Abstract
The mammalian Grf1 and Grf2 proteins are Ras guanine nucleotide exchange factors (GEFs) sharing a high degree of structural homology, as well as an elevated expression level in central nervous system tissues. Such similarities raise questions concerning the specificity and/or redundancy at the functional level between the two Grf proteins. grf1-null mutant mice have been recently described which showed phenotypic growth reduction and long-term memory loss. To gain insight into the in vivo function of Grf2, we disrupted its catalytic CDC25-H domain by means of gene targeting. Breeding among grf2+/− animals gave rise to viable grf2−/− adult animals with a normal Mendelian pattern, suggesting that Grf2 is not essential for embryonic and adult mouse development. In contrast to Grf1-null mice, analysis of grf2−/− litters showed similar size and weight as their heterozygous or wild-type grf2 counterparts. Furthermore, adult grf2−/− animals reached sexual maturity at the same age as their wild-type littermates and showed similar fertility levels. No specific pathology was observed in adult Grf2-null animals, and histopathological studies showed no observable differences between null mutant and wild-type Grf2 mice. These results indicate that grf2 is dispensable for mouse growth, development, and fertility. Furthermore, analysis of double grf1/grf2 null animals did not show any observable phenotypic difference with single grf1−/− animals, further indicating a lack of functional overlapping between the two otherwise highly homologous Grf1 and Grf2 proteins.
The activation of Ras proteins in mammals is mediated by specific guanine nucleotide exchange factors (GEFs) able to link the reception of upstream signals to formation of the active Ras/GTP complex via a process of GDP-GTP exchange.
Four main types of Ras-GEFs have been described in mammals to date: the highly conserved Grf1/Grf2 (8, 30) and Sos1/Sos2 (3, 5) families, GRP (11), and CNrasGEF (25). The Sos proteins activate Ras upon interaction with the adapter protein Grb2 through their SH3-binding domain (3, 5). Ras-GRP is a GEF coupling DAG and calcium signals to Ras activation in lymphoid tissues and the brain (11, 28). CNrasGEF is a novel exchanger which is able to activate Ras in response to cyclic AMP (cAMP) or cGMP (25). Within the Grf family, Grf1 and Grf2 share an overall homology close to 80% and are able to couple Ras protein activation to signal transduction processes mediated by Ca2+: CAM (13, 16), G proteins (18, 23, 29), cAMP (2), and receptor protein tyrosine kinases (9).
Besides the highly conserved REM (13, 21) and catalytic (CDC25-H) domains (7, 15, 24) present in all Ras-specific GEFs, the Grf proteins possess two pleckstrin homology domains and a Dbl homology domain. They also have a βγ-binding domain enabling signaling from G protein-coupled receptors to Ras (18, 29) and a calcium calmodulin-binding motif (IQ) able to couple calcium signals to Ras activation (13, 16). Ras-Grf2 contains also a cyclin destruction box apparently involved in ubiquitination and subsequent degradation of the full-length GEF by the 26S proteasome (10). Both Grf1 and Grf2 proteins are able to activate Ras and Rac through various pathways involving specific, differential contributions by their different structural domains (14, 19). Finally, Grf1 and Grf2 share a pattern of elevated expression in the central nervous system and brain (8, 13, 17), where they even appear to be able to form heterooligomers (1).
Inactivation of the Ras-Grf1 gene by means of targeted mutations in mice has been described (6, 20). The Grf1-null animals are viable and fertile, although they have defects in long-term memory consolidation (6), as well as reduced growth hormone and IGF-1 levels, showing a 15 to 25% lower body weight (20; unpublished observations). Like other paternally expressed imprinted genes, grf1 is implicated in growth stimulation, although it is the first imprinted gene described as exerting its effects after birth (20).
To gain insight into the functional specificity (or redundancy) of the highly homologous Grf1 and Grf2 proteins, we generated null mutant mice for the grf2 locus, as well as double-knockout (KO) grf1/grf2 animals. Analysis of our null mutant mice indicates that Grf2 is dispensable for normal development of mice to the adult stage and that there is no functional overlapping between the Grf1 and Grf2 proteins.
MATERIALS AND METHODS
Generation of mutant mice.
Lambda genomic clones containing several exons of the CDC25-H domain of Grf2 were isolated from a 129 SVJ mouse genomic library (Stratagene). Our GRF2 gene targeting vector was constructed by substituting an EcoRI/NcoI genomic fragment (including mGrf2 exon VI; see GenBank accession no. AF109311) for a neomycin cassette driven by the phosphoglycerate kinase 1 promoter (pGKneo), which was employed as a positive selectable marker. The pGK-thymidine kinase cassette was used as a negative selectable marker (27) in our vector. Electroporation of CJ7 embryonic stem (ES) cell line and selection of transfected clones was performed as described elsewhere (27). DNAs derived from G418/FIAU-resistant ES clones were screened by PCR and Southern blotting by means of EcoRV restriction enzyme digestion with a 5′ probe external to the targeting vector sequence as indicated in Fig. 1. Recombinant clones containing the predicted 6.6-kb rearranged band were obtained at a frequency of 1/33.
