Abstract
Recent studies have suggested that phosphorylation of human p53 at Ser20 is important for stabilizing p53 in response to DNA damage through disruption of the interaction between MDM2 and p53. To examine the requirement for this DNA damage-induced phosphorylation event in a more physiological setting, we introduced a missense mutation into the endogenous p53 gene of mouse embryonic stem (ES) cells that changes serine 23 (S23), the murine equivalent of human serine 20, to alanine (A). Murine embryonic fibroblasts harboring the p53S23A mutation accumulate p53 as well as p21 and Mdm2 proteins to normal levels after DNA damage. Furthermore, ES cells and thymocytes harboring the p53S23A mutation also accumulate p53 protein to wild-type levels and undergo p53-dependent apoptosis similarly to wild-type cells after DNA damage. Therefore, phosphorylation of murine p53 at Ser23 is not required for p53 responses to DNA damage induced by UV and ionizing radiation treatment.
The p53 gene is the most commonly mutated tumor suppressor gene in human cancers (20). Its role in tumor suppression is further highlighted by the creation of p53−/− mice, which are highly cancer prone and develop a large spectrum of tumors (15, 23). It has become clear that p53 plays several roles in regulating cellular events after DNA damage and other cellular stresses, including activating the arrest of cell cycle progression in G1 or initiating apoptosis (reviewed in references 24 and 32). These functions of p53, which depend in part on the cell type and nature of the DNA damage, protect the cellular genome from accumulating mutations and genome rearrangements and from passing these mutations to daughter cells, thus contributing to its tumor suppression activities. Structural and functional analyses of p53 have shown that p53 is a transcription factor with a central sequence-specific DNA-binding domain, a transcriptional activation domain at the N terminus, and a C-terminal domain that is involved in regulating p53 activity (24).
In response to DNA damage and other cellular stresses, p53 protein levels increase significantly and its DNA-binding activity is activated. p53 protein levels are regulated posttranscriptionally, and the increased levels observed following DNA damage are due primarily to increased protein stability (24). Degradation of p53 protein is mediated largely by the MDM2 oncoprotein, which targets p53 for ubiquitin-mediated degradation (18, 21, 25). p53 is phosphorylated at multiple sites in its N- and C-terminal domains after DNA damage, and it has become evident that phosphorylation of p53 plays important roles in regulating p53 stability and activity (reviewed in reference 2). In this context, the phosphorylation of human p53 at Ser15, -20, -33, and -37 is induced after cells are exposed to either UV light or ionizing radiation (IR), while the phosphorylation of Ser392 is induced by UV light but not by IR (2). A number of protein kinases have been found to phosphorylate human and murine p53 in vitro, including ATM, ATR, Chk1, Chk2, mitogen-activated protein kinase, Jun N-terminal kinase, protein kinase C, casein kinases I and II, double-stranded RNA activated protein kinase, and cyclin-dependent protein kinases (cdk), and several phosphorylation events are believed to be involved in p53 stabilization and activation (reviewed in references 2 and 30). The phosphorylation of human p53 at Ser15 (corresponding to Ser18 of mouse p53), which is mediated by the ATM family of kinases, is an early event following DNA damage and is reduced in ATM−/− cells after IR (4, 6). Reduced phosphorylation of Ser15 correlates with the reduced and delayed stabilization of p53 (38). In addition, we recently showed that a missense mutation introduced into the endogenous p53 gene of mouse cells that changed Ser18 to Ala impaired p53 stabilization after DNA damage (7). However, the defect caused by mutating the Ser18 codon was only partial, indicating that other phosphorylation events also must be involved in stabilizing and activating p53.
Ser20 lies directly within the region of the p53 transactivation domain that interacts with MDM2 (26, 40), and this interaction is required for MDM2-mediated degradation of p53. Several recent studies have suggested that phosphorylation of human p53 at Ser20 is important for stabilizing p53 after DNA damage (10, 39, 41). Since MDM2-mediated ubiquitination represents a major pathway for rapid p53 degradation, disruption of the MDM2-p53 interaction through phosphorylation of Ser20 could be important for stabilizing p53. The Chk1 and Chk2 kinases, which are activated by ATM after exposure to IR, phosphorylate human p53 at Ser20 in vitro (9, 37). Therefore, phosphorylation of human p53 at Ser20 by Chk1/2 kinases might represent another ATM-dependent pathway that stabilizes p53. Consistent with this notion, Chk2−/− mouse cells are defective in p53 stabilization and activation after IR (19).
To investigate the physiological roles of p53 phosphorylation at Ser20 in p53 responses to DNA damage, we introduced a missense mutation into the endogenous p53 gene of mouse embryonic stem (ES) cells that changes Ser23 (corresponding to Ser20 of human p53) to Ala. Various mutant primary cells, including mouse embryonic fibroblasts (MEFs) and thymocytes, were derived from the mutant ES cells and assayed for p53 responses to DNA damage. Surprisingly, the p53S23A mutation had no apparent effects on either the stability or the activity of p53 after DNA damage in mutant ES, MEFs, and thymocytes. Therefore, the Chk2-dependent stabilization and activation of p53 in mouse cells after IR must be mediated through pathways other than the phosphorylation of murine p53 at Ser23.
MATERIALS AND METHODS
Generation of mutant ES cells harboring p53S23A mutation.
