Abstract
Chromatin is the physiological template for many nuclear processes in eukaryotes, including transcription by RNA polymerase II. In vivo, chromatin is assembled from genomic DNA, core histones, linker histones such as histone H1, and nonhistone chromatin-associated proteins. Histone H1 is thought to act as a general repressor of transcription by promoting the compaction of chromatin into higher-order structures. We have used a biochemical approach, including an in vitro chromatin assembly and transcription system, to examine the effects of histone H1 on estrogen receptor α (ERα)-mediated transcription with chromatin templates. We show that histone H1 acts as a potent repressor of ligand- and coactivator-regulated transcription by ERα. Histone H1 exerts its repressive effect without inhibiting the sequence-specific binding of ERα to chromatin or the overall extent of targeted acetylation of nucleosomal histones by the coactivator p300. Instead, histone H1 acts by blocking a specific step in the ERα-dependent transcription process, namely, transcription initiation, without affecting transcription reinitiation. Together, our data indicate that histone H1 acts selectively to reduce the overall level of productive transcription initiation by restricting promoter accessibility and preventing the ERα-dependent formation of a stable transcription preinitiation complex.
Chromatin is the physiological template for many nuclear processes in eukaryotes, including transcription, replication, and repair of the genome. The assembly of genomic DNA into chromatin has important functional consequences for the regulation of RNA polymerase II (pol II)-encoding genes since chromatin acts as a general repressor of transcription. Sequence-specific DNA-binding activator proteins, ATP-dependent chromatin-remodeling complexes, and coactivators are required to overcome chromatin-mediated transcriptional repression (23). Transcriptional activator proteins target chromatin-remodeling complexes and coactivators to specific promoters. Chromatin-remodeling complexes use the energy from ATP to reorganize chromatin at the promoters, which allows access of the promoter DNA to the basal transcriptional machinery (reviewed in reference 26). Coactivators have at least two roles (reviewed in references 35 and 53). First, those coactivators with intrinsic histone acetyltransferase (HAT) activity acetylate nucleosomal histones at the promoter, which is thought to facilitate chromatin remodeling. Second, coactivators can make contacts with the basal transcriptional machinery and RNA pol II to facilitate the formation of transcription preinitiation complexes (PICs). The concerted actions of the activators, chromatin-remodeling complexes, and coactivators lead to the transcription of genes assembled into chromatin (reviewed in references 16, 23, 26, 34, and 35).
Chromatin is assembled in vivo from genomic DNA, core histones, linker histones, and nonhistone chromatin-associated proteins. The core histones form the structural core of the nucleosome (38), whereas the linker histones (e.g., histone H1) are thought to play a role in the compaction of chromatin and the formation of higher-order structures (reviewed in references 2, 9, 54, 55, 57, and 63). A number of studies have demonstrated an inhibitory effect of histone H1 on RNA pol II or pol III transcription (see, for example, references 3, 10, 11, 32, 37, and 51). Additional studies have suggested that the chromatin of transcriptionally active genes has reduced histone H1 content compared to that of repressed genes (reviewed in references 9 and 63). Although originally thought to be a general repressor of transcription, histone H1 has since been shown to have more specific effects on RNA pol II and pol III transcription (reviewed in references 9, 54, and 63). Cell-based studies in which the levels of histone H1 were artificially manipulated or genetically depleted indicate that, rather than global transcriptional effects, histone H1 has gene-specific effects (3, 5, 51). Although the mechanistic details of histone H1-mediated transcriptional inhibition are not clear, it is likely that the gene-specific effects of histone H1 are related to specific promoter architectures, as well as the specific subset of transcription factors (TFs) involved in the transcription of each gene.
In the present study, we have used a biochemical approach to examine the effects of histone H1 on the transcriptional activity of estrogen receptor α (ERα). ERα is a ligand-regulated, sequence-specific DNA-binding TF and a member of the nuclear hormone receptor superfamily (39). Previous studies have shown that chromatin plays an integral role in the ligand- and coactivator-dependent transcriptional activity of ERα (29). In the absence of chromatin (i.e., with naked DNA templates), spurious ligand-independent transcription by ERα is observed, the magnitude of ligand-dependent transcription is severely reduced, and coactivators such as p300 fail to enhance receptor-dependent transcriptional activity (29). Herein, we show that histone H1 acts as a potent repressor of ligand- and p300-regulated ERα transcriptional activity by inhibiting a specific step in the transcription process with ERα, namely, transcription initiation, without affecting transcription reinitiation.
MATERIALS AND METHODS
Plasmid templates and purified proteins.
The plasmid templates p2ERE-AdE4 and pERE contain two and four copies of the Xenopus vitellogenin A2 gene estrogen response element (ERE), respectively, located upstream of the adenovirus E4 core promoter in plasmid pIE0 (29, 30). His6-tagged human p300 and FLAG-tagged human ERα were expressed in Sf9 cells and purified by affinity chromatography as described previously (30). His6-tagged mouse SRC2(RID/PID), which contains the receptor and p300/CBP interaction domains of the protein (amino acids 624 to 1130) (25), was expressed in Escherichia coli by using a pET vector and purified by nickel-nitrilotriacetic acid affinity chromatography by using standard techniques.
Purification of histone H1.
Native calf thymus histone H1, purified by the method of Cole (8), was purchased from Gibco BRL and dissolved in H1 buffer (20 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 0.2 mM EDTA, 15% glycerol, 2 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) before use. Native Spodoptera frugiperda histone H1 was purified from cultured Sf9 cells by nonspecific association with anti-FLAG M2 affinity resin as follows. A 200-ml culture of Sf9 cells was grown to a density of approximately 1.5 × 106 cells per ml. The cells were pelleted, washed with ice cold phosphate-buffered saline, resuspended in 7 ml of lysis buffer (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 0.5% NP-40, 2 mM DTT, 1 mM PMSF, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml), and lysed by Dounce homogenization. The whole-cell lysate was centrifuged at 17,500 × g for 20 min at 4°C to pellet the insoluble material. The supernatant was recovered, diluted 1:1 with dilution buffer (20 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.5% NP-40, 2 mM DTT, 1 mM PMSF, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml), and incubated for 2 h at 4°C with 150 μl of a 50% slurry of anti-FLAG M2 affinity resin (Sigma). The resin was then washed four times with wash buffer (20 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 150 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.2% NP-40, 2 mM DTT, 1 mM PMSF). The histone H1 protein was eluted in batch with elution buffer (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 0.2 mM EDTA, 15% glycerol, 0.1% NP-40, 2 mM DTT, 1 mM PMSF, 0.5 mg of insulin per ml, 0.2 mg of FLAG peptide per ml). The identity of the purified histone H1 protein was confirmed by mass spectroscopic analysis of trypsin-digested samples (performed by the BioResource Center of Cornell University), as well as by Western blotting using a commercially available histone H1 antibody (Calbiochem).
