Abstract
UV lesions in the template strand block the DNA replication machinery. Genetic studies of the yeast Saccharomyces cerevisiae have indicated the requirement of the Rad6-Rad18 complex, which contains ubiquitin-conjugating and DNA-binding activities, in the error-free and mutagenic modes of damage bypass. Here, we examine the contributions of the REV3, RAD30, RAD5, and MMS2 genes, all of which belong to the RAD6 epistasis group, to the postreplication repair of UV-damaged DNA. Discontinuities, which are formed in DNA strands synthesized from UV-damaged templates, are not repaired in the rad5Δ and mms2Δ mutants, thus indicating the requirement of the Rad5 protein and the Mms2-Ubc13 ubiquitin-conjugating enzyme complex in this repair process. Some discontinuities accumulate in the absence of RAD30-encoded DNA polymerase η (Polη) but not in the absence of REV3-encoded DNA Polζ. We concluded that replication through UV lesions in yeast is mediated by at least three separate Rad6-Rad18-dependent pathways, which include mutagenic translesion synthesis by Polζ, error-free translesion synthesis by Polη, and postreplication repair of discontinuities by a Rad5-dependent pathway. We suggest that newly synthesized DNA possessing discontinuities is restored to full size by a “copy choice” type of DNA synthesis which requires Rad5, a DNA-dependent ATPase, and also PCNA and Polδ. The possible roles of the Rad6-Rad18 and the Mms2-Ubc13 enzyme complexes in Rad5-dependent damage bypass are discussed.
Replication of damaged DNA templates can occur by translesion synthesis. This process is usually mutagenic but can be error free as well (see below). In Escherichia coli, the umuC-encoded DNA polymerase V (PolV) promotes mutagenic translesion synthesis (29, 35). Error-prone translesion synthesis, however, accounts for only a minor portion of the damage bypass in E. coli, whereas much of the damage bypass occurs by means of error-free mechanisms. Error-free bypass mechanisms come into play when the replication machinery terminates synthesis at the site of a DNA lesion and replication restarts downstream of the lesion. This results in the formation of a gap in the newly synthesized strand across from the DNA lesion (30). In E. coli, this gap is filled in by RecA-dependent recombination mechanisms by which the DNA strand from the undamaged sister duplex is transferred to the gapped strand in the damaged duplex (31). In mammalian cells, a copy choice type of DNA synthesis has been invoked as the major mechanism for the filling in of gaps formed in the newly synthesized strand opposite DNA lesions (11). In this mechanism, the newly synthesized daughter strand of the undamaged complementary sequence is used as the template to bypass the lesion. Once the lesion is bypassed, DNA polymerase switches back to copying the damaged template strand.
Genetic studies of the yeast Saccharomyces cerevisiae have indicated the requirement of the RAD6 and RAD18 genes in error-free, as well as mutagenic, damage bypass processes (22, 28). rad6 and rad18 mutants exhibit a high degree of sensitivity to UV light and to other DNA-damaging agents, and they are defective in the postreplication repair of discontinuities that form in DNA synthesized from UV-damaged DNA templates (27) and in UV-induced mutagenesis (1, 5, 22). Rad6, a ubiquitin-conjugating (UBC) enzyme, exists in vivo in a complex with Rad18, a DNA-binding protein, and it has been suggested previously that Rad18 targets the Rad6 UBC activity to single-stranded DNA that results from blockage of DNA replication by DNA lesions (2, 3). However, how Rad6-Rad18-dependent protein ubiquitination promotes postreplication repair is not known.
In addition to RAD6 and RAD18, whose functions in damage bypass are expected to be regulatory, the products of the REV1, REV3, and REV7 genes play a direct role in mutagenic translesion synthesis. The Rev3 and Rev7 proteins together form DNA Polζ (26), which functions in translesion synthesis by extending from the nucleotide inserted opposite a DNA lesion by another DNA polymerase (10, 14, 19). For example, Polζ is highly inefficient at inserting nucleotides opposite the 3′ T of a cis-syn thymine-thymine (TT) dimer or a (6-4) TT photoproduct, but it is very efficient at extending from nucleotides inserted opposite the 3′ T of either lesion by another DNA polymerase (14, 19). The Rev1 protein has a deoxycytidyl transferase activity which transfers a dCMP residue to the 3′ end of the DNA primer (25). This activity, however, appears to have little or no role in promoting replication through UV-induced DNA lesions.
RAD30, another member of the RAD6 epistasis group, functions in the error-free replication of UV-damaged DNA (17, 24), and RAD30-encoded DNA Polη (16) replicates through a cis-syn TT dimer with the same efficiency and accuracy with which it replicates through undamaged T's (20, 39). In addition to inducing the formation of cyclobutane dimers at two adjacent thymines, UV light induces the formation of cis-syn cyclobutane dimers and (6-4) photoproducts at 5′-TC-3′ and 5′-CC-3′ sequences. In both yeasts and humans, UV-induced mutations occur primarily by a 3′ C-to-T transition, and the incidence of UV-induced mutations at TC and CC sites is elevated severalfold in the rad30Δ strain over that in the wild-type strain (41). These observations have implicated Polη in the error-free bypass of UV lesions formed at TC and CC sites.