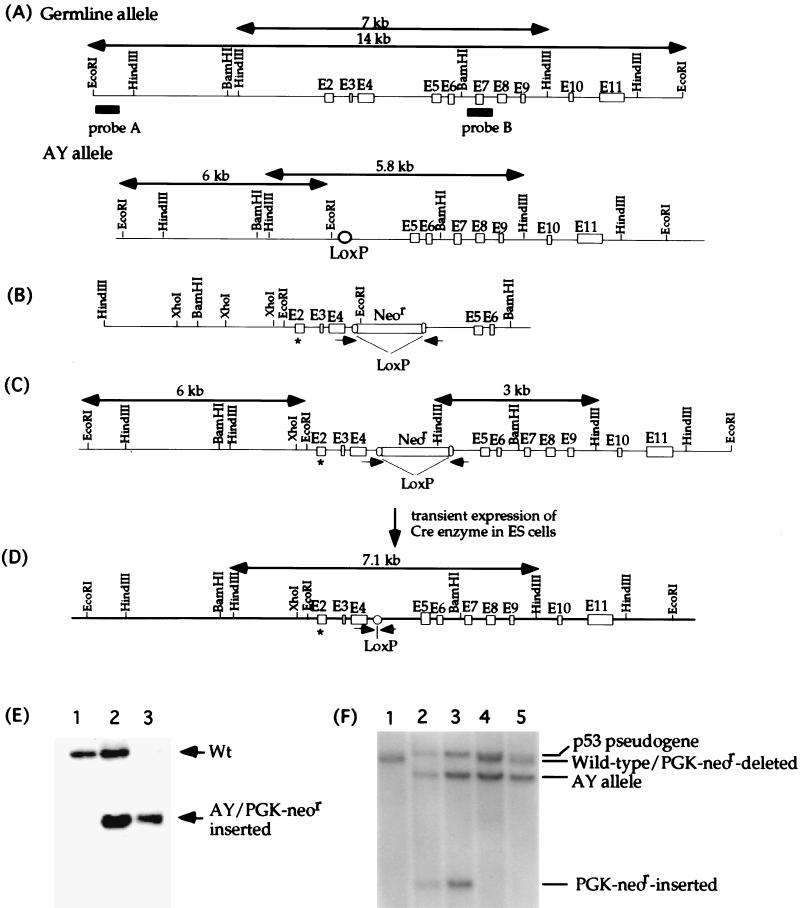
Ser23 is encoded by exon 2 of the mouse p53 gene. The strategy to introduce the Ser23Ala mutation was the same as that used previously to introduce a mutation into the endogenous p53 gene of murine ES cells that changed Ser18 to Ala (7). Briefly, site-directed mutagenesis was performed to introduce the Ser23Ala mutation into a mouse p53 genomic DNA fragment harboring exons 2 through 6. The knock-in vector was constructed by inserting the LoxP-flanked PGK-neor gene into an engineered unique SalI site within intron 4. An EcoRI site was introduced into intron 1 to assist in screening for a replacement of wild-type exon 2 by the targeting construct. The knock-in vector was electroporated into AY ES cells as previously described (7). AY ES cells are p53+/−, with exons 2 to 4 of one p53 allele deleted while the other allele remains wild type. The mutant p53 allele produced no truncated proteins, as confirmed by Western blot analysis (data not shown). Homologous recombination events between the wild-type p53 allele and the knock-in vector were screened by Southern blot analysis after EcoRI digestion of the DNAs and hybridization with probe A. AY ES cells displayed a 14-kbp wild-type allele and a 6-kbp AY mutant allele (Fig. 1A, B, and E). Under the same conditions, recombinant cells that had a mutant exon 2 and the introduced EcoRI site incorporated into the wild-type allele from the targeting vector displayed only a 6-kbp fragment, since both mutant alleles yielded a 6-kbp EcoRI fragment (Fig. 1A and C). To delete the PGK-neor gene from the p53 allele with the Ser23Ala mutation, 20 μg of a circular plasmid that drives expression of the CRE enzyme was transiently transfected into the mutant ES cells as described. ES cell transfectants were screened for the LoxP/Cre-mediated deletion by PCR as indicated in Fig. 1. Neor-deleted ES cells identified by PCR were subcloned, and the loss of the Neor cassette was confirmed by Southern blot analysis after HindIII digestion and hybridization with probe B, which revealed a 3-kbp fragment from the PGK-neor-inserted allele and a 7.1-kbp fragment from cells which had the PGK-neor cassette deleted (Fig. 1C and D). The wild-type allele yielded a 7-kbp fragment (Fig. 1A). Probe B also hybridized to a 7.8-kbp HindIII fragment derived from the p53 pseudogene.
FIG. 1.
Generation of p53S23A ES cells. (A) The endogenous configuration of the p53 gene in AY ES cells. AY ES cells have one wild-type p53 allele and one mutant p53 allele (AY allele) with p53 exons 2 through 4 deleted. The initiation codon, ATG, of the p53 gene is located in exon 2, and the AY allele does not produce a truncated p53 protein. Open boxes represent the p53 exons, and the filled bars represent probes for Southern blot analysis. The wild-type 14-kbp EcoRI and 7-kbp HindIII fragments are indicated by arrows, as are the mutant 6-kbp EcoRI and 5.8-kbp HindIII fragments on the AY allele. (B) The targeting construct. ∗, the Ser23-to-Ala mutation in exon 2. The PGK-neor gene flanked by LoxP sites was inserted into an engineered SalI site within intron 4. (C) Targeted p53 locus after homologous recombination between the wild-type p53 allele and the targeting vector. The positions of the PCR primer sets that were used to screen for LoxP/Cre-mediated deletion are shown by arrowheads. The sizes of the mutant EcoRI and HindIII fragments are indicated. (D) Mutant p53 allele after the PGK-neor gene was deleted. The size of the mutant HindIII fragment after the PGK-neor gene was deleted is indicated. (E) Southern blot analysis of genomic DNA derived from the wild type (lane 1), AY ES cells (lane 2), and targeted AY ES cells (lane 3), in which homologous recombination had occurred between the germ line allele and the targeting vector. Genomic DNA was digested with EcoRI and hybridized to probe A. Both the targeted allele and AY allele yielded the same 6-kbp mutant EcoRI fragment. The positions of both germ line and mutant alleles are indicated with arrowheads. (F) Southern blot analysis of genomic DNA derived from wild-type ES cells (lane 1), targeted AY ES cells with the PGK-neor gene inserted (lanes 2 and 3), AY ES cells (lane 4), and p53Ser23Ala ES cells (lane 5). Genomic DNA was digested with HindIII and hybridized to probed B. The 7-kbp wild-type band, 7.1-kbp PGK-neor gene deleted band, 5.8-kbp AY mutant band, and 3-kbp PGK-neor gene inserted band as well as the 7.8-kbp band derived from the p53 pseudogene are indicated.
