Abstract
Disruption of the peroxisome proliferator-activated receptor γ (PPARγ) gene causes embryonic lethality due to placental dysfunction. To circumvent this, a PPARγ conditional gene knockout mouse was produced by using the Cre-loxP system. The targeted allele, containing loxP sites flanking exon 2 of the PPARγ gene, was crossed into a transgenic mouse line expressing Cre recombinase under the control of the alpha/beta interferon-inducible (MX) promoter. Induction of the MX promoter by pIpC resulted in nearly complete deletion of the targeted exon, a corresponding loss of full-length PPARγ mRNA transcript and protein, and marked reductions in basal and troglitazone-stimulated expression of the genes encoding lipoprotein lipase, CD36, LXRα, and ABCG1 in thioglycolate-elicited peritoneal macrophages. Reductions in the basal levels of apolipoprotein E (apoE) mRNA in macrophages and apoE protein in total plasma and high-density lipoprotein (HDL) were also observed in pIpC-treated PPARγ-MXCre+ mice. Basal cholesterol efflux from cholesterol-loaded macrophages to HDL was significantly reduced after disruption of the PPARγ gene. Troglitazone selectively inhibited ABCA1 expression (while rosiglitazone, ciglitazone, and pioglitazone had little effect) and cholesterol efflux in both PPARγ-deficient and control macrophages, indicating that this drug can exert paradoxical effects on cholesterol homeostasis that are independent of PPARγ. Together, these data indicate that PPARγ plays a critical role in the regulation of cholesterol homeostasis by controlling the expression of a network of genes that mediate cholesterol efflux from cells and its transport in plasma.
Peroxisome proliferator-activated receptor γ (PPARγ) is a member of the nuclear receptor superfamily that has been implicated to play a possible role in adipocyte differentiation (34, 73, 82), insulin sensitization (42, 51, 61), and tumor suppression (11, 14, 25, 37, 43, 57, 77, 88). Ligands for PPARγ include modified fatty acids, members of the prostanoid family, linoleic acid derivatives (38, 39, 58), and synthetic insulin-sensitizing thiazolidinedione compounds (TZDs) such as BRL-49653 (rosiglitazone), pioglitazone, and troglitazone (28, 29, 38, 39, 58).
PPARγ is abundantly expressed in adipose tissue and at lower levels in T cells, neutrophils, epithelial cells, smooth muscle cells, and macrophages (19, 48, 81). The discovery of high levels of PPARγ protein expression in the foam cells of atherosclerotic lesions (53, 71) has led to intensive research efforts to identify possible roles of PPARγ in regulating cholesterol flux in macrophages and to better understand the possible effects of TZDs on the development of atherosclerosis. With regard to cholesterol uptake, the scavenger receptor, CD36, was demonstrated to be a direct target gene of PPARγ in macrophages (15, 56, 58, 87). Recent studies have shown, however, that neither treatment of macrophages with TZDs (56) nor mutation of the PPARγ gene (15) significantly alters oxLDL uptake. Likewise, the basal expression of other scavenger receptors believed to be involved in the uptake of cholesterol, including SR-A and SR-B1, were unaffected by deletion of the PPARγ gene in macrophages (15, 56).
ABCA1, a member of the ATP-binding cassette (ABC)-transporter family, is involved in the control of high-density lipoprotein (HDL) and apolipoprotein A1 (apoA1)-mediated cholesterol efflux from macrophages (12, 50, 63). Loss-of-function mutations in the ABCA1 gene in Tangier's patients is marked by severe accumulation of cholesterol in macrophages and other tissues, including the tonsils, liver, spleen, and mucosa (9, 12, 32, 75), suggesting that ABCA1 activity plays an integral role in influencing cellular cholesterol transport (64). ABCG1 may also play a role in cholesterol efflux, though its role is not well defined (40). PPARγ was recently shown to induce the expression of the cholesterol tranporter, ABCA1, in macrophages through a transcriptional cascade mediated by the nuclear receptor, LXRα (16, 21, 79, 91). Consistent with this PPARγ-LXRα-ABCA1 pathway, TZDs were shown to enhance cholesterol efflux from wild-type macrophages, while PPARγ-deficient macrophages were refractory to the effect of TZDs and also exhibited lowered basal levels of cholesterol efflux (16). Moreover, transplantation of PPARγ-null bone marrow into mice lacking low-density lipoprotein receptor (LDLr-null mice) resulted in a significant increase in atherosclerotic lesion size. The implication of these findings is that PPARγ exerts antiatherogenic effects by facilitating the removal of cholesterol from macrophages via cholesterol transporter proteins such as ABCA1.
Several lines of evidence indicate that apoE is also an important determinant of atherosclerosis and regulator of cholesterol efflux in macrophages (5, 6, 22, 24, 41, 54, 66, 96, 97). apoE-null macrophages exhibit markedly lowered cholesterol efflux to HDL3 and lipid-free apolipoproteins in mouse peritoneal macrophages (47). The transplantation of bone marrow derived from apoE-null mice into the LDLr-null background resulted in a significant increase in atherosclerotic lesion size compared to controls (26). Conversely, transplantation of bone marrow from wild-type mice into apoE-null mice reduced cholesterol levels in plasma and slowed the progression of atherosclerosis (10, 52). Likewise, macrophage-specific expression of human apoE in apoE-null mice reduces atherosclerosis (7). These results suggest that attenuation of apoE expression in bone marrow and differentiated cells derived from progenitor cells in bone marrow (including macrophages) is sufficient to cause the phenotype. Apart from effects on cholesterol flux, apoE overexpression in mice has also been shown to lower levels of isoprostane, an in vivo marker of oxidative stress (86), which is believed to contribute to the development of atherosclerosis (8, 69, 94).
A number of previous studies examining the role of PPARγ in regulating lipid flux in macrophages have been performed by using macrophage cancer cell lines and macrophages derived from embryonic stem (ES) cells. In the latter, wild-type and PPARγ-null ES cells were induced to differentiate into macrophages in vitro by using a two-stage process involving initial differentiation of ES cells into embryoid bodies and subsequent differentiation into monocytes and macrophages in the presence of interleukin-3 and macrophage colony-stimulating factor 1 (16, 56). To a similar end, macrophages have been isolated from chimeric mice that were generated by injection of PPARγ-null ES cells into wild-type blastocysts (56). In these mice, the contribution of PPARγ-deficient cells to the macrophage lineage was demonstrated, suggesting that PPARγ is not required for macrophage differentiation in vivo. Moreover, the expression of several macrophage-specific markers was shown to be similar in macrophages derived from both wild-type and chimeric mice (56). Despite this, it is noted that both the in vivo or in vitro differentiation of macrophages from ES cells occurs in the absence of the PPARγ gene. Given that a number of PPARγ target genes are expressed in macrophage precursor cells (27, 35, 85, 87), the possibility remains that ES cell-derived macrophage populations, while similar in terms of cell numbers, do not retain all of the phenotypic characteristics and physiological pathways (other than those tested) associated with normal macrophages. Indeed, the in vitro differentiation process itself may alter the expression of several key macrophage proteins involved in cholesterol flux, since macrophage colony-stimulating factor 1 was shown to regulate SR-A and CD36 expression (23, 95).
To circumvent such problems, a PPARγ conditional knockout mouse containing the Cre recombinase gene under the control of the MX promoter was developed. Despite some constitutive expression of the MX-Cre transgene, the majority of thioglycolate-elicited peritoneal macrophages from PPARγ-MXCre+ mice maintain an intact gene for PPARγ, suggesting that essentially normal development and/or differentiation of the myeloid lineage occurs. Induction of Cre recombinase and the ensuing loss of the targeted PPARγ exon, resulted in marked reductions in basal and troglitazone-stimulated expression of the genes encoding lipoprotein lipase (LPL), CD36, LXRα, and ABCG1 in thioglycolate-elicited peritoneal macrophages. Reductions in the basal levels of apoE mRNA in macrophages and apoE protein in total plasma and HDL were also observed in pIpC-treated PPARγ-MXCre+ mice. Further, basal cholesterol efflux from cholesterol-loaded macrophages to HDL was significantly reduced after disruption of the PPARγ gene. Surprisingly, and in sharp contrast to recent findings by using ES cell-derived macrophages (16, 18), troglitazone inhibited ABCA1 expression (whereas other TZDs, such as rosiglitazone, ciglitazone, and pioglitazone, had little effect) and cholesterol efflux in both PPARγ-deficient and control macrophages, indicating that troglitazone can exert paradoxical effects on cholesterol homeostasis that are independent of PPARγ. Together, these data indicate that PPARγ plays an important role in the regulation of cholesterol homeostasis by controlling the expression of a network of genes that mediate cholesterol efflux from cells and its transport in plasma.
MATERIALS AND METHODS
Reagents.
Oxysterols (Sigma, St. Louis, Mo.), dexamethasone (Sigma), troglitazone (Sankyo Research Laboratories, Fukuroi, Japan), rosiglitazone (Glaxo-SmithKline, Research Triangle Park, N.C.), ciglitazone (Biomol Research Laboratories, Plymouth Meeting, Pa.), and pioglitazone (Takeda Chemical Industries, Ltd., Osaka, Japan) were dissolved in dimethyl sulfoxide (DMSO; Sigma).
Targeting vector and generation of PPARγ conditional-null mice.
A mouse ES-129 P1 genomic library (Genome Systems, St. Louis, Mo.) was screened with the mouse PPARγ cDNA (17). A 4.5-kb HindIII fragment containing exons 1 and 2 of the mouse PPARγ gene was subcloned into a modified pGEM-3Zf vector (Promega, Madison, Wis.) in which the polylinker from EcoRI to SacI had been destroyed. The targeting vector was made by inserting the first loxP site into the EcoRI site of a region 1 kb upstream of exon 2. This exon encodes the DNA-binding domain of PPARγ. The first loxP site was inserted with a BamHI site in order to develop a means to genotype for the inclusion of this loxP site during homologous recombination. Insertion and orientation of the linker were confirmed by sequencing. A 2-kb SacI fragment containing a neomycin resistance cassette (neo) flanked by two loxP motifs was inserted into the SacI site located 0.75 kb downstream of exon 2. This fragment was cloned such that all three loxP sites were in the same orientation. The targeting vector contained 4-kb of homologous DNA upstream of the first loxP site and 2 kb of homologous DNA downstream of the loxP-neo-loxP cassette site. Finally, a thymidine kinase gene cassette was inserted into the SalI site of the polylinker. ES cells (RW4; Genome Systems) were electroporated with the linearized targeting construct and maintained on subconfluent embryonic fibroblasts. G418-resistant ES cell colonies were picked and expanded. The ES cells were scored for homologous recombination by Southern blotting. One (out of 264 ES cells screened) homologous recombinant was identified with a flanking probe (3′-probe; Fig. 1B). The targeted ES cells were injected into C57BL/6 blastocysts, and the blastocysts were transferred into pseudopregnant NIH Swiss mice. Highly chimeric male mice were bred with C57BL/6 females to generate heterozygous loxP-targeted/wild-type (t/+) mice. Deletion of the neomycin cassette in vivo was achieved by crossing the mice with an EIIaCre transgenic line (46). The neomycin cassette deleted heterozygous mice carrying one floxed (fl; for flanked by loxP) and one wild-type allele were bred with homozygous MX-Cre mice (44). These mice were kindly provided by Ralf Kühn and Klause Rajewsky (University of Cologne, Cologne, Germany). The heterozygous offspring for both the loxP-targeted PPAR γ gene (PPARγfl/+) and the MX-Cre transgene (PPARγfl/+ MXCre+) were then interbred with PPARγfl/+ littermates lacking Cre (PPARγfl/+ MXCre−) to generate mice hemizygous for the MX-Cre transgene and homozygous for the PPARγ-floxed allele (PPARγ-MXCre+) and littermate control mice, designated PPARγ-MXCre−. Subsequently, PPARγ-MXCre+ and PPARγ-MXCre− mice were interbred to generate littermates of the same genotypes.
