Abstract
The removal of interstrand cross-links (ICLs) from DNA in higher eucaryotes is not well understood. Here, we show that processing of psoralen ICLs in mammalian cell extracts is dependent upon the mismatch repair complex hMutSβ but is not dependent upon the hMutSα complex or hMlh1. The processing of psoralen ICLs is also dependent upon the nucleotide excision repair proteins Ercc1 and Xpf but not upon other components of the excision stage of this pathway or upon Fanconi anemia proteins. Products formed during the in vitro reaction indicated that the ICL has been removed or uncoupled from the cross-linked substrate in the mammalian cell extracts. Finally, the hMutSβ complex is shown to specifically bind to psoralen ICLs, and this binding is stimulated by the addition of PCNA. Thus, a novel pathway for processing ICLs has been identified in mammalian cells which involves components of the mismatch repair and nucleotide excision repair pathways.
DNA interstrand cross-linking agents have been widely used for more than 50 years as potent anticancer agents; nevertheless, the repair of the lesions created by these drugs in mammalian cells is not well understood. In Escherichia coli, two mechanisms for the removal of interstrand cross-links (ICLs) have been described as either recombination dependent or independent. The recombination-dependent pathway was first described by Cole and his colleagues and requires the products of the uvrA, uvrB, uvrC, uvrD, recA, and polA genes (17, 18), indicating that elements of both the nucleotide excision repair (NER) and homologous recombination pathways of E. coli were required for ICL repair. Cole (17) was unable to find evidence for a double-strand break (DSB) intermediate and therefore proposed a model for the removal of ICLs in E. coli in which repair is initiated by dual incisions in one strand on either side of the ICL. The resulting gap is then repaired by a recA-mediated recombination process using a homologous template as a donor. The remaining monoadduct is repaired by a second round of incision by the NER apparatus, and the resulting gap is filled in by the product of the polA gene (DNA polymerase I). Current findings suggest that this pathway is also largely conserved in Saccharomyces cerevisiae (36, 51), with the notable exception that DSBs have been observed during the repair of ICLs in yeast (31, 48, 50). Thus, components of both the RAD3 and RAD52 epistasis groups have been shown to be required for ICL repair in S. cerevisiae. A recombination-independent pathway of ICL repair has also been described for E. coli which requires the products of the uvr and polB genes (6, 7), suggesting that the gap created by NER is repaired by translesion synthesis. Recently a similar recombination-independent pathway requiring NER elements has been described for mammalian cells (61).
While NER plays a central role in repair of ICLs in E. coli and apparently yeast, the mammalian NER pathway does not appear to be required for recombination-dependent repair of ICLs. With the exception of mutants defective in ERCC1 and XPF, which are highly sensitive to ICL-inducing drugs, mammalian mutants defective in genes involved in NER are only mildly sensitive to these agents (3, 35). In fact, recent in vivo findings have shown that the Ercc1-Xpf heterodimer is required for the incision or “unhooking” steps of ICL repair but that other components of the early stages of mammalian NER are not involved (20). In addition, it has been inferred from in vitro studies that incisions that are formed by the NER pathway 5′ to an ICL lead to a futile cycle of incision and gap filling that does not result in repair (9, 52).
After the initial processing of ICLs in vivo, repair of these lesions appears to be funneled into the homologous recombination pathway, since mutants defective in the RAD51-related genes XRCC2 and XRCC3 are extremely sensitive to cross-linking agents (47). Both XRCC2 and XRCC3 have been shown to be required in a conservative pathway of recombination after the introduction of DSBs by the I-SceI endonuclease (37, 55). However, Ku or DNA-PKcs mutants, which are defective in the nonhomologous end joining pathway of DSB repair, do not exhibit an extreme sensitivity to ICL-inducing agents, suggesting that this pathway does not play a major role in the repair of ICLs (53).
While the evidence is compelling that Ercc1 and Xpf are required for the early stages of ICL repair in mammalian cells, other components of this pathway remain to be elucidated. In addition to their role in NER, Ercc1 and Xpf have also been shown to cooperate with mismatch repair (MMR) proteins in removal of insertion/deletion loops (IDLs), and in some pathways of recombination (16, 39, 40, 56). MMR proteins have also been shown to be involved in the recognition and excision repair of some types of alkylation damage (23, 24). These findings prompted us to examine the role of MMR proteins in the processing of ICLs in vitro. Here, we show that hMutSβ, but not hMutSα or hMlh1, is required for the initial processing of psoralen ICLs and that this processing leads to removal or uncoupling of the ICL. These findings identify a novel function for MutSβ and indicate a cooperation between this MMR complex and the Ercc1-Xpf heterodimer in the repair of ICLs in mammalian cells.
MATERIALS AND METHODS
Cell lines and proteins.
Human lymphoid cell lines were cultured in suspension in RPMI 1640 medium supplemented with 20% fetal calf serum (FCS). Human MMR deficient and hamster cell lines were cultured in minimal essential medium plus 10% FCS. HeLa cells were cultured in Joklit medium plus 10% FCS.
hMutSα and hMutSβ complexes were isolated from nuclear extracts prepared from HeLa cells (21). The extract was subjected to sequential chromatography on phosphocellulose and hydroxyapatite columns, and the fractions that contained hMutSα and hMutSβ were identified by Western blotting. hMutSα fractions were pooled and further purified by single-stranded DNA cellulose and monoQ chromatography as described previously (22). hMutSβ fractions were further purified by chromatography on heparin agarose, monoQ, CHT-1, and monoS columns. The Ercc1-Xpf complex was purified from HeLa nuclear extracts as described previously (1). Human PCNA was expressed from the PT7-hPCNA plasmid in E. coli BL21(DE3) and purified by sequential chromatography steps of phosphocellulose, Q-Sepharose, hydroxyapatite, and hydrophobic interaction chromatography essentially as described previously (66).
Substrate preparation.