Culture and treatment of ES cells.
Before UV radiation, ES cells were cultured without a feeder layer in Dulbecco’s minimal essential medium (DMEM) supplemented with 15% fetal calf serum (FCS), glutamine, nonessential amino acids, antibiotics, 100 μM β-mecaptoethanol, and recombinant lymphocyte inhibitory factor. For the study of p53 induction after UV treatment, ES cells were exposed to 60 J of UV light/m2 and harvested at different times after treatment. For the study of p53-dependent apoptosis after UV treatment, ES cells were treated with 10, 15, or 30 J of UV light/m2 and assayed for apoptosis 16 h after treatment as described below.
Derivation of MEFs and thymocytes from mutant ES cells.
We developed the Hprt-deficient blastocyst complementation approach to derive MEFs from mutant ES cells without a selectable marker (see Results). The MEFs were cultured in DMEM supplemented with 10% FCS, glutamine, antibiotics, 50 mM β-mecaptoethanol, and hypoxanthine-aminopterin-thymidine (HAT). After confirmation that all surviving MEFs were derived from ES cells, the MEFs were cultured in normal medium without HAT. Mouse thymocytes were derived from mutant ES cells by RAG2-deficient blastocyst complementation as previously described (11).
Radiation treatment of MEFs and thymocytes.
MEFs were treated with 60 J of UV light/m2 or 10 Gy of IR and harvested at different times after treatment for the analysis of p53 and p21 protein levels. Mouse thymocytes were resuspended in DMEM supplemented with 5% FCS and 25 mM HEPES, pH 7.4, at a density of 106 cells/ml. To assay for p53-dependent apoptosis after IR, thymocytes were exposed to 5, 10, or 20 Gy of IR. To assay for p53 induction after IR, thymocytes were exposed to 5 Gy of IR and harvested at the times indicated.
Western blot analysis of p53 and p21 and p53 Ser23 phosphospecific antibodies.
Protein extract from 4 × 105 ES or MEF cells or 107 mouse thymocytes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% (for Mdm2), 12% (for p53), or 15% (for p21) polyacrylamide gels and transferred to a nitrocellulose membrane. The membrane was blocked with 5% dry milk and probed with a monoclonal antibody against p53 (Pab240; Santa Cruz Biotechnology, Inc.) or a rabbit polyclonal antibody against p21 (Santa Cruz Biotechnology, Inc.) or a monoclonal antibody against Mdm2 (2A10; Oncogene Science). Then, the filter was incubated with horseradish peroxide-conjugated secondary antibody and developed with Enhanced Chemiluminescence PLUS (ECL PLUS; Amersham Pharmacia Biotech). To determine if the amounts of protein in each lane were comparable, the filter was stripped and probed with a rabbit polyclonal antibody against actin (Santa Cruz Biotechnology, Inc.).
Rabbit polyclonal antibody specific for murine p53 phosphorylated at Ser23 was prepared essentially as described previously for other sites (7, 8, 36, 35). Briefly, rabbits were immunized with the murine p53 phosphopeptide Ac-18-29(23P)Cys, i.e., Ac-SETFS(P)-GLWKLLC, coupled to keyhole limpet hemocyanin, and the resulting p53S23(P)-specific antibodies were purified from the hyperimmune sera by affinity chromatography using both the phosphospecific and the unphosphorylated peptides coupled with SulfoLink (Pierce Chemical Co.). The specificity of the purified antibodies was confirmed by enzyme-linked immunosorbent assay and immunoblot assays using a panel of p53 phosphorylated and unphosphorylated peptides as previously described (7, 35). Phosphorylation status was determined by Western immunoblot analysis of the immunoprecipitated p53.
p53-Mdm2 immunoprecipitation.
MEF cells with or without radiation treatment were lysed in the lysis buffer (20 mM Tris-HCl [pH 7.6], 170 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol [DTT], proteinase inhibitors). Cell lysates were incubated with anti-p53 antibody (pAb421) conjugated to agarose beads for 4 h at 4°C. After washing three times with washing buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40), p53 and Mdm2 in the immunoprecipitates were detected by Western blot analysis with monoclonal antibodies specific for p53 (pAb240) and Mdm2 (2A10).
Analysis of p53-dependent apoptosis in mouse ES cells and thymocytes.
ES cells were plated in six-well plates without a feeder layer at a density of 2 × 105 to 3 × 105 cells per well. Sixteen hours after UV treatment, ES cells were harvested and apoptotic cells were identified by staining with fluorescein isothiocyanate (FITC)-conjugated Annexin V (Pharmingen) as previously described (8). Thymocytes were recovered from p53−/− mice and AY-RAG2−/− and p53S23A-RAG2−/− chimeric mice, and IR-induced apoptosis was analyzed as previously described (27). The percentages of apoptotic thymocytes within the CD4+CD8+ and CD4+ thymocyte populations were determined at 10 h after IR by staining with phosphatidylethanolamine (PE)-conjugated anti-CD4 antibody and FITC-conjugated Annexin V (Pharmingen).