RAD5, MMS2, and UBC13 also belong to the RAD6 epistasis group (4, 12, 15). Rad5, a member of the SNF2/SWI2 family of ATPases, contains the seven consensus helicase motifs, and it has a C3HC4 RING finger motif, which is embedded in the middle of the helicase-like domain (15). Purified Rad5 displays a DNA-dependent ATPase activity, but it is devoid of any DNA helicase activity (18). Since UV-induced mutations still occur in the absence of RAD5 and a synergistic increase in UV sensitivity occurs in the absence of RAD5 and REV3, RAD5 is expected to promote damage bypass in an error-free manner (15). The mms2Δ mutation also results in proficiency in UV-induced mutagenesis, and it leads to a synergistic increase in UV sensitivity when it is combined with the rev3Δ mutation, implicating the involvement of MMS2 in error-free damage bypass as well (4). The Mms2 protein belongs to a family of noncanonical E2s, known as UBC enzyme variant proteins, that lack UBC (E2) activity because of the absence of an active-site cysteine residue (4). Mms2 forms a specific complex with the UBC13-encoded E2, and this complex assembles polyubiquitin chains linked through lysine 63 (12). A role for K63-linked ubiquitin chains has been previously suggested for RAD6-dependent damage bypass, since yeast cells carrying a mutated form of ubiquitin in which lysine 63 is replaced by arginine (ubiK63R) display a UV-sensitive phenotype and the rad6Δ mutation with the ubiK63R mutation results in epistasis (32). A ubc13Δ yeast strain also exhibits UV sensitivity, and UBC13, MMS2, and ubiquitin (ubiK63R) single, double, and triple mutants display comparable UV sensitivity phenotypes, suggesting that all these genes function in the same damage bypass pathway (12).
Here, we examine the roles of various RAD6 group genes, RAD5, MMS2, RAD30, and REV3, in the postreplication repair of DNAs synthesized from UV-irradiated DNA templates. We show that RAD5 and MMS2 make a major contribution to postreplication repair, and we suggest that conjugation of ubiquitin to Rad5 and/or associated proteins by the sequential action of the Rad6-Rad18 and Mms2-Ubc13 enzyme complexes promotes the assembly of Rad5 and associated proteins into the stalled replication machinery. Additionally, we discuss a model in which Rad5, together with PCNA and DNA Polδ, promotes postreplication repair by a copy choice type of DNA synthesis.
MATERIALS AND METHODS
Strains.
For postreplication repair studies, yeast strains were treated with ethidium bromide to obtain [rho0] derivatives lacking mitochondrial DNA. The following isogenic yeast strains, all derived from EMY74.7, MATa his3-Δ1 leu2-3,112 trp1Δ ura3-52, were used in these studies: YR1-65, rad1Δ [rho0]; YR1-118, rad1Δ rad5Δ [rho0]; YR1-77, rad1Δ rev3Δ [rho0]; YR1-120, rad1Δ rad5Δ rev3Δ [rho0]; YR1-206, rad1Δ rad30Δ [rho0]; YR1-205, rad1Δ rad5Δ rad30Δ [rho0]; YR1-218, rad1Δ mms2Δ [rho0]; and YR1-225, rad1Δ mms2Δ rad5Δ [rho0].
The following radΔ single mutant strains, used for determining UV sensitivity, were derived from EMY74.7 and the radΔ mms2Δ double mutant combinations were obtained from the EMY74.7 isogenic mms2Δ ura3 derivative strain YMMS2.6 by the gene replacement method. The strains and their relevant genotypes are as follows: YR1-62, rad1Δ; YR6-100, rad6Δ; YR5-62, rad52Δ; YR30.2, rad30Δ; YR5-50, rad5Δ; YR1-203, mms2Δ rad1Δ; YR6-190, mms2Δ rad6Δ; YR52-55, mms2Δ rad52Δ; YR30-19, mms2Δ rad30Δ; and YR5-53, mms2Δ rad5Δ.
Analysis of DNA synthesized from UV-irradiated templates by sedimentation in alkaline sucrose gradients.