Chromatin assembly and analysis.
Chromatin assembly reaction mixtures were prepared by using the S190 extract derived from Drosophila embryos as described previously (7, 24, 29, 30, 46). Briefly, purified Drosophila core histones, a plasmid DNA template, and an ATP-regenerating system were incubated with the S190 extract for 4 h at 27°C under buffer conditions described previously (28, 30). Purified recombinant ERα and 17β-estradiol (E2) were added during the chromatin assembly reactions. Purified histone H1 was added either during chromatin assembly or 15 to 20 min prior to the end of the chromatin assembly reaction. When included in the experiments, purified recombinant p300 was added after the chromatin assembly reactions were complete, followed by a 15-min incubation at 27°C to allow interaction with the chromatin and ERα. The amounts of the various factors included in the reaction mixtures are listed in the figure legends.
Micrococcal nuclease digestion analyses were performed as previously described (7, 24, 45, 46). After digestion and deproteinization, the DNA samples were separated on a 1% agarose gel and stained with ethidium bromide. DNase I primer extension footprint analyses of the chromatin templates were performed as previously described (44-46).
Glycerol gradient analysis of chromatin.
Glycerol gradient analyses were performed essentially as previously described (40), with slight modification. Briefly, histone H1 (estimated concentrations of 0, 50, and 100 nM) was added to the S190 chromatin assembly mixture 15 min prior to the end of the reaction. Each chromatin sample (400 μl containing 2 μg of plasmid DNA) was then applied to a 3.0-ml linear 15 to 40% glycerol gradient set on a 0.3-ml 100% glycerol cushion at the bottom of the tube. The gradients were centrifuged at 50,000 rpm in a Beckman SW60 rotor for 1 h at 4°C. After centrifugation, 335-μl fractions were removed from the top down for each gradient. Fifty microliters of each fraction was extracted with phenol-chloroform, ethanol precipitated, and subjected to 1% agarose gel electrophoresis in 1× Tris-borate-EDTA with ethidium bromide staining for DNA analysis. The remaining portion of each fraction was precipitated with 25% trichloroacetic acid, run on a sodium dodecyl sulfate (SDS)-15% acrylamide gel, transferred to nitrocellulose, and analyzed by Western blotting for core histones and histone H1 with an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech).
Restriction enzyme accessibility assays.
For the restriction enzyme accessibility assays, 20-μl aliquots of S190-assembled chromatin (containing 100 ng of plasmid DNA) were digested with 0, 2, 5, 10, and 20 U of the restriction endonuclease XbaI (New England Biolabs) for 30 min at 27°C. The digested chromatin samples were incubated for 15 min at 70°C to heat inactivate the enzyme and then treated sequentially with RNase A for 15 min at room temperature (RT) and proteinase K for 15 min at 37oC. The chromatin samples were extracted with phenol-chloroform, ethanol precipitated, simultaneously digested with the restriction endonucleases HindIII and EcoRI (New England Biolabs), and subjected to 1.75% agarose gel electrophoresis in 1× Tris-glycine buffer. The samples were transferred to nitrocellulose and analyzed by Southern blotting using an oligonucleotide probe that hybridizes between the XbaI and EcoRI sites (see schematic in Fig. 6).
FIG. 6.
Histone H1 inhibits localized increases in restriction enzyme accessibility induced by the binding of ERα. Specific structural changes at the promoter induced by histone H1 were analyzed by restriction enzyme accessibility assays using chromatin-assembled pERE. The experiment was performed under the conditions described for Fig. 3B (i.e., under conditions in which ERα-mediated transcription is inhibited by histone H1 but in the absence of the HeLa cell nuclear extract used for the transcription assays). (A) Restriction enzyme accessibility assay. (Top) Aliquots of chromatin, with or without added ERα or histone H1, were digested with increasing amounts of the restriction endonuclease XbaI, deproteinized, double digested with the restriction endonucleases HindIII and EcoRI, and subjected to Southern blot analysis with an end-labeled oligonucleotide probe that hybridizes to the XbaI/EcoRI fragment. The HindIII/EcoRI fragments (Uncut) and the XbaI-digested fragments (XbaI cut) are indicated by arrows. (Bottom) The schematic diagram denotes the locations of the EREs, TATA box, transcription start site, oligonucleotide probe, and restriction enzymes used in the assay. (B) Quantification of restriction enzyme accessibility assays. The signals in each lane in panel A were quantified with a PhosphorImager. The extent of digestion by XbaI under each condition was determined by dividing the signal from the XbaI-cut bands by the sum of the signals from the XbaI-cut and uncut bands. Each point represents the mean ± the standard error of the mean of three or more separate determinations.
In vitro transcription assays.
In vitro transcription reactions with the chromatin templates were performed as described previously (29, 30, 47, 52). Briefly, duplicate 15-μl aliquots of chromatin for each experimental condition (containing 75 ng of plasmid DNA) were mixed with 10 μl of HeLa cell nuclear extract (∼15 μg of extract protein) prepared by the method of Dignam et al. (12) in a 50-μl reaction mixture under buffer conditions described previously (30, 47, 52). For multiple-round transcription experiments, the chromatin was incubated with the HeLa cell nuclear extract for 15 min at RT, followed by the addition of ribonucleoside-5′-triphosphates (rNTPs) and an additional 30-min incubation at 30°C. Single-round transcription experiments were performed in a similar manner, except that the chromatin was incubated with the HeLa cell nuclear extract for 45 min at RT to allow a more complete formation of transcription PICs before the addition of rNTPs. The detergent Sarkosyl, which limits transcription to a single round (17, 18, 29), was added at a final concentration of 0.2% (wt/vol) about 15 s after the addition of the rNTPs. After the transcription reaction incubations were complete, the RNA samples were prepared for primer extension analyses as described previously (30, 47, 52). For most transcription experiments, histone H1 was added either during chromatin assembly or 15 min prior to the end of assembly. In the order of addition experiments, histone H1 was added at the time points described in the legend to Fig. 8. The data from the transcription experiments were analyzed with a Molecular Dynamics PhosphorImager. All transcription reactions were carried out in duplicate, and each experiment was performed at least two times to ensure reproducibility.
