to determine whether the other potential Gcn4 binding sites (i.e., Gcn4A and Gcn4B) also contribute to transcriptional activation and if the complete set of sites is necessary to achieve full activation of Ty1 transcription. Since the sequences of the five Gcn4 binding sites are relatively far from the consensus sequence recognized by Gcn4, it remains possible that other factors bind to these sites in the Ty1 promoter.
Gcn4 activates Ty1 transcription.
Gcn4 is a master regulator of gene expression under diverse stress conditions, such as amino acid, purine, nitrogen, or glucose limitation, hyperosmosis, and DNA damage (38). Several lines of evidence support the view that Gcn4 activates Ty1 transcription. First, elevated expression of Gcn4 caused an increase in Ty1 mRNA levels in cells grown under rich conditions (Fig. 4A). Second, we identified five potential Gcn4 binding sites in the 5′ LTRs of 8 out of 11 Ty1 elements expressed at high levels, and overproduction of Gcn4 activated the transcription of two of these. In contrast, overproduced Gcn4 did not activate the transcription of Ty1 copies lacking the Gcn4 binding sites (Fig. 4B). Third, Ty1 mRNA levels decreased in gcn4Δ cells grown under conditions of histidine or amino acid limitation, indicating that Gcn4 stimulates Ty1 transcription under these conditions (Fig. 5A and B). Fourth, under histidine starvation conditions, Gcn4 activated the expression of Ty1(PR1), which contains five Gcn4 binding sites. Finally, a nonconserved region of the 5′ LTR which contains the Gcn4E motif in highly expressed Ty1 copies conferred a Gcn4-dependent UAS activity on a reporter gene under conditions of histidine starvation. This region also interacts with Gcn4 in vitro. Taken together, these results indicate that the Ty1 retrotransposon could be a new target for the Gcn4 transcription activator. Several DNA binding factors contribute to Ty1 transcriptional activity, in addition to Gcn4 (see the introduction). From our data, we cannot exclude the possibility that Gcn4 cooperatively interacts with one of these factors to activate Ty1 transcription.
Targets of Gcn4 are normally activated upon amino acid limitation. Surprisingly, Ty1 mRNA levels were not increased in amino acid-starved wild-type cells. Likewise, overexpression of GCN4 did not raise Ty1 mRNA levels in wild-type cells grown under amino acid starvation conditions, although it did when wild-type cells were grown in rich medium. These peculiarities can be explained if the activation of Ty1 transcription by Gcn4 is counterbalanced by a higher rate of degradation of Ty1 mRNA when amino acids are limiting. As is generally the case, Ty1 mRNA is translated less efficiently in amino acid-starved cells than in nonstarved cells. Because the stability of Ty1 mRNA is probably the consequence of its interaction with Ty1A, a decrease in translation might affect Ty1 mRNA stability. Further analysis would be necessary to determine whether the half-life of Ty1 mRNA is effectively reduced under amino acid starvation.
A previous study indicated that the Ty2 retrotransposon contains in the TY2A open reading frame sequences that confer Gcn4-dependent activation on reporter genes (49). However, our finding is the first evidence that, besides activating genes involved in metabolic adaptation, Gcn4 activates transcription of a retroelement at its original location. Interestingly, we found that Ty1 retrotransposition increased sixfold when GCN4 was overexpressed (our unpublished data), indicating that stress conditions which activate GCN4 would induce not only Ty1 transcription but also Ty1 retrotransposition. Activation of Ty1 retrotransposition by stress has already been described and has been proposed to play a role in adaptive mutagenesis (34).
A model for activation of Ty1 transcription via recruitment of the Swi/Snf and SAGA chromatin-modifying complexes by Gcn4.
Some transcriptional regulators, such as Swi5, Ume6, Hir1, and Hir2, can recruit chromatin-modifying complexes to specific promoters (for a review, see reference 41). A similar function in targeting Swi/Snf and SAGA to specific promoters has been demonstrated for Gcn4 in order to activate transcription (29, 37, 39, 51, 56).