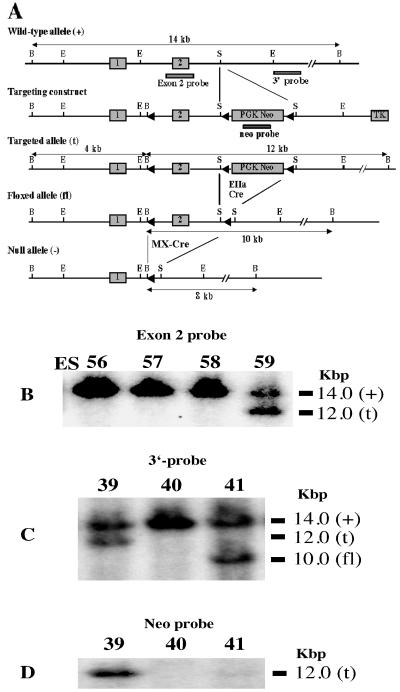
FIG. 1.
Gene targeting and conditional deletion of exon 2 of the PPARγ gene. (A) Restriction maps of the wild-type allele, targeting vector, targeted allele, floxed allele, and null allele. The indicated probes (3′-probe, exon 2, and neo probe) were used to assess recombination events. Open boxes represent exons and are numbered as indicated. PGK neomycin (PGK Neo) and thymidine kinase (TK) are positive and negative selection cassettes, respectively. Restriction sites: B, BamHI; E, EcoRI; S, SacI. (B) Southern blot analysis of homologous recombination in ES cells electroporated with the targeting vector. DNA derived from ES cells were digested with BamHI. Hybridizing fragments of wild-type (+) and targeted (t) alleles to the exon 2 probe and their respective sizes are indicated. Similar results were obtained with the 3′-probe (not shown). (C) Southern blot analysis of BamHI-digested tail DNA from pups derived from the cross between heterozygous (t/+) males and EIIaCre females (schematically shown in panel A). Hybridizing fragments of wild-type (+), targeted (t), and floxed (fl) alleles to the 3′-probe and their respective sizes are indicated. Recombination between loxP sites 2 and 3 was demonstrated with tail DNA derived from the mouse designated “41,” although other recombination events were observed. Similar results were obtained with the exon 2 probe (not shown). (D) Southern blot analysis of tail DNA used in panel C. A 12-kbp hybridizing fragment of the targeted (t) allele, but not wild-type (+) and floxed (fl) alleles, to the neo probe is indicated.
Isolation of macrophages.
mice were injected intraperitoneally with 250 μl of pIpC (2.5 mg/ml in isotonic saline; Sigma) on days 1, 4, and 7. On day 10, mice were injected intraperitoneally with 2 ml of 3% thioglycolate solution (Becton Dickinson, Cockeysville, Md.). On day 14, peritoneal macrophages were harvested by injecting 10 ml of ice-cold phosphate-buffered saline with an 18-gauge needle into the peritoneal cavity and withdrawing the fluid with a 21-gauge needle and then repeating the procedure. After centrifugation at 1,000 × g for 10 min, cells were resuspended in RPMI medium containing 10% fetal bovine serum and penicillin-streptomycin and then plated onto 150-mm petri dishes (Becton Dickinson Labware, Franklin Lakes, N.J.). After incubation of the cells at 37°C for 4 h, cells were further cultured in media containing the specified ligand or vehicle for 24 h.
Southern blot analysis.
Genomic DNA was isolated from peritoneal macrophages by using a Wizard genomic DNA purification kit (Promega) according to the manufacturer's protocol. It was then digested with BamHI, subjected to electrophoresis on a 0.6% agarose gel, and transferred to GeneScreen Plus Hybridization Transfer membranes (NEN Life Sciences, Boston, Mass.). A 0.6-kb DNA fragment (SalI-EcoRI) derived from intron 2 of the PPARγ gene (3′-probe) and DNA fragments derived from exon 2 and neo (exon 2 probe and neo probe, respectively) were used as probes.
RNase protection assay.
The RNA protection assay was performed by using RPAIII (Ambion, Austin, Tex.) according to the manufacturer's instructions. Antisense cRNA probes of PPARγ were prepared by using a MAXIscript kit from a clone containing exons 1 and 2 of the PPARγ gene. The clone was prepared by PCR with primers tagged with 5′-BamHI and 3′-EcoRI sites, and the PCR products were inserted into 5′-BamHI and 3′-EcoRI sites of the pTRiamp18 vector (Ambion). PCR was performed with the following primer sets: 5′-ACCTCTCCGTGATGGAAG-3′ (forward) and 5′-GGTGGAGATGCAGGTTC-3′ (reverse). Antisense riboprobes for β-actin were prepared from DNA provided with the RPAIII kit.
Northern blot analysis.
Total RNA was isolated from thioglycolate-elicited macrophages of mice by the acidic guanidine isocyanate-phenol-chloroform extraction method by using the Ultraspec RNA Isolation System (Biotecx Laboratories, Houston, Tex.). Total RNA was quantitated by optical densitometry at 260 nm and subjected to electrophoresis on a 1.0% agarose gel containing 7% formaldehyde in 20 mM morpholinepropanesulfonic acid-8 mM sodium acetate-1 mM EDTA buffer (pH 7.0) and transferred to GeneScreen Plus Hybridization Transfer membranes (NEN Life Sciences). The cDNA probes used for Northern blotting included PPARγ (17) and LPL (36). Probes for CD36, LXRα, ABCA1, ABCG1, and apoE cDNA were amplified from a mouse liver cDNA library by using gene-specific primers and cloned into pCR TOPO II (Stratagene, La Jolla, Calif.). The identity of the probes was confirmed by nucleotide sequencing. The intensities of hybridizing mRNAs were quantitated by using ImageQuant and normalized to levels of β-actin mRNA.
Western blot analysis.
Nuclear extracts from cultured macrophages were isolated according to the protocols provided with NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce, Rockford, Ill.). Hepa-1 cells (2 × 105) were transfected with 200 ng of an expression vector for PPARγ1, pSG5-PPARγ (provided by Walter Wahli, Université de Lausanne, Lausanne, Switzerland), by lipofection according to the protocols provided with LipofectAMINE Reagent (Gibco-BRL, Grand Island, N.Y.). Protein contained in the nuclear extracts of macrophages or total protein of Hepa-1 cells was assayed by the BCA protein assay (Pierce Chemical Co., Rockford, Ill.). Then, 10 μg of total protein from Hepa-1 cells and 10 μg of protein from nuclear extracts of macrophages were subjected to electrophoresis on a 4 to 15% Tris-HCl gradient gel (Bio-Rad, Hercules, Calif.), transferred to Immobilon-P membranes (Millipore, Bedford, Mass.), and probed according to the manufacturer's recommendations with anti-PPARγ antibodies (E-8 and H-100; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) as indicated. Detection of immunoreactive proteins was done by an enhanced chemiluminescence blot detection system (Amersham, Inc., Arlington Heights, Ill.).
Measurement of plasma lipids and lipoproteins.
Total cholesterol and triglyceride concentrations (Sigma), as well as free cholesterol and phospholipids (Wako, Osaka, Japan), were measured in 12-μl aliquots of plasma by using commercial kits and the Hitachi 911 automated chemistry analyzer (Boehringer Mannheim, Indianapolis, Ind.). Lipoproteins from pooled plasma samples (100 μl) were separated by gel filtration by using two Superose 6HR 10/30 columns in series (by fast-protein liquid chromatography [FPLC]; Pharmacia Biotech, Inc., Piscataway, N.J.). Mouse apoA-I, apoA-II, apoE, and apoB were identified by Western blotting as previously described (31).
Cholesterol loading and efflux.
Peritoneal macrophages were pretreated with the indicated ligands [troglitazone, 22(R)-hydroxycholesterol (22-HC), dexamethasone, or DMSO] and thereafter every 24 h in serum-free medium containing 0.1% albumin. The cells were cholesterol loaded by incubation with oxLDL (50 μg/ml containing [3H]cholesterol). After a 24-h incubation period, the cells were washed twice with phosphate-buffered saline, and fresh medium containing HDL (50 μg/ml) or not was added for 24 h. The radioactivity of 3H was measured in centrifuged media as well as in cellular lipids extracted with hexane-isopropanol. The total radioactivity in cells and media corresponds to the cellular loading of cholesterol from oxLDL. HDL-specific [3H]cholesterol efflux was measured as the fraction of total radiolabeled cholesterol appearing in the media in the presence of HDL after subtraction of values for HDL-free media (18).
RESULTS
Generation of a conditional allele of the PPARγ gene.
A targeting vector containing three loxP sites in the same orientation was constructed (Fig. 1A). The first loxP site (with an artificial BamHI site) was inserted into an EcoRI site of a region 1 kb upstream of exon 2 that encodes the DNA-binding domain of PPARγ. A 2-kb SacI fragment containing a neomycin resistance cassette (neo) flanked by two loxP motifs (fl-neo-fl) was inserted into the SacI site located 0.75 kb downstream of exon 2. Finally, a thymidine kinase gene cassette was inserted into the SalI site of the polylinker. After standard electroporation and culture of ES cells, one homologous recombinant (out of 264 ES cells screened; ES clone 59) was identified by Southern blotting of genomic DNA by using a probe derived from exon 2 (Fig. 1B). Similar results were obtained by using a flanking probe designated 3′-probe (data not shown). The targeted ES cells were injected into C57BL/6 blastocysts, which were then transferred into pseudopregnant NIH Swiss mice. Highly chimeric male mice were bred with C57BL/6 females to generate heterozygous loxP-targeted/wild-type (t/+) mice. Breeding of these offspring resulted in either wild-type (+/+) or t/+ mice, with no t/t mice, indicating that the neo cassette inactivated the PPARγ gene, resulting in embryonic lethality. This is in agreement with other studies showing that disruption of the PPARγ gene is lethal (3, 42, 73). To circumvent this problem, the neomycin cassette was removed in vivo by crossing the t/+ mice with a EIIaCre transgenic line (31, 46). Recombination events were analyzed by Southern blotting of the tail DNA with the 3′-probe (Fig. 1C) and exon 2 probe (identical results not shown). Deletion of the neomycin cassette in the same tail DNA samples was confirmed by Southern blotting with a neo probe (Fig. 1D). Mice that were heterozygous for the floxed PPARγ allele (PPARγfl/+) were bred with homozygous MXCre+ mice (44). Heterozygous offspring for the targeted PPARγ gene that were either hemizygous for the Cre transgene (PPARγfl/+ MXCre+) or lacking Cre (PPARγfl/+ MXCre−) were interbred. This breeding scheme yielded mice that were homozygous for the PPARγ-floxed allele and either hemizygous for or lacking the MX-Cre transgene (PPARγ-MXCre+ and PPARγ-MXCre−, respectively). These mice were subsequently interbred to generate littermates of the same genotypes. All breedings subsequent to the removal of the neo cassette resulted in a distribution of the genotypes of offspring that followed the predicted Mendelian frequency (data not shown), suggesting that neither the floxed PPARγ allele nor the MX-Cre transgene contributed to prenatal lethality.