Plasmids and psoralen interstrand cross-linked substrates for the cross-link induced-repair synthesis (CRS) assay were prepared as previously described (45). Briefly, two complementary oligonucleotides, 5′ GCTCTCGTCTGTACACCGAAG and 5′ GCTCTTCGGTGTACAGACGAG, were synthesized and phosphorylated. One hundred micrograms of this annealed substrate was added to 4,5′,8-trimethylpsoralen at a concentration of 5 μg/ml in a solution containing 10 mM Tris (pH 7.5), 0.5 mM EDTA, and 25 mM NaCl. The sample was irradiated with 365-nm UV light (10 min at 9 mW/cm2) to effect formation of the interstrand cross-link, and the cross-linked oligonucleotide was purified by denaturing polyacrylamide gel electrophoresis (PAGE). To insert the cross-linked oligonucleotide, plasmids were digested with HindIII, and a single deoxyadenosine residue was added to the 3′ end of the cleavage site by incubation with the Klenow fragment of DNA polymerase I. After ligation between the oligonucleotide and the linear plasmid, covalently closed cross-linked template (CLT) DNA was purified by CsCl-ethidium bromide gradient centrifugation. The control template (CT) and donor template (DT) plasmids were prepared in bulk quantities from host cells and purified by CsCl-ethidium bromide gradient centrifugation (see Fig. 1A).
FIG. 1.
Examination of extracts from hMsh2-defective cell lines in the CRS assay. (A) The CLT plasmid contains a single psoralen cross-link at the site identified by an “X.” The CT plasmid is identical to the CLT except for the presence of the cross-link. The DT (for donor template) plasmid is identical to the CT plasmid except for the presence of a short segment containing two SspI sites that replace the NcoI site. (B) Silver-stained gels of purified hMutSβ and hMutSα after sodium dodecyl sulfate-PAGE. (C) CRS assay with extracts prepared from HEC59 cells. The assays were performed as described in Materials and Methods. For the complementation assays, purified hMutSβ and hMutSα were added to a final concentration of 0.4 ng/ml. Subsequent to the incubation, the DNA was extracted, digested with NcoI and AvaII, subjected to agarose gel electrophoresis, and analyzed by autoradiography.
Substrates for electrophoretic mobility shift assays were prepared by digesting the CLT or CT with HincII and BamHI to release the 65-bp cross-linked (CL oligo) or non-cross-linked (NCL oligo) duplex DNA, respectively. The BamHI ends of the oligonucleotides were labeled and converted to blunt ends by treatment with the Klenow fragment of DNA polymerase I in the presence of dATP, dGTP, dTTP, and [α-32P]dCTP. The 65-bp substrates were purified by electrophoresis and extraction from an 8% polyacrylamide gel.
In vitro repair assays.
Mammalian whole-cell extracts were prepared by the method of Manley et al. (49). Where appropriate, before use in the in vitro cross-link assays, each extract was tested for competency in an in vitro NER assay (63). The incorporation assay (referred to as CRS) was performed as previously described (45). Briefly, 50-μl reaction mixtures contained (final concentration) 100 μg of extract, 30 ng of CLT or CT, 100 ng of DT, 45 mM HEPES-KOH (pH 7.8), 75 mM KCl, 7.4 mM MgCl2, 0.9 mM dithiothreitol, 0.4 mM EDTA, 2 mM ATP, 20 μM (each) dATP, dGTP, and TTP, and 8 μM dCTP, 2 μCi of [α-32P]dCTP (3,000 Ci/mmol), 40 mM phosphocreatine, 2.5 μg of creatine phosphokinase, 3% glycerol, and 18 μg of bovine serum albumin. The reactions were incubated for 3 h at 22°C. After incubation, the reactions were stopped by addition of EDTA and the samples were processed to remove RNA and protein. Plasmids were sequentially digested with the appropriate restriction enzymes and analyzed by agarose gel electrophoresis. After gel electrophoresis and staining with ethidium bromide, the gels were photographed and subsequently dried down for autoradiography.
To analyze the products that formed from the CLT during incubation in the mammalian extracts, CRS assays were performed as described above except that the extracted DNAs were digested with the indicated restriction enzymes and examined by 6% sequencing PAGE.
DNA sequencing.
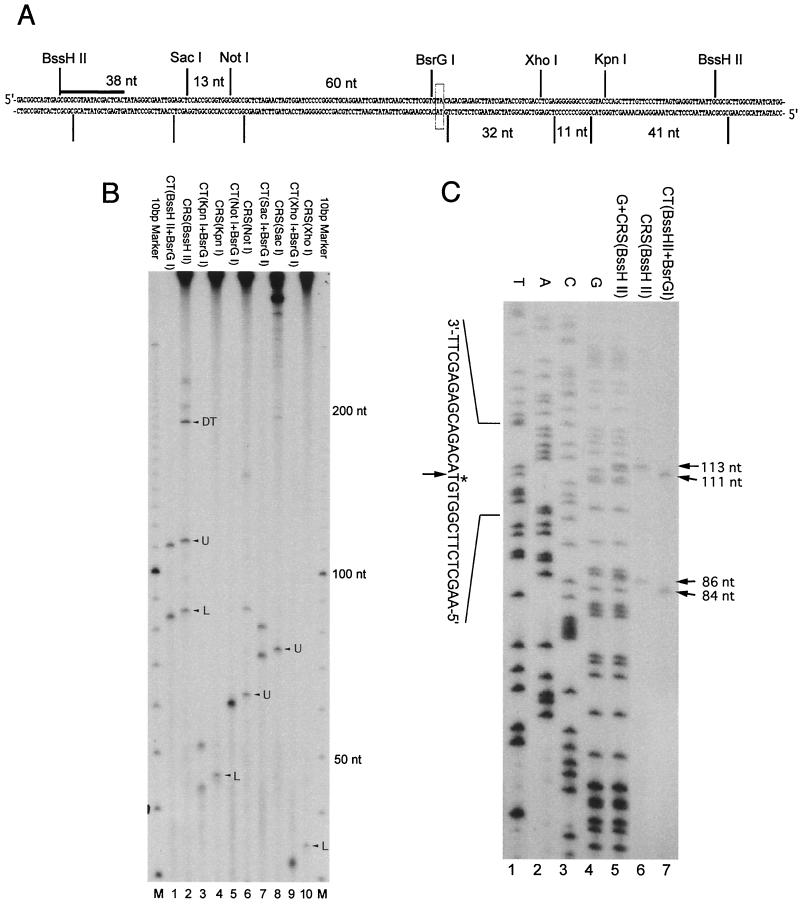
The primer 5′-CGCGCGTAATACGACTCACTAT, located at the leftmost BssHII site of the plasmid substrate (see Fig. 2A), was used for sequencing reactions carried out with the Sequenase Version 2.0 DNA sequencing kit from U.S. Biochemicals according to the manufacturer's recommendations. The CT plasmid was used as the sequencing template.