Asynchronously growing yeast cells were UV irradiated in logarithmic phase at a density of 0.5 × 107 to 1.0 × 107 cells per ml of synthetic complete medium lacking uracil but containing 5 μg of uridine/ml at room temperature with constant stirring in 150- by 20-mm petri dishes at a dose rate of 0.1 J/m2/s. All operations after irradiation were performed in yellow light to avoid photoreactivation. Cells were labeled with a radioisotope following UV irradiation and incubated for various times, followed by conversion to spheroplasts as described previously (27). Briefly, after UV irradiation, cells were collected by filtration and resuspended in fresh uridine medium at a density of 1 × 108 to 2 × 108 cells per ml. Pulse-labeling was achieved by the addition of 100 μCi of [3H]6′-uracil (20 to 25 Ci/mmol, 1 mCi/ml; Moravek Biochemicals and Radiochemicals, Brea, Calif.) to 1 ml of cells, followed by vigorous shaking for 15 min at 30°C. Cells were then washed, resuspended in synthetic complete medium containing 1.67 mg of uracil (high-uracil medium)/ml as described previously (27), and incubated for an additional 30 min or 6 h. Conversion of cells to spheroplasts, alkaline sucrose sedimentation, and processing of samples were done as described previously (36, 37).
UV sensitivity.
Cells were grown to midlogarithmic phase in yeast extract-peptone-dextrose medium, washed, sonicated to disperse cell clumps when necessary, and resuspended in sterile distilled water to a density of 2 × 108 cells per ml. Cell suspensions were diluted, spread onto yeast extract-peptone-dextrose medium, and irradiated at a dose rate of 1 J/m2/s. Plates were incubated in the dark, and colonies were counted after 3 to 5 days.
RESULTS
Impaired postreplication repair in the rad5Δ mutant.
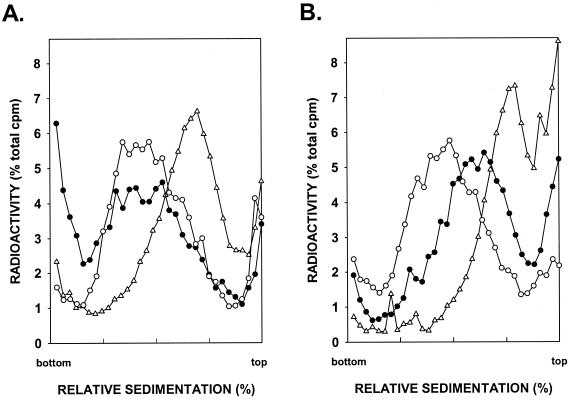
To determine if RAD5 was required for postreplication repair of UV-damaged DNA, we examined the sizes of newly synthesized DNA in UV-irradiated rad1Δ and rad1Δ rad5Δ cells. Because of the defect in the removal of UV-induced DNA damage, survival of UV-irradiated rad1Δ cells depends upon lesion bypass processes. The rad1Δ cells were UV irradiated at 2.5 J/m2, and the size of newly synthesized DNA formed from UV-damaged templates was examined by pulse-labeling of DNA with [3H]uracil for 15 min, followed by a chase for 30 min. DNA from rad1Δ cells obtained following this treatment sediments toward the top of the alkaline sucrose gradient, indicating the presence of discontinuities in the newly synthesized DNA (Fig. 1A). In unirradiated rad1Δ cells, the size of newly synthesized DNA following the 15-min pulse and 30-min chase periods was the same as in overnight-labeled cells (data not shown), indicating that this time interval is sufficient to reconstitute normal-size DNA in unirradiated cells. A previous study has indicated that the size of newly synthesized DNA in UV-irradiated rad1 cells correlates with the average distance between photoproducts present in the template strands (27), an expected result if replication is arrested at the site of UV lesions and a gap is formed in the newly synthesized strand across from the damage site.
FIG. 1.
Requirement of RAD5 for postreplication repair of UV-damaged DNA. rad1Δ (A) and rad1Δ rad5Δ (B) cells were UV irradiated at 2.5 J/m2 and then pulse-labeled with [3H]uracil for 15 min, followed by a 30-min (▵) or 6-h (•) chase in high-uracil medium, prior to conversion to spheroplasts and sedimentation of DNA in alkaline sucrose gradients. Synthesis of normal-sized DNA from unirradiated cells pulse-labeled with [3H]uracil for 15 min was followed by a 6-h chase (○). In the unirradiated rad1Δ and rad1Δ rad5Δ cells, normal-sized DNA reconstituted after a 15-min pulse with [3H]uracil and a 30-min chase. The total sedimentation counts was for cells pulse-labeled for 15 min followed by a 30-min or 6-h chase in high-uracil medium and for the unirradiated cells pulse-labeled for 15 min and then chased for 6 h were ∼11,100, 14,500, and 23,850, respectively.
In rad1Δ cells that were UV irradiated and allowed to have a 6-h repair period following the 15-min pulse, the size of daughter strands became the same as in unirradiated control cells, indicating proficient filling-in of daughter strand gaps (Fig. 1A). In the rad1Δ rad5Δ mutant strain, however, the efficiency of daughter strand gap filling was greatly reduced, as normal-size DNA was not reconstituted in this strain when cells were UV irradiated and allowed to have a 15-min pulse and a 6-h chase period (Fig. 1B). In the unirradiated rad1Δ rad5Δ cells, normal-sized DNA was reconstituted following a 15-min pulse with [3H]uracil and a 30-min chase (data not shown). These results indicate the requirement of RAD5 in daughter strand gap filling of UV-damaged DNA.