FIG. 8.
Histone H1 represses ERα-dependent transcription by selectively inhibiting the formation of a stable PIC. (A) Schematic representation of the order-of-addition, single-round transcription experiments with chromatin templates shown in panel B. The chromatin assembly and single-round transcription assays were performed under the conditions described in the legend to Fig. 7. Histone H1 was added at the following times: 1, directly to the chromatin-assembled template prior to the addition of the other reagents; 2, with the HeLa cell nuclear extract (HNE); 3, before the initiation of transcription (i.e., 1 min prior to addition of the rNTPs); 4, just after the initiation of transcription but prior to the addition of Sarkosyl to limit transcription to a single round. (B) Results of the order-of-addition, single-round transcription experiments. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The signals in each lane were quantified with a PhosphorImager.
Targeted histone acetylation assays.
The targeted histone acetylation assays were based on the method of Kim et al. (25). Briefly, the plasmid template pERE was assembled into chromatin by using the S190 extract in the presence or absence of 100 nM histone H1. The chromatin was subsequently purified by column chromatography with Sepharose CL-4B as described previously (43) to remove unincorporated histones. After purification, aliquots of chromatin containing approximately 750 ng of plasmid DNA were incubated in various combinations with ERα (350 ng), p300 (500 ng), SRC2(RID/PID) (100 ng), and E2 (to a final concentration of 100 nM) for 15 min at 27°C in a final volume of 35 μl to allow interaction of the factors with the chromatin template. Histone acetylation reactions were performed by using [3H]acetyl coenzyme A as described previously (30). The reaction mixtures were separated on an SDS-15% polyacrylamide gel and subjected to fluorography. The 3H-labeled histone bands were excised from the gel and quantified by liquid scintillation counting.
RESULTS
Histone H1 is efficiently incorporated into S190-assembled chromatin.
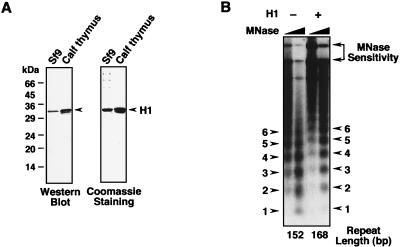
To examine the effects of histone H1 on various aspects of transcriptional activation by ERα with chromatin templates, we used native histone H1 purified from Sf9 cells or calf thymus (Fig. 1A) and an in vitro chromatin assembly and transcription system that faithfully recapitulates the known ligand-dependent functions of ERα (28, 29, 34). Incorporation of histone H1 into chromatin is known to increase the spacing between nucleosomes, also known as the nucleosomal repeat length (reviewed in reference 54). To ensure that exogenously added histone H1 was incorporated into the chromatin assembled with a Drosophila S190 chromatin assembly extract (7, 24), we performed micrococcal nuclease digestion of chromatin with or without histone H1 (Fig. 1B). Addition of histone H1 caused a discernible increase in the nucleosomal repeat length from 152 to 168 bp. Thus, the added histone H1 was incorporated into the chromatin.
FIG. 1.
Assembly of chromatin containing histone H1 by using a Drosophila chromatin assembly extract. (A) SDS-polyacrylamide gel electrophoresis and Western blotting analyses of histone H1 purified from Sf9 cells and calf thymus. The purified proteins were subjected to SDS-10% polyacrylamide gel electrophoresis with subsequent Western blotting with an anti-histone H1 antibody (left) or staining with Coomassie brilliant blue R-250 (right). (B) Addition of histone H1 to chromatin increases resistance to micrococcal nuclease and the nucleosomal repeat length. Chromatin was assembled by using the Drosophila S190 extract in the presence or absence of purified Sf9 cell histone H1 (100 nM) and then subjected to digestion with micrococcal nuclease (MNase). Note the increased resistance to micrococcal nuclease cleavage in the presence of histone H1 as indicated by increased amounts of higher-molecular-weight DNA species. The positions of the mono, di, and tri, etc., nucleosomal DNA fragments are indicated. Similar results were obtained with calf thymus histone H1 (not shown).
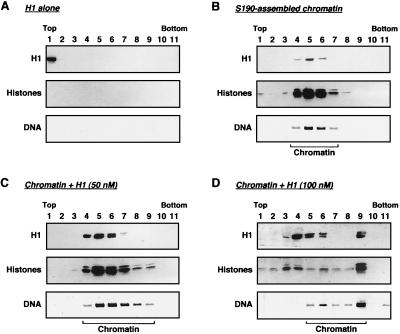
To determine the extent of incorporation of histone H1 into the chromatin assembled by using the S190 extract, we subjected the assembly reaction products to glycerol gradient analyses (Fig. 2). Chromatin was incubated with or without histone H1 (roughly 50 or 100 nM, as estimated from Coomassie-stained SDS-acrylamide gels with protein loading standards) and run through a 15 to 40% glycerol gradient. After centrifugation, fractions were removed from top to bottom and analyzed for histone H1 and core histone content by Western blotting and for DNA content by agarose gel electrophoresis with ethidium bromide staining. As shown in Fig. 2A, histone H1 in the absence of chromatin sedimented in fraction 1. When S190-assembled chromatin without exogenously added histone H1 was run on the gradient, the chromatin sedimented in fractions 4 through 7 (peak in fraction 5), as indicated by the presence of core histones and plasmid DNA (Fig. 2B). Note that trace amounts of histone H1 contributed by the S190 chromatin assembly extract comigrated in the same fractions. When ca. 50 nM histone H1 was added to the chromatin assembly reaction mixture, the sedimentation profile of the chromatin shifted to fractions 4 through 9 (peak in fractions 5 and 6), consistent with the incorporation of histone H1 (Fig. 2C). Complete incorporation of the exogenously added histone H1 was observed at the 50 nM concentration, as indicated by the shift of all of the histone H1 from fraction 1 to the chromatin-containing fractions (i.e., those fractions containing core histones and DNA). Finally, when ca . 100 nM histone H1 was added to the chromatin assembly reaction mixture, the sedimentation profile of the chromatin shifted to fractions 5 through 9, with the majority of the chromatin in fraction 9, which likely represents an H1-saturated chromatin fraction (Fig. 2D). At the 100 nM concentration, approximately 60 to 70% of the H1 was found in the chromatin-containing fractions (based on multiple experiments like those shown in Fig. 2D), with the remainder found comigrating with free core histones (i.e., without DNA) in fractions 3 and 4. From these results, we conclude that exogenously added histone H1 is efficiently incorporated into S190-assembled chromatin and that saturation occurs between histone H1 concentrations that we estimate to be between 50 and 100 nM.