FIG. 2.
Analysis of products created by processing of psoralen ICLs in HeLa extracts. CRS assays were performed as described in Materials and Methods, and extracted DNAs were digested with the indicated restriction enzymes and examined by denaturing PAGE. (A) The local sequence map of the region surrounding the defined ICL in the CLT. Cleavage sites of the appropriate restriction enzymes on both strands of the DNA are indicated by vertical bars. The numbers between the vertical bars indicate the distances in nucleotides between the cleavage sites. The boxed characters represent the thymine nucleotides that are cross-linked by the psoralen adduct. The horizontal bar represents the location of the sequencing primer. (B) Denaturing PAGE analysis of reaction products after digestion by the indicated restriction enzymes. DNAs (CLT and DT) recovered after incubation in HeLa extract were digested with the indicated enzymes (even-numbered lanes). U and L indicate the bands derived from the upper and lower strands, respectively. For size markers, the CT was digested with BsrGI and the appropriate restriction enzyme and radiolabeled by incubation with T4 polynucleotide kinase and [γ-32P]ATP (odd-numbered lanes). Lanes M indicate the 10-bp marker. (C) Determination by sequencing gel analysis of the 3′ terminus of the fragment resulting from incision and DNA synthesis occurring at the site of the psoralen ICL in HeLa extracts. Sequencing reactions (lanes 1 to 4) were performed with the CT plasmid as template and the indicated primer (seeMaterials and Methods). Assays were conducted with the CLT in the absence of the DT. After incubation, the recovered DNA was digested with BssHII, and loaded either alone (lane 6) or in combination with the “G” sequencing reaction (lane 5). Fragments resulting from digestion of the CT with BssHII and BsrGI are also shown as an additional marker (lane 7). The bands derived from the CLT and CT (lanes 6 and 7) are indicated by arrows on the right side of the panel. On the left side of the panel, the cross-linked thymine residue is indicated by an asterisk, and the 3′ end of the reaction product is indicated by an arrow.
Electrophoresis mobility shift assay.
Binding assays were performed in 20-μl volumes containing 25 mM HEPES-KOH (pH 7.8), 5 mM MgCl2, 80 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 1 mg/ml bovine serum albumin, and the indicated concentrations of hMutSβ at room temperature for 10 min. Carrier/competitor DNA consisted of 75 fmol of 195-bp unlabeled duplex DNA. Approximately 3.5 fmol of labeled 65-bp CL oligo or NCL oligo was added to each reaction. Reactions were separated by electrophoresis on 5% polyacrylamide gels at 4°C in TBE buffer (89 mM Tris borate, 2 mM EDTA). The gels were dried down for autoradiography, and quantification was performed by scanning autoradiograms with Adobe Photoshop software and determining band intensities with Intelligent Quantifier software (Bio Image).
RESULTS
hMutSβ is required for interstrand cross-link-induced DNA synthesis.
To understand the repair of ICLs in mammalian cells, we have developed an in vitro assay, referred to as CRS, using cell extracts, that measures DNA synthesis induced by a single psoralen ICL introduced at a defined site in a plasmid. The substrates for this assay are depicted in Fig. 1A. This single ICL was found previously to induce synthesis in the damaged plasmid and, interestingly, in an undamaged plasmid coincubated in the same extract (45), although the mechanism of incorporation in the undamaged plasmids remains unclear. We also showed that the undamaged plasmid was required only for stimulation of incorporation into the CLT at low levels of the latter plasmid, presumably due to a carrier effect in which the soluble concentration of inhibitory DNA binding proteins are reduced. The assay was also shown to be dependent upon Ercc1, Xpf, RPA, and PCNA but not upon other proteins involved in the disease xeroderma pigmentosum (XP) (45, 46, 65).
To identify additional factors involved in processing of psoralen ICLs, we focused on MMR proteins since they have been shown to cooperate with the Ercc1-Xpf heterodimer in other pathways of DNA repair (16, 39, 40, 56). We first used the CRS assay to examine extracts from the human tumor cell line HEC59. This cell line is defective in hMSH2 and thus deficient in both hMutSβ and hMutSα (11). As shown in Fig. 1C, extract prepared from HEC59 cells was defective in the ICL-induced DNA synthesis. To determine if this deficiency could be rectified by the addition of MMR proteins, we purified hMutSβ and hMutSα from HeLa nuclear extracts as described in Materials and Methods (Fig. 1B). Addition of purified hMutSβ, but not purified hMutSα, resulted in the complementation of the HEC59 extracts (Fig. 1C) and indicated that hMutSβ was specifically required for the observed ICL-induced DNA synthesis. Similar findings (data not shown) were also obtained with the hMSH2-defective human tumor cell line LoVo (11).
Psoralen ICLs are uncoupled during incubation in mammalian cell extracts.
The initial stages of ICL repair processing require the introduction of incisions at or near the site of cross-links, which is followed in vitro by the observed DNA synthesis. To examine the nature of the products formed during the reaction, we performed the CRS assay with HeLa extract, and after digestion with various restriction enzymes (Fig. 2A), the samples were analyzed by denaturing PAGE. For convenience sake, we will refer to this as the incision assay. As shown (Fig. 2B), digestion of the recovered plasmids with BssHII, SacI, NotI, XhoI, or KpnI all yielded unique bands that indicated that specific incisions had occurred at or near the site of the psoralen ICL. As an example, the digests with BssHII yielded two specific fragments that migrated just slightly more slowly than fragments produced from the CT by digestion with a combination of BssHII and BsrGI and labeling by incubation with T4 polynucleotide kinase and [γ-32P]ATP (Fig. 2B, lanes 1 and 2). An analysis of the bands created by the other restriction enzymes (Fig. 2B) indicated that these two bands represented the 5′ ends of the upper and lower strands shown in Fig. 2A. Again, as an example, consider the digest with SacI (Fig. 2B, lanes 7 and 8). Digestion of the CT with SacI and BsrGI yielded a fragment of 73 nucleotides, representing a portion of the upper strand (lane 7). (It also yielded a fragment of 81 nucleotides representing the lower strand). Digestion of the CLT with SacI after incubation and labeling in the HeLa extract yielded a fragment estimated at 75 nucleotides (lane 8). Since cleavage by BssHII and SacI results in 5′ and 3′ nucleotide overhangs, respectively, the only scenario that is consistent with the combined results from the BssHII, SacI, and NotI digestions is that the 5′ end of the upper strand is labeled during the assay and that the resulting 3′ end is located an apparent two nucleotides to the 3′ side of the BsrGI cleavage site. A similar analysis with KpnI and XhoI (Fig. 2B, lanes 3, 4, 9, and 10) indicates that the 5′ end of the lower strand is also labeled during the assay, with the resulting 3′ end also located to the 3′ side of the BsrGI site. Interestingly, the sites of these incisions appear to be located exactly 3′ to the ICL, which would indicate that the ICL must be uncoupled in order to release fragments of the observed sizes.