RESULTS
Introduction of Ser23Ala mutation into endogenous p53 gene in mouse ES cells.
Ser23 is encoded by exon 2 of the mouse p53 gene. A mouse genomic DNA fragment containing exons 2 to 6 was isolated, and site-directed mutagenesis was used to introduce a mutation (Ser23Ala) into exon 2 of the cloned p53 genomic DNA that changed serine 23 to alanine essentially as previously described (7, 8). The knock-in construct was generated by inserting the LoxP-flanked PGK-neor gene into intron 4 of the cloned p53 genomic DNA harboring the Ser23Ala mutation in exon 2 (Fig. 1B). To facilitate the mutagenesis processes, the knock-in construct was electroporated into a p53+/− ES cell line (AY ES cells), in which exons 2 to 4 of the p53 gene were replaced with a LoxP site on one allele, leaving the remaining p53 allele wild type (Fig. 1A). The mutant p53 allele in AY cells does not produce a truncated protein, which was confirmed by using polyclonal antibodies against p53 and a monoclonal antibody specific for the C terminus (data not shown). Therefore, the p53 genotype and phenotype of AY ES cells is p53+/−. Homologous recombination between the knock-in vector and the only germ line allele in AY ES cells replaced the wild-type exon 2 with the mutant exon 2 harboring the Ser23Ala mutation (Fig. 1C). Homologous recombination events were verified by Southern blot analysis of EcoRI-digested genomic DNAs with a fragment from p53 intron 1 (Fig. 1A, probe A). This probe detected a 14-kbp wild-type, germ line fragment and a 6-kbp fragment from the targeted allele and the AY mutant allele (Fig. 1C and E). The LoxP-flanked PGK-neor gene was deleted from the mutant ES cells through transient expression of the Cre enzyme as previously described (7, 8). The deletion event was screened by PCR with primers indicated in Fig. 1C and confirmed by Southern blot analysis with HindIII digestion and hybridization to probe B (Fig. 1E). ES cells with the PGK-neor gene deleted are denoted as p53S23A ES cells. p53 genomic DNA and cDNA from p53S23A ES cells were sequenced to confirm that the only p53 transcript present in these cells contained the Ser23Ala mutation without any accompanying mutations.
Phosphorylation of mouse p53 at Ser23 after DNA damage.
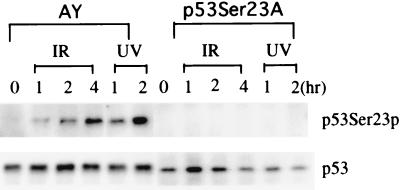
Phosphorylation of Ser20 of human p53 is induced in human cell lines exposed to UV light or IR treatment (10, 36, 37). Therefore, we analyzed whether murine p53 can be phosphorylated at Ser23, which is equivalent to Ser20 of human p53, after UV or IR treatment. The irradiated and untreated controls were cultured with proteosome inhibitor ALLN for 4 h before harvest to ensure that samples of all time points had similar amounts of p53 protein. Using an antibody specific for murine p53 phosphorylated at Ser23, we found that murine p53 was phosphorylated at Ser23 after the exposure of the ES cells to UV radiation or IR similarly to its human counterpart (Fig. 2).
FIG. 2.
Phosphorylation of murine p53 at Ser23 in ES cells at various times after IR treatment (5 Gy) or UV treatment (60 J/m2). Irradiated and untreated cells were treated with the proteosome inhibitor ALLN for 4 h before harvesting so that samples of all time points would have similar levels of p53 protein. Cell extracts from irradiated cells and untreated controls (0 h) were analyzed by Western blotting with antibodies specific for p53 or p53 phosphorylated at Ser23 (p53-S23p) as described previously (8, 33). The time after irradiation (in hours) is indicated at the top.
Derivation of mutant MEFs by complementation of Hprt-deficient blastocysts.
To derive MEFs from ES cells without a selectable marker, we developed a novel genetic approach denoted as Hprt-deficient blastocyst complementation (see Materials and Methods). ES cells without a selection marker were injected into blastocysts recovered from Hprt−/− mice, which were derived from Hprt-deficient ES cells (5). The injected blastocysts then were implanted into pseudopregnant females. At embryonic day 14, MEFs were recovered from the embryos and cultured under the selection of HAT (0.016 mg of hypoxanthine/ml, 0.01 mM aminopterin, 0.0048 mg of thymidine/ml). Because MEFs derived from Hprt-deficient blastocysts cannot survive in HAT-containing media, after two or three passages, essentially all surviving MEFs are derived from the Hprt-proficient ES cells. Using Hprt-deficient blastocyst complementation, we were able to routinely obtain pure populations of MEFs at passage 3 or 4 from AY and p53S23A ES cells as confirmed by Southern blot analysis of genomic DNA derived from these MEFs (Fig. 3). In addition to deriving MEFs from ES cells without a selectable marker, Hprt-deficient blastocyst complementation should be useful for obtaining any other primary cell types that can be selected in HAT medium.
FIG. 3.
Generation of AY and p53S23A MEFs by Hprt-deficient blastocyst complementation. Southern blot analysis of genomic DNA from passage 3 MEFs derived from embryos generated by injection of AY or p53S23A ES cells into Hprt-deficient blastocysts. Genomic DNA was digested with EcoRI and hybridized with probe A (see Fig. 1). Lane 1, wild-type MEFs; lane 2, AY MEFs; lane 3, p53S23A MEFs.
p53-mediated responses to DNA damage in p53S23A MEFs.