Conditional deletion of exon 2 of the PPARγ gene in peritoneal macrophages.
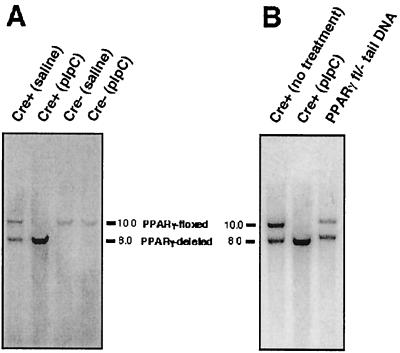
Treatment of PPARγ-MXCre+ mice with pIpC, which was shown to activate the MX promoter by stimulating the production of alpha and beta interferon (13, 83), results in a nearly complete deletion of the floxed exon 2 in thioglycolate-elicited peritoneal macrophages (Fig. 2). A partial deletion of the floxed exon 2 was detected in the macrophages of saline-treated PPARγ-MXCre+ mice. To test the possibility that spontaneous deletion resulting from constitutive expression of the MX-Cre transgene had occurred, Southern blot analysis of genomic DNA of macrophages from untreated PPARγ-MXCre+ mice was performed. Deletion of exon 2 was shown to occur in a portion of the macrophage population, as indicated by the presence of a 8-kb hybridizing band which represents a deleted PPARγ allele, suggesting that there is some spontaneous deletion or “leakiness” associated with the MX-Cre transgene (Fig. 2B). To determine the extent, the intensities of the hybridizing bands representing the PPARγ floxed and deleted alleles (10- and 8-kb hybridizing bands, respectively) were quantitated. Values were normalized to intensities of the hybridizing DNA bands with PPARγfl/− tail DNA, which represents a theoretical 1:1 ratio of floxed to deleted allele (Fig. 2B). The results show that the intensities of the PPARγ floxed and deleted allele are in a 3.4 to 1 ratio, respectively, indicating that the majority (77.2%) of thioglycolate-elicited macrophages from pretreated PPARγ-MXCre+ mice were differentiated in the presence of an intact gene for PPARγ. As expected, neither pIpC nor saline treatment resulted in recombination of the PPARγ gene in macrophages from PPARγ-MXCre− mice (Fig. 2A). Nearly complete disruption of the PPARγ gene was also demonstrated in the liver (not shown), a finding consistent with the results of earlier studies with the MX-Cre transgene in which a nearly complete pIpC deletion was shown in the liver (a tissue in which PPARγ is not well expressed) and macrophages (44, 68, 72). Based on these results, macrophages derived from pIpC-treated PPARγ-MXCre+ mice (designated as PPARγ-deficient macrophages) and pIpC-treated PPARγ-MXCre− mice (control macrophages) were used in the following studies.
FIG. 2.
(A) Southern blot analysis of BamHI-digested genomic DNA isolated from thioglycolate-elicited peritoneal macrophages from saline-treated PPARγ-MXCre+, pIpC-treated PPARγ-MXCre+, saline-treated PPARγ-MXCre−, and pIpC-treated PPARγ-MXCre− mice, as indicated. Hybridizing fragments of floxed and deleted alleles to the 3′-probe and their respective sizes are indicated. (B) Southern blot analysis of BamHI-digested genomic DNA isolated from macrophages of untreated PPARγ-MXCre+ mice, thioglycolate-elicited peritoneal macrophages from pIpC-treated PPARγ-MXCre+ mice, and tail of PPARγfl/− mice, as indicated. Hybridizing fragments of floxed and deleted alleles to the 3′-probe and their respective sizes are indicated.
Disruption of the PPARγ gene results in a truncated PPARγ mRNA transcript.
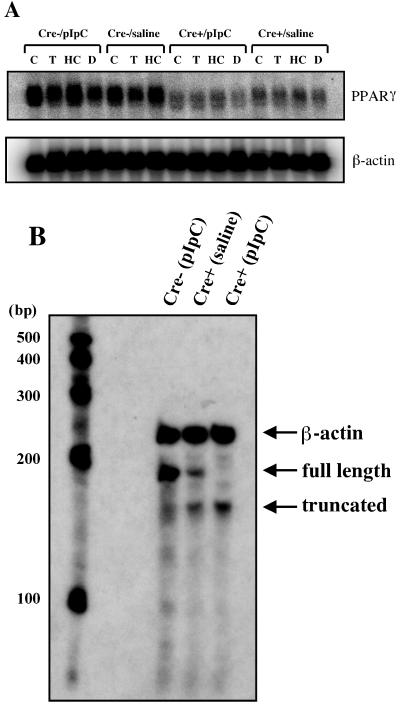
Northern blot analysis of total macrophage RNA with a PPARγ cDNA probe revealed a marked reduction in steady-state levels of a truncated PPARγ mRNA levels in macrophages derived from PPARγ-MXCre+ mice relative to control PPARγ-MXCre− mice (both pIpC and saline treatments) (Fig. 3A). It did not appear that treatment of either PPARγ-deficient and control macrophages with troglitazone, 22-HC, or dexamethasone in vitro significantly altered PPARγ mRNA levels. The loss of full-length PPARγ mRNA and the presence of a truncated PPARγ mRNA product, a finding consistent with the expected size based upon Cre recombination, was confirmed in PPARγ-deficient macrophages by using RNase protection assays (Fig. 3B). As expected, based on the partial deletion of the PPARγ gene in the macrophages of saline-treated PPARγ-MXCre+ mice (Fig. 2A), both full-length and truncated PPARγ transcripts were detected (Fig. 3B). Using a β-actin riboprobe to normalize for the amount of total RNA, PPARγ-deficient macrophages were shown to exhibit significantly lower levels of PPARγ transcript, thus suggesting the possibility that the truncated transcript is inherently less stable than wild-type PPARγ mRNA.
FIG. 3.
(A) Northern blot analysis of total RNA from thioglycolate-elicited peritoneal macrophages of PPARγ-MXCre− and PPARγ-MXCre+ mice treated with saline or pIpC, as indicated. Macrophages were treated with the indicated ligands [15 μM troglitazone (T), 2.5 μg of 22-HC/ml, 100 nM dexamethasone (D), and DMSO vehicle (C)] and total macrophage RNA (10 μg) was denatured and electrophoresed in formaldehyde-containing 1% agarose gels, blotted to nylon membranes, and probed with the indicated cDNA probes for PPARγ and β-actin. (B) RNase protection assay. Total RNA from thioglycolate-elicited peritoneal macrophages of pIpC-treated PPARγ-MXCre− (lane 1), saline-treated PPARγ-MXCre+ (lane 2), and pIpC-treated PPARγ-MXCre+ (lane 3) mice were hybridized with riboprobes for β-actin and PPARγ (as described in Materials and Methods) and subjected to digestion with RNase H. The products were then separated on a 5.0% polyacrylamide gel. Positions of the protected mRNA fragments for PPARγ and β-actin, as well as the expected size of the PPARγ truncated mRNA, are indicated.
Disruption of the PPARγ gene results in the loss of PPARγ protein in macrophages.
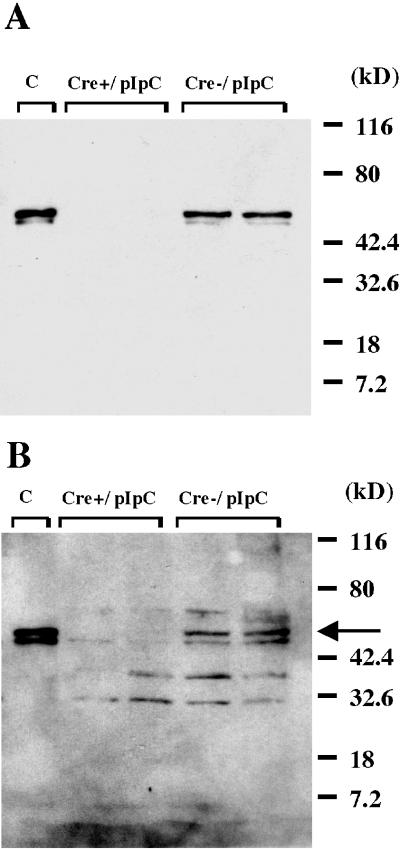
To determine whether disruption of the PPARγ gene results in a loss of PPARγ protein or the production of a truncated mutant form of the protein, nuclear extract proteins from peritoneal macrophages were analyzed by Western blotting. The results with a primary antibody raised against a peptide mapping at the carboxy terminus of PPARγ of human origin that is identical to the corresponding mouse sequence (E-8 PPARγ antibody) reveal that a 53-kDa protein product is expressed in thioglycolate-elicited macrophages from pIpC-treated PPARγ-MXCre− but not PPARγ-MXCre+ mice (Fig. 4A). This protein is consistent in size with the PPARγ1 protein and with the protein product expressed in Hepa-1 cells transfected with an expression vector for PPARγ1 (pSG5-PPARγ1 cDNA). As expected, the putative truncated PPARγ protein product, which has an expected size of 10 kDa, was not detected by the E-8 antibody (Fig. 4A). Similar results were obtained with the H-100 primary antibody, a PPARγ antibody that was raised against a recombinant protein corresponding to amino acids 6 to 105 of the PPARγ protein and is therefore expected to recognize both the wild-type and putative truncated form of this protein. A 53-kDa protein product that is consistent in size with the PPARγ protein was expressed in macrophages from pIpC-treated PPARγ-MXCre− mice but not in macrophages from pIpC-treated PPARγ-MXCre+ mice (Fig. 4B). Likewise, the expected translation product of PPARγ mRNA lacking exon 2, which is ca. 10 kDa, does not appear on Western blotting, perhaps due to increased instability of prematurely terminated proteins (Fig. 4B).
FIG. 4.