To determine precisely the sizes of the released fragments, and thus map the terminus of the 3′ ends, a sequencing gel analysis was performed. As shown (Fig. 2C), sequencing of the CT using the primer shown in Fig. 2A, and comparison to the products derived from the CLT after digestion with BssHII, demonstrated that these bands migrated at 113 and 86 nucleotides, indicating that the 3′ incisions resulting from incubation in HeLa extracts occurred precisely 3′ to the psoralen ICL on both strands. Taken together, our analyses of the products of the in vitro processing of psoralen ICLs indicate that DNA synthesis occurs 5′ to the cross-link in either strand and that the ICL is uncoupled during the reaction.
DSBs are created at the site of psoralen ICLs.
A direct prediction of the findings described above is that if these incisions occurred in both strands in the same molecule, a DSB would be created. DSBs have been observed in vivo in both yeast and mammalian cells during the repair of ICLs (5, 19, 20, 31, 50). To assay for DSBs, we performed the CRS assay and overexposed the film to allow easier visualization of products that would be produced if a DSB occurred at the site of the ICL. With the restriction enzymes used for linearization, these products would be predicted to migrate at approximately 1.7 and 0.7 kb. As shown (Fig. 3, top), bands migrating at these positions were observed after agarose gel electrophoresis, which is consistent with the formation of DSBs in the HeLa extract. In addition, the presence of these bands was stimulated by the addition of the DT plasmid, which is consistent with our previous results reported on the CRS assay (45). These results have also been confirmed with other extracts that are positive in the CRS and incision assays. We have also performed kinetic studies on the production of these bands, and the intensity of these bands increases with time of incubation, indicating that they are created as a result of exposure to the extract (data not shown). In addition, the two lower bands shown in Fig. 3, top panel, were purified from the agarose gel and examined by denaturing PAGE. This analysis showed that both bands migrated at the position expected for single-stranded DNA and that, therefore, the DNA in both fragments was not cross-linked (data not shown). This observation is consistent with the findings described above demonstrating that the ICL had been removed or uncoupled from the substrate. Nevertheless, it is possible that these fragments were derived from preexisting linear contaminants in our CLT preparations that become labeled during incubation in the HeLa extract. To rule out this possibility, we prepared a prelabeled cross-linked fragment by digestion of the CLT with BssHII and labeling of the ends with Klenow fragment. Incubation of this substrate with HeLa extract, in the absence of [α-32P]dNTPs, again yielded bands upon PAGE, consistent with the formation of DSBs at the site of the ICL, whereas no bands were observed upon mock incubation without extract, or in the same fragment derived from the CT (Fig. 3, bottom). Taken together, these results indicate that mammalian cell extracts can remove or uncouple psoralen ICLs and convert them into DSBs, consistent with in vivo results obtained in yeast and mammalian cells (5, 19, 20, 31, 50).
FIG. 3.
DSBs are created at the site of psoralen ICLs in HeLa cell extracts. Arrows to the left in each panel indicate fragments resulting from the formation of DSBs. Restriction enzyme maps are shown to the right of each gel figure, indicating the expected products from a DSB created at the site of the ICL, indicated by an X. Top, CRS assay demonstrating the formation of DSBs in vitro as a function of the concentration of the DT plasmid. After incubation in the extract, recovered DNAs were digested with NcoI and AvaII and examined by agarose gel electrophoresis followed by autoradiography. Bottom, DSB formation at the site of psoralen ICLs in a prelabeled cross-linked substrate requires the HeLa cell extract. CLT or CT plasmids were digested with BssHII and labeled by filling in the 3′ ends with Klenow fragment in the presence of [α-32P]dCTP. The labeled substrates were incubated with HeLa extract in the absence of the DT plasmid. Recovered DNAs were extracted and examined by PAGE followed by autoradiography.
Proteins involved in the NER and MMR pathways are required for induction of incisions at psoralen ICLs.
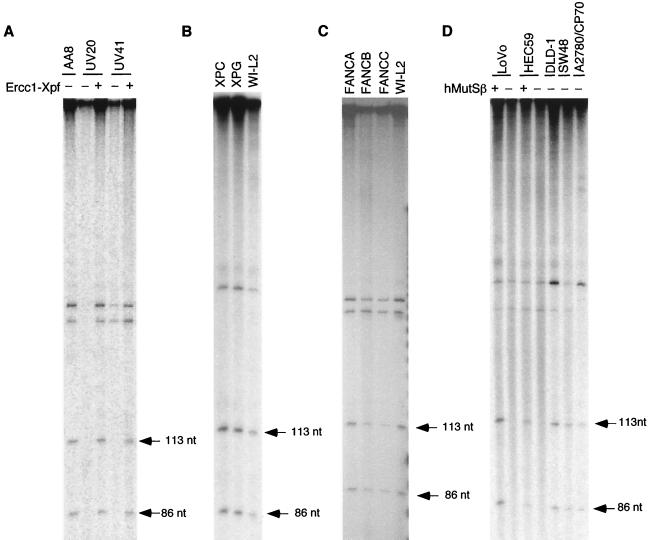
The Ercc1-Xpf heterodimer has been shown to be required for the incision of ICLs in vivo (20), and we have shown that it is required for the CRS assay in vitro (45, 65). We therefore examined extracts from the hamster mutant cell lines UV20 and UV41, which are defective in ERCC1 and XPF, respectively, in the incision assay as described above (Fig. 2B, lane 2). Extracts from both mutant cell lines were deficient in producing incisions at the site of the psoralen ICL, whereas extract from the parental cell line, AA8, was positive (Fig. 4A). Both mutant extracts were complemented by the addition of purified Ercc1-Xpf. Since Ercc1 and Xpf are components of the NER pathway, we next examined whether the observed incisions were due to NER (9, 52) or to a distinct mechanism. As shown (Fig. 4B), extracts from lymphoid cell lines derived from XP patients defective in XPC or XPG were as proficient in the incision assay as the repair-normal lymphoid cell line WI-L2. Consistent with previous results reported by others, extracts from each of the two XP cell lines were examined in the mammalian cell-free NER assay (63) and found to be negative (data not shown). These findings indicate than an intact NER pathway is not required for the observed ICL processing.