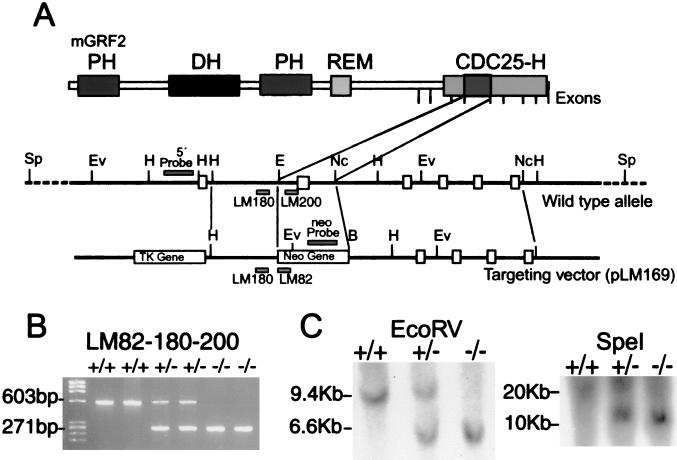
FIG. 1.
Targeted disruption of grf2 in ES cells and mice. (A) Schematic representation of the mouse grf2 genomic locus and the targeting vector used for replacement of an exon inside the CDC25-H domain. The positions of boundaries of the individual exons coding for the C-terminal portion of Grf2, including the CDC25-H domain, are indicated by the vertical marks. In the schematics for the wild-type allele and the targeting vector, filled boxes represent the exons, whereas open boxes represent the pgk-neo and pkg-tk selectable markers. Positions of the 5′ probe, neo probe, and primers used in routine genotyping experiments are also indicated. (B) Routine genotyping of mice was performed with a mixture of three oligonucleotides, allowing detection of the mutated and wild-type alleles. The pair LM200 and LM180 was able to amplify a 600-bp band from the wild-type grf2 gene, whereas the pair LM180 and LM82 was designed to amplify a 260-bp band from the mutated, targeted allele. (C) Proper homologous recombination of the targeting vector was checked by Southern blotting. Genomic DNA digested with EcoRV and hybridized to the indicated 5′ probe yielded an expected 9.5-kb band for the wild-type allele and a 6.5-kb fragment from the targeted gene. Digestion with SpeI and subsequent hybridization with the neo probe detected a unique 11-kb band only in heterozygous and KO mice, indicating that a single homologous recombination process occurred. Restriction enzymes: Ev, EcoRV; H, HindIII; E, EcoRI; Nc, NcoI; B, BamHI; Sp, SpeI.
Independent, targeted ES cell clones for the GRF2 gene injected into C57BL/6 blastocysts generated chimeras that transmitted the mutated allele to the progeny (4). Mice were bred in a specific-pathogen-free facility with food and water ad libitum. Breeding heterozygous animals obtained from chimera crossings gave rise to homozygous mutant grf2−/− mice at normal Mendelian proportions.
Genotyping of targeted ES cells and tail biopsies.
We extracted genomic DNA from ES cells, as well as tail biopsies, by the method described by Laird et al. (22). ES cells and tail tips were lysed, respectively, at 37 and 55°C for 4 h or overnight in lysis buffer (100 mM Tris-HCl [pH 8.0], 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl, 100 μg of proteinase K/ml). DNA was extracted with 1 volume of phenol-chloroform-isoamilic alcohol (25:24:1), and the upper phase was then precipitated with 2 volumes of absolute ethanol, washed with 70% ethanol, air dried, and resuspended in 200 μl of Tris-EDTA buffer. Genotyping of mice or ES cells was performed by PCR with the primers LM82 (5′-CTACCGGTGGATGTGGAATGTGTGCGA-3′), LM180 (5′-GGCAACCTCAAACAATATGGATGTCAG-3′), and LM200 (5′-GCCAGGCAGAGTCATCAAGGGTAGAG-3′), along with 2 μl of genomic DNA and 1.25 U of Taq polymerase (Roche Molecular Biochemicals). After a denaturing step for 10 min at 94°C, DNA was amplified for 30 cycles (94°C for 1 min, 62°C for 1 min, and 72°C for 1 min). PCR fragments were resolved on a 2.5% agarose gel (NuSieve 3:1 agarose; BioWhittaker Molecular Applications). LM180 and LM200 are primers specific for the grf2 gene designed to produce a 600-bp DNA fragment when amplifying the wild-type allele. Oligonucleotide LM82 (hybridizing to the neo-pgk promoter), together with LM180 are supposed to amplify a smaller 260-bp band from the KO allele. Oligonucleotides LM84, LM85, and LM82 were used for grf1 genotyping in the context of double grf1/grf2 KO generation. The pair LM84-LM85 amplifies a 824-bp band from the genomic copy of the gene, whereas the pair LM84-LM82 amplifies mutant genomic DNA, originating as a 615-bp band (LM84, 5′-AGT TGC TGA AGG GAG GAG GGT AGC GGC-3′; LM85, 5′-TGA CCA GGC CGT CCT CTG TGT AAT TGG-3′).
Southern blotting.