Western blot analysis of nuclear extracts from macrophages of pIpC-treated PPARγ-MXCre+ mice and similarly treated PPARγ-MXCre− mice. A total of 10 μg of protein from nuclear extracts of macrophages and 10 μg of total protein from Hepa-1 cells transfected with an expression vector for PPARγ1 (pSG5-PPARγ cDNA) were subjected to electrophoresis on a 4 to 15% Tris-HCl gradient gel (Bio-Rad), transferred to Immobilon-P membranes (Millipore), and probed as recommended by the manufacturer with anti-PPARγ antibodies (Santa Cruz Biotechnologies) specific for the N terminus (E-8, PPARγ antibody) (A) and the C terminus (H-100, PPARγ antibody) (B) of the PPARγ protein. Detection of immunoreactive proteins was done by using an enhanced chemiluminescence blot detection system (Amersham).
Disruption of the PPARγ gene results in the loss of PPARγ activity in macrophages.
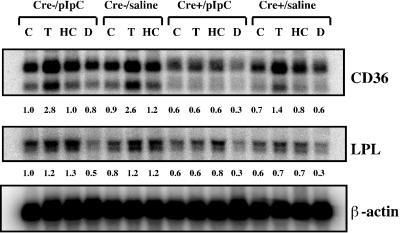
To evaluate whether disruption of the PPARγ gene results in a functional deficiency in PPARγ activity, the expression of the known PPARγ target gene, CD36, was examined in PPARγ-deficient and control macrophages. Consistent with previous findings (15, 56), the expression of the gene encoding CD36 was induced by troglitazone in control macrophages but not in PPARγ-deficient macrophages (Fig. 5). Basal levels of CD36 were also significantly lower in PPARγ-deficient macrophages relative to control macrophages. A gene dosage effect appeared to be present, since a less robust induction of CD36 mRNA by troglitazone was observed in macrophages derived from saline-treated PPARγ-MXCre+ mice, which have a partial deletion of the PPARγ gene (Fig. 5). Neither 22-HC nor dexamethasone significantly altered CD36 expression in both PPARγ-deficient or control macrophages. Together, these results indicate that targeted disruption of the PPARγ gene leads to a functional loss of PPARγ activity in primary peritoneal macrophages, which is consistent with the loss of PPARγ protein. Furthermore, the observation that troglitazone stimulated CD36 gene expression in control macrophages, which contain a homozygous floxed allele for the PPARγ gene, indicates that the presence of loxP sites within introns 1 and 2 of the PPARγ gene does not interfere with the activity of the PPARγ protein.
FIG. 5.
Northern blot analysis of total RNA from thioglycolate-elicited peritoneal macrophages of PPARγ-MXCre− and PPARγ-MXCre+ mice treated with saline or pIpC, as indicated. Macrophages were treated with the indicated ligands [15 μM troglitazone (T), 2.5 μg of 22-HC (HC)/ml, 100 nM dexamethasone (D), and DMSO vehicle (C)], and total macrophage RNA (10 μg) was denatured and electrophoresed in formaldehyde-containing 1% agarose gels, blotted onto nylon membranes, and probed with the indicated cDNA probes for CD36, LPL, and β-actin. The intensities of hybridizing mRNA bands were quantitated and normalized to β-actin mRNA. Numbers represent signal intensities relative to pIpC-treated Cre− macrophages (lane 1).
To examine whether PPARγ plays a role in regulating the expression of other scavenger receptors that bind modified lipoproteins, SR-A and SR-BI mRNA levels were determined in PPARγ-deficient and control macrophages. Consistent with previous studies (15, 16), however, the levels of SR-A and SR-BI mRNA were not substantially altered in response to the disruption of the PPARγ gene or by treatment with troglitazone, 22-HC, or dexamethasone in both PPARγ-deficient and control macrophages (data not shown).
PPARγ has a critical role in the regulation of the LPL gene in primary macrophages.
Associations between macrophage-derived LPL and the development of atherosclerosis have been described (1, 2, 90). This, coupled with the fact that the LPL gene is an established target of PPARγ in several other tissues, including liver and adipocytes (78), prompted us to examine LPL expression in primary macrophages. Similar to CD36, a striking reduction in basal and troglitazone-induced expression of the LPL gene was observed in PPARγ-deficient macrophages relative to controls (Fig. 5), suggesting that the LPL gene is a target of PPARγ in macrophages. Interestingly, 22-HC and dexamethasone were shown to enhance and suppress, respectively, the expression of LPL independent of the PPARγ gene disruption (Fig. 5).
Regulation of reverse cholesterol transport by PPARγ at the level of gene expression.
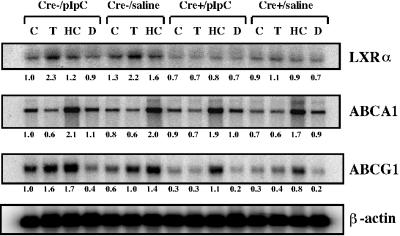
PPARγ was recently shown to induce the expression of the cholesterol transporter, ABCA1, via a transcriptional cascade involving LXRα (16, 21, 79, 91). Given the importance of the ABC family of transporter proteins in the regulation of cholesterol efflux in macrophages and its possible association with the development of foam cells of atherosclerotic plaques, components of the PPARγ-LXRα-ABCA1 pathway were examined at the level of gene expression in primary macrophages. In agreement with this pathway, troglitazone induced steady-state levels of LXRα mRNA in control macrophages, an effect which was absent in PPARγ-deficient macrophages (Fig. 6). Basal levels of LXRα mRNA expression were also lower in PPARγ-deficient macrophages, a finding consistent with reductions in basal levels of PPARγ expression. The LXRα ligand, 22-HC, slightly increased LXRα mRNA levels in control but not PPARγ-deficient macrophages (Fig. 6). To determine whether PPARγ-mediated regulation of LXRα extended to members of the ABC family of cholesterol transporters in macrophages, ABCG1 and ABCA1 gene expression levels were examined. Basal and troglitazone-inducible expression of the ABCG1 gene was found to be significantly inhibited in PPARγ-deficient macrophages relative to controls (Fig. 6), a finding consistent with the finding that lipid induction of ABCG1 (and ABCA1) expression in macrophages depends on LXRs (45, 70, 91, 92). Likewise, basal expression of ABCA1 was slightly reduced in PPARγ-deficient macrophages. Unexpectedly, however, troglitazone inhibited ABCA1 gene expression in both PPARγ-deficient and control macrophages, an effect which sharply contrasts the results of previous studies (16, 18) and the observed ABCG1 expression profiles (Fig. 5). The demonstration that 22-HC was capable of inducing ABCA1 and ABCG1 gene expression in PPARγ-deficient and control macrophages confirmed that the LXR signaling pathways downstream of PPARγ were intact in both macrophage populations.
FIG. 6.
Northern blot analysis of total RNA from thioglycolate-elicited peritoneal macrophages of PPARγ-MXCre− and PPARγ-MXCre+ mice treated with saline or pIpC, as indicated. Macrophages were treated with the indicated ligands [15 μM troglitazone (T), 2.5 μg of 22-HC (HC)/ml, 100 nM dexamethasone (D), and DMSO vehicle (C)], and total macrophage RNA (10 μg) was denatured and electrophoresed in formaldehyde-containing 1% agarose gels, blotted onto nylon membranes, and probed with the indicated cDNA probes for LXRα, ABCA1, ABCG1, and β-actin. The intensities of hybridizing mRNA bands were quantitated and normalized to β-actin mRNA. The numbers represent signal intensities relative to pIpC-treated Cre− macrophages (lane 1).
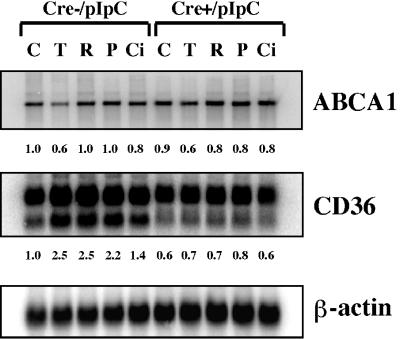
To determine whether the observed effects of troglitazone extend to other members of the TZD family of PPARγ agonists, the effect of rosiglitazone, ciglitazone, and pioglitazone on the expression of the gene encoding ABCA1 was examined. Unlike troglitazone, however, these TZDs were shown to have minimal effect on the expression of the gene encoding ABCA1 in both PPARγ-deficient and control macrophages (Fig. 7). To confirm the efficacy of these ligands, the expression of the gene encoding CD36 was also examined. As expected, each of these TZDs (rosiglitazone, ciglitazone, and pioglitazone) was capable of stimulating CD36 gene expression in control but not PPARγ-deficient macrophages (Fig. 7). It therefore appears that troglitazone has effects on ABCA1 gene expression that are unique for the TZD family of PPARγ agonists.
FIG. 7.
Northern blot analysis of total RNA from thioglycolate-elicited peritoneal macrophages of PPARγ-MXCre− and PPARγ-MXCre+ mice treated with pIpC, as indicated. Macrophages were treated with the indicated ligands [15 μM troglitazone (T), 5 μM rosiglitazone (R), 10 μM pioglitazone (P), 13 μM ciglitazone (Ci), and DMSO vehicle (C)], and the total macrophage RNA (10 μg) was denatured and electrophoresed in formaldehyde-containing 1% agarose gels, blotted onto nylon membranes, and probed with the indicated cDNA probes for ABCA1, CD36, and β-actin. The intensities of hybridizing mRNA bands were quantitated and normalized to β-actin mRNA. The numbers represent signal intensities relative to pIpC-treated Cre− macrophages (lane 1).
Reduction in basal levels of apoE gene expression in macrophages and plasma of pIpC-treated PPARγ-MXCre+ mice.
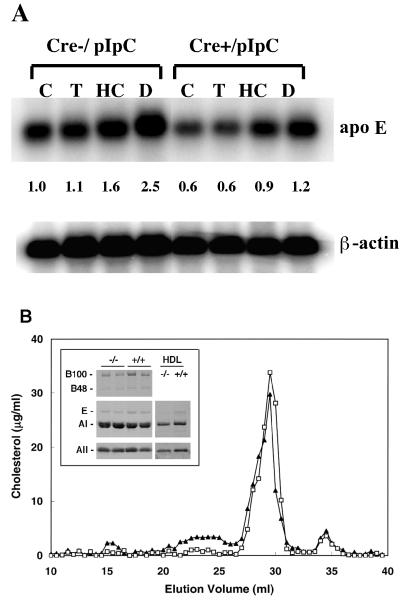
apoE may be an important determinant of atherosclerosis and a regulator of cholesterol efflux in macrophages (5, 6, 22, 24, 41, 54, 66, 96, 97). Consistent with the finding that the apoE gene is an LXRα target (45), basal expression of the gene encoding apoE was significantly attenuated in PPARγ-deficient macrophages relative to controls (Fig. 8A), while 22-HC was shown to induce apoE gene expression in a PPARγ-independent manner, an effect that was even more pronounced after dexamethasone treatment.
FIG. 8.