FIG. 2.
Histone H1 is efficiently incorporated into chromatin assembled in vitro. S190-assembled chromatin was incubated with calf thymus histone H1 at final concentrations of 0, 50, and 100 nM for 15 min at 27°C. After incubation, each sample was run on a 15 to 40% glycerol gradient. Fractions from the gradients were analyzed for histone H1 and core histones by Western blotting and for DNA by agarose gel electrophoresis with ethidium bromide staining. Note that the anti-core histone serum preferentially detects histones H2A and H2B (H2A > H2B > H3 > H4).
Previous studies have suggested that one molecule of histone H1 is incorporated into chromatin per nucleosome (1). We estimate that the concentration of nucleosomes in the chromatin used in our experiments is about 50 nM, which should also be the saturation point for histone H1 incorporation. However, we did not observe saturation of histone H1 incorporation at 50 nM but did observe saturation at a slightly higher histone H1 concentration (about 75 nM). Nonetheless, the histone H1 incorporation profiles shown in Fig. 2 are within the expected range given the amounts of histone H1 and nucleosomes in the chromatin assembly reaction mixtures. The differences from the predicted values are likely due to the fact that histone H1 concentrations are difficult to determine accurately (8), as well as the fact that the number of nucleosomes in the chromatin assembly reaction mixtures is an estimate.
Histone H1 is a potent repressor of ERα-mediated transcription at physiological concentrations.
ERα is an interesting transcriptional activator because it stimulates multiple steps in the transcription process (i.e., both transcription initiation and reinitiation; reference 29) and its activity is regulated by ligand binding (i.e., estrogens), activities that are limited to a subset of the known transcriptional activators. To examine the effects of histone H1 on ERα-mediated transcription with chromatin templates, a plasmid template containing two or four copies of a consensus ERE upstream of the adenovirus E4 promoter was assembled into chromatin by using the S190 extract. Purified ERα, ligand (i.e., E2), and histone H1 were added during chromatin assembly, and the chromatin templates were subsequently transcribed by using a HeLa cell nuclear extract as a source of the basal transcriptional machinery.
As shown in Fig. 3A, liganded ERα is a robust activator of transcription with chromatin using a template containing two EREs (compare lanes 1 and 2 and lanes 7 and 8) or four EREs (data not shown). The transcriptional activity of ERα in this assay system is ligand dependent (data not shown; reference 29). Addition of either Sf9 cell or calf thymus histone H1 during the chromatin assembly reaction reduced the amount of ERα-dependent transcription by about 85% at the highest concentration of histone H1 used (100 nM; compare lanes 2 and 6 and lanes 8 and 12). At a histone H1 concentration of 50 nM, which is below the saturation point for histone H1 incorporation in these assays (Fig. 2C), greater than 50% repression was observed (compare lanes 2 and 5 and lanes 8 and 11). Together, these results indicate that native histone H1, purified from either Sf9 cells or calf thymus, is a potent repressor of ERα-mediated transcription at physiological histone H1 concentrations. Note that nearly all of the remaining experiments were performed with both Sf9 cell and calf thymus histone H1, as well as with the two-ERE and four-ERE templates. In all cases, the results were essentially the same. Thus, for brevity, we will show only one case for each of the remaining experiments.
FIG. 3.
Histone H1 is a potent inhibitor of ERα-mediated transcription with chromatin templates. (A) Histone H1 inhibits ERα-mediated transcription with chromatin templates in a concentration-dependent manner. The plasmid template p2ERE-AdE4 was assembled into chromatin in the presence or absence of increasing amounts of purified Sf9 cell or calf thymus histone H1 (H1Sf9 and H1ct, respectively), as well as ERα (10 nM) and E2 (100 nM), as indicated. The concentrations of histone H1 in the chromatin assembly reaction mixtures were 12.5, 25, 50, and 100 nM. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The signals in each lane were quantified with a PhosphorImager. Similar results were obtained by using as a template pERE, which contains four EREs (not shown). (B) Histone H1 inhibits ERα-mediated transcription with chromatin templates when added either during or after chromatin assembly. The plasmid template p2ERE-AdE4 was assembled into chromatin in the presence or absence of purified Sf9 cell histone H1, ERα (10 nM), and E2 (100 nM), as indicated. Histone H1 was added at a concentration of 100 nM either during chromatin assembly or after chromatin assembly was complete. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The signals in each lane were quantified with a PhosphorImager. Similar results were obtained with calf thymus histone H1 (not shown).
In vivo, histone H1 interacts dynamically with preassembled chromatin (36, 42). Therefore, we determined whether histone H1 could repress ERα-mediated transcription when added to a fully assembled chromatin template. Nearly identical results were obtained when histone H1 was added at the beginning of chromatin assembly (Fig. 3B, During Assembly) or after chromatin assembly was complete (Fig. 3B, After Assembly). These results indicate that histone H1 can exert its repressive effects on ERα transcriptional activity when added to a fully assembled chromatin template.
Histone H1 does not inhibit sequence-specific binding of ERα to chromatin.