Proper insertion of the targeting construct was monitored by Southern blotting. Twenty-five micrograms of genomic DNA from tail biopsies was digested for 3 h at 37°C with EcoRV or SpeI. This DNA was loaded onto a 0.6% agarose gel in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA) (SeaKem ME agarose; BioWhittaker Molecular Applications) and run for 7 h at 50 V. After electrophoresis, DNA in the agarose gel was electrotransferred for 1 h at 500 mA to a nitrocellulose membrane previously equilibrated with 0.5× TBE for 30 min. DNA was then fixed to the membrane by cross-linking (UV Stratalinker 2400; Stratagene), washed with 2× SSPE (0.3 M NaCl, 23 mM NaH2PO4 [pH 7.4], 2.5 mM EDTA) and 0.5% SDS at 60°C for 30 min, and prehybridized with 10 ml of Hybrisol I (Intergen Company) at 50°C for 2 h. The 5′ probe (upstream of the insertion point) and the neo probe (inside the cassette) were labeled by random priming according to the manufacturer's instructions (Prime-It RmT kit; Stratagene). After hybridization at 50°C overnight, membranes were washed twice for 10 min at room temperature with 2× SSC (0.3 M NaCl, 30 mM sodium citrate; pH 7.0), once with 2× SSC, 0.5% SDS at 42°C, 10 min, and once with 0.5× SSC, 0.1% SDS at 42°C for 10 min. Film was exposed for 6 h at −80°C and developed.
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was extracted by adding 1 ml of TRIzol reagent (Gibco-BRL) to fresh or previously frozen tissues which were then homogenized with a mechanical cell disruptor (Ultra-Turrax T8 S8N-8G; IKA Labortechnik) for 30 s on ice. Homogenates were extracted with 0.2 ml of chloroform, and the aqueous phase was precipitated with 0.5 ml of isopropanol. The RNA was then washed with 70% ethanol, air dried, and resuspended in 20 μl of diethyl pyrocarbonate-H2O. cDNA was generated from 1.5 μg of RNA by using the SuperScript II RNase H-Reverse Transcriptase kit (Gibco-BRL) and oligo(dT) as a primer by the method described by the manufacturer. The pair LM112 (5′-GTG AGG GCC AGA AAG CTG TCT TTG ACG TCT-3′) and LM113 (5′-TCG GCT ACC TGT CCT CCA GGC CTG CCG ATT-3′), amplifying a coding region (nucleotides 2215 to 2478) (8) located upstream of the point of genomic disruption and including the cyclin destruction box domain, was used in PCR amplification to check for possible, partial grf2 locus expression in the KO mice. PCR conditions were as described above, analyzing the products on 2.5% NuSieve agarose 3:1 gels. grf1 cDNA amplification was performed as described above, with oligonucleotide pair LM90-LM91 for the PCR (LM90, 5′-AAC ATA CAC TCA CCC ATA TCT CCC TTC GGC-3′; LM91, 5′-TGC GGT TAT TGT CCA CCA GAT CAC GAC ACG-3′). Finally, 5′ and 3′ oligonucleotides from Clontech's Rat G3PDH Control Amplimer Set (catalog no. 5405-1) were used for PCR amplification of glyceraldehyde-3-phosphate dehydrogenase.
Western blot studies.
Tissues obtained from 5-week-old male mice were homogenized mechanically in 2 ml of radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 1% sodium deoxycholate; 0.1% SDS) and centrifuged for 30 min at 30,000 rpm, and 50 μg of supernatant protein was loaded onto a SDS-7.5% polyacrylamide gel. After electrophoresis, proteins were transferred onto nitrocellulose by using a semidry system (Transblot SD-Semidry Transfer Cell; Bio-Rad) for 30 min at 20 V. The membranes were blocked for 1 h with TTBS (10 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.05% Tween 20) plus 5% dry skim milk. Blots were then incubated for 3 h at room temperature with a rabbit polyclonal anti-Grf-2 antibody generated in our laboratory by immunization against an amino acid stretch (residues 800 to 1004) located immediately upstream of the CDC25-H domain of human Grf2. Antisera were diluted 1:1,000 with TTBS and 3% bovine serum albumin. Membranes were washed five times with TTBS for 5 min and incubated for 1 h at room temperature with bovine anti-rabbit horseradish peroxidase-conjugated secondary antibody (sc-2004; Santa Cruz Biotechnologies) diluted to 0.4 μg/ml in TTBS and 5% dry skim milk. Blots were washed again (four times for 10 min), and chemiluminescence was developed by using the ECL-Plus system as described by the manufacturer (Amersham Pharmacia Biotech). A Western blot for Grf1 was performed as described above with an antiserum obtained in our laboratory by inmunization against a fragment of the total mGrf1 protein stretching from amino acids 709 to 1034 lacking significant amino acid sequence homology with Grf2.
Immunocytochemistry.