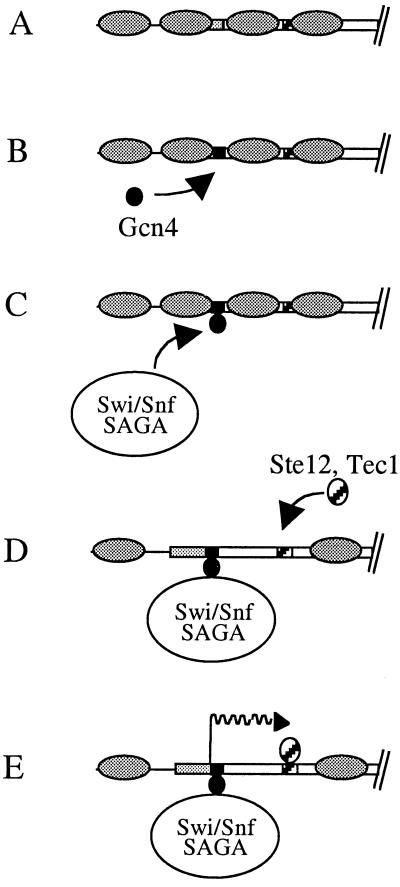
We showed that the overproduction of Gcn4 stimulates the transcription of Ty1 elements containing five Gcn4 binding sites. These elements require Swi/Snf and SAGA to be expressed at high levels. In contrast, the transcription of elements lacking Gcn4 binding sites was low and independent of Swi/Snf. These results strongly support a model in which Gcn4 recruits Swi/Snf and SAGA in order to activate Ty1 transcription. In this model, Ty1 elements lacking Gcn4 binding sites are expressed at low levels because Gcn4 does not bind to their sequences (Fig. 7A). As a result, Swi/Snf and SAGA are not targeted to these elements and do not remove repressive chromatin. In contrast, Ty1 elements containing Gcn4 binding sites are expressed at high levels because Gcn4 binds to their sequences and recruits Swi/Snf and SAGA (Fig. 7B to E). Another model is that chromatin remodeling might facilitate the binding of Gcn4, which in turn would activate Ty1 transcription. However, we disfavor this model because Gcn4 has already been shown to activate the transcription of several genes by recruiting Swi/Snf and SAGA to their promoter sequences. Ty1 transcription is not affected in gcn4Δ cells grown in rich medium (Fig. 5), suggesting that a transcriptional activator other than Gcn4 may also recruit Swi/Snf and SAGA to the Ty1 promoter. This factor might be Gcr1, as activation by Gcr1 at the his4-912δ promoter has been shown to depend on some components of the SAGA complex (13). Finally, we propose that Swi/Snf and SAGA alter the positions of nucleosomes along the Ty1 promoter and facilitate the access of other transcription factors, such as Ste12 and Tec1, to previously masked sequences. This hypothesis is supported by the fact that Ste12 and Tec1 are still required for Ty1 transcription when histones are depleted in hta1-htb1Δ cells.
FIG. 7.
Model for transcriptional activation of Ty1 elements. Shaded ovals, nucleosomes; shaded rectangles, Ty1 5′ LTR; open rectangles, transcriptional regulatory region of Ty1; solid box, Gcn4 binding site (only one site is shown for clarity); cross-hatched boxes, Ste12 and Tec1 binding sites. (A) Ty1 elements are contained within repressive chromatin, which is not removed at the Ty1 elements expressed at low levels because they lack the Gcn4 motifs. (B) Gcn4 binds to Ty1 sequences harboring Gcn4 binding sites. (C) Gcn4 recruits Swi/Snf and SAGA. (D) Swi/Snf and SAGA displace nucleosomes, allowing binding of Ste12 and Tec1 to their sites. (E) Ste12 and Tec1 activate Ty1 transcription (wavy arrow). Events B to E occur at the highly expressed elements.
At the HO promoter, the Swi5 DNA binding protein interacts first with the Swi/Snf complex, which in turn recruits SAGA (10), whereas at the HIS3 promoter, histone acetylation and chromatin remodeling act in parallel to activate transcription (37). Regarding Ty1, future investigations are necessary to determine whether the recruitment of one of the two complexes may alter nucleosomes in a manner that permits the association of the other complex, which will stabilize an “open” nucleosome state.
Acknowledgments
We are very grateful to P. Stragier for his hospitality to facilitate initiation of the project. We thank F. Winston and M. Proft for technical advice; B. Daignan-Fornier, A. Hinnebusch, S. Gangloff, M. Proft, and F. Winston for plasmids and strains; and F. Allemand for valuable help in the purification of His-tagged Gcn4 protein. Special thanks go to B. Daignan-Fornier, A. Gabriel, C. Condon, and P. Stragier for helpful comments on the manuscript.
This work was supported by a grant from the CNRS (UPR 9073). A.M. was a recipient of a Docteur Ingénieur fellowship from the CNRS and of a fellowship from ARC (Association pour la Recherche contre le Cancer).