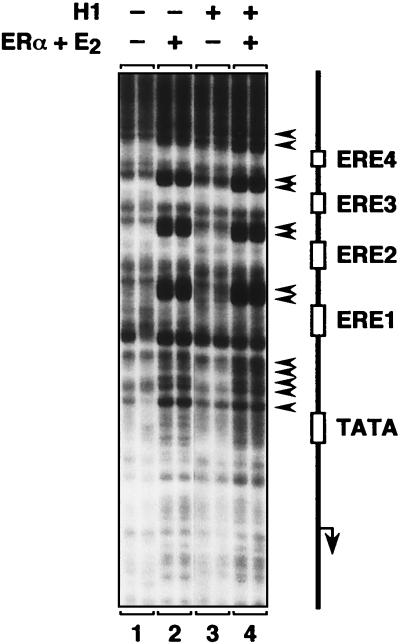
Next, we considered some possible mechanisms by which histone H1 might block ERα-mediated transcription. First, we examined the effects of histone H1 on the binding of ERα to chromatin. To do so, we subjected chromatin templates (assembled under the same conditions used for the transcription analyses in Fig. 3 but without addition of the HeLa cell nuclear extract used for the transcription reactions) to partial digestion with DNase I and resolved the cleavage patterns by primer extension analysis. As shown in Fig. 4, binding of liganded ERα to the chromatin templates results in a distinctive pattern of DNase I-hypersensitive sites surrounding the EREs (compare lanes 1 and 2), similar to what has been reported previously (29). Addition of histone H1 had no discernible effect on the pattern of DNase I cleavage in the presence of liganded ERα (compare lanes 2 and 4), indicating that binding of ERα to the chromatin templates was not affected.
FIG. 4.
Histone H1 does not inhibit sequence-specific binding of ERα to chromatin. The effect of histone H1 on the binding of liganded ERα to the EREs in the chromatin-assembled pERE template was analyzed by DNase I primer extension footprinting. The experiment was performed under the conditions described for Fig. 3B (i.e., under conditions in which ERα-dependent transcription is inhibited by histone H1 but in the absence of the HeLa cell nuclear extract used for the transcription assays). The schematic diagram denotes the locations of the EREs, TATA box, and transcription start site for the pERE template (arrow). Binding of ERα to the chromatin template results in regions of DNase I hypersensitivity, which are indicated by arrowheads. Each sample was run in duplicate.
Histone H1 inhibits ERα-mediated transcription in the presence of exogenously added p300 without blocking the targeted acetylation of nucleosomal histones by p300.
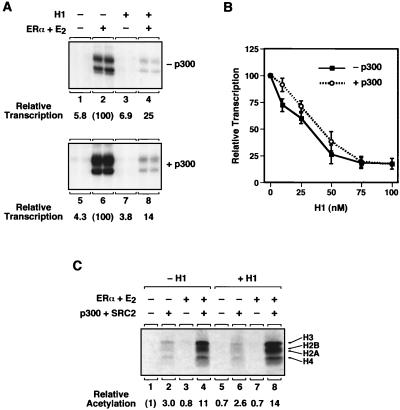
The transcriptional activity of ERα is enhanced by a variety of coactivator proteins, including p300/CBP and members of the steroid receptor coactivator (SRC) family of proteins (reviewed in references 34 and 41). The SRC proteins interact simultaneously with both ERα and p300/CBP, functioning to recruit p300/CBP to the DNA-bound receptor at estrogen-regulated promoters. In the next set of experiments, we examined the possibility that histone H1 might inhibit ERα-mediated transcription, at least in part, by blocking coactivator activity. In particular, we focused on the activity of p300, which has been shown previously to function as a potent transcriptional coactivator for ERα in vitro with chromatin templates (29, 30). As shown in Fig. 5A, addition of purified recombinant human p300 to the transcription reaction mixtures enhanced ERα-mediated transcription four- to fivefold, on average (compare lanes 2 and 6). Histone H1 was a potent repressor of ERα-mediated transcription, even in the presence of exogenously added p300 (compare lanes 6 and 8). However, the fold enhancement of ERα-mediated transcription by p300 was similar with or without histone H1 (i.e., about four- to fivefold, on average). In dose-response experiments, histone H1 inhibited ERα-mediated transcription similarly in the presence or absence of exogenously added p300 (Fig. 5B). The inhibition by histone H1 was dose dependent and occurred at physiological histone H1-to-nucleosome ratios. Thus, histone H1 reduces the absolute levels of transcription in the presence of exogenously added p300 but the fold enhancement by p300 remains the same.
FIG. 5.
Histone H1 inhibits ERα-mediated transcription in the presence of exogenously added p300 but not the targeted acetylation of nucleosomal histones by p300. (A) Histone H1 inhibits ERα-mediated transcription in the presence of added p300. In vitro chromatin assembly and transcription experiments were performed with the plasmid template p2ERE-AdE4 as described in the legend to Fig. 3B. Purified p300 was added to the assembled chromatin at a concentration of 60 nM. (B) Histone H1 inhibits ERα-mediated transcription in the presence of added p300 in a concentration-dependent manner. In vitro chromatin assembly and transcription experiments were performed with the plasmid template p2ERE-AdE4 and increasing concentrations of histone H1 as described in the legend to Fig. 3A. Purified p300 was added to the assembled chromatin at a concentration of 60 nM. Each point represents the mean ± the standard error of the mean of three or more separate determinations. (C) Histone H1 does not inhibit the targeted acetylation of nucleosomal histones by p300. The plasmid template pERE was assembled into chromatin with or without histone H1 and subsequently purified by column chromatography with Sepharose CL-4B to remove traces of unincorporated (i.e., free) histones. ERα, E2, p300, and SRC2(RID/PID) were added, and histone acetylation reactions were performed in the presence of [3H]acetyl coenzyme A as described in Methods and Materials. The reaction mixtures were separated on an SDS-15% polyacrylamide gel and subjected to fluorography. The 3H-labeled histone bands were excised from the gel and quantified by liquid scintillation counting. The results are expressed as relative acetylation compared to the control sample.
p300 has potent HAT activity, which is required for ERα-mediated transcription with chromatin templates (30). Thus, we explored the possibility that addition of histone H1 reduces the maximal levels of transcription achieved in the presence of exogenously added p300 by blocking the targeted acetylation of nucleosomal histones by p300. For the assay, an ERE-containing template was assembled into chromatin in the presence or absence of a saturating amount of histone H1 and traces of free core histones and unincorporated histone H1 were subsequently removed from the templates by size exclusion chromatography. ERα, E2, p300, and a fragment of the coactivator SRC2 containing the receptor and p300/CBP interaction domains [i.e., SRC(RID/PID)] were added in various combinations to the chromatin templates (Fig. 5C). In this assay, the simultaneous binding of the SRC2 protein fragment to p300 and chromatin-bound ERα in the presence of ligand recruits p300 HAT activity to the chromatin template, resulting in the acetylation of nucleosomal histones (compare lanes 1 and 4). Maximal acetylation by p300 is dependent on the presence of ERα, E2, and the SRC2 protein fragment (Fig. 5C; reference 25). Addition of histone H1 to the chromatin template had no discernible effect on ERα-dependent targeted acetylation of nucleosomal histones by p300 (Fig. 5C, compare lanes 4 and 8). Note, however, that this type of activator-dependent HAT recruitment assay measures acetylation events that occur over a range of at least several nucleosomes in the location of the DNA-binding sites (13, 31) and thus might preclude the detection of more subtle effects of histone H1 on the acetylation of histones in a single nucleosome in the promoter region. Nonetheless, these assays indicate that under conditions in which histone H1 reduces the maximal levels of transcription in the presence of exogenously added p300, it does not interfere with the overall extent of targeted acetylation of nucleosomal histones by p300.