Adult, male C57BL/6 mice were anesthesized with chloral hydrate (5% [wt/vol] in saline, 10 μl/g of body weight [given intraperitoneally]) and perfused through the aorta, first with heparinized saline (5 IU/ml; Byk Leo, Madrid, Spain) and then with a solution containing 4% (wt/vol) depolymerized paraformaldehyde and 0.2% (wt/vol) picric acid in 0.1 M phosphate buffer (pH 7.4) (PB) for 20 min. All animal manipulations adhered to the directives of the European Communities Council (no. 86/609/EEC) and conformed to National Institues of Health guidelines. Brains were dissected and postfixed in the same solution for 2 h at 4°C. For cryoprotection, brains were immersed in 30% (wt/vol) sucrose in PB until they sank. Thirty-micrometer-thick parasagittal sections were obtained with a cryostat (Leica Jung, Nussloch, Germany), collected in cold PB, and sequentially incubated in (i) either anti-human-Grf1 or anti-human-Grf2 rabbit polyclonal primary antibodies diluted 1:100 in PB containing 10% normal goat serum and 0.03% Triton X-100 for 48 h at 4°C; (ii) biotinylated anti-rabbit immunoglobulin G (Vector Laboratories, Burlingame, Calif.) diluted 1:250 in PB for 2 h at room temperature; and (iii) horseradish peroxidase-coupled avidin complex (1:200 in PB; Vector) for 2 h at room temperature. Peroxidase was visualized with 0.07% 3,3′-diaminobenzidine and 0.003% hydrogen peroxide in 0.1 M Tris-HCl buffer (pH 7.6). Sections were stored in the dark until analyzed. Bright-field digital images were taken with an Olympus Provis AX microscope and a digital camera of 1.34 Mpixels (Apogee Instruments, Inc., Tucson, Ariz.). The objective lenses used were an Olympus ×1.25 (PlanApo N.A. 0.04) and UPlanFl ×4 (N.A. 0.13), ×20 (N.A. 0.50), and ×40 (N.A. 0.75). The images were captured with Adobe Photoshop 6.0 software (Adobe Systems, Inc., San Jose, Calif.) connected to a trichromatic sequential filter (Cambridge Research & Instrumentation, Inc., Boston, Mass.). After conversion into black and white, the sharpness, contrast, and brightness were adjusted to reflect the appearance of the labeling seen through the microscope.
RESULTS AND DISCUSSION
To advance our understanding of the in vivo function(s) of the Ras-Grf2 protein, we designed a gene-targeting approach aimed at inactivating the grf2 locus in mice. For this purpose we targeted its CDC25-H catalytic domain (residues 952 to 1189) expecting to disrupt the GEF activity of Grf2 in a similar approach to that used to target other GEFs such as Sos1, Sos2, or Grf1 (6, 12, 26).
Our analysis of the grf2 mouse gene by means of Southern blot analyses and genome partial sequencing (GenBank numbers AF109309 to AF109312) allowed the mapping of seven exons coding for specific contiguous regions of the catalytic domain of the protein (Fig. 1A). On the basis of these data we decided to target a very conserved region of the CDC25-H domain of Ras-Grf2 stretching from amino acids 1005 to 1070. Our targeting vector pLM169 was used to substitute this fragment for a neo cassette, including a pgk promoter and the neomycin resistance gene (Fig. 1A). The vector included also the herpes simplex virus tk gene, which was used as a negative selection marker of the homologous recombination process (Fig. 1A).
The targeting vector pLM169 was used for ES cell electroporation and subsequent generation of chimeras, as described in Materials and Methods. Mating of chimeras with wild-type C57BL/6 animals yielded grf2+/− heterozygous animals, which were then interbred to produce grf2−/− mutant animals according to the Mendelian expected frequency of 1/4. Genotyping of animals resulting from the crosses was first obtained by routine PCR, with oligonucleotides hybridizing to the 5′ end of the pgk-neo cassette (LM82), as well as to regions located immediately before (LM180) or after (LM200) the neo insertion site in the wild-type allele. The presence of mutant grf2 alleles in adult animals was clearly identified by the amplification of a diagnostic, expected band of 260 bp (Fig. 1B). Proper homologous recombination was checked by Southern blot hybridization. Thus, with a probe hybridizing to the 5′ region of the disruption (5′ probe; Fig. 1A), an expected 6.5-kb fragment was detected in mutant animals when their genomic DNA was digested with EcoRV (Fig. 1C). Another probe, hybridizing inside the neo cassette (neo probe; Fig. 1A) was used to discard the possibility of multiple copy insertion. A single 11-kb band was detected in animals by using this probe on SpeI-digested genomic DNA (Fig. 1C), indicating a unique insertion by homologous recombination of the mutated allele.
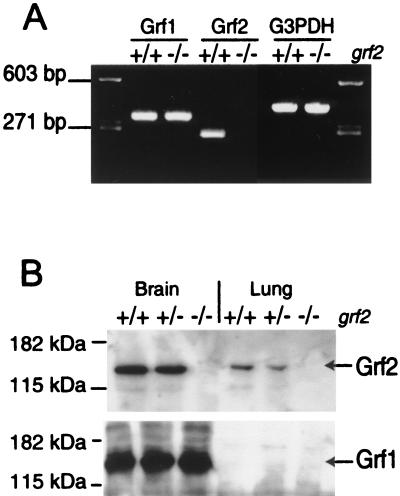
To confirm our generation of a null mutation of the grf2 locus, we analyzed the expression of this gene in normal and mutant mice (Fig. 2). Since Grf2 has been reported to show good expression levels in brain and lung (8, 13), we selected here samples from these tissues for our analysis (Fig. 2). Thus, RT-PCR analysis of RNA extracted from brains of wild-type and mutant animals confirmed the complete absence in tissues of grf2−/− animals of a 263-bp diagnostic band which was otherwise easily detected in the corresponding tissues of wild-type animals (Fig. 2A). In contrast, control RT-PCR assays of the grf1 and G3PDH loci showed similar levels of amplified, diagnostic bands in wild-type and KO grf2 animals (Fig. 2A). Since the oligonucleotides used for grf2 (LM112 and LM113; see Materials and Methods) anneal to a region of this gene located upstream of the neo cassette insertion site, these results also indicate that no shorter or partial grf2 transcripts are generated in our null mutants as a consequence of the disruption strategy used here.