FIG. 4.
Incisions and DNA synthesis during psoralen ICL processing in vitro are dependent upon Ercc1-Xpf and hMutSβ. Assay were performed as described in the legend to Fig. 2 using the BssHII restriction enzyme, which generates fragments of 113 and 86 nucleotides (see Fig. 2B, lane 2). (A) Assay of extracts from Chinese hamster ovary cells defective in either ERCC1 or XPF. UV20 and UV41 were derived from the repair-normal cell line AA8 used as a control. For complementation assays, purified Ercc1-Xpf was added to a final concentration of 0.5 ng/ml. (B) Assay of extracts from XP lymphoid cell lines defective in XPC (XP1BE) or XPG (XPG83). WI-L2 is a repair-normal human lymphoid cell line used as a control. (C) Assay of extracts from Fanconi anemia lymphoid cell line defective in FANCA (GM13022A), FANCB (GM13071), or FANCC (GM13020). (D) Assay of MMR-deficient human tumor cell lines defective in hMSH2 (HEC59, LoVo), hMSH6 (DLD1), or hMLH1 (SW48, A2780/CP70). For complementation assays, hMutSβ or hMutSα was added to a final concentration of 0.4 ng/ml.
Fanconi anemia (FA) cells exhibit a pronounced hypersensitivity to cross-linking agents, suggesting that FA proteins may play a role in ICL repair (13, 27). We therefore examined extracts from cells representing FA complementation groups A, B, and C in the incision assay, and we found that all three were proficient (Fig. 4C) and are therefore not required for the early stages of processing psoralen ICLs in vitro.
To determine whether MMR proteins play a role in the introduction of the observed incisions at psoralen ICLs, we examined extracts from the two hMSH2-defective cell lines described above, LoVo and HEC59 (11). We also examined a third line, DLD1, which is defective in hMSH6 (11) and, therefore, deficient in hMutSα but not hMutSβ (Fig. 4D). Of these three cell lines, LoVo and HEC59 extracts were negative in the incision assay but could be complemented by the addition of purified hMutSβ, whereas the DLD1 extract was positive in the incision assay. These findings confirm the results described above and indicate that hMutSβ, but not hMutSα, has a unique role in the processing of psoralen ICLs in vitro. We also examined extracts from two human tumor cell lines, SW48 (11) and A2780/CP70 (60), defective in hMLH1. Extracts from both were positive in the incision assay, indicating that hMlh1 was not required for this activity (Fig. 4D).
MutSβ recognizes psoralen ICLs.
The demonstrated role of MutSβ in MMR is to recognize small IDLs and thus initiate MMR repair processing (29, 33, 54, 62). To determine whether hMutSβ plays the same role in lesion recognition of psoralen ICLs, we performed gel mobility shift studies using a psoralen cross-linked duplex oligonucleotide as a substrate. As shown (Fig. 5A), hMutSβ induced the formation of two specific bands (S1 and S2) and one nonspecific (NS) band. Also, addition of unlabeled cross-linked template reduced binding to the labeled cross-linked probe, indicating that the binding is specific to the ICL (data not shown). These results indicate that hMutSβ can recognize and bind a psoralen ICL with a specificity similar to that previously observed for IDLs.
FIG. 5.
Binding of hMutSβ complex to psoralen DNA ICLs as determined by EMSA. Binding assays were performed as described in Materials and Methods. The positions of the specific (S1 and S2) and nonspecific (NS) complexes are indicated (arrows). (A) hMutSβ binds specifically to DNA containing psoralen ICLs. (B) Effect of ADP and ATP on binding of hMutSβ to psoralen ICLs. (C) PCNA stimulates binding of hMutSβ to psoralen ICLs. A nonspecific band created by incubation of the CL oligo or NCL oligo with PCNA alone is indicated (asterisk). (D) Quantitation of the gel shown in panel C. The S and NS complexes in each lane were quantified by densitometry.
Binding of MutS proteins to mismatched DNAs is reduced upon the addition of ATP, although the interpretation of this finding has proved to be controversial (10, 30, 38). To determine if this binding pattern is maintained in the recognition of psoralen ICLs by hMutSβ, we performed a gel mobility shift assay in the presence of ATP and observed that formation of the S complexes was dramatically reduced (Fig. 5B). Furthermore, addition of ADP reversed this effect, as has been observed for the binding of hMutSβ to mismatched DNA (29). (For a discussion of current theories on the roles of ADP and ATP in MMR see references 14, 38, and 41).
PCNA has been shown to interact with both Msh3 and Msh6 and to act at a step prior to DNA synthesis by enhancing mismatch recognition by MutS proteins (15, 26, 32, 58). To determine if PCNA affected the binding of hMutSβ to psoralen ICLs, we again performed gel mobility shift assays with the psoralen cross-linked duplex oligonucleotide. As shown (Fig. 5C and D), addition of PCNA produced approximately 11-fold and 2-fold increases in the formation of the S1 and S2 complexes, respectively. Taken together, the mobility shift assay results indicate that hMutSβ efficiently discriminated between psoralen cross-linked DNA and undamaged DNA and responded to the addition of nucleotide factors and PCNA similarly to the previously observed binding of MutS proteins to mismatched DNA.