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874-880. [DOI] [PubMed]
- 3.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., D. J. Garfinkel, C. A. Styles, and G. R. Fink. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491-500. [DOI] [PubMed] [Google Scholar]
- 6.Boeke, J. D., and S. B. Sandmeyer. 1991. Yeast transposable elements, p. 193-261. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the Rdn1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed]
- 9.Ciriacy, M., K. Freidel, and C. Lohning. 1991. Characterization of trans-acting mutations affecting Ty and Ty-mediated transcription in Saccharomyces cerevisiae. Curr. Genet. 20:441-448. [DOI] [PubMed] [Google Scholar]
- 10.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 11.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curcio, M. J., A. M. Hedge, J. D. Boeke, and D. J. Garfinkel. 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 220:213-221. [DOI] [PubMed] [Google Scholar]
- 13.Dudley, A. M., L. J. Gansheroff, and F. Winston. 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912Δ promoter in Saccharomyces cerevisiae. Genetics 151:1365-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder, R. T., T. P. St. John, D. T. Stinchcomb, R. W. Davis, S. Scherer, and R. W. Davis. 1981. Studies on the transposable element Ty1 of yeast. Cold Spring Harbor Symp. Quant. Biol. 45:581-591. [DOI] [PubMed] [Google Scholar]
- 15.Errede, B. 1993. MCM1 binds to a transcriptional control element in Ty1. Mol. Cell. Biol. 13:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Errede, B., M. Company, and C. A. Hutchison III. 1987. Ty1 sequence with enhancer and mating-type-dependent regulatory activities. Mol. Cell. Biol. 7:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulton, A. M., P. D. Rathjen, S. M. Kingsman, and A. J. Kingsman. 1988. Upstream and downstream transcriptional control signals in the yeast retrotransposon, TY. Nucleic Acids Res. 16:5439-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garfinkel, D. J. 1992. Retroelements in microorganisms, p. 107-158. In J. A. Levy (ed.), The Retroviridae, vol. 1. Plenum Press, New York, N.Y.
- 19.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 20.Gray, W. M., and J. S. Fassler. 1996. Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol. Cell. Biol. 16:347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray, W. M., and J. S. Fassler. 1993. Role of Saccharomyces cerevisiae Rap1 protein in Ty1 and Ty1-mediated transcription. Gene Expr. 3:237-251. [PMC free article] [PubMed] [Google Scholar]
- 22.Happel, A. M., M. S. Swanson, and F. Winston. 1991. The SNF2, SNF5 and SNF6 genes are required for Ty transcription in Saccharomyces cerevisiae. Genetics 128:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, D. E., I. A. Hope, J. P. Macke, and K. Struhl. 1986. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science 234:451-457. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, A. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in S. cerevisiae, p. 319-415. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6:2288-2298. [DOI] [PubMed] [Google Scholar]
- 26.Hull, M. W., J. Erickson, M. Johnston, and D. R. Engelke. 1994. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 14:1266-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel, and D. F. Voytas. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8:464-478. [DOI] [PubMed] [Google Scholar]
- 29.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 30.Laloux, I., E. Dubois, M. Dewerchin, and E. Jacobs. 1990. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol. Cell. Biol. 10:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurent, B. C., I. Treich, and M. Carlson. 1993. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 7:583-591. [DOI] [PubMed] [Google Scholar]
- 32.Madison, J. M., A. M. Dudley, and F. Winston. 1998. Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (δ) in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maftahi, M., C. Gaillardin, and J. M. Nicaud. 1996. Sticky-end polymerase chain reaction method for systematic gene disruption in Saccharomyces cerevisiae. Yeast 12:859-868. [DOI] [PubMed] [Google Scholar]
- 34.Morillon, A., M. Springer, and P. Lesage. 2000. Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5766-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moye-Rowley, W. S., K. D. Harshman, and C. S. Parker. 1989. Yeast YAP1 encodes a novel form of the Jun family of transcriptional activator proteins. Genes Dev. 3:283-292. [DOI] [PubMed] [Google Scholar]
- 36.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 37.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 38.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcription profiling shows that Gcn4 is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 40.Pascual-Ahuir, A., R. Serrano, and M. Proft. 2001. The Sko1p repressor and Gcn4p activator antagonistically modulate stress-regulated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:16-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 42.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 46.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St John, T. P., and R. W. Davis. 1981. The organization and transcription of the galactose gene cluster of Saccharomyces. J. Mol. Biol. 152:285-315. [DOI] [PubMed] [Google Scholar]
- 48.Tice-Baldwin, K., G. R. Fink, and K. T. Arndt. 1989. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246:931-935. [DOI] [PubMed] [Google Scholar]
- 49.Turkel, S., and P. J. Farabaugh. 1993. Interspersion of an unusual GCN4 activation site with a complex transcriptional repression site in Ty2 elements of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2091-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turkel, S., X. B. Liao, and P. J. Farabaugh. 1997. Gcr1-dependent transcriptional activation of yeast retrotransposon Ty2-917. Yeast 13:917-930. [DOI] [PubMed] [Google Scholar]
- 51.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 52.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winston, F. 1992. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast, p. 1271-1293. In S. L. McKnight and K. R. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed]
- 55.Winston, F., K. J. Durbin, and G. R. Fink. 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675-682. [DOI] [PubMed] [Google Scholar]
- 56.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]