Effect of the rev3Δ and rad30Δ mutations on postreplication repair.
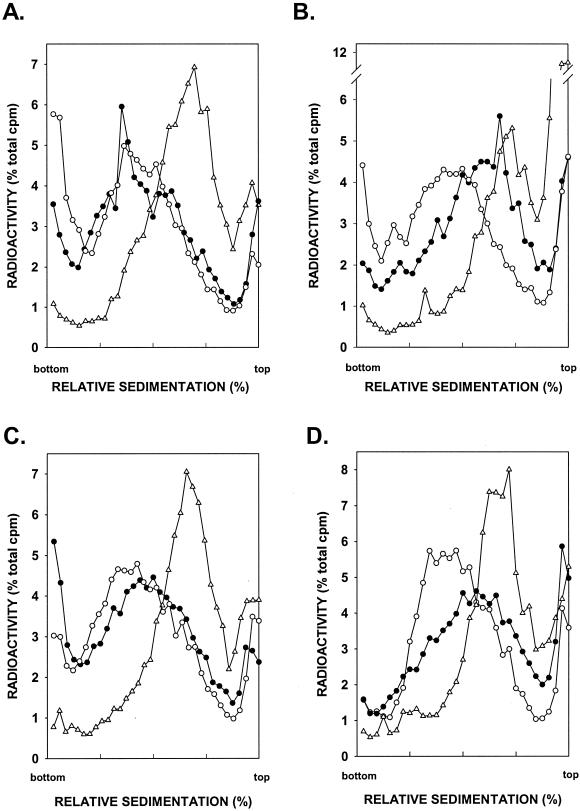
The REV3- and RAD30-encoded DNA polymerases function in mutagenic and error-free modes of translesion DNA synthesis, respectively. Here, we examine the contribution of these polymerases to the reconstitution of normal-sized DNA following the replication of UV-damaged DNA. A previous study had indicated that postreplicative gap filling of UV-damaged DNA was not affected in the rad1 rev3 mutant (27). However, since the rev3 mutation used in that study was a missense allele, the possibility that REV3 function was not entirely inhibited by this mutation could not be excluded. As shown in Fig. 2A, however, the rad1Δ rev3Δ mutant strain was also fully proficient in postreplicative gap filling of UV-damaged DNA. Since the UV sensitivity of the rev3Δ rad5Δ mutant is much greater than that of the rev3Δ or rad5Δ single mutant (15), we examined the ability of the rad1Δ rev3Δ rad5Δ mutant strain to carry out gap filling. As shown in Fig. 2B, the sedimentation profile of DNA from rad1Δ rev3Δ rad5Δ cells that had been UV irradiated and allowed a 6-h repair period following a 15-min pulse period was the same as the size of DNA from rad1Δ rad5Δ cells (compare Fig. 1B and 2B). Thus, the rev3Δ mutation does not enhance the postreplication repair defect of the rad1Δ rad5Δ mutant strain. From these observations, we infer that Polζ makes no significant contribution to the filling in of postreplicative gaps formed opposite UV lesions.
FIG. 2.
Effect of the rev3Δ and rad30Δ mutations on postreplication repair of UV-damaged DNA. rad1Δ rev3Δ (A), rad1Δ rad5Δ rev3Δ (B), rad1Δ rad30Δ (C), and rad1Δ rad5Δ rad30Δ (D) cells were UV irradiated at 2.5 J/m2 and pulse-labeled with [3H]uracil for 15 min followed by a 30-min (▵) or 6-h (•) chase in high-uracil medium. Cells were converted to spheroplasts, and the size of nuclear DNA was examined by sedimentation in alkaline sucrose gradients. Sedimentation patterns of DNA from unirradiated cells pulse-labeled with [3H]uracil for 15 min followed by a 6-h chase (○) are also shown.
The rad30Δ mutation impaired to some degree the restoration of normal-sized DNA in UV-irradiated cells. The rad1Δ rad30Δ mutant strain was not quite as proficient in the repair of discontinuities formed in the newly synthesized DNA strand following UV irradiation as the rad1Δ mutant (compare Fig. 1A and 2C), and the defectiveness in the repair of discontinuities was also somewhat greater in the rad1Δ rad30Δ rad5Δ triple mutant than that in the rad1Δ rad5Δ double mutant (compare Fig. 1B and 2D). Thus, in the absence of translesion synthesis by Polη, some discontinuities accumulate in the DNAs synthesized from UV-damaged templates.
Epistasis of RAD5 to MMS2.