DISCUSSION
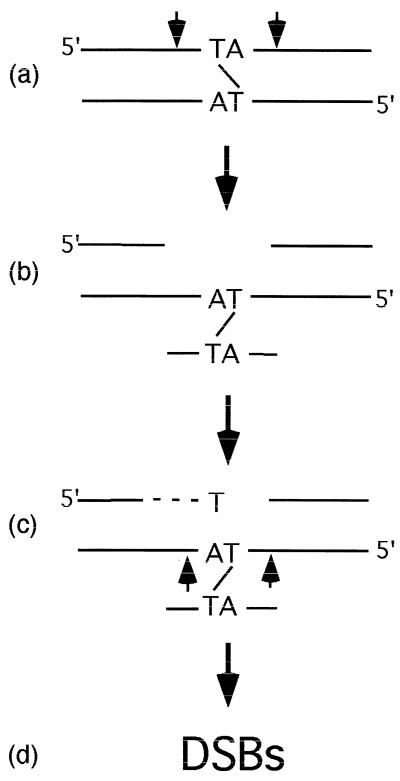
To summarize our findings, we have shown that processing of psoralen ICLs in mammalian cell extracts results in specific products that are produced as a result of incisions near the ICL followed by extension or gap-filling DNA synthesis. The formation of these products also requires that the ICL be uncoupled or removed from the substrate, and this conclusion is confirmed by our finding that DSBs are also formed during the in vitro reaction. A putative model explaining our findings is shown in Fig. 6. Initially, incisions are formed on both sides of the ICL, although the exact sites of these incisions remains to be determined. DNA synthesis then fills in the resulting gap until it reaches the adducted thymine in the template strand, where the polymerase is stalled from further progression, thus giving rise to the fragments we identified after restriction enzyme digestion. Processing of both strands in the same manner or the induction of a single nick at either of the indicated locations (Fig. 6c) would give rise to DSBs, as we have observed. These DSBs and/or the gapped structure (Fig. 6b) may be intermediates that are further processed by homologous recombination pathways to yield fully repaired chromosomes in vivo. DSBs have been observed as intermediates of ICL repair in both yeast and mammalian cells, although the process by which they are produced has not been identified (5, 19, 20, 31, 50). Some support for this model comes from a recent report in which a substrate containing a psoralen ICL placed in a duplex four to six bases from a junction with unpaired DNA was incised in the duplex region on either side of the adduct by purified Ercc1-Xpf (43). Such a structure could arise during replication or it could be formed by recognition and processing of ICLs by repair factors.
FIG. 6.
Model for processing of a psoralen DNA ICLs in mammalian cell extracts. An ICL in duplex DNA stimulates cleavage (indicated by arrows) on both sides of the cross-link in one strand (a), creating a gapped intermediate (b). DNA synthesis (dashed line) fills in the gap but is blocked at the adducted thymine in the template strand from further progression (c). A second incision(s) in the opposite strand at either or both of the indicated points (arrows) would result in the formation of DSBs (d).
We have also identified a number of proteins that are required for the in vitro processing of psoralen ICLs. Previous in vivo and in vitro findings (3, 20, 35, 45, 65) have demonstrated that the Ercc1-Xpf heterodimer, but not other components of the NER pathway, is required for the initial stages of the major pathway of ICL repair in mammalian cells. However, it should be noted that NER has been shown in vivo to participate in an apparently minor, but mutagenic, pathway of ICL repair that presumably entails translesion bypass synthesis (61). Our results confirm the role of Ercc1-Xpf and also implicate hMutSβ and PCNA as proteins involved in the recognition step of ICL processing. These conclusions are based on both the lack of activity in Msh2-deficient cell extracts, which can be complemented by addition of purified MutSβ but not by MutSα, and the demonstration of direct binding of psoralen ICLs by MutSβ which is stimulated by PCNA. Interestingly, our findings rule out hMlh1 as being required for the incisions observed at psoralen ICLs. The precise function of Mlh1 in MMR repair is not completely understood, but it is required for the repair of both base-base mismatches and IDLs (14, 41). The lack of a requirement for hMlh1 and the involvement of Ercc1-Xpf suggest that the initial processing of psoralen ICLs represents a pathway distinct from the archetypal MMR pathway that repairs DNA mismatches. The yeast homologs of Ercc1 and Xpf (Rad10 and Rad1, respectively) and Msh proteins have been shown to function together in several biochemical pathways. The repair of large IDLs has been shown to require Msh2 and Rad1 (40), and the removal of nonhomologous ends during some types of recombination requires the Rad1-Rad10 heterodimer and MutSβ (16, 56). Both Rad1 and Rad10 have also been shown to physically interact with Msh2 (8). Our findings indicate that the Ercc1-Xpf heterodimer, hMutSβ, and PCNA also cooperate in a novel pathway involved in the early stages of processing of psoralen ICLs in mammalian cells. Also, recent in vivo results have shown that cell cycle arrest and repair of psoralen ICLs in primary human fibroblasts requires passage through S phase (2). A requirement for DNA replication for removal of ICLs would be consistent with an involvement of MMR components, since current models suggest a direct association between these proteins and the replication machinery.
Our results indicate that proteins involved in FA complementation groups A, B, and C are not required for the observed in vitro processing of psoralen ICLs. Although there have been reports of an involvement of FA proteins in forming incisions at sites of ICLs (42), their function still remains unclear (13). However, the recent cloning and characterization of the FANCD2 gene has resulted in the discovery of a signal transduction pathway that responds to DNA damage by the mono-ubiquitination of the FANCD2 protein (28, 57). This ubiquitination pathway is dependent upon other FA proteins, and it targets FANCD2 to nuclear foci containing BRCA1. Our findings described here suggest that the ultimate target of this pathway does not involve the initial recognition and processing of ICLs. However, it should be noted that FA proteins may comprise a separate pathway of ICL repair that does not involve the Ercc1-Xpf heterodimer.
MMR proteins have been shown to be involved in the cellular response to DNA damage caused by alkylating agents (12). Defects in MSH2, MSH6, MLH1, and PMS2 in mammalian cells lead to an increased resistance to compounds such as N-methyl-N′-nitro-N-nitrosoguanidine and N-methyl-N-nitrosourea of more than 100-fold compared to wild-type cell lines (44, 59). This increased resistance is apparently due to a defect in both cell cycle arrest and apoptotic responses in these MMR-deficient cells. In wild-type cells this response is mediated by MutSα and MutLα (24, 34, 64). Interestingly, treatment of MMR-defective cells with bifunctional alkylating agents, such as 1,3-bis(2-chloroethyl)-1-nitrosourea, 1-(2-chloroethyl)-3-cyclohexyl-nitrosourea, and mitomycin C results in resistance that is typically somewhat less than that of wild-type cells (4, 25, 34). This phenotype suggests the possibility that MMR proteins are involved in the apoptotic response in the case of monoalkylation but are involved in both apoptosis and DNA repair in the case of ICLs induced by alkylating agents. In this situation, the lack of both apoptosis and DNA repair may effectively offset each other, resulting in the observed level of sensitivity to these drugs that is less than that of wild-type cells. Current findings indicate that MutSβ is not involved in these signaling pathways for O6-alkylguanine lesions (24, 34); however, since we have shown that hMutSβ binds to psoralen ICLs, it is possible that it may also play a dual role in DNA repair and signal transduction.