(A) Northern blot analysis of total RNA from thioglycolate-elicited peritoneal macrophages of pIpC-treated PPARγ-MXCre− or PPARγ-MXCre+ mice. Macrophages were treated with the indicated ligands [15 μM troglitazone (T), 2.5 μg of 22-HC (HC)/ml, 100 nM dexamethasone (D), and DMSO vehicle (C)], and total macrophage RNA (10 μg) was denatured and electrophoresed in formaldehyde-containing 1% agarose gels, blotted to nylon membranes, and probed with the indicated cDNA probes for apoE and β-actin. The intensities of the hybridizing mRNA bands were quantitated and normalized to β-actin mRNA. The numbers represent signal intensities relative to pIpC-treated Cre− macrophages (lane 1). (B) Lipid profiles in serum of pIpC-treated PPARγ-MXCre− and pIpC-treated PPARγ-MXCre+ mice. Lipoproteins were separated from 100 μl of pooled mouse plasma samples by FPLC (n = 4 for each group). Profiles from pIpC-treated PPARγ-MXCre− (control; ▴) and pIpC-treated PPARγ-MXCre+ (□) mice are shown. Concentrations of cholesterol are indicated on the y axis. (Inset) Immunoblot analysis of apolipoproteins B, E, A-I, A-II, and Cs in plasma and in HDL fraction from pIpC-treated PPARγ-MXCre− (control) and pIpC-treated PPARγ-MXCre+ mice, as indicated.
It is also noteworthy that pIpC-treated PPARγ-MXCre+ mice exhibited a significant reduction of apoE (−30% by densitometric analysis) in plasma, mostly due to decreased HDL apoE levels (Fig. 8B, inset). The total cholesterol, phospholipid, triglyceride, and cholesterol ester levels in plasma were similar in pIpC-treated PPARγ-MXCre+ mice and pIpC-treated controls (not shown), but in pIpC-treated PPARγ-MXCre+ mice, FPLC analysis revealed a decrease in LDL-cholesterol accompanied by a modest increase in HDL-cholesterol (Fig. 8B). The decrease in LDL-cholesterol was paralleled by a concomitant decrease in plasma apoB levels (Fig. 8B, inset). The possibility that PPARγ deficiency in tissues other than macrophages may have contributed to changes in the serum lipid profiles cannot be ruled out, since a nearly complete disruption of the PPARγ gene was also detected in liver (not shown). Indeed, the loss of even low levels of endogenous PPARγ expression in liver may play a significant role in altering the FPLC profile, given the importance of this organ in regulating lipoprotein metabolism in serum.
Disruption of the PPARγ gene results in reduced basal levels of cholesterol efflux from macrophages.
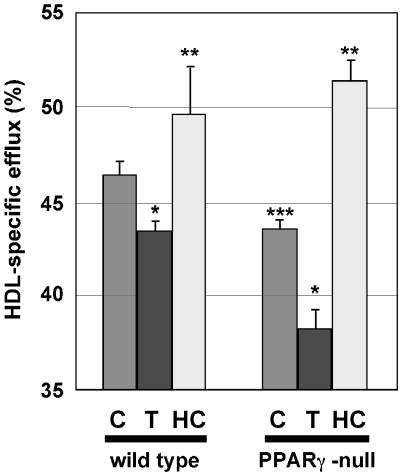
To examine possible roles of PPARγ in regulating cholesterol efflux in primary macrophages, cholesterol efflux studies were performed. After disruption of the PPARγ gene, basal cholesterol efflux from cholesterol-loaded macrophages to HDL was significantly reduced (Fig. 9). Paradoxically, and in sharp contrast to the results of previous cholesterol efflux studies (15, 18, 56), troglitazone lowered cholesterol efflux to HDL in both PPARγ-deficient and control macrophages (Fig. 7). On the other hand, 22-HC was shown to stimulate cholesterol efflux in both PPARγ-deficient and control macrophages. In fact, the levels of cholesterol efflux from PPARγ-deficient and control macrophages induced by 22-HC treatment were similar, indicating that the effects of LXRα signaling on cholesterol efflux can compensate for a lack of PPARγ activity. These results are in agreement with our observation that troglitazone inhibited, whereas 22-HC induced, the expression of the gene encoding the cholesterol transporter ABCA1 in both macrophage populations. Together, these results point to a critical role of PPARγ in the regulation of basal cholesterol efflux in macrophages and imply that troglitazone may have effects on cholesterol homeostasis that are independent of PPARγ.
FIG. 9.
Thioglycolate-elicited peritoneal macrophages from pIpC-treated PPARγ-MXCre− and pIpC-treated PPARγ-MXCre+ mice were plated in 24-well plates and incubated for 24 h in RPMI 1640 supplemented with 10% fetal bovine serum and [3H]cholesterol in the absence or presence of 15 μM troglitazone or 2.5 μg of 22-HC (HC)/ml as indicated. HDL-dependent cholesterol efflux to the medium was determined as described in Materials and Methods. Data are presented as a percentage (± the standard error) of the total radioactivity in the cells and the medium. Each point is a numerical average of quadruplicate experiments. The PPARγ-independent effects of troglitazone and 22-HC, as well as the reduction of cholesterol efflux in PPARγ-deficient macrophages, are statistically significant at P values of <0.05 and are indicated by the asterisks (★, ★★, and ★★★, respectively).
DISCUSSION
This report describes the development of a viable PPARγ conditional gene knockout mouse that circumvents the embryonic lethality associated with disruption of the PPARγ gene. Introduction of the MX-Cre transgene into this mouse line allowed for the pIpC-inducible deletion of the targeted PPARγ exon 2 in primary peritoneal macrophages and corresponding loss of the full-length PPARγ mRNA transcript and protein. Significant reductions in both basal and troglitazone-inducible expression of the known PPARγ macrophage target genes, CD36 (87) and LXRα (16), were shown, indicating a loss of PPARγ function and suggesting that these macrophages may be useful in studying possible roles of PPARγ in macrophage functions relevant to atherosclerosis. The changes in basal expression of PPARγ target genes in PPARγ-deficient mice also imply the production of endogenous ligands for PPARγ and its heterodimeric partner, RXRα. Stimulation of PPARγ target genes by troglitazone alone provides further evidence for the presence of sufficient levels of endogenous RXR ligands to support PPARγ signaling in primary macrophages.
The demonstration that PPARγ is required for basal and troglitazone-inducible expression of the gene encoding LPL represents a novel finding suggesting that the LPL gene (which contains a PPRE in its promoter) is a PPARγ target in macrophages, as has been shown in other tissues, including liver and adipose tissue (78). The PPARγ-independent induction of LPL gene expression by 22-HC and suppression by dexamethasone also represent novel findings that may have important implications. Indeed, the transplantation of LPL-null bone marrow into wild-type C57BL/6 mice (90) or LDLr-null mice (1) reduced atherosclerotic lesion size, whereas the in vivo expression of macrophage-derived LPL promotes foam cell formation and atherosclerosis (1, 2, 90).
The marked reductions in basal and troglitazone-stimulated expression of the gene encoding ABCG1 in PPARγ-deficient macrophages corresponded well with the LXRα expression profile and was consistent with previous studies (with LXR knockout mice) showing that LXR signaling pathways modulate ABCG1 (and ABCA1) expression in macrophages (45). Based on these results, it is proposed that a PPARγ-LXRα-ABCG1 pathway is likely to present in macrophages, although the ABCG1 gene has yet to be established as a direct target of LXRα. In addition to a direct transcriptional effect of PPARγ on LXRα, PPARγ may also regulate ABC gene expression by enhancing the uptake of modified lipoproteins containing LXRα ligands through CD36, a possibility which remains to be examined. In contrast to the results of ABCG1, however, troglitazone inhibited the expression of the gene encoding ABCA1 in both PPARγ-deficient and control macrophages, a reproducible result that was evident in the same RNA samples in which other PPARγ targets were shown to be induced. This result was rather surprising in light of the proposed PPARγ-LXRα-ABCA1 pathway and previous findings showing that PPARγ agonists were capable of inducing ABCA1 gene expression. However, other TZDs, including rosiglitazone, ciglitazone, and pioglitazone, have minimal effect on the expression of the gene encoding ABCA1 in both PPARγ-deficient and control primary macrophages. It therefore appears that troglitazone has effects on ABCA1 gene expression that are unique for the TZD family of PPARγ agonists. Given that the expression of the ABCA1 gene is regulated by a number of factors, including cyclic AMP, serum deprivation in vitro, 9-cis-retinoic acid, and various cytokines (76), troglitazone may exert specific effects on ABCA1 gene expression through one or more of these factors, possibilities which remain to be examined. For example, troglitazone was shown to inhibit phorbol ester-induced tumor necrosis factor alpha release in a monocyte cell line, THP-1, whereas neither pioglitazone nor rosiglitazone had any effect (59). The demonstration that 22-HC induced both ABCA1 and ABCG1 gene expression in both PPARγ-deficient and control macrophages indicates that the suppression of ABCA1 by troglitazone is not likely to be the result of defective LXR signaling pathways.
The demonstration that basal levels of cholesterol efflux to HDL were lower in PPARγ-deficient macrophages than control macrophages points to a critical role of PPARγ in modulating constitutive levels of cholesterol efflux in primary macrophages. The presence of functional LXR signaling pathways was again indicated by the fact that 22-HC not only induced both ABCA1 and ABCG1 gene expression but also promoted cholesterol efflux in both PPARγ-deficient and control macrophages. The suppression of ABCA1 and HDL-specific cholesterol efflux by troglitazone in both PPARγ-deficient and control primary macrophages was an unexpected finding that sharply contrasts with the results of previous cholesterol efflux studies with ES-cell derived macrophages (16). Although the reasons for these differences are not entirely clear, it is noted that costimulation with the LXRα ligand, 22-HC, was required in that study to achieve maximal effects of PPARγ agonists on ABCA1 and ABCG1 gene expression, thus suggesting that endogenous ligands for LXRα in ES cell-derived macrophages were not in sufficient concentration to fully support the proposed PPARγ-LXRα-ABCA1 regulatory pathway. Moreover, differences in these results may be attributable to the use of primary macrophages in the present study. Indeed, others have reported that LXR or RXR ligands, but not the PPARγ ligand, rosiglitazone, induced ABCA1 expression and enhanced cholesterol efflux (20) in primary murine macrophages. Effects of PPARγ agonists on macrophages may also be species specific since they were shown to induce ABCA1 gene expression and enhance cholesterol removal from human primary macrophages (18). An important consideration may pertain to the experimental conditions under which independent studies were performed. In the above study (18), mononuclear cells from the blood of healthy normolipidemic donors were cultured for a period of 10 days, enough time to induce differentiation into macrophages. Since the expression of CD36 and apoE, two proteins believed to affect lipid flux in macrophages, is highly upregulated during the conversion of monocytes to macrophages (4, 33), the effects of PPARγ ligand stimulation on ABC expression and cholesterol efflux must be considered in the context of in vitro differentiated primary macrophages from monocytes. Other PPARγ-independent effects of troglitazone have been reported (62). Our results with troglitazone thus add to the growing list of PPARγ-independent effects of PPARγ agonists, highlighted by the prostaglandin metabolite, 15d-PGJ2 (74, 84, 89, 93).