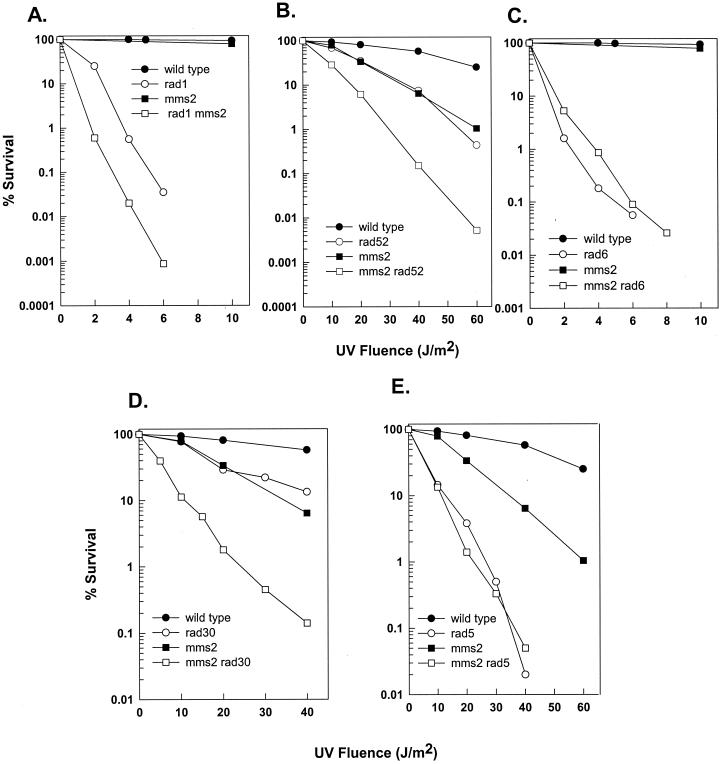
Based on the UV survival curves of single and double mutants, MMS2 has been assigned to the RAD6 epistasis group. However, from these studies, it was not clear if MMS2 also contributed to nucleotide excision repair (NER), since levels of survival after UV irradiation of the rad4 mutant defective in NER and the rad4 mms2 double mutant were not very different (4). Therefore, we have reexamined the epistasis relationships between a strain with the mms2Δ mutation and strains with the rad1Δ, rad52Δ, and rad6Δ mutations, which are defective in NER, recombinational repair, and damage bypass, respectively. We found a synergistic enhancement in UV sensitivity when the mms2Δ mutation was combined with the rad1Δ mutation (Fig. 3A) or with the rad52Δ mutation (Fig. 3B), whereas the UV sensitivity of the mms2Δ rad6Δ strain was no greater than that of the rad6Δ strain (Fig. 3C). These observations provide confirmatory evidence for the involvement of MMS2 in RAD6-dependent damage bypass.
FIG. 3.
Epistasis analysis of the mms2Δ mutation. Shown is survival after UV irradiation of wild-type strain EMY74.7, its isogenic mms2Δ derivative strain YMMS2.6, and EMY74.7 strains carrying deletion mutations of different RAD genes constructed by the gene replacement method. Survival curves present results from an average of approximately three experiments for each strain.
MMS2 functions in the error-free pathway of RAD6-dependent damage bypass, since UV-induced mutagenesis was not affected in the mms2Δ mutant and a synergistic enhancement in UV sensitivity occurs when the mms2Δ mutation is combined with the rev3Δ mutation (4). In contrast to the requirement of REV3-encoded Polζ for error-prone damage bypass, error-free bypass of UV lesions is mediated by two alternate pathways that involve RAD30 and RAD5, respectively. This conclusion stems from observations that the rad5Δ rad30Δ double mutant strain displays a synergistic increase in UV sensitivity compared to the levels of sensitivity of the rad5Δ and rad30Δ single mutants and that the incidence of UV-induced mutations is much higher in the double mutant than in the single mutants (17, 24). The UV sensitivities of strains with the rad5Δ and rad30Δ mutations in combination with the mms2Δ mutation have been examined previously by two independent groups. While in one study, rad5 was shown to be epistatic to mms2 and additive with rad30 (38), in another study, rad5 showed additivity with mms2 (40). Because of these conflicting observations, we reexamined the epistasis relationships of mms2 with rad5 and rad30. We found that, compared to the UV sensitivities of the mms2Δ and rad30Δ single mutants, the UV sensitivity of the mms2Δ rad30Δ double mutant was synergistically enhanced (Fig. 3D) but that the UV sensitivity of the mms2Δ rad5Δ double mutant was the same as that of the rad5Δ mutant (Fig. 3E). These observations support a role for MMS2 in RAD5-dependent error-free postreplication repair.
Defective postreplication repair in the mms2Δ mutant.