FIG. 2.
Grf2 expression in brain and lung of wild-type, heterozygous, and KO animals. (A) RT-PCR amplification of mouse grf1, grf2, and G3PDH loci. Total RNA samples extracted from grf2-null animals and their wild-type +/+ counterparts were assayed with specific pairs of oligonucleotides as described in Materials and Methods. Oligonucleotides LM112 and LM113, which can amplify a grf2-specific fragment, failed to detect any amplified band in RNA from the grf2−/− animals, whereas a 265-bp band was amplified from the RNA extracted from wild-type tissues. In contrast, the grf1- and G3PDH-specific oligonucleotide pairs amplified their specific bands at similar levels in the wild-type and Grf2 KO brain samples. (B) Western immunoblot analysis of protein extracts (see Materials and Methods) allowed detection in wild-type and heterozygous tissues of an expected 135-kDa Grf2 protein band that was totally absent in the tissue extracts from KO animals. The levels of the 140-kDa Grf1 protein were not affected in the KO mice, indicating that there is no compensatory increase of expression of this protein in the absence of Grf2.
Furthermore, we confirmed the absence of expression of Grf2 protein in the KO animals by means of Western immunoblotting with antibodies generated in our laboratory that readily discriminate the Grf1 and the Grf2 proteins (Fig. 2B). Thus, our specific antiserum against Grf2 detected a 135-kDa protein band present at similar levels in tissues of wild-type and heterozygous animals but totally absent in the tissues of KO mice (Fig. 2B). Since the antibodies used in this experiment are directed against a Grf2 region located upstream of the disruption site and no smaller band was detected in the grf2-null samples, we can also exclude the synthesis of any truncated form of the Grf2 protein in our KO animals. Interestingly, control with our Grf1-specific antibodies showed the presence of similar levels of the 140-kDa Grf1 protein in brain samples of wild-type and mutant animals (Fig. 2B), indicating that there is no overexpression of Grf1 to compensate for the absence of Grf2 in the KO animals. Similar results were obtained with commercially available antibodies against the Grf proteins (not shown).
We have kept the grf2−/− animals in our lab for over 18 months. Both males and females are as fertile as their wild-type and heterozygous counterparts, producing similar numbers of pups per litter. In contrast to the reduced size of grf1−/− animals (20; unpublished observations), the grf2−/− mutant mice showed growth rates and weights similar to those of their wild-type and heterozygous controls (data not shown). Finally, histopathological studies of various tissues in adult animals of different ages showed no observable anatomical defects or histopathological changes of any kind.
Taken together, these data show that Grf2-dependent guanine nucleotide exchange activity is dispensable for mouse growth, fertility, and somatic development to the adult stage. Animals lacking the Grf2 protein show a normal phenotype and do not appear to develop any physical illness due to the null mutation. This is in clear contrast to the phenotypes (reduced size, long-term memory defects) observed in KO animals lacking the highly homologous Grf1 protein (6, 20).
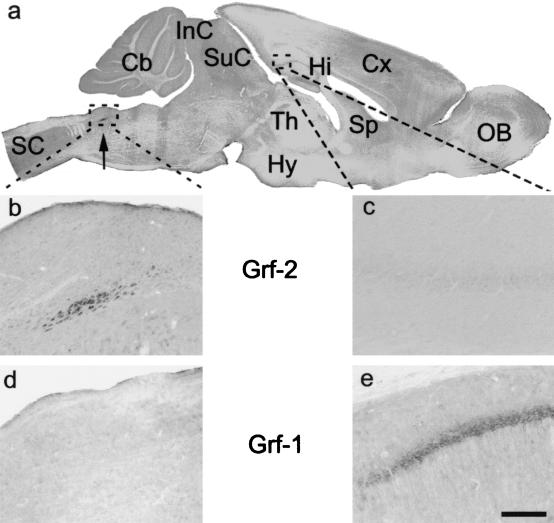
These results suggest that, despite their high sequence similarity, the functions of the Grf1 and Grf2 proteins must be, at least in part, different. Consistent with that notion, we demonstrated a clearly different pattern of expression of both proteins in adult mouse brain, an organ where expression of both proteins has been reported to be highest (8, 13). Thus, our immunohistochemical study of adult mouse brain with our specific anti-Grf1 and anti-Grf2 antibodies showed (Fig. 3) that Grf2 expression is located in the nucleus of the solitary tract, a region implicated in control of breathing and oxytocin synthesis during lactation. In contrast, no Grf2 expression was observed in the hippocampus, where Grf1 was mainly expressed (6) (Fig. 3), suggesting that Grf2 is not implicated in LTP (6). On the other hand, Grf2 animals have a normal weight development during lactation, and no obvious breathing problems have been observed, suggesting that Grf2 absence in the solitary nuclei does not affect their normal functions in mice.