Histone H1 decreases promoter accessibility without inhibiting gross ERα-induced alterations in chromatin structure.
Next, we examined possible effects of histone H1 on promoter-proximal nucleosome remodeling induced by ERα. In nucleosomal array disruption assays that measure remodeling over a range of five to six nucleosomes (45, 46), binding of ERα caused a localized disruption of the nucleosomal array near the promoter that was not inhibited by addition of histone H1 (data not shown). To examine the possibility that histone H1 might cause more subtle or specific changes that are not detected by nucleosomal array disruption assays, we performed restriction enzyme accessibility assays (Fig. 6). Briefly, S190-assembled chromatin, with or without liganded ERα or histone H1, was digested with increasing amounts of the restriction endonuclease XbaI, which cuts at −52 in pERE (relative to the most 3′ transcription initiation site), between the EREs and the TATA box. The samples were deproteinized, digested with the restriction endonucleases HindIII and EcoRI to set 5′ and 3′ boundaries surrounding the promoter, and analyzed by Southern blotting with a probe that hybridizes between the XbaI and EcoRI sites (see the schematic at the bottom of Fig. 6A). The extent of XbaI digestion under each condition was quantified and plotted graphically (Fig. 6B). The extent of XbaI digestion increased upon the addition of liganded ERα by about three- to fourfold when analyzed in the presence of subsaturating amounts of XbaI (see, for example, 2 and 5 U in Fig. 6B). Interestingly, the extent of XbaI digestion decreased upon the addition of histone H1 by about three- to fourfold either in the presence or in the absence of liganded ERα. Collectively, the results of the nucleosomal array disruption assays and the restriction enzyme accessibility assays indicate that although histone H1 does not substantially inhibit gross alterations in chromatin structure induced by ERα, it does reduce the extent of localized promoter accessibility. Thus, it is possible that histone H1 inhibits ERα-dependent transcription, in part, by decreasing the frequency of promoter opening and the accessibility of the promoter to DNA-binding components of the basal transcriptional machinery (e.g., TFIID).
Histone H1 inhibits ERα-mediated transcription by blocking transcription initiation but not reinitiation.
In previous experiments, ERα was shown to have a dual role in the transcription process with chromatin templates, stimulating both transcription initiation and reinitiation (29). To determine the steps in the transcription process at which histone H1 acts to inhibit transcription by ERα, we examined the inhibitory activity of histone H1 in a single round of transcription. The detergent Sarkosyl, which inhibits the assembly of transcription PICs and transcription initiation but not elongation by transcriptionally engaged RNA pol II (17, 18), was used to limit transcription to a single round under various experimental conditions. Limiting transcription to a single round allows one to study events associated with transcription initiation. As shown in Fig. 7A, histone H1 is a potent inhibitor of ERα-mediated transcription in a single round (compare lanes 6 and 8), achieving approximately the same level of inhibition as that observed with histone H1 in multiple rounds. These results indicate that histone H1 inhibits ERα-mediated transcription by reducing the efficiency of transcription initiation.
FIG. 7.
Histone H1 inhibits transcription initiation, but not reinitiation, by liganded ERα. (A) Histone H1 inhibits ERα-mediated transcription in a single round of transcription. In vitro chromatin assembly and transcription experiments were performed with the plasmid template p2ERE-AdE4 as described for Fig. 3B. For the single-round transcription experiments, Sarkosyl was added after initiation of transcription by addition of rNTPs. Under these conditions, Sarkosyl inhibits transcription reinitiation but not elongation and, thus, a single round of transcription is obtained. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The signals in each lane were quantified with a PhosphorImager. (B) Histone H1 does not inhibit ERα-mediated transcription reinitiation. The number of rounds of transcription in experiments like the one shown in panel A was determined by dividing the amount of transcription in the absence of Sarkosyl by the amount of transcription in the corresponding sample in the presence of Sarkosyl. Each bar represents the mean ± the range of two separate determinations.
By dividing the amount of transcription observed in the multiple-round experiment by the amount of transcription for the corresponding sample in the single-round experiment, we were able to determine the total number of rounds of transcription obtained under each condition. As shown in Fig. 7B, the addition of liganded ERα caused a 10-fold increase in the number of rounds of transcription, a fold increase similar to what has been reported previously (29). The addition of histone H1, however, had no effect on the number of rounds of transcription stimulated by liganded ERα. Thus, histone H1 acts selectively to inhibit a specific step in the transcription process with liganded ERα. That is, histone H1 inhibits ERα-mediated transcription initiation but not transcription reinitiation.
To explore the effects of histone H1 on ERα-mediated transcription initiation with chromatin templates in further detail, we performed single-round, order-of-addition transcription experiments, adding histone H1 at various time points during the transcription process (the experimental setup is described in Fig. 8A). Histone H1 efficiently inhibited ERα-mediated transcription (i.e., a threefold reduction) when it was added prior to the addition of the HeLa cell nuclear extract and the assembly of transcription PICs (Fig. 8B, condition 1). Histone H1 also inhibited ERα-mediated transcription when it was added simultaneously with the HeLa cell nuclear extract but prior to the assembly of PICs, although it did so less efficiently (an about twofold reduction in ERα-mediated transcription; condition 2). Addition of histone H1 after the assembly of PICs, before or after initiation of transcription by the addition of rNTPs, had little or no effect on ERα-mediated transcription (conditions 3 and 4). Together with the results of the restriction enzyme accessibility assays (Fig. 6), the results of the single-round and order-of-addition transcription experiments suggest that histone H1 inhibits the process of ERα-mediated transcription initiation by blocking the formation of stable transcription PICs.