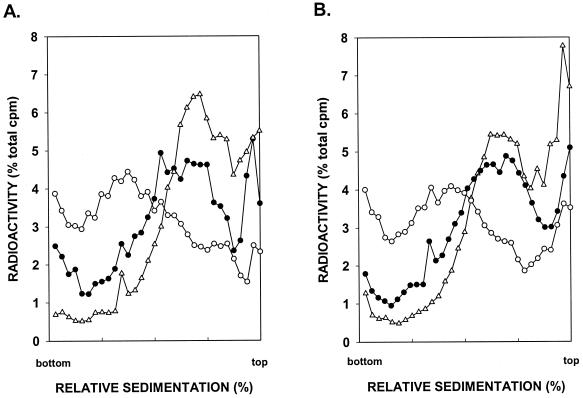
Next, we examined the effect of the mms2Δ mutation on postreplication repair. The mms2Δ mutation was combined with the rad1Δ mutation, and the double mutant was UV irradiated at 2.5 J/m2. The size of newly synthesized DNA was then examined by centrifugation in alkaline sucrose gradients. As shown in Fig. 4A, the efficiency of postreplication repair was greatly reduced by the mms2Δ mutation, since there was only a small increase in the size of newly synthesized DNA even after a 6-h repair period in the rad1Δ mms2Δ strain. Also, little, if any, postreplication repair occurred in the rad1Δ mms2Δ rad5Δ mutant strain (Fig. 4B). The requirement of MMS2 for postreplication repair implies that ubiquitin conjugation by the Mms2-Ubc13 complex is indispensable for this repair process.
FIG. 4.
Requirement of MMS2 for postreplication repair of UV-damaged DNA. rad1Δ mms2Δ (A) and rad1Δ mms2Δ rad5Δ (B) cells were UV irradiated at 2.5 J/m2 and then pulse-labeled with [3H]uracil for 15 min, followed by a 30-min (▵) or 6-h (•) chase in high-uracil medium, prior to conversion to spheroplasts and sedimentation of DNAs in alkaline sucrose gradients. Synthesis of normal-sized DNA from unirradiated cells pulse-labeled with [3H]uracil for 15 min followed by a 6-h chase (○) is also shown.
DISCUSSION
Roles of REV3, RAD30, and RAD5 in the replication of UV-damaged DNA.
Here, we show that the discontinuities formed in the strands synthesized from UV-irradiated DNA templates are repaired with a much-reduced efficiency in the presence of the rad5Δ mutation but that the rev3Δ mutation has little effect on this repair process and that some discontinuities accumulate in the absence of translesion synthesis by Polη. These observations indicate a major role for RAD5 in the postreplication repair of UV-damaged DNA, whereas Polζ and Polη contribute to damage bypass by translesion DNA synthesis.
On its own, Polζ bypasses UV lesions quite poorly. This is so because Polζ is highly inefficient at inserting nucleotides opposite a cis-syn TT dimer or a (6-4) TT photoproduct. However, Polζ efficiently extends from nucleotides inserted opposite the 3′ T of these lesions by another DNA polymerase. For both lesions, Polζ efficiently extends from a G opposite the 3′ T (14, 19), and that accounts for the T-to-C transitions that occur at these lesion sites.
Polη promotes highly efficient and error-free bypass of cis-syn TT dimers, and genetic studies have indicated a role for Polη in the error-free bypass of cyclobutane dimers formed at 5′-TC-3′ and 5′-CC-3′ sites. Thus, we expect Polη to play a prominent role in the bypass of cyclobutane dimers formed at different dipyrimidine sites. Polη may also promote replication through some of the (6-4) dipyrimidine photoproducts (also see below) since it is able to insert a G, from which Polζ subsequently extends, opposite the 3′ T of the (6-4) TT lesion (14).
In contrast to the involvement of Polζ and Polη in translesion DNA synthesis, the Rad5-dependent bypass pathway functions in the postreplication repair of discontinuities that form in the DNA synthesized from UV-damaged templates. These discontinuities may arise primarily opposite the (6-4) dipyrimidine lesions present in the template DNA. In contrast to a cis-syn cyclobutane pyrimidine dimer (CPD), which has only a modest effect on DNA structure, a (6-4) lesion induces a large structural distortion, leading to a 44° bend in the DNA helix, and the 3′ nucleotide of the lesion is held perpendicular to the 5′ nucleotide (21). Consequently, while Polη can efficiently replicate through a CPD, it is unable to bypass a (6-4) lesion, and although Polη can insert a G opposite the 3′ T of the (6-4) TT lesion, it does so with a reduced efficiency (14). Thus, because of the less efficient insertion of nucleotides opposite (6-4) lesions, we expect translesion synthesis to promote the bypass of only a fraction of these lesions. Consequently, the Rad5-dependent postreplication repair pathway may be the major means for bypassing the (6-4) dipyrimidine lesions.
Although Polη and Rad5 may primarily promote the bypass of CPDs and (6-4) photoproducts, respectively, we expect some overlap in their lesion bypass abilities. This overlap is suggested by the facts that the UV sensitivity of the rad5Δ rad30Δ double mutant is greater and its level of mutagenesis is higher than those of the rad5Δ and rad30Δ single mutants (17, 24). Thus, in the absence of RAD30, we expect the RAD5-dependent pathway to promote the bypass of CPDs as well.