FIG. 3.
Immunohistochemical analysis of adult mouse brain with specific anti-Grf1 and anti-Grf2 antibodies. (a) Panoramic view of a parasagittal section of the mouse brain and cervical spinal cord after Grf2 immunocytochemistry. Immunostained cells were observed in the nucleus of the solitary tract (arrow). (b) Magnification of the Grf2 immunoreactive neurons of the nucleus of the solitary tract outlined in panel a. (c) Magnification of the caudal hippocampus outlined in panel a. No labeling is observed after Grf2 immunocytochemistry. (d) Magnification of the nucleus of the solitary tract after Grf1 immunocytochemistry. No immunoreaction is observed. (e) Magnification of the caudal hippocampus after Grf1 immunocytochemistry. Pyramidal cells of the hippocampus were immunolabeled, whereas they were not stained after Grf2 immunocytochemistry (see panels a and c). Scale bar: a, 1 mm; b and d, 20 μm; c and e, 50 μm. Abbreviations: Cb, cerebellum; Cx, cerebral cortex; Hi, hippocampus; Hy, hypothalamus; InC, inferior colliculus; OB, olfactory bulb; SC, spinal cord; SuC, superior colliculus; Sp, septum; Th, thalamus.
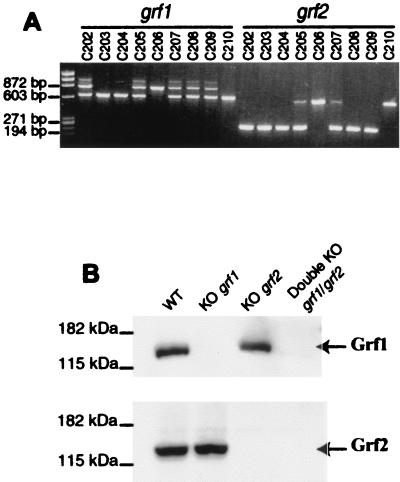
In order to further test the possibility of some degree of functional overlapping between Grf1 and Grf2, we carried out cross-breeding between the grf2−/− animals described above and grf1−/− mice also generated in our laboratory. These crosses yielded double-heterozygous grf1/grf2 mice, which were then crossed among themselves in an effort to obtain double grf1/grf2 KO animals. Genotypic analysis of the grf1 and grf2 loci in the offspring from such crosses demonstrated that adult, double-null mutant grf1/g rf2 mice were perfectly viable (see, for example, animals C203 and C204 in Fig. 4A). Furthermore, we confirmed the absence of both Grf1 and Grf2 proteins in these adult mice by Western immunoblotting with our specific antibodies (Fig. 4B). We observed that Grf2 protein levels were normal in grf1 KO animals and that Grf1 levels were also unchanged in grf2 KO animals (Fig. 4B), showing no compensatory oversynthesis of either protein in the simple KOs. Moreover, both proteins were completely absent in the double-KO mice (Fig. 4B).
FIG. 4.
Analysis of double-mutant mice deficient for grf1 and grf2. (A) DNA genotyping of representative offspring resulting from crosses among double heterozygous grf1/grf2 mice. The oligonucleotides used were as described in Materials and Methods. LM82, LM180, and LM200 were used for the grf2 gene, and LM82, LM84, and LM85 were used for the grf1 gene. Wild-type and mutant grf1 loci originate at bands of 824 and 615 bp, respectively. Likewise, our wild-type and null mutant grf2 genes amplify bands of 600 and 260 bp, respectively. Note that mice C203 and C204 harbor double homozygous null mutations for both grf1 and grf2 genes. (B) Western immunoblot of brain extracts from representative wild-type, single KO grf1 and grf2, and double-KO grf1/grf2 mice. With specific anti-Grf1 antibodies, the upper panel shows the absence of Grf1 protein in Grf1- and Grf1/Grf2-KO mice. Similarly, with specific anti-Grf2 antibodies, the lower panel shows a total absence of Grf2 protein in brain extracts of single grf2 KO and double grf1/grf2 KO mice.
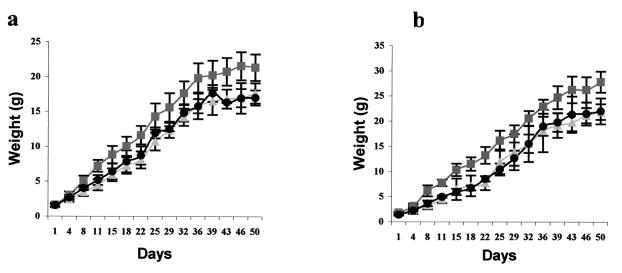
Analysis of the offspring resulting from crosses among heterozygous animals showed that the double-null grf1/grf2 mutants were born at expected Mendelian ratios (Table 1). Preliminary observations of the grf1/grf2 double-KO mice showed that these animals are perfectly viable and fertile, reaching sexual maturity at the same age as their wild-type control counterparts and producing normal-size litters. Five-month-old double-null mutant mice kept in our laboratory have not shown so far any obvious developmental or anatomical defect specifically associated with the double null genotype. On the other hand, the grf1/grf2 double-KO animals keep the same weight defect observed in single grf1-null mice (20; unpublished observations), with no further change in this phenotype due to the addition of the grf2 mutation (Fig. 5).