It is becoming increasingly clear that a number of proteins not associated with the PPARγ-LXRα-ABCA1 pathway also play an important role in regulating cholesterol efflux in macrophages. Indeed, a recent study has provided evidence for the existence of a transcriptional repressor of ABCA1 gene expression, the zinc-finger protein 202, that significantly reduces HDL-mediated lipid efflux in macrophages (67), although effects of troglitazone on this repressor have yet to be examined. Numerous studies have also shown that lack of macrophage-derived apoE is sufficient to inhibit cholesterol efflux from macrophages and promote atherosclerosis (10, 26, 47, 52). It is thus likely that the reduction in basal levels of expression of the gene encoding apoE contributed to the attenuation of cholesterol efflux in PPARγ-deficient macrophages. This result is consistent with the finding that the apoE gene is a direct target of LXRα and LXRβ in macrophages (45), since constitutive levels of LXRα expression were reduced in PPARγ-deficient macrophages. apoE insufficiency may have also contributed to the significant increase in atherosclerotic lesion size shown in LDLr-null mice transplanted with PPARγ-null bone marrow (16), a possibility which remains to be examined. The lack of a robust induction of apoE by troglitazone in either PPARγ-deficient or control macrophages may be related to a sharing of the regulatory control of apoE expression by both LXRα and LXRβ since oxysterol induction of apoE was only partially compromised in LXRα−/− mice relative to LXRα−/− LXRβ−/− double knockout mice (45). Interestingly, dexamethasone was also shown to strongly stimulate apoE gene expression in macrophages in a PPARγ-independent manner, pointing to a potential therapeutic use for glucocorticoids in protecting against the development of atherosclerosis.
It is also noteworthy that pIpC-treated PPARγ-MXCre+ mice exhibited a significant reduction of apoE (−30% by densitometric analysis) in plasma, mostly due to decreased HDL apoE levels (Fig. 8B, inset). The attenuation of apoE expression in serum may be the result of lowered macrophage LPL expression, since transplantation of LPL−/− bone marrow into wild-type mice was shown to result in a 50% reduction in apoE expression in serum relative to control mice (90). Although plasma lipid levels in pIpC-treated PPARγ-MXCre+ mice were similar relative to pIpC-treated control mice, FPLC analysis revealed a decrease in LDL-cholesterol and apoB accompanied by a modest increase in HDL-cholesterol. Since the PPARγ gene was also disrupted in the liver (not shown), an organ which plays an important role in the regulation of serum lipoproteins, it must be emphasized that the possibility that PPARγ deficiency in the liver contributed to the observed changes in lipoprotein-associated cholesterol levels cannot be ruled out, despite the fact that PPARγ is expressed at very low levels in the liver. These results are consistent with the fact that little changes in plasma lipid levels were observed in studies in which the human apoE transgene was expressed in the macrophages of apoE−/− mice (7) and in which apoE−/− macrophage stem cells were transplanted into apoE+/+ mice (26), despite major changes in atherosclerotic plaque lesion size.
Interest in the pleiotropic effects of TZDs stems largely from their widespread use in diabetic patients. The suppressive effects of troglitazone on ABCA1 expression and cholesterol efflux carries with it the implication that troglitazone promotes the formation of foam cells associated with atherosclerotic plaques by inhibiting cholesterol removal from macrophages independently of the expression of PPARγ. The PPARγ-independent inhibitory effect of troglitazone on ABCA1 expression probably masks or reduces PPARγ-dependent effects that are likely to be clinically beneficial. It will therefore be important to determine whether these effects are seen in human cells and whether PPARγ-independent effects on cholesterol efflux are observed for other classes of PPARγ agonists. Clearly, the development of PPARγ ligands that do not have this property would be desirable from a clinical standpoint. Numerous whole-body effects of troglitazone on insulin sensitivity, blood pressure, hyperlipidemia, and oxidation of LDL have been reported (30, 60). Further, TZDs were shown to have beneficial effects on local events in the artery wall, including reducing carotid thickness in humans, neointimal formation following balloon injury in rats, and macrophage recruitment to the artery wall in atherosclerosis-susceptible apoE-deficient mice (49, 55, 65, 80). These effects preclude us from making accurate conclusions regarding the long-term effects of TZDs on atherosclerosis in animals and humans based solely on cholesterol flux in macrophages. The introduction of the PPARγ conditional allele into atherogenic backgrounds including LDLr-null and apoE-null mouse models should further aid in our understanding of the effects of PPARγ signaling and TZDs on atherogenesis. In particular, an apoE-null background may help to separate the putative effects of apoE from other PPARγ-mediated effects on cholesterol efflux or atherosclerosis.
Taken together, this study describes the development of a conditional PPARγ knockout mouse. Based on this in vivo model of PPARγ deficiency, evidence is provided here indicating that PPARγ plays a critical role in controlling the expression of a network of genes that mediate cholesterol efflux from cells and cholesterol transport in plasma.
Acknowledgments
T.E.A. and S.S. contributed equally to this study.
M.R. was supported by Beginning Grant-In-Aid American Heart Association Western Affiliate.
REFERENCES
- 1.Babaev, V. R., S. Fazio, L. A. Gleaves, K. J. Carter, C. F. Semenkovich, and M. F. Linton. 1999. Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in vivo. J. Clin. Investig. 103:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babaev, V. R., M. B. Patel, C. F. Semenkovich, S. Fazio, and M. F. Linton. 2000. Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in low density lipoprotein receptor-deficient mice. J. Biol. Chem. 275:26293-26299. [DOI] [PubMed] [Google Scholar]
- 3.Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, K. R. Chien, A. Koder, and R. M. Evans. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4:585-595. [DOI] [PubMed] [Google Scholar]
- 4.Basheeruddin, K., C. Rechtoris, and T. Mazzone. 1992. Transcriptional and post-transcriptional control of apolipoprotein E gene expression in differentiating human monocytes. J. Biol. Chem. 267:1219-1224. [PubMed] [Google Scholar]
- 5.Basu, S. K., J. L. Goldstein, and M. S. Brown. 1983. Independent pathways for secretion of cholesterol and apolipoprotein E by macrophages. Science 219:871-873. [DOI] [PubMed] [Google Scholar]
- 6.Basu, S. K., Y. K. Ho, M. S. Brown, D. W. Bilheimer, R. G. Anderson, and J. L. Goldstein. 1982. Biochemical and genetic studies of the apoprotein E secreted by mouse macrophages and human monocytes. J. Biol. Chem. 257:9788-9795. [PubMed] [Google Scholar]
- 7.Bellosta, S., R. W. Mahley, D. A. Sanan, J. Murata, D. L. Newland, J. M. Taylor, and R. E. Pitas. 1995. Macrophage-specific expression of human apolipoprotein E reduces atherosclerosis in hypercholesterolemic apolipoprotein E-null mice. J. Clin. Investig. 96:2170-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berliner, J. A., M. Navab, A. M. Fogelman, J. S. Frank, L. L. Demer, P. A. Edwards, A. D. Watson, and A. J. Lusis. 1995. Atherosclerosis: basic mechanisms. Oxidation, inflammation, and genetics. Circulation 91:2488-2496. [DOI] [PubMed] [Google Scholar]
- 9.Bodzioch, M., E. Orso, J. Klucken, T. Langmann, A. Bottcher, W. Diederich, W. Drobnik, S. Barlage, C. Buchler, M. Porsch-Ozcurumez, W. E. Kaminski, H. W. Hahmann, K. Oette, G. Rothe, C. Aslanidis, K. J. Lackner, and G. Schmitz. 1999. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22:347-351. [DOI] [PubMed] [Google Scholar]
- 10.Boisvert, W. A., J. Spangenberg, and L. K. Curtiss. 1995. Treatment of severe hypercholesterolemia in apolipoprotein E-deficient mice by bone marrow transplantation. J. Clin. Investig. 96:1118-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockman, J. A., R. A. Gupta, and R. N. Dubois. 1998. Activation of PPARγ leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology 115:1049-1055. [DOI] [PubMed] [Google Scholar]
- 12.Brooks-Wilson, A., M. Marcil, S. M. Clee, L. H. Zhang, K. Roomp, M. van Dam, L. Yu, C. Brewer, J. A. Collins, H. O. Molhuizen, O. Loubser, B. F. Ouelette, K. Fichter, K. J. Ashbourne-Excoffon, C. W. Sensen, S. Scherer, S. Mott, M. Denis, D. Martindale, J. Frohlich, K. Morgan, B. Koop, S. Pimstone, J. J. Kastelein, and M. R. Hayden. 1999. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22:336-345. [DOI] [PubMed] [Google Scholar]
- 13.Chang, K. C., G. Goldspink, and J. Lida. 1990. Studies in the in vivo expression of the influenza resistance gene Mx by in-situ hybridization. Arch. Virol. 110:151-164. [DOI] [PubMed] [Google Scholar]
- 14.Chang, T. H., and E. Szabo. 2000. Induction of differentiation and apoptosis by ligands of peroxisome proliferator-activated receptor gamma in non-small cell lung cancer. Cancer Res. 60:1129-1138. [PubMed] [Google Scholar]
- 15.Chawla, A., Y. Barak, L. Nagy, D. Liao, P. Tontonoz, and R. M. Evans. 2001. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat. Med. 7:48-52. [DOI] [PubMed] [Google Scholar]
- 16.Chawla, A., W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans, and P. Tontonoz. 2001. A PPARγ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7:161-171. [DOI] [PubMed] [Google Scholar]
- 17.Chen, F., S. W. Law, and B. W. O'Malley. 1993. Identification of two mPPAR-related receptors and evidence for the existence of five subfamily members. Biochem. Biophys. Res. Commun. 196:671-677. [DOI] [PubMed] [Google Scholar]