DISCUSSION
Histone H1 has been shown previously to function as a gene-specific repressor of transcription (reviewed in references 9, 54, and 63). However, the detailed mechanisms of histone H1-mediated repression remain unclear. Herein, we show that histone H1 is a potent repressor of ERα-mediated transcription in the context of chromatin. Although previous studies with naked DNA templates have demonstrated an inhibitory effect of histone H1 on transcription initiation (10, 37), we show that histone H1 acts selectively to block transcription initiation without affecting transcription reinitiation. Our data indicate that histone H1 reduces the overall level of productive transcription initiation by restricting the access of the transcriptional machinery to promoters assembled into chromatin and preventing the formation of a stable transcription PIC. However, those templates that do initiate transcription in the presence of histone H1 are refractory to its repressive effects in subsequent rounds of transcription. Together, our results suggest a stochastic process in which the activating effects of ERα are countered by the repressive effects of histone H1. Histone H1 is likely to act by promoting the formation of higher-order chromatin structures that restrict the access of the transcriptional machinery to the promoter (reviewed in references 2, 9, 54, 55, 57, 58, and 63). The result is that, in the presence of histone H1, the number of transcriptionally active templates is reduced but the total number of rounds of transcription on each active template is unaffected.
How might histone H1 act selectively to repress transcription initiation without affecting transcription reinitiation? One possibility relates to the fact that transcription initiation and reinitiation are mechanistically distinct (reviewed in reference 15). Initiation requires recruitment of the entire set of factors that make up the transcription PIC to the promoter, a subset of which is left behind after initiation occurs (49, 50, 61, 62). The factors remaining at the promoter, which include TFIID, other basal TFs, and the Mediator complex (49, 50, 61, 62), form a platform for the assembly of a second transcription complex (i.e., a reinitiation platform). Therefore, the formation of the PIC, which involves the rate-limiting step of TFIID binding to the promoter (27, 60), is biochemically different from the formation of the reinitiation complex. These distinct processes are likely to have different sensitivities to the repressive effects of histone H1 and, thus, be differentially regulated by histone H1. A second possibility is that histone H1 acts to counter transcription initiation until it is displaced from the chromatin upon the successful formation of a transcription PIC. Subsequent rounds of reinitiation would then proceed in an uninhibited manner in the absence of histone H1. Such a model is consistent with the observation that the chromatin of transcriptionally active genes has a reduced histone H1 content compared to that of repressed genes (reviewed in references 9 and 63). In this regard, ERα might facilitate the formation of transcription PICs by promoting the displacement of histone H1 from the chromatin template, as has been reported previously for nuclear receptors (4, 6, 33, 59). Whether the displacement of histone H1, subsequent chromatin remodeling, and the establishment of a stable transcription PIC would necessarily be coupled or independent events is not clear. In future studies, it will be interesting to explore these possibilities further.
Interestingly, we find that histone H1 elicits its repressive effects without blocking certain aspects of ERα-mediated transcription. For example, our results show that histone H1 does not appear to inhibit the sequence-specific binding of ERα to chromatin (Fig. 4) or the overall extent of targeted acetylation of nucleosomal histones by p300 (Fig. 5C). In this regard, our findings stand in contrast to those of previous studies showing inhibitory effects of histone H1 on activator binding to chromatin (21, 22) and nucleosomal histone acetylation (14, 19). However, with respect to activator binding to chromatin, previous reports have shown that not all activators are affected by the presence of histone H1 (21, 59), suggesting that the type of DNA-binding domain may play a role. The lack of an inhibitory effect of histone H1 on these particular aspects of ERα-mediated transcription underscores the selective manner in which histone H1 inhibits ERα activity. The results of our restriction enzyme accessibility assays are consistent with previously reported inhibitory effects of histone H1 on nucleosome mobility and chromatin remodeling (20, 48, 56).
In summary, we find that histone H1 exerts its repressive effects on ERα-mediated transcription by selectively inhibiting transcription initiation without affecting reinitiation. Thus, histone H1, which was originally thought to be a general repressor of transcription, can act at a specific step in the transcription process. Together, our results suggest an important role for the selective inhibitory effects of histone H1, as well as higher-order chromatin structure, in regulating ERα-mediated transcription.
Acknowledgments
We thank John Lis, Jeff Roberts, Erik Andrulis, and Mari Acevedo for critical reading of the manuscript. We are grateful to Tory Manning for the p300 protein and to Mi Young Kim for the SRC2(RID/PID) protein and help with the acetylation assays.
This work was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a grant from the National Institutes of Health (DK58110) to W.L.K. and a postdoctoral fellowship from the Susan G. Komen Breast Cancer Foundation to E.C.