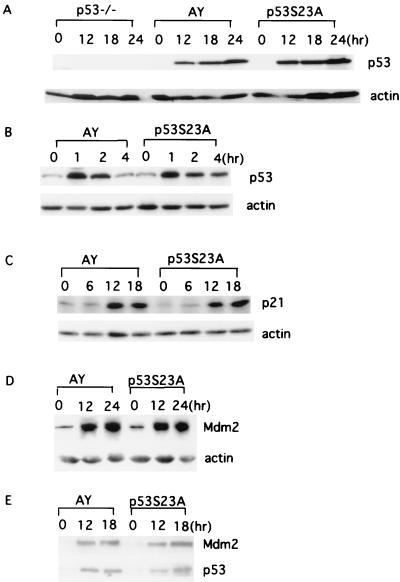
p53 protein levels are significantly but transiently increased in MEFs following exposure to DNA damage-inducing agents. The activation of p53 subsequently induces the expression of a number of genes, including p21CIP/WAF1. Employing Hprt-deficient blastocyst complementation, we derived MEFs from AY and p53S23A ES cells and examined the DNA damage responses produced by exposure to IR or UV in these MEFs. p53 protein levels at different times following exposure to UV light or IR were evaluated by Western blot analyses of whole-cell extracts from p53−/−, AY, and p53S23A MEFs. As expected, no p53 protein was present before or after DNA damage in p53−/− MEFs (Fig. 4A); however, p53 protein levels increased dramatically and similarly after UV (Fig. 4A) or IR (Fig. 4B) treatment in both AY and p53S23A MEFs, indicating that phosphorylation of mouse p53 at Ser23 does not significantly affect p53 stability after DNA damage. To test whether p53 activity was normal in p53S23A MEFs after exposure to DNA damage, we also analyzed p21CIP/WAF1 and Mdm2 protein levels. Expression of both proteins are activated in a p53-dependent manner after UV radiation. The kinetics and extent of p21 and Mdm2 induction after UV radiation was similar between AY and p53S23A MEFs, indicating that p53 is activated as a transcription factor in a normal manner after DNA damage in p53S23A MEFs (Fig. 4C and D). Since previous publications suggested that phosphorylation of human p53 at Ser20 could stablize p53 through disruption of the p53-MdM2 interaction (10, 39, 41), we tested the p53-Mdm2 interaction with or without DNA damage in p53S23A and control MEFs by immunoprecipitation and Western blot analysis. Similar ratios of Mdm2 versus p53 were observed in the p53 immunoprecipitates from p53S23A and control MEFs at various time points after UV radiation, indicating that phosphorylation of mouse p53 at Ser23 does not significantly affect the p53-Mdm2 interaction in MEF cells (Fig. 4E).
FIG. 4.
Induction of p53 and p21 in AY and p53S23A MEFs after DNA damage. Cell extracts were prepared from AY and p53S23A MEFs at the times indicated and analyzed for p53 expression by Western immunoblot analysis after exposure to 60 J of UV light/m2 (A) or 10 Gy of IR (B). The times after treatment (in hours) and genotypes are labeled at the top of the lanes. p53 and actin are indicated on the right. Shown is the induction of p21 (C) and Mdm2 (D) proteins in AY and p53S23A MEFs after 60- and 30-J/m2 UV treatment, respectively. The genotype and time points are indicated on top. p21, Mdm2, and actin are indicated on the right. (E) Immunoprecipitation and Western blot analysis of the p53-Mdm2 interaction in AY and p53S23A MEFs with or without UV radiation (30 J/m2). The genotypes and time points after radiation are labeled on top. p53 and Mdm2 are indicated on the right.
p53 induction and p53-dependent apoptosis in p53S23A ES cells after UV radiation.
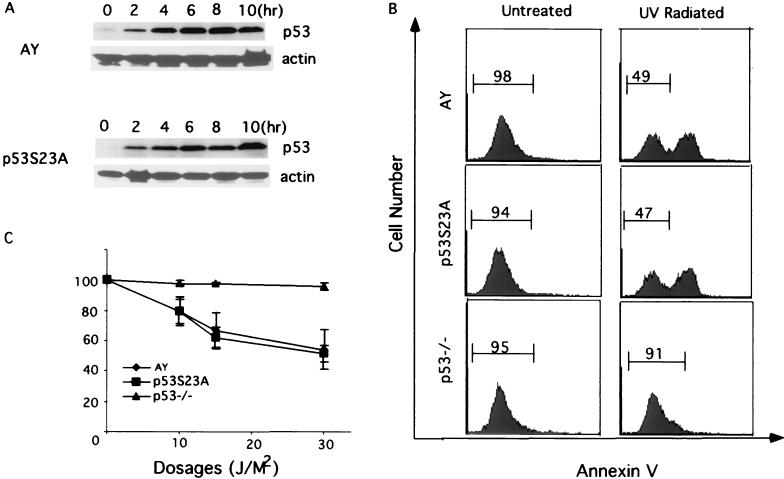
Murine ES cells undergo a typical induction of p53 accumulation following exposure to UV light (8, 13, 34). To confirm that the induction of p53 accumulation was normal in p53S23A ES cells after DNA damage, we analyzed p53 levels in AY and p53S23A ES cells by Western blot analysis at different times after exposure to UV light. Consistent with the findings in MEFs, p53 accumulated at similar rates and to similar levels in both AY and p53S23A ES cells (Fig. 5A and B).
FIG. 5.