TABLE 1.
Analysis of offspring resulting from crossings between mice carrying double heterozygous null mutations of grf1 and grf2a
| Genotype (grf1 | grf2) from offspring of crossesb | No. (%) of animals | Expected Mendelian ratio (%) |
|---|---|---|
| +/+ | +/+ | 12 (9) | 6 |
| +/+ | +/− | 28 (20) | 12 |
| +/+ | −/− | 4 (3) | 6 |
| +/− | +/+ | 14 (10) | 12 |
| +/− | +/− | 33 (24) | 25 |
| +/− | −/− | 17 (12) | 12 |
| −/− | +/+ | 7 (5) | 6 |
| −/− | +/− | 12 (9) | 12 |
| −/− | −/− | 11 (8) | 6 |
Analysis of genotypic distribution of a total of 138 newborn animals obtained in 16 separate litters resulting from breeding three males with seven females of the indicated genotypes. The numbers in parentheses indicate the percentages of the total numbers of animals. Genotypes were determined at weaning as described in Materials and Methods. The number of double-KO animals obtained corresponded to the expected ratio if normal Mendelian segregation occurred.
(Male) grf1+/− grf2+/− × (female) grf1+/− grf2+/−.
FIG. 5.
Postnatal growth curve of wild-type (▪), single grf1 KO (▴), and double grf1/grf2 KO (•) mice. (a) Growth in females; (b) weight change in males. Each value point plotted represents the average weight and standard deviation (in grams) of a total of 5 to 10 comparable littermates of the same age. Single grf1 KO and double grf1/grf2 KO mice weights were ca. 70% of their wild-type littermates.
Taken together, our data suggest that Grf2 is dispensable for normal mouse growth development and that there is no functional overlap between Grf2 and its highly homologous Grf1 counterpart.
Acknowledgments
This work was supported by FEDER grants 1FD97-1735 and 1FD97-1678 and CICYT grant SAF00-0069 from MCYT (Spain).
A.F.-M. and L.M.E. contributed equally to this work.
REFERENCES
- 1.Anborgh, P. H., X. Qian, A. G. Papageorge, W. C. Vass, J. E. DeClue, and D. R. Lowy. 1999. Ras-specific exchange factor GRF: oligomerization through its Dbl homology domain and calcium-dependent activation of Raf. Mol. Cell. Biol. 20:4611-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baouz, S., E. Jacquet, K. Accorsi, C. Hountondji, M. Balestrini, R. Zippel, E. Sturani, and A. Parmeggiani. 2001. Sites of phosphorylation by protein kinase A in CDC25Mm/Grf1, a guanine nucleotide exchange factor for Ras. J. Biol. Chem. 276:1742-1749. [DOI] [PubMed] [Google Scholar]
- 3.Bonfini, L., C. A. Karlovich, C. Dasgupta, and U. Banerjee. 1992. The Son of sevenless gene product: a putative activator of Ras. Science 255:603-606. [DOI] [PubMed] [Google Scholar]
- 4.Bonin, A., S. W. Reid, and L. Tessarollo. 2001. Isolation, microinjection, and transfer of mouse blastocysts. Methods Mol. Biol. 158:121-134. [DOI] [PubMed] [Google Scholar]
- 5.Bowtell, D., P. Fu, M. Simon, and P. Senior. 1992. Identification of murine homologues of the Drosophila son of sevenless gene: potential activators of ras. Proc. Natl. Acad. Sci. USA 89:6511-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brambilla, R., N. Gnesutta, L. Minichiello, G. White, A. J. Roylance, C. E. Herron, M. Ramsey, D. P. Wolfer, V. Cestari, C. Rossi-Arnaud, S. G. N. Grant, P. F. Chapman, H. P. Lipp, E. Sturani, and R. Klein. 1997. A role for the Ras signaling pathway in synaptic transmission and long-term memory. Nature 390:281-286. [DOI] [PubMed] [Google Scholar]
- 7.Camonis, J. H., M. Kalekine, B. Gondre, H. Garreau, E. Boy-Marcotte, and M. Jacquet. 1986. Characterization, cloning, and sequence analysis of the CDC25 gene which controls the cyclic AMP level of Saccharomyces cerevisiae. EMBO J. 5:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., L. J. Zhang, P. Greer, P. S. Tung, and M. F. Moran. 1993. A murine CDC25/ras-GRF-related protein implicated in Ras regulation. Dev. Genet. 14:339-346. [DOI] [PubMed] [Google Scholar]
- 9.Darznin, B., L. Chang, J. W. Leitner, Y. Takata, and J. M. Olefsky. 1993. Insulin activates p21Ras and guanine nucleotide releasing factor in cells expressing wild-type and mutant insulin receptors. J. Biol. Chem. 268:19998-20001. [PubMed] [Google Scholar]