- 18.Chinetti, G., S. Lestavel, V. Bocher, A. T. Remaley, B. Neve, I. P. Torra, E. Teissier, A. Minnich, M. Jaye, N. Duverger, H. B. Brewer, J. C. Fruchart, V. Clavey, and B. Staels. 2001. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 7:53-58. [DOI] [PubMed] [Google Scholar]
- 19.Clark, R. B., D. Bishop-Bailey, T. Estrada-Hernandez, T. Hla, L. Puddington, and S. J. Padula. 2001. The nuclear receptor PPAR gamma and immunoregulation: PPAR gamma mediates inhibition of helper T cell responses. J. Immunol. 164:1364-1371. [DOI] [PubMed] [Google Scholar]
- 20.Claudel, T., M. D. Leibowitz, C. Fievet, A. Tailleux, B. Wagner, J. J. Repa, G. Torpier, J. M. Lobaccaro, J. R. Paterniti, D. J. Mangelsdorf, R. A. Heyman, and J. Auwerx. 2001. Reduction of atherosclerosis in apolipoprotein E knockout mice by activation of the retinoid X receptor. Proc. Natl. Acad. Sci. USA 98:2610-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costet, P., Y. Luo, N. Wang, and A. R. Tall. 2000. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 275:28240-28245. [DOI] [PubMed] [Google Scholar]
- 22.Cullen, P., A. Cignarella, B. Brennhausen, S. Mohr, G. Assmann, and A. von Eckardstein. 1998. Phenotype-dependent differences in apolipoprotein E metabolism and in cholesterol homeostasis in human monocyte-derived macrophages. J. Clin. Investig. 101:1670-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Villiers, W. J., I. P. Fraser, D. A. Hughes, A. G. Doyle, and S. Gordon. 1994. Macrophage-colony-stimulating factor selectively enhances macrophage scavenger receptor expression and function. J. Exp. Med. 180:705-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dory, L. 1989. Synthesis and secretion of apoE in thioglycolate-elicited mouse peritoneal macrophages: effect of cholesterol efflux. J. Lipid Res. 30:809-816. [PubMed] [Google Scholar]
- 25.Elstner, E., C. Muller, K. Koshizuka, E. A. Williamson, D. Park, H. Asou, P. Shintaku, J. W. Said, D. Heber, and H. P. Koeffler. 1998. Ligands for peroxisome proliferator-activated receptor gamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc. Natl. Acad. Sci. USA 95:8806-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazio, S., V. R. Babaev, A. B. Murray, A. H. Hasty, K. J. Carter, L. A. Gleaves, J. B. Atkinson, and M. F. Linton. 1997. Increased atherosclerosis in mice reconstituted with apolipoprotein E null macrophages. Proc. Natl. Acad. Sci. USA 94:4647-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng, J., J. Han, S. F. Pearce, R. L. Silverstein, A. M. J. Gotto, D. P. Hajjar, and A. C. Nicholson. 2000. Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-γ. J. Lipid Res. 41:688-696. [PubMed] [Google Scholar]
- 28.Forman, B. M., J. Chen, and R. M. Evans. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. USA 94:4312-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forman, B. M., P. Tontonoz, J. Chen, R. P. Brun, B. M. Spiegelman, and R. M. Evans. 1995. 15-Deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 83:803-812. [DOI] [PubMed] [Google Scholar]
- 30.Ghazzi, M. N., J. E. Perez, T. K. Antonucci, J. H. Driscoll, S. M. Huang, B. W. Faja, R. W. Whitcomb, and the Troglitazone Study Group. 1997. Cardiac and glycemic benefits of troglitazone treatment in NIDDM. Diabetes 46:433-439. [DOI] [PubMed] [Google Scholar]
- 31.Hayhurst, G. P., Y. H. Lee, G. Lambert, J. M. Ward, and F. J. Gonzalez. 2001. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21:1393-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobbs, H. H., and D. J. Rader. 1999. ABC1: connecting yellow tonsils, neuropathy, and very low HDL. J. Clin. Investig. 104:1015-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh, H. Y., S. F. Pearce, L. M. Yesner, J. L. Schindler, and R. L. Silverstein. 1996. Regulated expression of CD36 during monocyte-to-macrophage differentiation: potential role of CD36 in foam cell formation. Blood 87:2020-2028. [PubMed] [Google Scholar]
- 34.Imai, T., M. Jiang, P. Chambon, and D. Metzger. 2001. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc. Natl. Acad. Sci. USA 98:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kintscher, U., S. Goetze, S. Wakino, S. Kim, S. Nagpal, R. A. Chandraratna, K. Graf, E. Fleck, W. A. Hsueh, and R. E. Law. 2000. Peroxisome proliferator-activated receptor and retinoid X receptor ligands inhibit monocyte chemotactic protein-1-directed migration of monocytes. Eur. J. Pharmacol. 401:259-270. [DOI] [PubMed] [Google Scholar]
- 36.Kirchgessner, T. G., K. L. Svenson, A. J. Lusis, and M. C. Schotz. 1987. The sequence of cDNA encoding lipoprotein lipase. A member of a lipase gene family. J. Biol. Chem. 262:8463-8466. [PubMed] [Google Scholar]
- 37.Kitamura, S., Y. Miyazaki, Y. Shinomura, S. Kondo, S. Kanayama, and Y. Matsuzawa. 1999. Peroxisome proliferator-activated receptor gamma induces growth arrest and differentiation markers of human colon cancer cells. Jpn. J. Cancer Res. 90:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kliewer, S. A., J. M. Lenhard, T. M. Willson, I. Patel, D. C. Morris, and J. M. Lehmann. 1995. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell 83:813-819. [DOI] [PubMed] [Google Scholar]
- 39.Kliewer, S. A., S. S. Sundseth, S. A. Jones, P. J. Brown, G. B. Wisely, C. S. Koble, P. Devchand, W. Wahli, T. M. Willson, J. M. Lenhard, and J. M. Lehmann. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA 94:4318-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klucken, J., C. Buchler, E. Orso, W. E. Kaminski, M. Porsch-Ozcurumez, G. Liebisch, M. Kapinsky, W. Diederich, W. Drobnik, M. Dean, R. Allikmets, and G. Schmitz. 2000. ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc. Natl. Acad. Sci. USA 97:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruth, H. S., S. I. Skarlatos, P. M. Gaynor, and W. Gamble. 1994. Production of cholesterol-enriched nascent high density lipoproteins by human monocyte-derived macrophages is a mechanism that contributes to macrophage cholesterol efflux. J. Biol. Chem. 269:24511-24518. [PubMed] [Google Scholar]
- 42.Kubota, N., Y. Terauchi, H. Miki, H. Tamemoto, T. Yamauchi, K. Komeda, S. Satoh, R. Nakano, C. Ishii, T. Sugiyama, K. Eto, Y. Tsubamoto, A. Okuno, K. Murakami, H. Sekihara, G. Hasegawa, M. Naito, Y. Toyoshima, S. Tanaka, K. Shiota, T. Kitamura, T. Fujita, O. Ezaki, S. Aizawa, and T. Kadowaki. 1999. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell 4:597-609. [DOI] [PubMed] [Google Scholar]
- 43.Kubota, T., K. Koshizuka, E. A. Williamson, H. Asou, J. W. Said, S. Holden, I. Miyoshi, and H. P. Koeffler. 1998. Ligand for peroxisome proliferator-activated receptor gamma (troglitazone) has potent antitumor effect against human prostate cancer both in vitro and in vivo. Cancer Res. 58:3344-3352. [PubMed] [Google Scholar]
- 44.Kuhn, R., F. Schwenk, M. Aguet, and K. Rajewsky. 1995. Inducible gene targeting in mice. Science 269:1427-1429. [DOI] [PubMed] [Google Scholar]
- 45.Laffitte, B. A., J. J. Repa, S. B. Joseph, D. C. Wilpitz, H. R. Kast, D. J. Mangelsdorf, and P. Tontonoz. 2001. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. USA 98:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langer, C., Y. Huang, P. Cullen, B. Wiesenhutter, R. W. Mahley, G. Assmann, and A. von Eckardstein. 2000. Endogenous apolipoprotein E modulates cholesterol efflux and cholesteryl ester hydrolysis mediated by high-density lipoprotein-3 and lipid-free apolipoproteins in mouse peritoneal macrophages. J. Mol. Med. 78:217-227. [DOI] [PubMed] [Google Scholar]
- 48.Law, R. E., S. Goetze, X. P. Xi, S. Jackson, Y. Kawano, L. Demer, M. C. Fishbein, W. P. Meehan, and W. A. Hsueh. 2000. Expression and function of PPARγ in rat and human vascular smooth muscle cells. Circulation 101:1311-1318. [DOI] [PubMed] [Google Scholar]
- 49.Law, R. E., W. P. Meehan, X. P. Xi, K. Graf, D. A. Wuthrich, W. Coats, D. Faxon, and W. A. Hsueh. 1996. Troglitazone inhibits vascular smooth muscle cell growth and intimal hyperplasia. J. Clin. Investig. 98:1897-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawn, R. M., D. P. Wade, M. R. Garvin, X. Wang, K. Schwartz, J. G. Porter, J. J. Seilhamer, A. M. Vaughan, and J. F. Oram. 1999. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J. Clin. Investig. 104:R25-R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann, J. M., L. B. Moore, T. A. Smith-Oliver, W. O. Wilkison, T. M. Willson, and S. A. Kliewer. 1995. An antidiabetic thiazolidinedione is a high-affinity ligand for peroxisome proliferator-activated receptor gamma (PPARγ). J. Biol. Chem. 270:12953-12956. [DOI] [PubMed] [Google Scholar]
- 52.Linton, M. F., J. B. Atkinson, and S. Fazio. 1995. Prevention of atherosclerosis in apolipoprotein E-deficient mice by bone marrow transplantation. Science 267:1034-1037. [DOI] [PubMed] [Google Scholar]
- 53.Marx, N., G. Sukhova, C. Murphy, P. Libby, and J. Plutzky. 1998. Macrophages in human atheroma contain PPARγ: differentiation-dependent peroxisomal proliferator-activated receptor gamma (PPARγ) expression and reduction of MMP-9 activity through PPARγ activation in mononuclear phagocytes in vitro. Am. J. Pathol. 153:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazzone, T., and C. Reardon. 1994. Expression of heterologous human apolipoprotein E by J774 macrophages enhances cholesterol efflux to HDL3. J. Lipid Res. 35:1345-1353. [PubMed] [Google Scholar]
- 55.Minamikawa, J., M. Yamauchi, D. Inoue, and H. Koshiyama. 1998. Another potential use of troglitazone in noninsulin-dependent diabetes mellitus. J. Clin. Endocrinol. Metab. 83:1041-1042. [DOI] [PubMed] [Google Scholar]
- 56.Moore, K. J., E. D. Rosen, M. L. Fitzgerald, F. Randow, L. P. Andersson, D. Altshuler, D. S. Milstone, R. M. Mortensen, B. M. Spiegelman, and M. W. Freeman. 2001. The role of PPAR-γ in macrophage differentiation and cholesterol uptake. Nat. Med. 7:41-47. [DOI] [PubMed] [Google Scholar]
- 57.Mueller, E., P. Sarraf, P. Tontonoz, R. M. Evans, K. J. Martin, M. Zhang, C. Fletcher, S. Singer, and B. M. Spiegelman. 1998. Terminal differentiation of human breast cancer through PPARγ. Mol. Cell 1:465-470. [DOI] [PubMed] [Google Scholar]