Induction of apoptosis in p53−/−, AY, and p53S23A ES cells after UV treatment. (A) p53 protein levels in AY and p53S23A ES cells at different times after UV treatment. Time after treatment is indicated at the top, genotypes are indicated on the left, and p53 and actin are indicated on the right. (B) Flow cytometric analysis of AY, p53−/−, and p53S23A ES cells harvested 16 h after exposure to 30 J of UV light/m2. Cell number is plotted as a function of the intensity of staining for Annexin V. Cells staining positive for Annexin V are apoptotic. The percentages of nonapoptotic cells are shown. (C) The percentage ratio of nonapoptotic cells in irradiated AY, p53−/−, and p53S23A ES cells relative to nonapoptotic cells in unirradiated controls 16 h after exposure to 10, 20, or 30 J of UV light/m2. Mean values from four independent experiments are presented with error bars. Percentages (y axis) were determined as follows: (no. of nonapoptotic cells in irradiated ES cells/no. of nonapoptotic cells in untreated control) × 100%.
ES cells also undergo p53-dependent apoptosis after UV radiation. To determine if the Ser23Ala mutation affected the ability of p53 to activate apoptosis, we analyzed p53−/−, AY, and p53S23A ES cells for Annexin V expression, which is a sensitive indicator of apoptosis, after treatment with UV light. Consistent with previous findings that UV-induced apoptosis in ES cells is p53 dependent, little apoptosis was observed in p53−/− ES cells exposed to UV light at a dosage of 10 to 30 J/m2 (Fig. 5B and C). In contrast, exposure of AY and p53S23A ES cells resulted in strong staining with Annexin V as determine by flow cytometry; at all UV dosages tested, little if any difference in the extent of apoptosis between the two cell lines was observed (Fig. 5B and C). Therefore, phosphorylation of mouse p53 at Ser23 is not required either for stabilizing or activating p53 in response to DNA damage induced by UV radiation.
p53 induction and p53-dependent apoptosis in p53S23A thymocytes after IR.
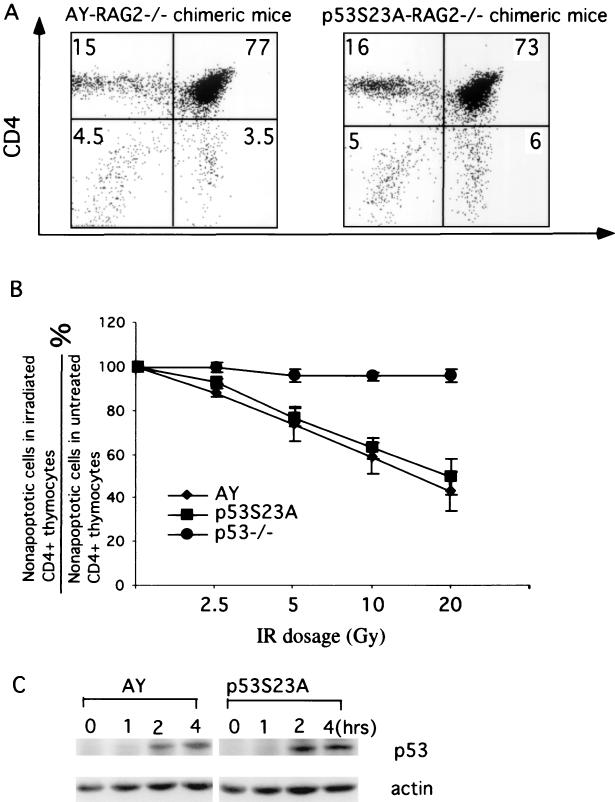
Mouse thymocytes undergo p53-dependent apoptosis after IR (12, 27). To further confirm the absence of an effect of p53Ser23Ala mutation on p53 stability and activity, we also examined p53 accumulation and p53-dependent apoptosis in mouse thymocytes derived from AY and p53S23A ES cells. Mouse thymocytes were derived from AY and p53S23A ES cells by RAG2-deficient blastocyst complementation as previously described (8). Because thymocyte development is blocked at the CD4−CD8− stage in RAG2-deficient mice, all CD4+CD8+ double-positive and CD4+ or CD8+ single-positive thymocytes in the chimeric mice, which together constitute more than 90% of thymus cellularity, were derived from the injected ES cells (Fig. 6A). To test the IR-induced apoptotic responses in mouse thymocytes, thymocytes recovered from p53−/− mice as well as AY-RAG2−/− and p53S23A-RAG2−/− chimeric mice were treated with increasing dosages of IR and analyzed for apoptotic cells by Annexin V staining 10 h later. Because CD4+CD8+ thymocytes are the ones undergoing p53-dependent apoptosis after IR (14, 27) and to prevent contamination by CD4−CD8− thymocytes derived from the RAG2−/− blastocysts, we analyzed only CD4+ thymocytes. As expected, p53−/− thymocytes showed little IR-induced apoptosis (Fig. 6B). The overlap of the error bars for the percentage of apoptotic cells in AY and p53S23A thymocytes indicated that IR-induced p53-dependent apoptosis is similar in p53S23A thymocytes and AY thymocytes (Fig. 6B). Consistent with these findings, the kinetics and extent of p53 induction in p53S23A thymocytes were very similar to those in AY thymocytes. Together, these findings indicated that phosphorylation of mouse p53 at Ser23 plays no significant role in p53 stability or activity in mouse thymocytes after IR.
FIG. 6.
Induction of p53 and apoptosis in p53−/−, AY, and p53S23A mouse thymocytes after IR. (A) Mouse thymocytes were recovered from AY-RAG2−/− and p53S23A-RAG2−/− chimeric mice, stained for CD4 and CD8, and analyzed by flow cytometry. Cells residing in the lymphocyte gate were analyzed, and the percentages of total cells in a particular gate are indicated. (B) The percentage ratio of nonapoptotic CD4+ thymocytes in AY, p53−/−, and p53S23A thymocytes treated with 2.5, 5, 10, 15, and 20 Gy of IR to the nonapoptotic CD4+ thymocytes from untreated controls. Mean values from four independent experiments are presented with error bars. (C) p53 protein levels in AY and p53S23A thymocytes at different times after IR treatment were determined by Western blot analysis. Time after treatment and genotypes are indicated at the top; p53 and actin are indicated on the right.
p53−/− mice mainly develop thymic lymphomas and sarcomas by 6 months of age (15, 23). Therefore, we examined the chimeric mice for sarcomas and analyzed thymocytes derived from eight 6- to 8-month-old p53S23A-RAG2−/− chimeric mice. No tumors could be identified in these chimeric mice, suggesting that p53 tumor suppression activity is not significantly affected in p53S23A cells.