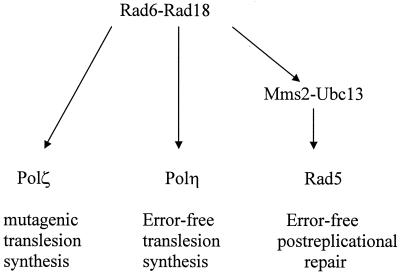
In summary, the bypass of UV lesions in S. cerevisiae is mediated via three separate Rad6-Rad18-dependent pathways: Polζ-dependent mutagenic translesion synthesis, Polη-dependent error-free translesion synthesis through CPDs, and Rad5-dependent repair of postreplicative gaps which presumably are formed in the newly synthesized strand opposite the highly distorting (6-4) dipyrimidine lesions (Fig. 5).
FIG. 5.
Rad6-Rad18-dependent pathways for the replication of UV-damaged DNA in yeast. Whereas Polη primarily carries out error-free translesion synthesis through CPDs and the Rad5-dependent error-free postreplication repair pathway is proposed to primarily promote the bypass of (6-4) dipyrimidine lesions by a copy choice type of DNA synthesis, Polζ contributes to the mutagenic bypass of both these UV lesions. The Mms2-Ubc13 protein complex is proposed to function specifically in the Rad5-dependent postreplication repair pathway. Subsequent to the attachment of ubiquitin to Rad5 and/or associated proteins by the Rad6-Rad18 complex, the Mms2-Ubc13 complex may attach additional ubiquitin moieties to these proteins, and that may be important for the assembly of these repair proteins into the replication machinery.
Requirement of the Mms2-Ubc13 UBC enzyme complex for Rad5-dependent postreplication repair.
In agreement with previous observations (38), we find epistasis of the mms2Δ mutation with rad5Δ but that UV sensitivity is greatly enhanced when the mms2Δ mutation is combined with the rad30Δ mutation (Fig. 3) or the rev3Δ mutation (4). Consistent with the epistasis of rad5Δ and mms2Δ mutations, we find a drastic inhibition in postreplication repair of UV-damaged DNA in the absence of the Mms2 protein. From these observations, we infer the requirement of the Mms2-Ubc13 enzyme complex for Rad5-dependent postreplication repair. Curiously, even though the mms2Δ strain is not as UV sensitive as the rad5Δ strain (Fig. 3E), it is much more defective in postreplication repair than the rad5Δ strain (compare Fig. 1B and 4A). Thus, while the Mms2-Ubc13 complex is almost indispensable for postreplication repair, perhaps, in the absence of Rad5, another protein can partially substitute for it in this repair process. The higher UV sensitivity of the rad5Δ strain than that of the mms2Δ strain may suggest that, in addition to having a role in postreplication repair, Rad5 promotes the repair of UV lesions by some other means as well but that the Mms2-Ubc13 function is primarily dedicated to the filling in of postreplicative gaps.
Possible roles of Rad6-Rad18 and Mms2-Ubc13 enzyme complexes in Rad5-dependent postreplication repair.
Rad5 physically interacts with both Rad18 and Ubc13 (38), suggesting either that Rad5 is a target of ubiquitin conjugation by the Rad6-Rad18 and Mms2-Ubc13 enzyme complexes or that Rad5 serves to recruit these UBC complexes for the ubiquitination of other postreplication repair proteins. Thus, the Rad6-Rad18 and Mms2-Ubc13 complexes may either act on Rad5 directly or coordinate the ubiquitination of some Rad5-associated protein(s) via their interactions with Rad5. Since Rad6 attaches ubiquitin directly to a protein substrate with or without the help of a ubiquitin ligase (7, 13, 33, 34) and the Mms2-Ubc13 complex assembles unanchored polyubiquitin chains linked through lysine 63 (12), following the Rad6-Rad18-mediated attachment of ubiquitin to Rad5 or to Rad5-associated proteins, the Mms2-Ubc13 complex may attach additional ubiquitin moieties linked through lysine 63. Since lysine 63-linked ubiquitin chains are not substrates for proteolytic degradation (6, 8), they may facilitate the assembly of Rad5 and/or its associated proteins into the replication machinery.
Model for Rad5 action in postreplication repair.
Previously, it was shown that the pol30-46 mutation in PCNA results in a defect in the error-free postreplication repair of UV-damaged DNA in yeast (37) and that postreplication repair is also severely inhibited by the yeast pol3-3 mutation, which results in a temperature-sensitive defect in Polδ (36). The requirement of Rad5, PCNA, and Polδ for postreplication repair suggests that these proteins function together in this process.