- 58.Nagy, L., P. Tontonoz, J. G. Alvarez, H. Chen, and R. M. Evans. 1998. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell 93:229-240. [DOI] [PubMed] [Google Scholar]
- 59.Naitoh, T., M. Kitahara, and N. Tsuruzoe. 2000. The effect of activation of peroxisome proliferator-activated receptor gamma (PPARγ) on human monocyte function: PPARγ ligands do not inhibit tumor necrosis factor-alpha release in human monocytic cell line THP-1. Cell Biol. Toxicol. 16:131-135. [DOI] [PubMed] [Google Scholar]
- 60.Noguchi, N., H. Sakai, Y. Kato, J. Tsuchiya, Y. Yamamoto, E. Niki, H. Horikoshi, and T. Kodama. 1996. Inhibition of oxidation of low-density lipoprotein by troglitazone. Atherosclerosis 123:227-234. [DOI] [PubMed] [Google Scholar]
- 61.Okuno, A., H. Tamemoto, K. Tobe, K. Ueki, Y. Mori, K. Iwamoto, K. Umesono, Y. Akanuma, T. Fujiwara, H. Horikoshi, Y. Yazaki, and T. Kadowaki. 1998. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J. Clin. Investig. 101:1354-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okura, T., M. Nakamura, Y. Takata, S. Watanabe, Y. Kitami, and K. Hiwada. 2000. Troglitazone induces apoptosis via the p53 and Gadd45 pathway in vascular smooth muscle cells. Eur. J. Pharmacol. 407:227-235. [DOI] [PubMed] [Google Scholar]
- 63.Oram, J. F., R. M. Lawn, M. R. Garvin, and D. P. Wade. 2000. ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J. Biol. Chem. 275:34508-34511. [DOI] [PubMed] [Google Scholar]
- 64.Oram, J. F., and A. M. Vaughan. 2000. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr. Opin. Lipidol. 11:253-260. [DOI] [PubMed] [Google Scholar]
- 65.Pasceri, V., H. D. Wu, J. T. Willerson, and E. T. Yeh. 2000. Modulation of vascular inflammation in vitro and in vivo by peroxisome proliferator-activated receptor-γ activators. Circulation 101:235-238. [DOI] [PubMed] [Google Scholar]
- 66.Plump, A. S., J. D. Smith, T. Hayek, K. Aalto-Setala, A. Walsh, J. G. Verstuyft, E. M. Rubin, and J. L. Breslow. 1992. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell 71:343-353. [DOI] [PubMed] [Google Scholar]
- 67.Porsch-Ozcurumez, M., T. Langmann, S. Heimerl, H. Borsukova, W. E. Kaminski, W. Drobnik, C. Honer, C. Schumacher, and G. Schmitz. 2001. The zinc finger protein 202 (ZNF202) is a transcriptional repressor of ATP binding cassette transporter A1 (ABCA1) and ABCG1 gene expression and a modulator of cellular lipid efflux. J. Biol. Chem. 276:12427-12433. [DOI] [PubMed] [Google Scholar]
- 68.Raabe, M., M. M. Veniant, M. A. Sullivan, C. H. Zlot, J. Bjorkegren, L. B. Nielsen, J. S. Wong, R. L. Hamilton, and S. G. Young. 1999. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J. Clin. Investig. 103:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajagopalan, S., X. P. Meng, S. Ramasamy, D. G. Harrison, and Z. S. Galis. 1996. Reactive oxygen species produced by macrophage-derived foam cells regulate the activity of vascular matrix metalloproteinases in vitro. Implications for atherosclerotic plaque stability. J. Clin. Investig. 98:2572-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Repa, J. J., S. D. Turley, J. A. Lobaccaro, J. Medina, L. Li, K. Lustig, B. Shan, R. A. Heyman, J. M. Dietschy, and D. J. Mangelsdorf. 2000. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289:1524-1529. [DOI] [PubMed] [Google Scholar]
- 71.Ricote, M., J. Huang, L. Fajas, A. Li, J. Welch, J. Najib, J. L. Witztum, J. Auwerx, W. Palinski, and C. K. Glass. 1998. Expression of the peroxisome proliferator-activated receptor gamma (PPARγ) in human atherosclerosis and regulation in macrophages by colony-stimulating factors and oxidized low-density lipoprotein. Proc. Natl. Acad. Sci. USA 95:7614-7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rohlmann, A., M. Gotthardt, R. E. Hammer, and J. Herz. 1998. Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. J. Clin. Investig. 101:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosen, E. D., P. Sarraf, A. E. Troy, G. Bradwin, K. Moore, D. S. Milstone, B. M. Spiegelman, and R. M. Mortensen. 1999. PPAR γ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4:611-617. [DOI] [PubMed] [Google Scholar]
- 74.Rossi, A., P. Kapahi, G. Natoli, T. Takahashi, Y. Chen, M. Karin, and M. G. Santoro. 2000. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IκB kinase. Nature 403:103-108. [DOI] [PubMed] [Google Scholar]
- 75.Rust, S., M. Rosier, H. Funke, J. Real, Z. Amoura, J. C. Piette, J. F. Deleuze, H. B. Brewer, N. Duverger, P. Denefle, and G. Assmann. 1999. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 22:352-355. [DOI] [PubMed] [Google Scholar]
- 76.Santamarina-Fojo, S., A. T. Remaley, E. B. Neufeld, and H. B. J. Brewer. 2001. Regulation and intracellular trafficking of the ABCA1 transporter. J. Lipid Res. 42:1339-1345. [PubMed] [Google Scholar]
- 77.Sarraf, P., E. Mueller, D. Jones, F. J. King, D. J. DeAngelo, J. B. Partridge, S. A. Holden, L. B. Chen, S. Singer, C. Fletcher, and B. M. Spiegelman. 1998. Differentiation and reversal of malignant changes in colon cancer through PPARγ. Nat. Med. 4:1046-1052. [DOI] [PubMed] [Google Scholar]
- 78.Schoonjans, K., J. Peinado-Onsurbe, A. M. Lefebvre, R. A. Heyman, M. Briggs, S. Deeb, B. Staels, and J. Auwerx. 1996. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15:5336-5348. [PMC free article] [PubMed] [Google Scholar]
- 79.Schwartz, K., R. M. Lawn, and D. P. Wade. 2000. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem. Biophys. Res. Commun. 274:794-802. [DOI] [PubMed] [Google Scholar]
- 80.Shinohara, E., S. Kihara, N. Ouchi, T. Funahashi, T. Nakamura, S. Yamashita, K. Kameda-Takemura, and Y. Matsuzawa. 1998. Troglitazone suppresses intimal formation following balloon injury in insulin-resistant Zucker fatty rats. Atherosclerosis 136:275-279. [DOI] [PubMed] [Google Scholar]
- 81.Spiegelman, B. M. 1997. Peroxisome proliferator-activated receptor gamma: a key regulator of adipogenesis and systemic insulin sensitivity. Eur. J. Med. Res. 2:457-464. [PubMed] [Google Scholar]
- 82.Spiegelman, B. M., E. Hu, J. B. Kim, and R. Brun. 1997. PPARγ and the control of adipogenesis. Biochimie 79:111-112. [DOI] [PubMed] [Google Scholar]
- 83.Staeheli, P., P. Danielson, O. Haller, and J. G. Sutcliffe. 1986. Transcriptional activation of the mouse Mx gene by type I interferon. Mol. Cell. Biol. 6:4770-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Straus, D. S., G. Pascual, M. Li, J. S. Welch, M. Ricote, C. H. Hsiang, L. L. Sengchanthalangsy, G. Ghosh, and C. K. Glass. 2000. 15-Deoxy-Δ12,14-prostaglandin J2 inhibits multiple steps in the NF-κB signaling pathway. Proc. Natl. Acad. Sci. USA 97:4844-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suzuki, H., Y. Kurihara, M. Takeya, N. Kamada, M. Kataoka, K. Jishage, O. Ueda, H. Sakaguchi, T. Higashi, T. Suzuki, Y. Takashima, Y. Kawabe, O. Cynshi, Y. Wada, M. Honda, H. Kurihara, H. Aburatani, T. Doi, A. Matsumoto, S. Azuma, T. Noda, Y. Toyoda, H. Itakura, Y. Yazaki, and T. Kodama. 1997. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature 386:292-296. [DOI] [PubMed] [Google Scholar]
- 86.Tangirala, R. K., D. Pratico, G. A. FitzGerald, S. Chun, K. Tsukamoto, C. Maugeais, D. C. Usher, E. Pure, and D. J. Rader. 2001. Reduction of isoprostanes and regression of advanced atherosclerosis by apolipoprotein E. J. Biol. Chem. 276:261-266. [DOI] [PubMed] [Google Scholar]
- 87.Tontonoz, P., L. Nagy, J. G. Alvarez, V. A. Thomazy, and R. M. Evans. 1998. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93:241-252. [DOI] [PubMed] [Google Scholar]
- 88.Tontonoz, P., S. Singer, B. M. Forman, P. Sarraf, J. A. Fletcher, C. D. Fletcher, R. P. Brun, E. Mueller, S. Altiok, H. Oppenheim, R. M. Evans, and B. M. Spiegelman. 1997. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc. Natl. Acad. Sci. USA 94:237-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vaidya, S., E. P. Somers, S. D. Wright, P. A. Detmers, and V. S. Bansal. 1999. 15-Deoxy-Δ12,14-prostaglandin J2 inhibits the β2 integrin-dependent oxidative burst: involvement of a mechanism distinct from peroxisome proliferator-activated receptor gamma ligation. J. Immunol. 163:6187-6192. [PubMed] [Google Scholar]
- 90.Van Eck, M., R. Zimmermann, P. H. Groot, R. Zechner, and T. J. Van Berkel. 2000. Role of macrophage-derived lipoprotein lipase in lipoprotein metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 20:E53-E62. [DOI] [PubMed] [Google Scholar]
- 91.Venkateswaran, A., B. A. Laffitte, S. B. Joseph, P. A. Mak, D. C. Wilpitz, P. A. Edwards, and P. Tontonoz. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXRα. Proc. Natl. Acad. Sci. USA 97:12097-12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Venkateswaran, A., J. J. Repa, J. M. Lobaccaro, A. Bronson, D. J. Mangelsdorf, and P. A. Edwards. 2000. Human white/murine ABC8 mRNA levels are highly induced in lipid-loaded macrophages. A transcriptional role for specific oxysterols. J. Biol. Chem. 275:14700-14707. [DOI] [PubMed] [Google Scholar]
- 93.Willoughby, D. A., A. R. Moore, and P. R. Colville-Nash. 2000. Cyclopentenone prostaglandins-new allies in the war on inflammation. Nat. Med. 6:137-138. [DOI] [PubMed] [Google Scholar]
- 94.Witztum, J. L. 1994. The oxidation hypothesis of atherosclerosis. Lancet 344:793-795. [DOI] [PubMed] [Google Scholar]
- 95.Yesner, L. M., H. Y. Huh, S. F. Pearce, and R. L. Silverstein. 1996. Regulation of monocyte CD36 and thrombospondin-1 expression by soluble mediators. Arterioscler. Thromb. Vasc. Biol. 16:1019-1025. [DOI] [PubMed] [Google Scholar]
- 96.Zhang, S. H., R. L. Reddick, J. A. Piedrahita, and N. Maeda. 1992. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science 258:468-471. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, W. Y., P. M. Gaynor, and H. S. Kruth. 1996. Apolipoprotein E produced by human monocyte-derived macrophages mediates cholesterol efflux that occurs in the absence of added cholesterol acceptors. J. Biol. Chem. 271:28641-28646. [DOI] [PubMed] [Google Scholar]