Our findings indicate a previously unidentified role for hMutSβ in combination with Ercc1-Xpf and possibly PCNA in the recognition and processing of psoralen ICLs. If these findings are applicable to lesions induced by other types of bifunctional alkylating compounds, they could have important implications for their use as chemotherapeutic agents.
Acknowledgments
Nianxiang Zhang and Xiaoyan Lu contributed equally to this work.
We thank Stephen Chaney, Tom Kunkel, and Marsha Frazier for providing cell lines.
This work was supported by grants CA52461 and CA75160 from the National Institutes of Health.
REFERENCES
- 1.Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin, V. Podust, M. Protic, U. Hubscher, J. M. Egly, and R. D. Wood. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859-868. [DOI] [PubMed] [Google Scholar]
- 2.Akkari, Y. M., R. L. Bateman, C. A. Reifsteck, S. B. Olson, and M. Grompe. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 20:8283-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, B. S., T. Sadeghi, M. J. Siciliano, R. Legerski, and D. Murray. 1996. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosphamide analogs. Cancer Chemother. Pharmacol. 38:406-416. [DOI] [PubMed] [Google Scholar]
- 4.Aquilina, G., S. Ceccotti, S. Martinelli, R. Hampson, and M. Bignami. 1998. N-(2-chloroethyl)-N′-cyclohexyl-N-nitrosourea sensitivity in mismatch repair-defective human cells. Cancer Res. 58:135-141. [PubMed] [Google Scholar]
- 5.Averbeck, D., M. Dardalhon, and N. Magana-Schwencke. 1990. Repair of furocoumarin-plus-UVA-induced damage and mutagenic consequences in eukaryotic cells. J. Photochem. Photobiol. 6:221-236. [DOI] [PubMed] [Google Scholar]
- 6.Berardini, M., P. L. Foster, and E. L. Loechler. 1999. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 181:2878-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berardini, M., W. Mackay, and E. L. Loechler. 1997. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry 36:3506-3513. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand, P., D. X. Tishkoff, N. Filosi, R. Dasgupta, and R. D. Kolodner. 1998. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl. Acad. Sci. USA 95:14278-14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessho, T., D. Mu, and A. Sancar. 1997. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol. Cell. Biol. 17:6822-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackwell, L. J., D. Martik, K. P. Bjornson, E. S. Bjornson, and P. Modrich. 1998. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 273:32055-32062. [DOI] [PubMed] [Google Scholar]
- 11.Boyer, J. C., A. Umar, J. I. Risinger, J. R. Lipford, M. Kane, S. Yin, J. C. Barrett, R. D. Kolodner, and T. A. Kunkel. 1995. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. J. Biol. Chem. 55:6063-6070. [PubMed] [Google Scholar]
- 12.Branch, P., G. Aquilina, M. Bignami, and P. Karran. 1993. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature 362:652-654. [DOI] [PubMed] [Google Scholar]
- 13.Buchwald, M., and E. Moustacchi. 1998. Is Fanconi anaemia caused by a defect in the processing of DNA damage? Mutat. Res. 408:75-90. [DOI] [PubMed] [Google Scholar]
- 14.Buermeyer, A. B., S. M. Deschenes, S. M. Baker, and R. M. Liskay. 1999. Mammalian DNA mismatch repair. Annu. Rev. Genet. 33:533-564. [DOI] [PubMed] [Google Scholar]
- 15.Clark, A. B., F. Valle, K. Drotschmann, R. K. Gary, and T. A. Kunkel. 2000. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 275:36498-36501. [DOI] [PubMed] [Google Scholar]
- 16.Colaiacovo, M. P., F. Paques, and J. E. Haber. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151:1409-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole, R. S. 1973. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl. Acad. Sci. USA 70:1064-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, R. S., D. Levitan, and R. R. Sinden. 1976. Removal of psoralen interstrand cross-links from DNA of Escherichia coli: mechanism and genetic control. J. Mol. Biol. 103:39-59. [DOI] [PubMed] [Google Scholar]
- 19.Dardalhon, M., and D. Averbeck. 1988. Induction and removal of DNA interstrand cross-links in V-79 Chinese hamster cells measured by hydroxylapatite chromatography after treatments with bifunctional furocoumarins. Int. J. Radiat. Biol. 54:1007-1020. [DOI] [PubMed] [Google Scholar]
- 20.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond, J. T., G. M. Li, M. J. Longley, and P. Modrich. 1995. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268:1909-1912. [DOI] [PubMed] [Google Scholar]
- 23.Duckett, D. R., J. T. Drummond, A. I. H. Murchie, J. T. Reardon, A. Sancar, D. M. J. Lilley, and P. Modrich. 1996. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. USA 93:6443-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duckett, D. R., S. M. Bronstein, Y. Taya, and P. Modrich. 1999. hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl. Acad. Sci. USA 96:12384-12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiumicino, S., S. Martinelli, C. Colussi, G. Aquilina, C. Leonetti, M. Crescenzi, and M. Bignami. 2000. Sensitivity to DNA cross-linking chemotherapeutic agents in mismatch repair-defective cells in vitro and in xenografts. Int. J. Cancer. 85:590-596. [DOI] [PubMed] [Google Scholar]
- 26.Flores-Rozas, H., D. Clark, and R. D. Kolodner. 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 26:375-378. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Higuera, I., Y. Kuang, and A. D. D'Andrea. 1999. The molecular and cellular biology of Fanconi anemia. Curr. Opin. Hematol. 6:83-88. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 29.Genschel, J., S. J. Littman, J. T. Drummond, and P. Modrich. 1998. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 273:19895-19901. [DOI] [PubMed] [Google Scholar]
- 30.Gradia, S., S. Acharya, and R. Fishel. 1997. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91:995-1005. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg, R. B., M. Alberti, J. E. Hearst, M. A. Chua, and W. A. Saffran. 2001. Recombinational and mutagenic repair of psoralen interstrand crosslinks in Saccharomyces cerevisiae. J. Biol. Chem. 276:31551-31560. [DOI] [PubMed] [Google Scholar]