DISCUSSION
Human p53 is phosphorylated at several N-terminal serines, including Ser20, following DNA damage, and murine p53 is similarly phosphorylated at an equivalent residue (2). Several biochemical studies utilizing purified glutathione S-transferase (GST)-p53 fusion protein or in vivo studies employing transient transfection of wild-type or mutant p53 into cell lines have concluded that the phosphorylation of human p53 at Ser20 is important for p53 stabilization after DNA damage through disruption of the p53-MDM2 interaction (10, 39, 41). In addition, Ser20 phosphorylation has been shown to affect the apoptotic activity of the p53 protein and is correlated with the induction of p21 and MDM2 after DNA damage in human tumor cell lines (22, 42). However, another study showed no such effect by expression of p53 lacking all N-terminal phosphorylation sites in human tumor cell lines through transient transfection (3). It is known that high levels of the p53 protein may dictate the apoptotic response of the cell (33); therefore, one possible explanation for these different results is that different expression levels of the transfected proteins are present in the cells. In addition, two reports provided biochemical evidence suggesting that phosphorylation of human p53 Ser20 did not directly interrupt p53-MDM2 interaction (22, 36). To elucidate the physiological importance of phosphorylation of p53 at Ser23 in p53 responses to DNA damage, we introduced a missense mutation (Ser23 to Ala) into the endogenous p53 gene of mouse embryonic stem cells through homologous recombination and LoxP/Cre-mediated deletion. These mutant cells express a p53 from their endogenous gene that cannot be phosphorylated at Ser23. Extensive analysis of the p53 responses to DNA damage in the mutant ES cells and other primary cells derived from them, including MEFs and thymocytes, showed no apparent defects in p53 stabilization or activity. Therefore, we conclude that phosphorylation of murine p53 at Ser23 is not required for these classical p53-dependent responses to DNA damage. The possibility that Ser23 phosphorylation is required for more subtle DNA damage responses or for p53 responses to other forms of stress will await the derivation of knock-in mice that harbor this mutation.
The apparent discrepancy between our conclusion and those from previous publications could be explained by several possibilities. First, it is possible that the roles of p53 phosphorylation in regulating p53 stability and activity are different between mice and humans. Several lines of evidence argue against this possibility, however. The N-terminal amino acid sequences of human and mouse p53 are highly conserved with residues Pro13 to Pro27 (human numbering) being identical. The patterns of N-terminal phosphorylation after DNA damage as well as signaling pathways involved also are highly conserved. Furthermore, knock-in mice in which the mouse p53 gene was largely replaced with the human p53 sequence have normal p53 responses to DNA damage and p53 tumor suppression activity (28). Therefore, the DNA damage-activated signaling pathways leading to p53 activation must be conserved between humans and the mouse. In support of this notion, phosphorylation of human p53 at Ser15 and at the murine p53 equivalent site (Ser18) is required for efficient p53 responses to DNA damage (7, 16). Second, phosphorylation of mouse p53 at Ser23 and at other sites may serve redundant functions in regulating p53 stability and activity. Therefore, in the absence of Ser23 phosphorylation, perhaps other p53 phosphorylation events activate p53 responses to the normal level. In this context, several phosphorylation events, including phosphorylation of human p53 at Ser15 and Thr18, also have been shown to activate p53 activity and cause disruption of the p53-MDM2 interaction (14, 36, 38). Nevertheless, this scenario would require that when the previous studies of p53-MDM2 interactions were performed, the hypothesized redundant pathways were not operative, thus revealing the importance of Ser20 phosphorylation. Third, most previous in vivo assay systems involved overexpression of various mutant p53s in tumor cell lines. It is possible that certain signaling pathways leading to p53 phosphorylation at other N-terminal sites are defective in these tumor cells, contributing to the more apparent requirement of p53 phosphorylation at Ser20 in p53 stabilization after DNA damage or that overexpression of p53 somehow enhances an effect of Ser20 phosphorylation on p53 stability.
Phosphorylation of human p53 at Ser20 after DNA damage is believed to be mediated by the Chk1 and Chk2 protein kinases, which are activated in response to DNA damage by ATM family kinases (1, 29, 31). Chk2−/− mouse thymocytes do not accumulate p53 protein after IR treatment, consistent with the notion that Chk2-dependent phosphorylation of p53 at Ser20 might be involved in p53 stabilization after DNA damage (19). Our findings that p53 stabilization and activity are normal in p53S23A thymocytes after IR do not argue against the notion that Chk2-dependent pathways are required for p53 stabilization in mouse thymocytes after IR. However, our findings indicated that Chk2 functions to activate p53 through pathways that are independent of Ser23 phosphorylation.
Hprt-deficient blastocyst complementation.