- 10.De Hoog, C. L., J. A. Koehler, M. D. Goldstein, P. Taylor, D. Figeys, and M. F. Moran. 2001. Ras binding triggers ubiquitination of the Ras exchange factor Ras-Grf2. Mol. Cell. Biol. 21:2107-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebinu, J. O., D. A. Bottorff, E. Y. Chan, S. L. Stang, R. J. Dunn, and J. C. Stone. 1998. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science 280:1082-1086. [DOI] [PubMed] [Google Scholar]
- 12.Esteban, L. M., A. Fernández-Medarde, E. López, K. Yienger, C. Guerrero, J. M. Ward, L. Tessarollo, and E. Santos. 2000. Ras-guanine exchange factor Sos2 is dispensable for mouse growth and development. Mol. Cell. Biol. 20:6410-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fam, N. P., W. Fan, Z. Wang, L. Zhang, H. Chen, and M. F. Moran. 1997. Cloning and characterization of Ras-GRF2, a novel guanine nucleotide exchange factor for Ras. Mol. Cell. Biol. 17:1396-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, W., C. A. Koch, C. L. de Hoog, N. P. Fam, and M. F. Moran. 1998. The exchange factor Ras-Grf2 activates Ras-dependent and Rac-dependent mitogen-activated protein kinase pathways. Curr. Biol. 8:935-938. [DOI] [PubMed] [Google Scholar]
- 15.Feig, L. A. 1994. Guanine nucleotide exchange factors: a family of positive regulators of Ras and related proteins. Curr. Opin. Cell Biol. 6:204-211. [DOI] [PubMed] [Google Scholar]
- 16.Freshney, N. W., S. D. Goonesekera, and L. A. Feig. 1997. Activation of the exchange factor Ras-GRF by calcium requires an intact Dbl homology domain. FEBS Lett. 407:111-115. [DOI] [PubMed] [Google Scholar]
- 17.Guerrero, C., J. M. Rojas, M. Chedid, L. M. Esteban, D. B. Zimonjic, N. C. Popescu, J. Font de Mora, and E. Santos. 1996. Expression of alternative forms of Ras exchange factors GRF and SOS1 in different human tissues and cell lines. Oncogene 12:1097-1107. [PubMed] [Google Scholar]
- 18.Inglese, J., J. Koch, K. Touhara, and R. J. Lefkowitz. 1995. Gβγ interactions with PH domains and Ras-MAPK signaling pathways. Trends Biochem. Sci. 20:151-156. [DOI] [PubMed] [Google Scholar]
- 19.Innocenti, M., R. Zippel, R. Brambilla, and E. Sturani. 1999. CDC25Mm/Ras-Grf1 regulates both Ras and Rac signaling pathways. FEBS Lett. 460:357-362. [DOI] [PubMed] [Google Scholar]
- 20.Itier, J., G. L. Tremp, J. Léonard, M. Multon, G. Ret, F. Schweighoffer, B. Tocqué, M. Bluet-Pajot, V. Cormier, and F. Dautry. 1998. Imprinted gene in postnatal growth role. Nature 393:125-126. [DOI] [PubMed] [Google Scholar]
- 21.Lai, C. C., M. Boguski, D. Broek, and S. Powers. 1993. Influence of guanine nucleotides on complex formation between Ras and CDC25 proteins. Mol. Cell. Biol. 13:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattingly, R. R., and I. G. Macara. 1996. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature 382:268-272. [DOI] [PubMed] [Google Scholar]
- 24.Overbeck, A. F., T. R. Brtva, A. D. Cox, S. M. Graham, S. Y. Huff, R. Khosravi-Far, L. A. Quilliam, P. A. Solski, and C. J. Der. 1995. Guanine nucleotide exchange factors: activators of Ras superfamily proteins. Mol. Reprod. Dev. 42:468-476. [DOI] [PubMed] [Google Scholar]
- 25.Pham, N., I. Cheglakov, C. A. Koch, C. L. de Hoog, M. F. Moran, and D. Rotin. 2000. The guanine exchange factor CNrasGEF activates Ras in response to cAMP and cGMP. Curr. Biol. 10:555-558. [DOI] [PubMed] [Google Scholar]
- 26.Qian, X., L. Esteban, W. C. Vass, C. Upadhyaya, A. G. Papageorge, K. Yienger, J. M. Ward, D. R. Lowry, and E. Santos. 2000. The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. EMBO J. 19:642-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tessarollo, L. 2001. Manipulating mouse embryonic stem cells. Methods Mol. Biol. 158:47-63. [DOI] [PubMed] [Google Scholar]
- 28.Tognon, C. E., H. E. Kirk, L. A. Passmore, I. P. Whitehead, C. J. Der, and R. J. Kay. 1998. Regulation of RasGRP via a phorbol ester-responsive C1 domain. Mol. Cell. Biol. 18:6995-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touhara, K., J. Inglese, J. A. Pitcher, G. Shaw, and R. J. Lefkowitz. 1994. Binding of G protein beta gamma-subunits to pleckstrin homology domains. J. Biol. Chem. 269:10217-10220. [PubMed] [Google Scholar]
- 30.Wolfman, A., and I. G. Macara. 1990. A cytosolic protein catalyzes the release of GDP from p21ras. Science 248:67-69. [DOI] [PubMed] [Google Scholar]