- 32.Gu, L., Y. Hong, S. McCulloch, H. Watanabe, and G. M. Li. 1998. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 26:1173-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habraken, Y., P. Sung, L. Prakash, and S. Prakash. 1996. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 6:1185-1187. [DOI] [PubMed] [Google Scholar]
- 34.Hickman, M. J., and L. Samson. 1999. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl. Acad. Sci. USA 96:10764-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoy, C. A., L. H. Thompson, C. L. Mooney, and E. P. Salazar. 1985. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 45:1737-1743. [PubMed] [Google Scholar]
- 36.Jachymczyk, W. J., R. C. von Borstel, M. R. A. Mowat, and P. J. Hastings. 1981. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol. Gen. Genet. 182:196-205. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, R. D., N. Liu, and M. Jasin. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401:397-399. [DOI] [PubMed] [Google Scholar]
- 38.Junop, M. S., G. Obmolova, K. Rausch, P. Hsieh, and W. Yang. 2001. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7:1-12. [DOI] [PubMed] [Google Scholar]
- 39.Kirkpatrick, D. T. 1999. Roles of the DNA mismatch repair and nucleotide excision repair proteins during meiosis. Cell. Mol. Life Sci. 55:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirkpatrick, D. T., and T. D. Petes. 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387:929-931. [DOI] [PubMed] [Google Scholar]
- 41.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 42.Kumaresan, K. R., B. Hang, and M. W. Lambert. 1995. Human endonucleolytic incision of DNA 3′ and 5′ to a site-directed psoralen monoadduct and interstrand cross-link. J. Biol. Chem. 270:30709-30716. [DOI] [PubMed] [Google Scholar]
- 43.Kuraoka, I., W. R. Kobertz, R. R. Ariza, M. Biggerstaff, J. M. Essigmann, and R. D. Wood. 2000. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 275:26632-26636. [DOI] [PubMed] [Google Scholar]
- 44.Li, G. M. 1999. The role of mismatch repair in DNA damage-induced apoptosis. Oncol. Res. 11:393-400. [PubMed] [Google Scholar]
- 45.Li, L., C. A. Peterson, X. Lu, P. Wei, and R. J. Legerski. 1999. Interstrand cross-links induce DNA synthesis in damaged and undamaged plasmids in mammalian cell extracts. Mol. Cell. Biol. 19:5619-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, L., C. A. Peterson, X. Zhang, and R. J. Legerski. 2000. Requirement for PCNA and RPA in interstrand crosslink-induced DNA synthesis. Nucleic Acids Res. 28:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783-793. [DOI] [PubMed] [Google Scholar]
- 48.Magana-Schwencke, N., J. A. Henriques, R. Chanet, and E. Moustacchi. 1982. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA 79:1722-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manley, J. L., A. Fire, A. Cano, P. A. Sharp, and M. L. Gefter. 1980. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl. Acad. Sci. USA 77:3855-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McHugh, P. J., W. R. Sones, and J. A. Hartley. 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, R. D., L. Prakash, and S. Prakash. 1982. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol. Cell. Biol. 2:939-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu, D., T. Bessho, L. V. Nechev, D. J. Chen, T. M. Harris, J. E. Hearst, and A. Sancar. 2000. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol. 20:2446-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller, C., P. Calsou, and B. Salles. 2000. The activity of the DNA-dependent protein kinase (DNA-PK) complex is determinant in the cellular response to nitrogen mustards. Biochimie 82:25-28. [DOI] [PubMed] [Google Scholar]
- 54.Palombo, F., I. Iaccarino, E. Nakajima, M. Ikejima, T. Shimada, and J. Jiricny. 1996. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6:1181-1184. [DOI] [PubMed] [Google Scholar]
- 55.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Gene Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmers, C., T. Taniguchi, J. Hejna, C. Reifsteck, L. Lucas, D. Bruun, M. Thayer, B. Cox, S. Olson, A. D. D'Andrea, R. Moses, and M. Grompe. 2001. Positional cloning of a novel fanconi anemia gene, FANCD2. Mol. Cell 7:241-248. [DOI] [PubMed] [Google Scholar]
- 58.Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65-73. [DOI] [PubMed] [Google Scholar]
- 59.Umar, A., M. Koi, J. I. Risinger, W. E. Glaab, K. R. Tindall, R. D. Kolodner, C. R. Boland, J. C. Barrett, and T. A. Kunkel. 1997. Correction of hypermutability, N-methyl-N′-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 57:3949-3955. [PubMed] [Google Scholar]
- 60.Vaisman, A., M. Varchenko, A. Umar, T. A. Kunkel, J. I. Risinger, J. C. Barett, T. C. Hamilton, and S. G. Chaney. 1998. The role of hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin resistance: correlation with replicative bypass of platinum-DNA adducts. Cancer Res. 58:3579-3585. [PubMed] [Google Scholar]
- 61.Wang, X., C. A. Peterson, H. Zheng, R. S. Nairn, R. J. Legerski, and L. Li. 2001. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell. Biol. 21:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson, T., S. Guerrette, and R. Fishel. 1999. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J. Biol. Chem. 274:21659-21664. [DOI] [PubMed] [Google Scholar]
- 63.Wood, R. D., P. Robins, and T. Lindahl. 1988. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell 53:97-106. [DOI] [PubMed] [Google Scholar]
- 64.Wu, J., L. Gu, H. Wang, N. E. Geacintov, and G. M. Li. 1999. Mismatch repair processing of carcinogen-DNA adducts triggers apoptosis. Mol. Cell. Biol. 19:8292-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, N., X. Zhang, C. Peterson, L. Li, and R. Legerski. 2000. Differential processing of UV mimetic and interstrand crosslink damage by XPF cell extracts. Nucleic Acids Res. 28:4800-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, P., S. J. Zhang, Z. Zhang, J. F. Woessner, Jr., and M. Y. Lee. 1995. Expression and physicochemical characterization of human proliferating cell nuclear antigen. Biochemistry 34:10703-10712. [DOI] [PubMed] [Google Scholar]