The RAD6, RAD18, and RAD5 genes do not function in genetic recombination, and mutational inactivation of these genes results in a hyper-recombination phenotype (23, 28). A plausible way by which the Rad5 pathway may promote replication of UV-damaged templates is through a copy choice type of DNA synthesis. Upon stalling of the replication fork at the damage site, ubiquitination of Rad5 and/or associated proteins by the Rad6-Rad18 and Mms2-Ubc13 complexes may promote the assembly of these proteins into the stalled replication machinery. Although Rad5 has no DNA helicase activity (18), such an activity may appear in Rad5 upon its ubiquitination or an as yet unidentified DNA helicase may unwind the 3′ end of the nascent strand, which would then anneal with the homologous strand of the undamaged sister duplex. An assembly of PCNA, Polδ, Rad5, and additional, as yet unidentified proteins may perform limited DNA synthesis through the undamaged sister duplex. Once the 3′ end of the invading strand has been copied past the site of DNA lesion, Polδ would switch back to copying the original template strand. A copy choice type of mechanism has been proposed to account for replication across from damage sites in mammalian cells (11), and a purified system consisting of DNA polymerase holoenzyme, gene 32 protein, dda DNA helicase, and uvsx protein supports a copy choice type of DNA synthesis in bacteriophage T4 (9).
Acknowledgments
This work was supported by National Institutes of Health grant GM19261.
REFERENCES
- 1.Armstrong, J. D., D. N. Chadee, and B. A. Kunz. 1994. Roles of the yeast RAD18 and RAD52 DNA repair genes in UV mutagenesis. Mutat. Res. 315:281-293. [DOI] [PubMed] [Google Scholar]
- 2.Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. [DOI] [PubMed] [Google Scholar]
- 3.Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. [DOI] [PubMed] [Google Scholar]
- 4.Broomfield, S., B. L. Chow, and W. Xiao. 1998. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 95:5678-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassier-Chauvat, C., and F. Fabre. 1991. A similar defect in UV-induced mutagenesis conferred by the rad6 and rad18 mutations of Saccharomyces cerevisiae. Mutat. Res. 254:247-253. [DOI] [PubMed] [Google Scholar]
- 6.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 7.Dohmen, R. J., K. Madura, B. Bartel, and A. Varshavsky. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 88:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finley, D., S. Sadis, B. P. Monia, P. Boucher, D. J. Ecker, S. T. Crooke, and V. Chau. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Formosa, T., and B. M. Alberts. 1986. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47:793-806. [DOI] [PubMed] [Google Scholar]
- 10.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. J. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 13.Jentsch, S., J. P. McGrath, and A. Varshavsky. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329:131-134. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, R. E., L. Haracska, S. Prakash, and L. Prakash. 2001. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21:3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, R. E., S. T. Henderson, T. D. Petes, S. Prakash, M. Bankmann, and L. Prakash. 1992. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12:3807-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 274:15975-15977. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R. E., S. Prakash, and L. Prakash. 1994. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem. 269:28259-28262. [PubMed] [Google Scholar]
- 19.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J.-K., D. Patel, and B.-S. Choi. 1995. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem. Photobiol. 62:44-50. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, C. W. 1982. Mutagenesis in Saccharomyces cerevisiae. Adv. Genet. 21:173-254. [DOI] [PubMed] [Google Scholar]
- 23.Liefshitz, B., R. Steinlauf, A. Friedl, F. Eckardt-Schupp, and M. Kupiec. 1998. Genetic interactions between mutants of the ′error-prone' repair group of Saccharomcyes cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407:135-145. [DOI] [PubMed] [Google Scholar]
- 24.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382:729-731. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 27.Prakash, L. 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 28.Prakash, S., P. Sung, and L. Prakash. 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27:33-70. [DOI] [PubMed] [Google Scholar]
- 29.Reuven, N. B., G. Arad, A. Maor-Shoshani, and Z. Livneh. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763-31766. [DOI] [PubMed] [Google Scholar]
- 30.Rupp, W. D., and P. Howard-Flanders. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 31:291-304. [DOI] [PubMed] [Google Scholar]
- 31.Rupp, W. D., C. E. I. Wilde, D. L. Reno, and P. Howard-Flanders. 1971. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 61:25-44. [DOI] [PubMed] [Google Scholar]
- 32.Spence, J., S. Sadis, A. L. Haas, and D. Finley. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung, P., E. Berleth, C. Pickart, S. Prakash, and L. Prakash. 1991. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 10:2187-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung, P., S. Prakash, and L. Prakash. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2:1476-1485. [DOI] [PubMed] [Google Scholar]
- 35.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres-Ramos, C. A., S. Prakash, and L. Prakash. 1997. Requirement of yeast DNA polymerase δ in post-replicational repair of UV-damaged DNA. J. Biol. Chem. 272:25445-25448. [DOI] [PubMed] [Google Scholar]
- 37.Torres-Ramos, C. A., B. L. Yoder, P. M. J. Burgers, S. Prakash, and L. Prakash. 1996. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proc. Natl. Acad. Sci. USA 93:9676-9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, W., B. L. Chow, S. Broomfield, and M. Hanna. 2000. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics 155:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, S.-L., R. E. Johnson, S. Prakash, and L. Prakash. 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]