The knock-in ES cell technology, which allows the introduction of subtle mutations into the endogenous genes in mice so that the mutant gene is expressed under the control of its own promoter and regulatory elements, has become a powerful tool for investigating gene functions. Because the PGK-neor gene, which is uniformly used as the selection marker for ES cell transfectants, can affect chromatin remodeling and thus transcription through the locus, it is necessary to excise it from the genome after homologous recombination to allow expression of the mutant gene under physiological control (17). Therefore, the mutant ES cells with the knock-in mutation will not have a selection marker. While mice with germ line transmission of the mutant gene can be generated to study its effects, often it is worthwhile to first assay for effects by using chimeric mice generated from the ES cells. This approach is particularly useful when the mutation leads to embryonic lethality in germ line-transmitted mice. While RAG2-deficient blastocyst complementation can be employed to derive lymphocytes from mutant knock-in mutant ES cells, the derivation of other primary cells from chimeric mice with the mutant ES cells is difficult because the mutant ES cells do not have a selection marker to distinguish them from blastocyst-derived cells. The Hprt-deficient blastocyst complementation method we described here can solve this problem, and it enabled us to efficiently select for primary embryonic fibroblasts derived from mutant ES cells. This approach potentially can be used to derive any other type of primary cells that can be grown in a cell culture system.
Acknowledgments
This work was supported by an American Cancer society grant (ACS RPG-99-170-01-CCG) to Yang Xu. C.W.A. was supported in part by a CRADA funded by the Laboratory Technology Research Program in the Office of Science of the U.S. Department of Energy.
REFERENCES
- 1.Ahn, J. Y., J. K. Schwarz, H. Piwnica-Worms, and C. E. Canman. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60:5934-5936. [PubMed] [Google Scholar]
- 2.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft, M., M. H. Kubbutat, and K. H. Vousden. 1999. Regulation of p53 function and stability by phosphorylation. Mol. Cell Biol. 19:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 5.Bronson, S. K., E. G. Plaehn, K. D. Kluckman, J. R. Hagaman, N. Maeda, and O. Smithies. 1996. Single-copy transgenic mice with chosen-site integration. Proc. Natl. Acad. Sci. USA 93:9067-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 7.Chao, C., S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2000a. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl. Acad. Sci. USA 97:11936-11941. [DOI] [PMC free article] [PubMed]
- 8.Chao, C., S. Saito, J. Kang, C. W. Anderson, E. Appella, and Y. Xu. 2000b. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. Embo J. 19:4967-4975. [DOI] [PMC free article] [PubMed]
- 9.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 10.Chehab, N. H., A. Malikzay, E. S. Stavridi, and T. D. Halazonetis. 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA 96:13777-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., R. Lansford, V. Stewart, F. Young, and F. W. Alt. 1993. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. USA 90:4528-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, A. R., C. A. Purdie, D. J. Harrison, R. G. Morris, C. C. Bird, M. L. Hooper, and A. H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849-852. [DOI] [PubMed] [Google Scholar]
- 13.Corbet, S. W., A. R. Clarke, S. Gledhill, and A. H. Wyllie. 1999. P53-dependent and -independent links between DNA-damage, apoptosis and mutation frequency in ES cells. Oncogene 18:1537-1544. [DOI] [PubMed] [Google Scholar]
- 14.Craig, A. L., L. Burch, B. Vojtesek, J. Mikutowska, A. Thompson, and T. R. Hupp. 1999. Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem. J. 342:133-141. [PMC free article] [PubMed] [Google Scholar]
- 15.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 16.Fiscella, M., S. J. Ullrich, N. Zambrano, M. T. Shields, D. Lin, S. P. Lees-Miller, C. W. Anderson, W. E. Mercer, and E. Appella. 1993. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene 8:1519-1528. [PubMed] [Google Scholar]
- 17.Gorman, J. R., and F. W. Alt. 1998. Regulation of immunoglobulin light chain isotype expression. Adv. Immunol. 69:113-181. [DOI] [PubMed] [Google Scholar]
- 18.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 19.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 20.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 21.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 22.Jabbur, J. R., P. Huang, and W. Zhang. 2000. DNA damage-induced phosphorylation of p53 at serine 20 correlates with p21 and Mdm-2 induction in vivo. Oncogene 19:6203-6208. [DOI] [PubMed] [Google Scholar]
- 23.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 25.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 26.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 27.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 28.Luo, J. L., Q. Yang, W. M. Tong, M. Hergenhahn, Z. Q. Wang, and M. Hollstein. 2001. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 20:320-328. [DOI] [PubMed] [Google Scholar]
- 29.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meek, D. W. 1999. Mechanisms of switching on p53: a role for covalent modification? Oncogene 18:7666-7675. [DOI] [PubMed] [Google Scholar]
- 31.Melchionna, R., X. B. Chen, A. Blasina, and C. H. McGowan. 2000. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2:762-765. [DOI] [PubMed] [Google Scholar]
- 32.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 33.Ronen, D., D. Schwartz, Y. Teitz, N. Goldfinger, and V. Rotter. 1996. Induction of HL-60 cells to undergo apoptosis is determined by high levels of wild-type p53 protein whereas differentiation of the cells is mediated by lower p53 levels. Cell Growth Differ. 7:21-30. [PubMed] [Google Scholar]
- 34.Sabapathy, K., M. Klemm, R. Jaenisch, and E. F. Wagner. 1997. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 16:6217-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakaguchi, K., S. Saito, Y. Higashimoto, S. Roy, C. W. Anderson, and E. Appella. 2000. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 275:9278-9283. [DOI] [PubMed] [Google Scholar]
- 37.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. (Erratum, 14:760.) [PMC free article] [PubMed] [Google Scholar]
- 38.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 39.Shieh, S. Y., Y. Taya, and C. Prives. 1999. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 18:1815-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uesugi, M., and G. L. Verdine. 1999. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. USA 96:14801-14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unger, T., T. Juven-Gershon, E. Moallem, M. Berger, R. Vogt Sionov, G. Lozano, M. Oren, and Y. Haupt. 1999. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unger, T., R. V. Sionov, E. Moallem, C. L. Yee, P. M. Howley, M. Oren, and Y. Haupt. 1999. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene 18:3205-3212. [DOI] [PubMed] [Google Scholar]