REFERENCES
- 1.Bates, D. L., and J. O. Thomas. 1981. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 9:5883-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belikov, S., and V. Karpov. 1998. Linker histones: paradigm lost but questions remain. FEBS Lett. 441:161-164. [DOI] [PubMed] [Google Scholar]
- 3.Bouvet, P., S. Dimitrov, and A. P. Wolffe. 1994. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 8:1147-1159. [DOI] [PubMed] [Google Scholar]
- 4.Bresnick, E. H., M. Bustin, V. Marsaud, H. Richard-Foy, and G. L. Hager. 1992. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 20:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. T., A. Gunjan, B. T. Alexander, and D. B. Sittman. 1997. Differential effect of H1 variant overproduction on gene expression is due to differences in the central globular domain. Nucleic Acids Res. 25:5003-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruhat, A., and J. P. Jost. 1996. Phosphorylation/dephosphorylation of the repressor MDBP-2-H1 selectively affects the level of transcription from a methylated promoter in vitro. Nucleic Acids Res. 24:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulger, M., and J. T. Kadonaga. 1994. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5:241-262. [Google Scholar]
- 8.Cole, R. D. 1989. Purification and analysis of H1 histones. Methods Enzymol. 170:524-532. [DOI] [PubMed] [Google Scholar]
- 9.Crane-Robinson, C. 1999. How do linker histones mediate differential gene expression? Bioessays 21:367-371. [DOI] [PubMed] [Google Scholar]
- 10.Croston, G. E., L. A. Kerrigan, L. M. Lira, D. R. Marshak, and J. T. Kadonaga. 1991. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science 251:643-649. [DOI] [PubMed] [Google Scholar]
- 11.Croston, G. E., P. J. Laybourn, S. M. Paranjape, and J. T. Kadonaga. 1992. Mechanism of transcriptional antirepression by GAL4-VP16. Genes Dev. 6:2270-2281. [DOI] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-Driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell 6:1049-1058. [DOI] [PubMed] [Google Scholar]
- 14.Gunjan, A., D. B. Sittman, and D. T. Brown. 2000. Core histone acetylation is regulated by linker histone stoichiometry in vivo. J. Biol. Chem. 276:3635-3640. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, S. 1998. Activation and the role of reinitiation in the control of transcription by RNA polymerase II. Cold Spring Harbor Symp. Quant. Biol. 63:181-188. [DOI] [PubMed] [Google Scholar]
- 16.Hampsey, M., and D. Reinberg. 1999. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev. 9:132-139. [DOI] [PubMed] [Google Scholar]
- 17.Hawley, D. K., and R. G. Roeder. 1987. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem. 262:3452-3461. [PubMed] [Google Scholar]
- 18.Hawley, D. K., and R. G. Roeder. 1985. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. Biol. Chem. 260:8163-8172. [PubMed] [Google Scholar]
- 19.Herrera, J. E., K. L. West, R. L. Schiltz, Y. Nakatani, and M. Bustin. 2000. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol. Cell. Biol. 20:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, D. A., and A. N. Imbalzano. 2000. Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry 39:11649-11656. [DOI] [PubMed] [Google Scholar]
- 21.Juan, L. J., R. T. Utley, C. C. Adams, M. Vettese-Dadey, and J. L. Workman. 1994. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 13:6031-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juan, L. J., R. T. Utley, M. Vignali, L. Bohm, and J. L. Workman. 1997. H1-mediated repression of transcription factor binding to a stably positioned nucleosome. J. Biol. Chem. 272:3635-3640. [DOI] [PubMed] [Google Scholar]
- 23.Kadonaga, J. T. 1998. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92:307-313. [DOI] [PubMed] [Google Scholar]
- 24.Kamakaka, R. T., M. Bulger, and J. T. Kadonaga. 1993. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 7:1779-1795. [DOI] [PubMed] [Google Scholar]
- 25.Kim, M. Y., S. J. Hsiao, and W. L. Kraus. 2001. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 27.Klein, C., and K. Struhl. 1994. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science 266:280-282. [DOI] [PubMed] [Google Scholar]
- 28.Kraus, W. L., and J. T. Kadonaga. 1999. Ligand- and cofactor-regulated transcription with chromatin templates, p. 167-189. In D. Picard (ed.), Steroid/nuclear receptor superfamily: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 29.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 32.Laybourn, P. J., and J. T. Kadonaga. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254:238-245. [DOI] [PubMed] [Google Scholar]
- 33.Lee, H. L., and T. K. Archer. 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 17:1454-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, K. C., and W. L. Kraus. 2001. Nuclear receptors, coactivators and chromatin: new approaches, new insights. Trends Endocrinol. Metab. 12:191-197. [DOI] [PubMed] [Google Scholar]
- 35.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 36.Lever, M. A., J. P. Th'ng, X. Sun, and M. J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873-876. [DOI] [PubMed] [Google Scholar]
- 37.Levine, A., A. Yeivin, E. Ben-Asher, Y. Aloni, and A. Razin. 1993. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J. Biol. Chem. 268:21754-21759. [PubMed] [Google Scholar]
- 38.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 39.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev 20:321-344. [DOI] [PubMed]
- 42.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 43.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 44.Pazin, M. J., J. W. Hermann, and J. T. Kadonaga. 1998. Promoter structure and transcriptional activation with chromatin templates assembled in vitro. A single Gal4-VP16 dimer binds to chromatin or to DNA with comparable affinity. J. Biol. Chem. 273:34653-34660. [DOI] [PubMed] [Google Scholar]
- 45.Pazin, M. J., and J. T. Kadonaga. 1998. Transcriptional and structural analysis of chromatin assembled in vitro, p. 173-194. In H. Gould (ed.), Chromatin: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 46.Pazin, M. J., R. T. Kamakaka, and J. T. Kadonaga. 1994. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science 266:2007-2011. [DOI] [PubMed] [Google Scholar]
- 47.Pazin, M. J., P. L. Sheridan, K. Cannon, Z. Cao, J. G. Keck, J. T. Kadonaga, and K. A. Jones. 1996. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 10:37-49. [DOI] [PubMed] [Google Scholar]
- 48.Pennings, S., G. Meersseman, and E. M. Bradbury. 1994. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl. Acad. Sci. USA 91:10275-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, S. G., B. Choy, S. S. Walker, Y. S. Lin, and M. R. Green. 1995. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr. Biol. 5:508-516. [DOI] [PubMed] [Google Scholar]
- 50.Sandaltzopoulos, R., and P. B. Becker. 1998. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol. Cell. Biol. 18:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, X., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86:475-483. [DOI] [PubMed] [Google Scholar]
- 52.Sheridan, P. L., C. T. Sheline, K. Cannon, M. L. Voz, M. J. Pazin, J. T. Kadonaga, and K. A. Jones. 1995. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 9:2090-2104. [DOI] [PubMed] [Google Scholar]
- 53.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, J. O. 1999. Histone H1: location and role. Curr. Opin. Cell Biol. 11:312-317. [DOI] [PubMed] [Google Scholar]
- 55.Travers, A. 1999. The location of the linker histone on the nucleosome. Trends Biochem. Sci. 24:4-7. [DOI] [PubMed] [Google Scholar]
- 56.Ura, K., J. J. Hayes, and A. P. Wolffe. 1995. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 14:3752-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolffe, A. P. 1997. Histone H1. Int. J. Biochem. Cell Biol. 29:1463-1466. [DOI] [PubMed] [Google Scholar]
- 58.Wolffe, A. P., S. Khochbin, and S. Dimitrov. 1997. What do linker histones do in chromatin? Bioessays 19:249-255. [DOI] [PubMed] [Google Scholar]
- 59.Wong, J., Q. Li, B. Z. Levi, Y. B. Shi, and A. P. Wolffe. 1997. Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J. 16:7130-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao, H., J. D. Friesen, and J. T. Lis. 1995. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. Cell. Biol. 15:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 62.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general TFs during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 63.Zlatanova, J., P. Caiafa, and K. Van Holde. 2000. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 14:1697-1704. [DOI] [PubMed] [Google Scholar]